Abstract
Adeno-associated virus (AAV) type 2 and 5 proteins Rep52 and Rep40 were polyubiquitinated during AAV-adenovirus type 5 (Ad5) coinfection and during transient transfection in either the presence or absence of Ad5 E4orf6 and E1b-55k. Polyubiquitination of small Rep proteins via lysine 48 (K48) linkages, normally associated with targeting of proteins for proteasomal degradation, was detected only in the presence of E4orf6. The small Rep proteins were ubiquitinated via lysine 63 (K63) following transfection in either the presence or absence of E4orf6 or following coinfection with Ad5. E4orf6/E1b-55k-dependent K48-specific polyubiquitination of small Rep proteins could be inhibited using small interfering RNA (siRNA) to cullin 5.
Together, adenovirus type 5 (Ad5) early gene products E1a, E1b-55k, E2a, E4orf6, and virus-associated (VA) RNA can support efficient replication of adeno-associated virus (AAV) (4, 31). E4orf6 and E1b-55k are known to interact with cellular cullin 5 (cul5), elongins B and C, and the ring box protein Rbx1 to form an E3 ubiquitin ligase complex that specifically targets a small population of cellular proteins for degradation by the proteasome (1, 7, 21, 22, 24, 27). This property has been implicated in a number of functions presumed to be required for both Ad and AAV replication (3, 8-10, 17, 23, 24, 34, 35).
Previously, only p53, Mre11, DNA ligase IV, and integrin α3 had been shown to be substrates of the Ad5 E3 ubiquitin ligase complex (1, 7, 21, 22, 24, 27); however, we have recently shown (16, 17) that the small Rep proteins and capsid proteins of AAV5 are also degraded in the presence of Ad E4orf6 and E1b-55k in a proteasome-dependent manner. These proteins were restored to levels required during infection by the action of VA RNA (17). The targeting for degradation of AAV5 protein by the E4orf6/E1b-55k E3 ubiquitin ligase complex required functional BC-box motifs in E4orf6 and could be inhibited by depletion of the scaffolding protein cullin 5 using directed small interfering RNA (siRNA) (16). In addition, the degradation of AAV5 protein was partially prevented by overexpression of pUBR7, a plasmid that generates a dominant-negative ubiquitin (16). The role this targeted degradation plays in the life cycle of AAV has not yet been clarified; however, E4orf6 mutants that cannot function in this regard do not support AAV replication as well as wild-type E4orf6 (R. Nayak and D. J. Pintel, unpublished data). Degradation of Mre11 by the Ad5 E3 ligase has also been implicated in allowing efficient Ad5 and AAV replication (24). Ubiquitination of AAV Rep proteins during viral infection, however, has not previously been reported.
AAV5 Rep proteins are polyubiquitinated during coinfection with Ad5. During transient transfection, AAV5 small Rep proteins are polyubiquitinated in the presence or absence of E4orf6.
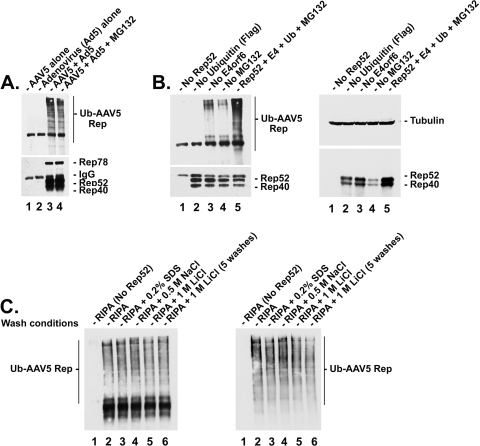
Figure 1 demonstrates that AAV5 Rep52/40 proteins are ubiquitinated during AAV5-Ad coinfection. Immunoblots, using an anti-ubiquitin antibody (P4D1; #sc-8017 [Santa Cruz Biotechnology, Inc., Santa Cruz, CA]) and previously published procedures (13, 14, 25, 26, 33, 36), of Rep proteins immunoprecipitated with a pan-specific anti-Rep antibody (76.3; #03-61073 [ARP, Belmont, MA]), displayed a specific, high-molecular-weight laddering typical of ubiquitin-conjugated proteins (Fig. 1A, lanes 3 and 4). For Rep immunoprecipitations (IPs), cells were lysed by boiling in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 2 mM EDTA, 1% NP-40) plus 1% SDS, extracts were diluted to 0.2% SDS, and antibody was added, and following capture on protein G beads (#10-1243; Invitrogen, Carlsbad, CA), the cells were subsequently washed in RIPA buffer. Results were confirmed with more stringent washes as described below. The use of a pan-specific Rep antibody prevented the determination of which Rep proteins were ubiquitinated in this experiment; however, characterization of individually expressed Rep52/40 proteins as described below revealed that they undergo this modification. (Specific analysis of the large Rep proteins is currently ongoing.) The accumulation of ubiquitinated Rep proteins in this experiment was not significantly increased in the presence of the proteasome inhibitor MG132 (10 μM; #474791 [Calbiochem, Gibbstown, NJ]) (Fig. 1A, compare lanes 3 and 4), likely because, as we have previously shown, the E4orf6-targeted degradation of AAV5 small Rep proteins is compensated for by the action of Ad5-expressed VA RNA (17), which serves to enhance translation of AAV proteins. Additionally, as shown below, the spectrum of ubiquitinated proteins detected also included a component that likely did not lead to degradation.
FIG. 1.
AAV5 Rep proteins are polyubiquitinated during coinfection with Ad. (A) Postimmunoprecipitation (IP) Western blot analysis of AAV5 Rep protein ubiquitination during AAV5-Ad coinfection. Western blots were performed as previously described (16). Briefly, cells were infected with AAV5 (MOI, 10) and/or Ad5 (MOI, 5) for 42 h. Where applicable, cells were treated after 36 h with either MG132 or equivalent amounts of dimethyl sulfoxide (DMSO) as a vehicle control. Cells were lysed with RIPA buffer containing 1% SDS and boiled to disassociate any interactions between Rep protein and other potentially ubiquitinated (Ub) cellular proteins. Lysates were diluted in RIPA buffer to a final SDS concentration of 0.2% and precleared once with protein G beads prior to immunoprecipitation. AAV5 Rep proteins were immunoprecipitated from 293-infected lysates with anti-Rep antibody and subjected to Western analysis using the anti-Rep (bottom) or anti-ubiquitin (top) antibodies. 293 cells were infected with AAV5 alone (lane 1), Ad5 alone (lane 2), or both AAV and Ad (lanes 3 and 4) in the absence (lane 3) or presence (lane 4) of the proteasome inhibitor MG132 (10 μM). The presence of ubiquitinated Rep proteins is indicated by the appearance of high-molecular-weight isoforms of the protein, observed as a laddering or smearing effect. (B) Western blot analysis of AAV5 small Rep protein ubiquitination in 293 cells in the absence or presence of adenovirus E4orf6 during transient transfection. Lysates were isolated 36 to 42 h posttransfection with vectors expressing AAV5 small Rep proteins plus a construct expressing E4orf6 and another expressing a Flag-tagged ubiquitin in the absence or presence of MG132 (added 6 h prior to cell harvest). Cells were transfected with each of the constructs except where noted: no Rep (lane 1), no Flag-ubiquitin (lane 2), no E4orf6 (lane 3), or no MG132 (lane 4). (Left panel) Post-IP Western blot following immunoprecipitation with an anti-Rep antibody, with an immunoblot using an anti-Rep (bottom) or anti-Flag (top) antibody. (Right panel) Pre-IP Western blot of the same samples as in the left panel, with anti-tubulin (top) or anti-Rep (bottom) antibodies. (C) Post-IP Western blot analysis of AAV5 small Rep protein ubiquitination in 293 cells in the presence of E4orf6 and MG132 during transient transfection following various stringent washing conditions. Cells were transfected with Flag-Ub and E4orf6 and treated with MG132 in the absence (lane 1) or presence (lanes 2 to 6) of AAV5 Rep52/40. Following immunoprecipitation, Rep/antibody-conjugated beads were washed twice with either RIPA buffer alone (lanes 1 and 2), RIPA buffer plus 0.2% SDS (lane 3), RIPA buffer plus 0.5 M NaCl (lane 4), or RIPA buffer plus 1 M LiCl (lane 5) or washed five times with RIPA buffer plus 1 M LiCl (lane 6). Samples were either blotted with anti-Rep antibody and exposed with high-sensitivity Femto reagent (left) or blotted with anti-ubiquitin antibody and exposed with Pico reagent (right). Antibody dilutions were as follows: anti-Flag (Sigma-Aldrich; #F1804), 1:5,000; anti-Rep (ARP; #03-61073), 1:200; anti-ubiquitin (Santa Cruz Biotechnology, Inc.; #sc-8017), 1:500; and goat anti-mouse horseradish peroxidase (HRP) conjugate (Sigma-Aldrich; #A5278) secondary antibody, 1:5,000.
Because we had previously demonstrated the E4orf6- and ubiquitin-dependent degradation of transiently expressed AAV5 and AAV2 Rep52/40 (16), we chose next to further characterize the ubiquitination of AAV small Rep proteins following transient transfection of Ad E1a- and E1b-55k-containing 293 cells, performed as previously described (16). This also afforded us the opportunity to characterize the role of E4orf6 in this process, an analysis complicated during coinfection studies by the requirement of E4orf6 for AAV genome replication (9, 24, 30, 31). Following immunoprecipitation with anti-Rep antibody, immunoblots using anti-Flag antibody (#F1804; Sigma-Aldrich, St. Louis, MO) demonstrated significant levels of high-molecular-weight ubiquitinated isoforms of AAV5 Rep52/40 proteins in the presence of cotransfected Flag-tagged ubiquitin (with a plasmid that was a gift from M. Hannink), AAV5 Rep52/40 (expressed from cytomegalovirus Rep52 [CMV-Rep52], as described previously [16]), CMV-E4orf6 (plasmid gift from T. Shenk), and added MG132 (Fig. 1B, left panel, lane 5). As expected, a marked decrease in the levels of ubiquitinated substrate was observed in the absence of MG132 (Fig. 1B, left panel, lane 4). Examination by immunoblotting of the overall levels of AAV5 small Rep proteins in this experiment confirmed, as previously reported (16), that the accumulated levels of AAV5 small Rep proteins were reduced in the presence of E4orf6 and the absence of MG132 in 293 cells (Fig. 1B, right panel, compare lane 4 to lanes 2, 3, and 5). This reduction was not apparent in immunoprecipitated samples, likely because of subsaturating immunoprecipitation of the Rep proteins in these experiments (Fig. 1B, left panel, lanes 2 to 5).
Surprisingly, significant, albeit lower, levels of ubiquitinated Rep were also observed in the absence of E4orf6 (Fig. 1B, left panel, lane 3). These results suggested that AAV5 small Rep proteins were ubiquitinated in 293 cells through both E4orf6-dependent and E4orf6-independent mechanisms. As mentioned above, this may also have partially explained the high accumulated levels of polyubiquitinated Rep protein observed during coinfection in the absence of MG132 (Fig. 1A, lane 3). The gels shown here and in Fig. 2 are representative examples of multiple experiments, which include those in which levels of E4orf6 were confirmed to be as expected.
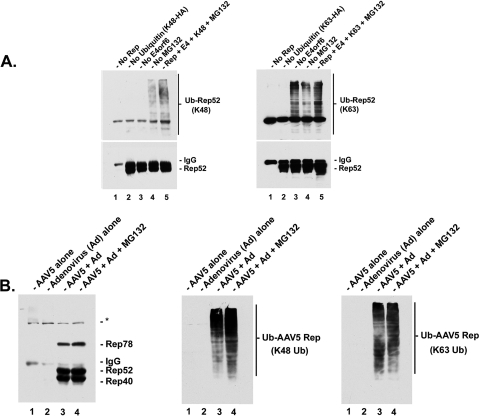
FIG. 2.
AAV5 small Rep proteins exhibit different E4orf6-dependent and -independent patterns of polyubiquitination in 293 cells during transient transfection. (A) Post-IP Western blot analysis of AAV5 small Rep protein K48-specific or K63-specific polyubiquitination in 293 cells in the absence or presence of adenovirus E4orf6 during transient transfection. Lysates were isolated 36 to 42 h posttransfection with vectors expressing myc-tagged AAV5 small Rep proteins plus a construct expressing E4orf6 and another expressing either HA-tagged K48-specific (left panel) or HA-tagged K63-specific (right panel) ubiquitin in the absence or presence of MG132 or equivalent amounts of DMSO. Cells were transfected with each of the constructs, except where noted: no Rep (lane 1), no HA-ubiquitin (lane 2), no E4orf6 (lane 3), or no MG132 (lane 4). (Left panel) Post-IP Western blot of K48-specific polyubiquitination following immunoprecipitation with an anti-Rep antibody, with an immunoblot using an anti-Myc (bottom) or anti-HA (top) antibody. (Right panel) Post-IP Western blot of K63-specific polyubiquitination following immunoprecipitation with an anti-Rep antibody, with an immunoblot using an anti-Myc (bottom) or anti-HA (top) antibody. (B) Post-IP Western blot analysis of AAV5 Rep protein wild-type, K48-specific, and K63-specific polyubiquitination during AAV5-Ad5 coinfection. AAV5 Rep proteins were immunoprecipitated from 293-infected lysates with anti-Rep antibody and subjected to Western anaylsis using the anti-Rep (left panel), anti-K48-specific ubiquitin (center panel), or anti-K63-specific ubiquitin (right panel) antibody. 293 cells were infected with AAV5 alone (lane 1), Ad5 alone (lane 2), or both AAV and Ad5 (lanes 3 and 4) in the absence (lane 3) or presence (lane 4) of MG132. Antibody dilutions were as follows: anti-HA (Sigma-Aldrich; #H9658), 1:5,000; anti-Rep (ARP; #03-61073), 1:200; anti-c-Myc (Santa Cruz Biotechnology, Inc.; #sc-40), 1:500; anti-ubiquitin (Santa Cruz Biotechnology, Inc.; #sc-8017), 1:500; anti-K48- and anti-K63-ubiquitin antibodies, which recognize only K48 or K63 polyubiquitin chains, as described previously (18), 1:400; and goat anti-mouse-HRP (Sigma-Aldrich; #A5278) secondary antibody, 1:5,000.
Multiple high-molecular-weight forms of Rep could be directly demonstrated by immunoblotting, using anti-Rep antibody following immunoprecipitation of Rep, when highly sensitive detection methods were used. 293 cells were transfected with Flag-tagged ubiquitin and E4orf6 in the presence of MG132 and in either the presence (Fig. 1C, lanes 2 to 6) or absence (Fig. 1C, lane 1) of AAV5 Rep52/40. Following immunoprecipitation as performed for the experiments shown in Fig. 1B, samples were either blotted with anti-Rep antibody (Fig. 1C, left panel) and exposed with high-sensitivity Femto reagent (#34095; Thermo Scientific, Rockford, IL) or blotted with anti-ubiquitin antibody (Fig. 1C, right panel) and exposed with Pico luminol reagent (#34080; Thermo Scientific, Rockford, IL), per the manufacturer's instructions. In addition, to confirm direct conjugation of ubiquitin to Rep, Rep/antibody-conjugated beads were washed under conditions of increasing stringency, predicted to disrupt nonspecific interactions between Rep and other, potentially ubiquitinated, cellular proteins. Higher-molecular-weight forms detected by both anti-Rep and anti-ubiquitin antibodies were similarly resistant to two 1-ml washes each with RIPA buffer alone (as done for Fig. 1A and 1B) (shown here in Fig. 1C, lanes 2) or RIPA buffer containing 0.2% SDS (Fig. 1C, lanes 3), 0.5 M NaCl (Fig. 1C, lanes 4), and 1 M LiCl (Fig. 1C, lanes 5) or following five 1-ml washes with RIPA buffer containing 1 M LiCl (Fig. 1C, lanes 6). These results demonstrate that the high-molecular-weight protein complexes detected in Fig. 1A to C are comprised of ubiquitinated, high-molecular-weight isoforms of AAV5 Rep proteins. RIPA buffer washes were thus deemed adequate for subsequent experiments.
In addition, high levels of high-molecular-weight ubiquitinated isoforms of AAV2 Rep52—expressed from a CMV-AAV2Rep52(HA [hemagglutinin]) plasmid previously described (16)—could be detected in the presence or absence of E4orf6 in 293 cells (data not shown).
AAV5 small Rep proteins exhibit different E4orf6-dependent and -independent patterns of polyubiquitination in 293 cells.
Two important types of polyubiquitination include conjugation through ubiquitin lysine 48 (K48), which is associated with proteasome-mediated protein degradation, and conjugation through ubiquitin lysine 63 (K63), a less-well-characterized nonproteolytic modification associated with effects on protein characteristics such as trafficking and protein-protein interaction (5, 11, 12, 20, 28). Because we have observed ubiquitination of Rep52/40 both in the absence and in the presence of E4orf6, we investigated whether the small Rep proteins might undergo more than one type of ubiquitination in 293 cells, perhaps dependent upon the presence of E4orf6.
For these experiments, we utilized vectors expressing two specific HA-tagged ubiquitin mutants, as has been done by others in other systems (6, 14, 15, 19). Each of the ubiquitins expressed by these vectors contains multiple mutations in which all the ubiquitin lysines were changed to arginine, except either the lysine at position 48 (K48-HA Ub) or that at position 63 (K63-HA Ub), thus allowing visualization of these specific types of polyubiquitination. As expected, substantial levels of the degradation-associated K48-specific ubiquitination of Myc-tagged Rep52/40 were detected in the presence of E4orf6, with higher levels observable with added MG132 (Fig. 2A, left panel). Significantly lower levels of K48 ubiquitination were detected in the absence of E4orf6 (Fig. 2A, left panel, compare lanes 3 through 5). Interestingly, substantial levels of K63-specific ubiquitination of Rep52/40 were detected in the absence (and presence) of E4orf6 (Fig. 2A, right panel, compare lanes 3 and 5). Expression of an HA-tagged K29-specific ubiquitin mutant did not result in significant levels of K29-specific polyubiquitination of the small Rep proteins (data not shown).
The recent availability of antibodies that specifically recognize K48- and K63-linked polyubiquitin chains (gift from Genentech, Inc. [18]) has allowed us to confirm that both K48- and K63-linked polyubiquitination of AAV5 Rep proteins occurs during AAV5-Ad coinfection. Immunoblots using these antibodies demonstrated significant levels of both K48-linked (Fig. 2B, center panel, lanes 3 and 4) and K63-linked (Fig. 2B, right panel, lanes 3 and 4) polyubiquitinated isoforms of AAV5 Rep proteins during coinfection. These results demonstrate that the AAV2 and AAV5 Rep52/40 proteins underwent at least two forms of polyubiquitination during transient transfection as well as coinfection with Ad5.
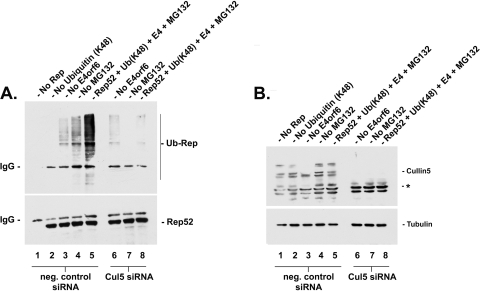
The levels of K48-linked polyubiquitinated forms of AAV5 small Rep proteins are significantly reduced in the presence of siRNA against cullin 5.
We have previously demonstrated that the addition of siRNAs to cullin 5 was capable of preventing degradation of AAV5 capsid proteins in the presence of E4orf6 and E1b-55k (16). To determine whether cullin 5 participates in the Ad E4orf6/E1b-55k ubiquitin ligase that specifically targets AAV5 small Rep proteins for K48-linked polyubiquitination, we pretreated 293 cells with two applications of siRNAs against cullin 5 (cocktail of #SI00052360, #SI00052367, #SI00052374, #SI00052381, or negative control #SI03650325; Qiagen, Inc., Valencia, CA) as per the manufacturer's protocols. In support of our previous data, the depletion of cullin 5 by siRNAs against cullin 5 (Fig. 3A and B, lanes 6 to 8), and not the negative control siRNA (Fig. 3A and B, lanes 1 to 5), significantly decreased the levels of K48-linked polyubiquitinated AAV5 small Rep proteins during transient transfection with the HA-tagged K48-specific ubiquitin construct in the presence of E4orf6 and E1b-55k (Fig. 3A, compare lanes 5 and 8). This data confirmed that the Ad5 E4orf6/E1b-55k/Cul5 E3 ubiquitin ligase complex specifically targets AAV proteins for proteasomal degradation by the addition of K48-linked polyubiquitin chains. We were unable detect a significant difference in the (low) levels of E4orf6-independent K48-linked polyubiquitination of Rep in the presence of siRNAs against cullin 5 (Fig. 3A, compare lanes 3 and 6), indicating that there may be an additional cellular E3 ubiquitin ligase responsible for a basal level of K48-linked ubiquitination of AAV5 small Rep proteins. Experiments regarding the effects of Cul5 siRNA on the ubiquitination of the large Rep proteins during infection are currently ongoing.
FIG. 3.
The levels of K48-linked polyubiquitinated forms of AAV5 small Rep proteins are significantly reduced in the presence of siRNA against cullin 5. Shown is Western blot analysis of AAV5 small Rep protein K48-specific polyubiquitination in 293 cells in the presence of cullin 5 siRNA (lanes 6 to 8) or negative control siRNA (lanes 1 to 5) during transient transfection. Lysates were isolated 36 to 42 h posttransfection with vectors expressing myc-tagged AAV5 small Rep proteins plus a construct expressing E4orf6 and another expressing HA-tagged K48-specific ubiquitin in the presence of MG132 or equivalent amounts of DMSO. Cells were transfected with each of the constructs, except where noted: no Rep (lane 1); no HA-ubiquitin (lane 2); no E4orf6 (lanes 3 and 6); and no MG132 (lanes 4 and 7). (A) Post-IP Western blot following immunoprecipitation with an anti-Myc antibody, with an immunoblot using an anti-Myc (bottom) or anti-HA (top) antibody. The accumulation (or lack thereof) of polyubiquitinated isoforms of AAV5 small Rep proteins is indicated by the presence of high-molecular-weight laddering or a smearing effect. (B) Pre-IP immunoblot of same samples as in panel A with anti-tubulin and anti-cullin 5 antibodies, showing depletion of cullin 5. The bands marked with an asterisk (*) are nonspecific and of unknown origin. Antibody dilutions were as follows: anti-HA (Sigma-Aldrich; #H9658), 1:5,000; anti-cullin 5 (Santa Cruz Biotechnology, Inc.; #sc-13014), 1:500; anti-c-Myc (Santa Cruz Biotechnology, Inc.; #sc-40), 1:500; goat anti-mouse-HRP (Sigma-Aldrich; #A5278); and goat anti-rabbit-HRP (Sigma-Aldrich; #A9169) secondary antibodies, 1:5,000.
AAV5 small Rep proteins could be coimmunoprecipitated with Ad5 E1b-55k in the presence or absence of E4orf6 (data not shown). This finding supported previous studies suggesting that E1b-55k interacts with its protein substrate and then acts as a chaperone to bring the substrate to the rest of the Cul5 E3 ubiquitin ligase complex, where it and the substrate (in this case, Rep52) are anchored to the ubiquitin ligase through E4orf6 and its interactions with the elongins B and C (2). It is unknown at this time whether Rep/Cul5 complexes formed in the absence of E4orf6 also contain other components normally associated with the Ad5 E3 ubiquitin ligase complex. Experiments more fully characterizing the composition of this complex are currently under way.
Degradation via K48-linked polyubiquitination of the AAV small Rep proteins by E4orf6 and E1b-55k may seem counterintuitive, given the well-established requirement of E4orf6 and E1b-55k for AAV replication (9, 24, 30, 31). It may be that E4orf6/E1b-55k-dependent degradation of AAV5 proteins is a secondary effect of its role in targeting the degradation of cellular proteins required for viral replication. We have previously shown that the enhancement of translation provided by adenovirus VA RNA as part of its helper function is necessary to restore AAV5 protein to levels necessary for viral replication (17); however, this scenario would predict that only the required levels of AAV5 proteins, and not a cellular target whose degradation is required for viral replication, become restored by VA RNA activity. In addition, we have found that AAV5 protein degradation targeted by the Ad5 E3 ubiquitin ligase complex is reduced in the presence of high levels of AAV protein. This suggests another possible consequence of AAV ubiquitination: that the cellular degradative machinery (either components of the E3 ubiquitin ligase or the proteasome) could be saturated or overwhelmed by AAV protein during infection such that efficient degradation of certain cellular targets could not be achieved (16). Alternatively, because AAV replication represses Ad5 replication during coinfection (29), perhaps degradation of AAV Rep proteins as targeted by Ad E1b-55k and E4orf6 has evolved as a defense mechanism by Ad5 for the maintenance of Ad5 replicative fitness in the presence of AAV coinfection.
In this report, we also identify ubiquitination of the small Rep proteins in a manner not associated with protein degradation. K63-linked polyubiquitination modulates the function of a number of cellular proteins. It has been implicated in governing protein localization, trafficking, protein-protein interactions, and other functions (5, 11, 12, 20, 28). It thus may be that the polyubiquitination of AAV small Rep proteins via K63 conjugation is required for its function as well. Additionally, work in other systems has suggested that the same lysine on a protein substrate may be differentially polyubiquitinated by K48-linked and K63-linked polyubiquitin chains as a method of controlling the levels of the protein, depending on the cellular conditions (32). Whatever its role in the function or stability of the structural and nonstructural proteins, polyubiquitination of AAV Rep proteins is likely an important facet of AAV biology.
Acknowledgments
We thank Lisa Burger for excellent technical help and Ramnath Nayak, Mark Hannink, Mick Petris, and Rick Sachdev for advice and reagents.
This work was supported by PHS grants AI46458 and AI56310 from NIAID to D.J.P.
Footnotes
Published ahead of print on 4 November 2009.
REFERENCES
- 1.Baker, A., K. J. Rohleder, L. A. Hanakahi, and G. Ketner. 2007. Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J. Virol. 81:7034-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanchette, P., C. Y. Cheng, Q. Yan, G. Ketner, D. A. Ornelles, T. Dobner, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2004. Both BC-box motifs of adenovirus protein E4orf6 are required to efficiently assemble an E3 ligase complex that degrades p53. Mol. Cell. Biol. 24:9619-9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanchette, P., K. Kindsmuller, P. Groitl, F. Dallaire, T. Speiseder, P. E. Branton, and T. Dobner. 2008. Control of mRNA export by adenovirus E4orf6 and E1B55K proteins during productive infection requires E4orf6 ubiquitin ligase activity. J. Virol. 82:2642-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowles, D. E., J. E. Rabinowitz, and R. J. Samulski. 2006. The genus Dependovirus. Hodder Arnold, London, United Kingdom.
- 5.Buchberger, A. 2002. From UBA to UBX: new words in the ubiquitin vocabulary. Trends Cell Biol. 12:216-221. [DOI] [PubMed] [Google Scholar]
- 6.Conze, D. B., C. J. Wu, J. A. Thomas, A. Landstrom, and J. D. Ashwell. 2008. Lys63-linked polyubiquitination of IRAK-1 is required for interleukin-1 receptor- and Toll-like receptor-mediated NF-kappaB activation. Mol. Cell. Biol. 28:3538-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dallaire, F., P. Blanchette, P. Groitl, T. Dobner, and P. E. Branton. 2009. Identification of integrin alpha3 as a new substrate of the adenovirus E4orf6/E1B 55-kilodalton E3 ubiquitin ligase complex. J. Virol. 83:5329-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan, D., P. Sharma, L. Dudus, Y. Zhang, S. Sanlioglu, Z. Yan, Y. Yue, Y. Ye, R. Lester, J. Yang, K. J. Fisher, and J. F. Engelhardt. 1999. Formation of adeno-associated virus circular genomes is differentially regulated by adenovirus E4 ORF6 and E2a gene expression. J. Virol. 73:161-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrari, F. K., T. Samulski, T. Shenk, and R. J. Samulski. 1996. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J. Virol. 70:3227-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez, R. A., and S. J. Flint. 2002. Effects of mutations in the adenoviral E1B 55-kilodalton protein coding sequence on viral late mRNA metabolism. J. Virol. 76:4507-4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hicke, L., and R. Dunn. 2003. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 19:141-172. [DOI] [PubMed] [Google Scholar]
- 12.Hochstrasser, M. 2000. Evolution and function of ubiquitin-like protein-conjugation systems. Nat. Cell Biol. 2:E153-E157. [DOI] [PubMed] [Google Scholar]
- 13.Ley, R., K. Balmanno, K. Hadfield, C. Weston, and S. J. Cook. 2003. Activation of the ERK1/2 signaling pathway promotes phosphorylation and proteasome-dependent degradation of the BH3-only protein, Bim. J. Biol. Chem. 278:18811-18816. [DOI] [PubMed] [Google Scholar]
- 14.Lim, K. L., K. C. Chew, J. M. Tan, C. Wang, K. K. Chung, Y. Zhang, Y. Tanaka, W. Smith, S. Engelender, C. A. Ross, V. L. Dawson, and T. M. Dawson. 2005. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J. Neurosci. 25:2002-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim, K. L., V. L. Dawson, and T. M. Dawson. 2006. Parkin-mediated lysine 63-linked polyubiquitination: a link to protein inclusions formation in Parkinson's and other conformational diseases? Neurobiol. Aging 27:524-529. [DOI] [PubMed] [Google Scholar]
- 16.Nayak, R., K. D. Farris, and D. J. Pintel. 2008. E4Orf6-E1B-55k-dependent degradation of de novo-generated adeno-associated virus type 5 Rep52 and capsid proteins employs a cullin 5-containing E3 ligase complex. J. Virol. 82:3803-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nayak, R., and D. J. Pintel. 2007. Positive and negative effects of adenovirus type 5 helper functions on adeno-associated virus type 5 (AAV5) protein accumulation govern AAV5 virus production. J. Virol. 81:2205-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newton, K., M. L. Matsumoto, I. E. Wertz, D. S. Kirkpatrick, J. R. Lill, J. Tan, D. Dugger, N. Gordon, S. S. Sidhu, F. A. Fellouse, L. Komuves, D. M. French, R. E. Ferrando, C. Lam, D. Compaan, C. Yu, I. Bosanac, S. G. Hymowitz, R. F. Kelley, and V. M. Dixit. 2008. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell 134:668-678. [DOI] [PubMed] [Google Scholar]
- 19.Olzmann, J. A., L. Li, M. V. Chudaev, J. Chen, F. A. Perez, R. D. Palmiter, and L. S. Chin. 2007. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J. Cell Biol. 178:1025-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pickart, C. M. 2004. Back to the future with ubiquitin. Cell 116:181-190. [DOI] [PubMed] [Google Scholar]
- 21.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Querido, E., R. C. Marcellus, A. Lai, R. Charbonneau, J. G. Teodoro, G. Ketner, and P. E. Branton. 1997. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J. Virol. 71:3788-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samulski, R. J., and T. Shenk. 1988. Adenovirus E1B 55-Mr polypeptide facilitates timely cytoplasmic accumulation of adeno-associated virus mRNAs. J. Virol. 62:206-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwartz, R. A., J. A. Palacios, G. D. Cassell, S. Adam, M. Giacca, and M. D. Weitzman. 2007. The Mre11/Rad50/Nbs1 complex limits adeno-associated virus transduction and replication. J. Virol. 81:12936-12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi, D., M. S. Pop, R. Kulikov, I. M. Love, A. L. Kung, and S. R. Grossman. 2009. CBP and p300 are cytoplasmic E4 polyubiquitin ligases for p53. Proc. Natl. Acad. Sci. U. S. A. 106:16275-16280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sourisseau, T., C. Desbois, L. Debure, D. D. Bowtell, A. C. Cato, J. Schneikert, E. Moyse, and D. Michel. 2001. Alteration of the stability of Bag-1 protein in the control of olfactory neuronal apoptosis. J. Cell Sci. 114:1409-1416. [DOI] [PubMed] [Google Scholar]
- 27.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348-352. [DOI] [PubMed] [Google Scholar]
- 28.Sun, L., and Z. J. Chen. 2004. The novel functions of ubiquitination in signaling. Curr. Opin. Cell Biol. 16:119-126. [DOI] [PubMed] [Google Scholar]
- 29.Timpe, J. M., K. C. Verrill, and J. P. Trempe. 2006. Effects of adeno-associated virus on adenovirus replication and gene expression during coinfection. J. Virol. 80:7807-7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ward, P. 2006. Replication of adeno-associated virus DNA. Hodder Arnold, London, United Kingdom.
- 31.Weitzman, M. D. 2006. The parvovirus life cycle: an introduction to molecular interactions important for infection. Hodder Arnold, London, United Kingdom.
- 32.Wertz, I. E., K. M. O'Rourke, H. Zhou, M. Eby, L. Aravind, S. Seshagiri, P. Wu, C. Wiesmann, R. Baker, D. L. Boone, A. Ma, E. V. Koonin, and V. M. Dixit. 2004. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 430:694-699. [DOI] [PubMed] [Google Scholar]
- 33.Weston, C. R., K. Balmanno, C. Chalmers, K. Hadfield, S. A. Molton, R. Ley, E. F. Wagner, and S. J. Cook. 2003. Activation of ERK1/2 by deltaRaf-1:ER* represses Bim expression independently of the JNK or PI3K pathways. Oncogene 22:1281-1293. [DOI] [PubMed] [Google Scholar]
- 34.Woo, J. L., and A. J. Berk. 2007. Adenovirus ubiquitin-protein ligase stimulates viral late mRNA nuclear export. J. Virol. 81:575-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yew, P. R., and A. J. Berk. 1992. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature 357:82-85. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, D. D., S. C. Lo, J. V. Cross, D. J. Templeton, and M. Hannink. 2004. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 24:10941-10953. [DOI] [PMC free article] [PubMed] [Google Scholar]