Abstract
Mokola virus (MOKV) is a nonsegmented, negative-sense RNA virus that belongs to the Lyssavirus genus and Rhabdoviridae family. MOKV phosphoprotein P is an essential component of the replication and transcription complex and acts as a cofactor for the viral RNA-dependent RNA polymerase. P recruits the viral polymerase to the nucleoprotein-bound viral RNA (N-RNA) via an interaction between its C-terminal domain and the N-RNA complex. Here we present a structure for this domain of MOKV P, obtained by expression of full-length P in Escherichia coli, which was subsequently truncated during crystallization. The structure has a high degree of homology with P of rabies virus, another member of Lyssavirus genus, and to a lesser degree with P of vesicular stomatitis virus (VSV), a member of the related Vesiculovirus genus. In addition, analysis of the crystal packing of this domain reveals a potential binding site for the nucleoprotein N. Using both site-directed mutagenesis and yeast two-hybrid experiments to measure P-N interaction, we have determined the relative roles of key amino acids involved in this interaction to map the region of P that binds N. This analysis also reveals a structural relationship between the N-RNA binding domain of the P proteins of the Rhabdoviridae and the Paramyxoviridae.
Like rabies virus (RABV), Mokola virus (MOKV) belongs to the Lyssavirus genus and the Rhabdoviridae family. Lyssaviruses often cause lethal encephalitis and are a significant global health risk. RABV alone is responsible for an estimated 55,000 deaths a year, primarily in poor rural areas in Africa and Asia (26), in spite of the availability of effective vaccines. In addition, poor surveillance and the emergence of novel variants that appear to be insensitive to traditional rabies vaccines (such as MOKV) mean that the true risk that these viruses pose remains underappreciated (44). Lyssaviruses belong to the order Mononegavirales and have a nonsegmented, negative-sense (NNS) RNA genome that contains five common genes in the following order: N (nucleoprotein), P (phosphoprotein), M (matrix protein), G (glycoprotein), and L (large protein) (9). During all stages of the virus life cycle, their RNA genome and RNA antigenome are encapsidated by N, which forms a large helical polymer known as the nucleocapsid (13, 22). The replication/transcription complex is formed by L, P, and N. The L protein harbors the RNA-dependent RNA polymerase (RdRP) catalytic activity but also caps and polyadenylates the viral mRNA as shown for vesicular stomatitis virus (VSV) (1, 33). The P protein fulfils a crucial role during RNA synthesis, as it is an essential cofactor for the RdRP activity of L. P contains a dimerization domain located between residues 91 and 131 (RABV P numbering) (15, 16, 23). Both structural and biochemical analyses suggest that RABV P forms elongated dimers in solution, which is in line with other evidence suggesting an important role for dimerization in replication (15). In addition, P binds two distinct forms of N, i.e., No and N-RNA; No refers to a soluble form of N that is not associated with RNA molecules, and N-RNA corresponds to the multimerized form of N associated with genomic RNA. There is evidence that N° interacts with two distinct regions of P located at the N terminus (RABV P residues 4 to 40) (7, 31) and C terminus (RABV P residues 185 to 297) of the phosphoprotein (7, 14). In contrast, only the C-terminal domain of P interacts with N when it is bound to genomic RNA in the nucleocapsid (16). The lyssavirus phosphoprotein is also phosphorylated (20). Experimental evidence obtained for VSV further suggests that the differential phosphorylation of the N and C termini of P may be involved in regulating viral transcription and replication (8). Conversely, N is also phosphorylated, but the exact role of phosphorylation of these two proteins in regulating their binding remains controversial (29, 45). Binding of P to N-RNA probably involves the C-terminal region of N (RABV N residues 376 to 450) (38). A recent model proposes that during replication the L protein forms a complex with P, which in turn binds to the N-RNA polymer and acts as a bridge to allow access of L to the RNA. The model further suggests that the P-N0 complex may bind to the replicating L-P-N-RNA complex and feed the newly formed RNA strand with uncomplexed N for immediate encapsidation (1). However, it is important to note that many of the details of the interactions between L, P, and N in the context of the replication or transcription complexes are not well understood. This is due primarily to the lack of high-resolution structural data about these proteins and their complexes. To date the structures of the VSV and RABV N-RNAs have been determined (2, 19), as well as individual domains of the VSV and RABV P proteins (specifically the dimerization domain of VSV P and the N-RNA binding domains of VSV and RABV P) (10, 30, 36). Very recently the structure of VSV N-RNA complexed with the C-terminal domain of VSV P has been determined, which reveals that P binds in the cleft between two adjacent N molecules in the nucleocapsid (18) primarily via helix α4 and residues in β2. In contrast, no high-resolution structural data are available for L. The structure of the RABV P N-RNA binding domain in combination with previous studies of P-N complexes has led to a tentative model for the interaction between RABV N-RNA and P (1, 37). However, the lack of structural data means that the exact nature of the interactions remains uncertain.
Because the functional analysis of P in the L-P-N-RNA complex remains incomplete, we crystallized full-length MOKV P to better define the structure of this protein. Here we present a structure of the C-terminal domain of MOKV P, which is involved in the interaction with N, obtained by expression of full-length P in Escherichia coli, which was subsequently truncated during crystallization. The structure shows a high degree of homology with the RABV P. The details of the contacts between molecules of P in the crystal reveal that the C-terminal acidic tail specific to MOKV P and absent in RABV P packs against a positively charged surface area of P that may represent the N-RNA binding site. On the basis of this structure, we tested a number of amino acid residues potentially involved in P-N-RNA interaction, by yeast two-hybrid analysis. Using this strategy, we have been able to define a number of residues in the C-terminal domain of P, mutations of which block binding to N.
MATERIALS AND METHODS
Cloning.
For in-fusion cloning, the cDNA of the gene encoding the phosphoprotein of Mokola virus was obtained by reverse transcription-PCR (RT-PCR) from total RNA isolated from infected BSR cells (a clone of BHK-21) using hexamer primers (Roche Boehringer). PCR was carried out with the primers 5′-AGGAGATATACCATGAGCAAAGATTTGGTGCATCCTAGTC-3′ and 5′-GTGATGGTGATGTTTCTCTGCCTCCTCGAGCCGGGC-3′, where the gene-specific sequence is indicated in bold. The PCR product was cloned into pOPINE using the in-fusion cloning methodology (6), yielding a full P construct with a C-terminal 6×His tag.
For cloning of the target and bait proteins for the yeast two-hybrid experiments the same cDNA was also used to amplify by PCR the DNA sequence encoding full-length N and PΔ176, which corresponds to P deleted of the first 176 amino acids. The following pairs of primers were used: 5′-GGGGACAACTTTGTACAAAAAAGTTGGCATGGAGTCTGACAAGATTGTG-3′ and 5′-GGGGACAACTTTGTACAAGAAAGTTGGTTATGATCCATCTGCATACAC-3′ for N and 5′-GGGGACAACTTTGTACAAAAAAGTTGGCATGCACACACCCGATCACGATG-3′ and 5′-GGGGACAACTTTGTACAAGAAAGTTGGTTACTCTGCCTCCTCGAGC-3′ for PΔ176. The gene-specific sequence is indicated in bold. PCR products were cloned into pDONR207 (Invitrogen) using Gateway.
Site-directed mutagenesis was performed with the QuikChange site-directed mutagenesis kit (Stratagene) following the manufacturer's instructions, using PΔ176 cloned into pDON207 as a template for PCR. Primers used for site-directed mutagenesis were the following: PΔ176(F210A), 5′-GCCCATCAGGTAGCAGAAAGCGCTTCAAAGAAATACAAGTTCCC-3′ and 5′-GGGAACTTGTATTTCTTTGAAGCGCTTTCTGCTACCTGATGGGC-3′; PΔ176(S211A), 5′-CATCAGGTAGCAGAAAGCTTTGCAAAGAAATACAAGTTCCCTT-3′ and 5′-AAGGGAACTTGTATTTCTTTGCAAAGCTTTCTGCTACCTGATG-3′; PΔ176(K212E), 5′-CATCAGGTAGCAGAAAGCTTTTCAGAGAAATACAAGTTCCCTT-3′ and 5′-AAGGGAACTTGTATTTCTCTGAAAAGCTTTCTGCTACCTGATG-3′; PΔ176(K212A), 5′-CATCAGGTAGCAGAAAGCTTTTCAGCGAAATACAAGTTCCCTTCTAGATC-3′ and 5′-GATCTAGAAGGGAACTTGTATTTCGCTGAAAAGCTTTCTGCTACCTGATG-3′; PΔ176(K213E), 5′-GGTAGCAGAAAGCTTTTCAAAGGAATACAAGTTCCCTTCTAGATC-3′ and 5′-GATCTAGAAGGGAACTTGTATTCCTTTGAAAAGCTTTCTGCTACC-3′; PΔ176(K213A), 5′-GGTAGCAGAAAGCTTTTCAAAGGCATACAAGTTCCCTTCTAGATCC-3′ and 5′-GGATCTAGAAGGGAACTTGTATGCCTTTGAAAAGCTTTCTGCTACC-3′; PΔ176(Y214E), 5′-CAGGTAGCAGAAAGCTTTTCAAAGAAAGAAAAGTTCCCTTCTAGATCCTC-3′ and 5′-GAGGATCTAGAAGGGAACTTTTCTTTCTTTGAAAAGCTTTCTGCTACCTG-3′; PΔ176(Y214A), 5′-CCATCAGGTAGCAGAAAGCTTTTCAAAGAAAGCCAAGTTCCCTTCTAGAT-3′ and 5′-ATCTAGAAGGGAACTTGGCTTTCTTTGAAAAGCTTTCTGCTACCTGATGG-3′; PΔ176(K215E), 5′-AGCAGAAAGCTTTTCAAAGAAATACGAGTTCCCTTCTAGATCC-3′ and 5′-GGATCTAGAAGGGAACTCGTATTTCTTTGAAAAGCTTTCTGCT-3′; PΔ176(K215A), 5′-GGTAGCAGAAAGCTTTTCAAAGAAATACGCGTTCCCTTCTAGATCCTCG-3′ and 5′-CGAGGATCTAGAAGGGAACGCGTATTTCTTTGAAAAGCTTTCTGCTACC-3′; PΔ176(I223E), 5′-CCCTTCTAGATCCTCGGGAGAATTCTTGTGGAACTTTGAG-3′ and 5′-CTCAAAGTTCCACAAGAATTCTCCCGAGGATCTAGAAGGG-3′; PΔ176(L225E), 5′-CTAGATCCTCGGGAATATTCGAGTGGAACTTTGAGCAGCTTA-3′ and 5′-TAAGCTGCTCAAAGTTCCACTCGAATATTCCCGAGGATCTAG-3′; and PΔ176(Y295F), 5′-CCAAGAAGATATCAACAGTTTCATGGCCCGGCTCG-3′ and 5′-CGAGCCGGGCCATGAAACTGTTGATATCTTCTTGG-3′.
Expression and purification.
The P protein was expressed and purified using standard methods as described previously (35). Briefly, E. coli expressing MOKV P was grown in 0.5 L of TB Overnight Express (Novagen) for 5 to 6 h at 37°C followed by 20 h at 25°C. The cell pellet was harvested and frozen at −80°C. For protein purification, the pellet was defrosted on ice and resuspended in buffer A (25 mM Tris [pH 7.5], 500 mM NaCl, 30 mM imidazole) with 0.1% Tween 20, protease inhibitors, and DNase I added. The suspension was passed through a cell disruptor at 30 kpsi and then centrifuged for 30 min at 30,000 × g at 4°C. The soluble fraction was transferred to an Aktä Express equipped with an Ni-nitrilotriacetic acid (NTA) column connected in-line to a Hiload 16/60 Superdex 75 gel filtration column (GE Healthcare). The Ni-NTA column was washed using buffer A, eluted using buffer B (25 mM Tris [pH 7.5], 250 mM NaCl, 250 mM imidazole) before transfer to the gel filtration column equilibrated in buffer C (20 mM Tris [pH 7.5], 200 mM NaCl). Peak fraction collection was performed using the Aktä Express software. Peak fractions were pooled, and the protein was concentrated to 12 mg/ml. The integrity of the protein was verified by electrospray ionization (ESI) mass spectroscopy as described previously (35).
Crystallization, data collection, and structure determination.
The concentrated protein was subjected to the OPPF automated sitting-drop vapor diffusion crystallization pipeline as described previously (48). Crystallization trials were attempted at 20.5°C in sitting drops containing 100 nl protein and 100 nl precipitant solution equilibrated against 95-μl reservoirs in 96-well plates. In total, 576 conditions were tested. Diffraction-quality crystals appeared after 15 days in a condition containing 30% (vol/vol) pentaerythritol ethoxylate (15/4 EO/OH), 50 mM ammonium sulfate, and 100 mM bis-Tris, pH 6.5 (Hampton Index screen).
Crystals were cryoprotected by brief immersion in reservoir solution supplemented with 20% (vol/vol) glycerol before flash-cryocooling in a stream of cold (100 K) nitrogen gas. The diffraction quality of the crystal was significantly improved by annealing, i.e., blocking the cryogenic gas stream with a credit card for 5 s (3). Diffraction data were recorded from a single crystal at the I03 Diamond beamline (λ = 0.890 Å) on a Quantum 315 detector (Area Detector Systems Corporation) and processed using HKL2000. The structure was solved by molecular replacement using the structure of the RABV P N-RNA binding domain (Protein Data Bank [PDB] accession no. 1VYI) with Molrep (46) and TLS+ restrained refinement at 2.1 Å (0.21 nm) completed using REFMAC (32). Refinement statistics are shown in Table 1. Structure-based superpositions were performed using SHP (40), and sequence alignments carried out with ClustalX (43) or MUSCLE (12). Graphics were produced using PYMOL (http://www.pymol.org/).
TABLE 1.
X-ray data collection and refinement statistics
| Parameter | Value |
|---|---|
| Data collection | |
| X-ray source | Diamond I03 |
| Wavelength (Å) | 0.890 |
| Space group | P212121 |
| Unit cell (Å) | a = 25.84, b = 48.87, c = 85.95 |
| Resolution range (Å) | 30.0-2.10 (2.18-2.10) |
| Unique reflections | 6,812 (653) |
| Completeness (%) | 99.6 (97.9) |
| Redundancy | 5.5 (3.3) |
| Avg I/σI | 9.9 (1.7) |
| Rmerge | 0.173 (0.585) |
| Refinement statistics | |
| Resolution range (Å) | 30.0-2.10 (2.18-2.10) |
| No. of reflections (working/test) | 6,454/325 |
| R factora (Rwork/Rfree) | 0.220/0.266 |
| No. of atoms (protein/water) | 851/59 |
| Bond length RMSD (Å) | 0.007 |
| Bond angle RMSD (°) | 0.9 |
| Mean B factor (protein/water; Å2) | 17/32 |
Rwork and Rfree are defined by R = Σhkl¦¦Fobs ¦ − ¦Fcalc¦¦/Σhkl ¦Fobs¦, where h, k, and l are the indices of the reflections (used in refinement for Rwork; 5%, not used in refinement, for Rfree) and Fobs and Fcalc are the structure factors deduced from measured intensities and calculated from the model, respectively. Numbers in parentheses are for the highest-resolution shell.
Yeast two-hybrid procedure.
Yeast culture media were prepared as previously described (47). PΔ176 or the appropriate mutants were transferred by in vitro recombination from pDONR207 in a Gateway-compatible pDEST32 yeast expression vector (Invitrogen) to be fused with the DNA binding domain of Gal4 (Gal4DB). N protein was transferred from pDONR207 into pPC86 (Invitrogen) to be fused to the activation domain of Gal4 (Gal4AD). These constructs were transformed in yeast strain AH109 (Clontech). Yeast cells were plated on a selective medium lacking histidine and supplemented or not with the indicated amount of 3-aminotriazole (3-AT) (Sigma-Aldrich) to test the interaction-dependent transactivation of the HIS3 reporter gene.
Protein structure accession number.
The final refined coordinates and structure factors have been deposited with the PDB under accession number 2wzl.
RESULTS
Crystallization of MOKV P and structure determination.
We expressed full-length MOKV P in E. coli and, following purification, subjected it to nanodrop crystallization trials. Diffraction-quality crystals formed within 15 days, and the 2.1-Å structure was determined by molecular replacement using the structure of the RABV P N-RNA binding domain (PDB accession no. 1VYI [30]) and refined with residuals Rwork/Rfree of 0.220/0.266. The crystal structure belongs to space group P212121, with cell dimensions a = 25.8 Å, b = 48.9 Å, and c = 86.0 Å, and reveals one copy of a truncated form of P in the asymmetric unit. The region of P crystallized is the C-terminal, N-RNA binding domain (residues 197 to 303), and the experimental electron density shows a sharp cutoff at residue 197, suggesting that this is where the truncation occurred.
The refined model for residues 197 to 303 is complete and has good stereochemistry (Table 1), with more than 90% of residues in the most favored region of the Ramachandran plot (28). The data show that P had been truncated during the crystallization process, as mass spectrometry analysis of the protein immediately before the crystallization trials showed the protein to be intact at that point (data not shown). It is interesting to note that the previously published structure of the rabies virus C-terminal domain of P was obtained by expression of full-length P in insect cells, which was subsequently also truncated during the crystallization process (30).
Analysis of MOKV P structure.
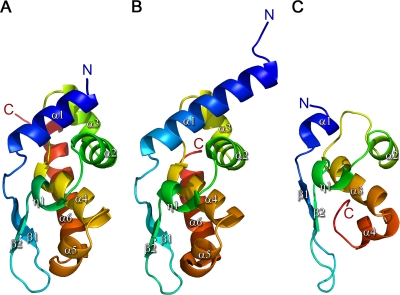
The structure of the MOKV P C-terminal domain is formed by six tightly folded α helices, a 310 helix, and a small β sheet consisting of two β strands (Fig. 1), with a morphology previously described as resembling a sliced pear, with a flat and curved face. It shows a high degree of structural homology with RABV P (PDB accession no. 1VYI) (Fig. 1B) (101 amino acids aligned with a root mean square deviation [RMSD] of 0.66 Å using SHP [40]), consistent with the high degree of sequence identity (68%). As the first residue visible in the electron density is residue 197, this means that the MOKV P structure is 10 amino acids shorter at the N terminus than the available RABV P structure (which starts at residue 187), yielding a shorter α1 helix. In addition, there are five extra residues at the C terminus compared to RABV P, leading to an extended α6 helix. Apart from the N- and C-terminal extremities of the P structures, the MOKV and RABV P proteins have very similar structures (Fig. 1) with similar surface charge properties. As observed for RABV P, a clear positively charged cluster is present on the round face of MOKV P, which is implicated in binding N (23), whereas for the flat face the charge is more evenly distributed. The so-called “W hole,” present on the flat face, which has been implicated in binding N (although no direct evidence exists to support this [30]), is formed by three residues (C262, F266, and I288 in MOKV P and C261, W265, and M287 in RABV P). In the Mokola virus structure, however, this hole is not as pronounced as it is in the RABV P structure, where W186, from a neighboring molecule, packs into the hole and displaces W265. The role of this hydrophobic region, if any, remains unclear.
FIG. 1.
Side-by-side comparison of rhabdovirus P structures: (A) MOKV P; (B) RABV P (PDB accession no. 1VYI); (C) VSV P (PDB accession no. 2K47). The structures are rainbow ramped from blue (N terminus) to red (C terminus).
There is also clear structural similarity to P of VSV, although the fragment of VSV P solved by nuclear magnetic resonance (NMR) is considerably smaller. In this case SHP aligns 57 residues out of 70 in VSV, with an RMSD of 2.73 Å, despite the aligned residues sharing less than 11% sequence identity.
Mapping the P-N binding site.
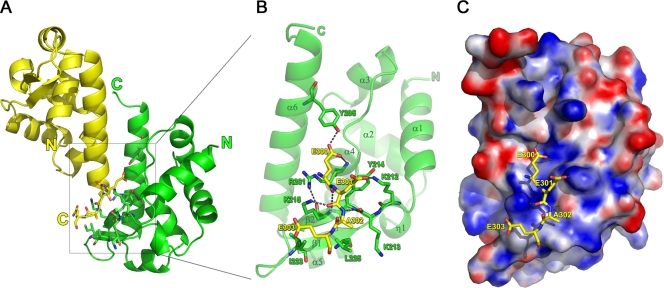
Analysis of the crystal structure of MOKV P using PISA (27) reveals that the major crystal packing interface involves the positively charged cluster implicated in N-RNA binding (23). Specifically, the interface is established primarily by the interactions of the C-terminal α6 helices of two symmetry-related monomers (symmetry operator: x−1/2,−y+3/2,−z), orientated in an inverted fashion relative to each other (Fig. 2A). The C-terminal four residues (E300, E301, A302, and E303) of one monomer (drawn in yellow in Fig. 2) interact, via a combination of both polar and hydrophobic contacts, with a positively charged region consisting of residues located on β1, β2, α4, and α6 (K212, K213, Y214, K215, I223, L225, R261, and Y295) in the other monomer (drawn in green in Fig. 2A and B). K212, K213, K215, R261, and Y295 engage in both main- and side-chain polar contacts with the acidic tail, with Y214, K215, I223, L225, and R261 contributing to additional hydrophobic interactions (Table 2). This interaction probably does not occur under physiological conditions because the interaction surface is small, with a total buried surface area between the two monomers of 757.4 Å2 and 392.3 Å2 contributed by the acidic tail. This is further supported by the observation that the acidic extension of MOKV P is poorly conserved among the lyssaviruses. A total of 384 GenBank sequences of P that encompass at least the motif described here and represent all the described genotypes of lyssaviruses were aligned using ClustalX (available on request). Of all lyssaviruses characterized to date, only MOKV (L-E-E/D-A-D/E), Lagos bat virus (L-E-E/D-A/V/I-D), and raccoon rabies virus (R-Q-L-N-L) P proteins contain a similar extension.
FIG. 2.
Details of the major crystal packing interaction interface. (A) View of the interaction between the two neighboring monomers. (B) Close-up of the interactions between the positively charged cluster located on one monomer (green) with the acidic C terminus of the second monomer (yellow). (C) The same view as in panel B but with the green monomer shown as an electrostatic surface representation, generated using the PDB2PQR server (using the AMBER force field [11]) in combination with APBS tools (4) and pymol (http://www.pymol.org/).
TABLE 2.
PISA analysis of the MOKV P-P interface
| Structure | Residue | Bondsa | No. of bonds | Accessible surface area (Å2) | Buried surface area (Å2) | ΔiG (kcal/M)b | Yeast two-hybrid resultsc |
|---|---|---|---|---|---|---|---|
| 1 | Lys 212 | HS | 1 | 113.9 | 35.5 | −0.72 | ++ |
| Lys 213 | H | 1 | 107.97 | 23.99 | −0.12 | − | |
| Tyr 214 | 36.74 | 27.25 | 0.44 | ++ | |||
| Lys 215 | HS | 3 | 111.52 | 84.8 | −1.07 | ++ | |
| Ile 223 | 109.32 | 28.72 | 0.46 | − | |||
| Leu 225 | 95.81 | 28.48 | 0.46 | + | |||
| Arg 261 | H | 1 | 55.12 | 46.99 | −0.64 | ++ | |
| Tyr 295 | H | 1 | 72.9 | 37.33 | 0.02 | − | |
| 2 | Glu 300 | H | 3 | 141.8 | 133.95 | −0.46 | ND |
| Glu 301 | HS | 2 | 104.71 | 56.01 | −0.1 | ND | |
| Ala 302 | H | 1 | 107.22 | 54.08 | 0.69 | ND | |
| Glu 303 | S | 1 | 215.04 | 62.45 | −-0.19 | ND |
H, hydrogen bond; S, salt bridge.
ΔiG, solvation energy gain upon formation of the interface.
−, no effect; +, weak effect; ++, strong effect; ND, not determined.
Strikingly, K212, K213, Y214, and K215 were previously identified as being involved in the interaction with N-RNA (23). Moreover, the acidic tail bears a striking resemblance to the acidic region in N (DEED in MOKV N versus EEAE in MOKV P) immediately following S389, whose phosphorylation status may modulate the interaction of N with P (45).
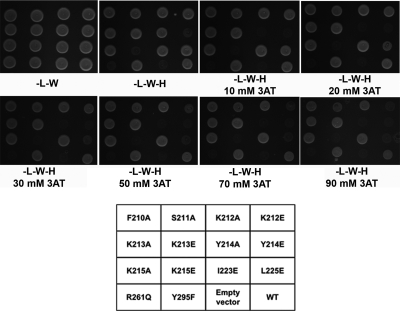
To precisely map which amino acid residues of the MOKV P domain are involved in this interaction with N, we mutated the eight amino acids (K212 to 215, I223, L225, R261, and Y295) implicated in the interaction with the acidic tail of P. F210 and S211, which were previously postulated to play a role in P-N interaction, were also tested (23). Yeast cells were transformed with Gal4DB-P and Gal4AD-N, and the absence of toxicity when expressing these two proteins was assessed on synthetic medium without leucine and tryptophan (−L−W). P-N interaction was monitored by measuring yeast growth on the same medium but without histidine (−L−W−H) and supplemented or not with 3-AT to increase stringency. The interaction between P and N was tested by yeast two-hybrid experiments between full-length MOKV N and truncated MOKV P lacking amino acids 1 to 176 (PΔ176) to remove both the P-N° interaction site in the N-terminal part of P and the dimerization domain (7, 16, 31). This approach is similar to the experiments conducted previously by Jacob et al. (23) but with the important difference that we introduced specific, single mutations whereas Jacob et al. used a random mutagenesis approach yielding constructs with multiple substitutions.
As shown in Fig. 3, mutations of Y214, K215, and R261 resulted in the most significant growth defects of the yeast strain, while for K212 and L225 higher-stringency conditions were necessary to reveal an effect on N binding. The number of bonds formed and the reduction in accessible surface area upon binding the acidic tail of each of the residues shows a good overall correspondence with the Y2H results (Table 2). These results suggest that these residues are involved in the interaction between MOKV P and N. However, it should be noted that it is distinctly possible that the Y214E mutation causes incorrect protein folding, given the packing of this aromatic side chain in the core of the molecule. Table 2 shows that the accessible surface area of Y214 in the unbound state is low (36.74Å2) and is reduced only slightly upon binding the acidic tail (27.25Å2), indicative of a weak interaction with the tail and burial of a significant portion into the core of the protein. Similarly, increasing the stringency by the addition of 3-AT revealed that the Y214A mutation did impair yeast growth, but whether this is due to the effects on P-N complex stability or destabilization of P remains to be seen.
FIG. 3.
Mapping the P-N interaction by yeast two-hybrid analysis. Yeast cells expressing Gal4 DNA binding domain (Gal4DB) fused to wild-type PΔ176 (WT) or the indicated mutants were cotransformed with a plasmid encoding the Gal4 transactivation domain (AD) alone (empty vector) or fused to N. Yeast cells were plated on selective medium lacking leucine and tryptophan to validate yeast growth after transformation (−L−W). Selective medium lacking histidine and supplemented with indicated concentrations of 3-aminotriazole (3-AT) were used to test the interaction-dependent transactivation of the HIS3 reporter gene.
For residues K212, K215, L225, and R261 the MOKV and RABV P structures show that the effects observed are not likely due to a defect in P folding but are likely due to disruption of the interaction with N. Figure 3 clearly reveals the importance of the electrostatic environment in the interaction, as mutations of these residues to glutamic acid had a much more substantial effect on disrupting the interaction with N than alanine substitution. This further supports the idea that the interaction observed for P in the crystal with its acidic tail mimics the interaction with N, as the crystal structure showed that the interaction with the acidic tail was largely electrostatically driven and involved primarily main-chain atoms of the acidic tail.
DISCUSSION
We have crystallized and solved the structure of the N-RNA binding domain of Mokola virus P via expression of full-length P which was subsequently truncated during the crystallization process. The MOKV P structure shows a high degree of structural similarity with the homologous domain of RABV P, consistent with the high degree of sequence identity (68%). Due to the longer sequence of MOKV P compared to RABV P, the C-terminal alpha helix (α6) is six residues longer than that of RABV P. Whether the MOKV P α1 in the intact protein is longer than observed here remains to be established, as the sequence prior to residue 197 is poorly conserved between the RABV and MOKV P proteins and secondary structure prediction methods do not predict the presence of a helix even for RABV P in this region.
Analysis of the crystal packing interactions revealed that a positively charged region of P, previously shown to be involved in the interaction with N, interacts with four acidic C-terminal amino acids in a symmetry-related P molecule that may resemble part of the putative P binding region in N. The interaction is electrostatically driven and, together with the small buried surface area, suggests that the interacting regions in P and/or N are likely to be larger than can be ascertained on the basis of the MOKV P crystal structure alone, consistent with the recent structure of VSV N-RNA in complex with P indicating that the opposing face of the domain in P, involving helices α4 and α5, is involved in the interaction with N (18, 37, 38). The involvement of the positively charged region in P in binding N was further confirmed by a combination of site-directed mutagenesis and yeast two-hybrid analysis to study the effects of the mutations on the interaction of MOKV P with MOKV N. The analysis revealed that out of 10 amino acids in P potentially involved in binding N, five negatively affected the P-N interaction. As previously demonstrated (23), we confirmed that K212, K215, and perhaps Y214 are involved in P-N binding. We further show that a positive charge at positions 212 and 215 is crucial for P-N interaction. Two other positions not previously described, L225 and R261, were also shown to be necessary for the P-N interaction to occur. However, our work suggests that the role of amino acids 210, 211, and 213, recognized as potentially involved in P-N binding, is less significant (or at least they contribute less to the interaction with N), as their mutation did not alter the interaction. This further reinforces the hypothesis of potential core packing destabilization (30) of some mutations when generated at random since, only a small number of the residues of P identified show here a clear effect on N binding in the yeast two-hybrid screen when tested in isolation. The high degree of conservation of these positions in lyssaviruses despite the high variability of this region of the P further supports their importance in the interaction with N. The observation that at all sites alanine substitutions are tolerated in the interaction with N indicates that the acidic tail-positive cluster interactions are probably weak and therefore mutations in this region may be compensated for by the presence of other interacting regions in P and/or N.
The region of N interacting with P probably involves residues in the vicinity of S389, as its phosphorylation state may be important for the interaction with P (45) and trypsin removal of RABV N residues 376 to 450 abolishes P binding (16, 38). S389 is followed by four acidic residues with a high degree of similarity with the acidic C-terminal extension of MOKV P (DEED in MOKV N versus EEAE in MOKV P). However, mutation of all four acidic residues in MOKV N (D390A, E391A, E392A, and D393A) did not affect binding of N to PΔ176 in a yeast two-hybrid screen (not shown). Another acidic region in N is also highly conserved among N proteins (ELEE in MOKV N, positions 371 to 374). Here again mutations E373A and E374A did not alter P-N binding in yeast two-hybrid experiments (not shown). One or both of these sites may still interact with P, since it has recently been shown that measles virus (MeV) N has two boxes of amino acid residues that are involved in P-N binding (5). However, MOKV N mutated at both sites (residues 373 to 374 and 390 to 393) still binds to MOKV PΔ176 (not shown). This lack of effect on binding of P by mutating individually or in combination the two acidic regions of N may reflect the fact that these regions are simply not involved in P-N binding or that the P binding region involves more than just the acidic residues mutated in this study, as suggested by the structure of the VSV N-RNA P complex (18). Given that, in our model, much of the molecular interaction of the negatively charged tail involves main-chain atoms, it remains possible that replacement of even the entire acidic region with alanine is not sufficient to destabilize the interaction with P, as a significant number of main-chain contacts with P are likely to remain in place, as shown for VSV (18).
None of the MOKV P residues identified in this study as being important for the interaction with MOKV N appear to be conserved in the VSV P structure, suggesting that VSV P interacts differently with N than the lyssavirus P. In addition, biochemical data also indicate that P-N and P-L interactions are likely to be different (41, 42). This has been confirmed by the structure of VSV N-RNA complexed with the C-terminal domain of VSV P, which reveals that those residues of VSV P that are involved in binding lie close to but do not map directly onto the residues that we predict are involved in MOKV N-P binding (18). This lack of agreement is probably due primarily to differences in the sizes of the P proteins (the C-terminal domain of MOKV P is 50% larger than the equivalent C-terminal domain of VSV).
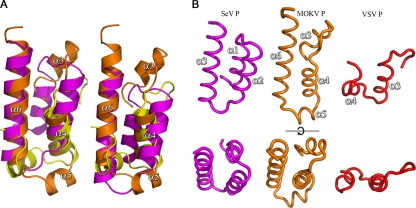
Alongside the structures of the N-RNA binding domains from P proteins of members of the Rhabdoviridae (MOKV and RABV for the Lyssavirus genus and VSV P for the Vesiculovirus genus), the structures of functionally equivalent domains of the P proteins from other members of the order Mononegavirales have been solved. The structures of the N-RNA binding domains of Sendai virus (SeV) and measles virus (MeV) P proteins consist of an antiparallel triple-helix bundle, and, as for the Rhabdoviridae, the N-RNA binding domains among paramyxoviruses are also structurally conserved in spite of low sequence conservation (5, 21, 24, 25). Careful analysis of the structures reveals a structural relationship between the N-RNA binding domains of the P proteins of the Rhabdoviridae and the Paramyxoviridae, where the three-helix bundles of the SeV and MeV P proteins align with helices α3, α4, and α6 of MOKV (and RABV) (Fig. 4B). Structural superposition using SHP (40) shows that 43 and 40 residues of MOKV P (out of 107) can be aligned with SeV P and MeV P, with RMSDs of 3.3 Å and 3.2 Å, respectively, over the aligned residues, despite 14% and 0% sequence identity. The similarity between VSV P and the paramyxovirus and lyssavirus P proteins is much poorer due to a shorter C-terminal helix α4 and the absence of a helix equivalent to MOKV α3 in VSV P, although the alignment suggests that at least VSV P α3 (α4 in MOKV P) is conserved. This low but detectable degree of structural similarity would suggest that the P proteins of Rhabdoviridae and Paramyxoviridae originated from a common ancestor in spite of the high degree of amino acid divergence.
FIG. 4.
Comparison of the structures of Rhabdovirus and Paramyxovirus N-RNA binding domains of P. (A) Superposition of the conserved elements of MOKV P (orange), VSV P (yellow) (PDB accession no. 2K47), and SeV (magenta) (PDB accession no. 1R4G). Helix numbering is as for MOKV P. The left-hand drawing is based on SHP alignment of the Cα's (40) whereas the right hand drawing is based on the sequence alignment. (B) Side-by-side comparison of the conserved elements of the SeV, MOKV, and VSV P proteins.
Finally, it is of interest to note that the MOKV P positively charged cluster (K212, K213, and K215) in combination with R261 was recently identified as a functional nuclear localization signal (NLS) (34). Although the possible role of P nuclear localization remains to be established, the structures of the RABV and MOKV P proteins show that these regions do not conform to the classic NLS structure (39), as they are not part of a disordered or flexible loop but rather are part of an ordered region that includes beta sheet β1, the (ordered) loop separating α1 and β1, and helix α4. As noted by Pasdeloup et al. (34), the overlap with the NLS suggests that the N-RNA binding region in P may additionally act to mask the NLS upon binding to N. If verified, this would constitute another example of the modulation of the biological activities achieved by a lyssavirus protein by sequestering a biologically active protein interface during morphogenesis and/or the viral life cycle, as already postulated for the matrix protein (17).
Acknowledgments
We thank the staff of Diamond for assistance, Christian Siebold for help with synchrotron data collection, and Joanne Nettleship for the mass spectrometry analysis.
The OPPF is supported by the United Kingdom Medical Research Council (MRC) and the Biotechnology and Biological Sciences Research Council. R.A. and O.D. are supported by the EU-funded FP6 Vizier program (LSHG-CT-2004-511960) and J.M.G. by Spine2Complexes (LSGH-CT-2006-031220).
Footnotes
Published ahead of print on 11 November 2009.
REFERENCES
- 1.Albertini, A. A., G. Schoehn, W. Weissenhorn, and R. W. Ruigrok. 2008. Structural aspects of rabies virus replication. Cell Mol. Life Sci. 65:282-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albertini, A. A., A. K. Wernimont, T. Muziol, R. B. Ravelli, C. R. Clapier, G. Schoehn, W. Weissenhorn, and R. W. Ruigrok. 2006. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science 313:360-363. [DOI] [PubMed] [Google Scholar]
- 3.Assenberg, R., O. Delmas, S. C. Graham, A. Verma, N. Berrow, D. I. Stuart, R. J. Owens, H. Bourhy, and J. M. Grimes. 2008. Expression, purification and crystallization of a lyssavirus matrix (M) protein. Acta Crystallogr. F Struct. Biol. Crystallogr. Commun. 64:258-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker, N. A., D. Sept, S. Joseph, M. J. Holst, and J. A. McCammon. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 98:10037-10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernard, C., S. Gely, J. M. Bourhis, X. Morelli, S. Longhi, and H. Darbon. 2009. Interaction between the C-terminal domains of N and P proteins of measles virus investigated by NMR. FEBS Lett. 583:1084-1089. [DOI] [PubMed] [Google Scholar]
- 6.Berrow, N. S., D. Alderton, S. Sainsbury, J. Nettleship, R. Assenberg, N. Rahman, D. I. Stuart, and R. J. Owens. 2007. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 35:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chenik, M., K. Chebli, Y. Gaudin, and D. Blondel. 1994. In vivo interaction of rabies virus phosphoprotein (P) and nucleoprotein (N): existence of two N-binding sites on P protein. J. Gen. Virol. 75:2889-2896. [DOI] [PubMed] [Google Scholar]
- 8.Das, S. C., and A. K. Pattnaik. 2004. Phosphorylation of vesicular stomatitis virus phosphoprotein P is indispensable for virus growth. J. Virol. 78:6420-6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delmas, O., E. C. Holmes, C. Talbi, F. Larrous, L. Dacheux, C. Bouchier, and H. Bourhy. 2008. Genomic diversity and evolution of the lyssaviruses. PLoS One 3:e2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding, H., T. J. Green, S. Lu, and M. Luo. 2006. Crystal structure of the oligomerization domain of the phosphoprotein of vesicular stomatitis virus. J. Virol. 80:2808-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dolinsky, T. J., J. E. Nielsen, J. A. McCammon, and N. A. Baker. 2004. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32:W665-W667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edgar, R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finke, S., and K. K. Conzelmann. 2005. Replication strategies of rabies virus. Virus Res. 111:120-131. [DOI] [PubMed] [Google Scholar]
- 14.Fu, Z. F., Y. Zheng, W. H. Wunner, H. Koprowski, and B. Dietzschold. 1994. Both the N- and the C-terminal domains of the nominal phosphoprotein of rabies virus are involved in binding to the nucleoprotein. Virology 200:590-597. [DOI] [PubMed] [Google Scholar]
- 15.Gerard, F. C., A. Ribeiro Ede, Jr., A. A. Albertini, I. Gutsche, G. Zaccai, R. W. Ruigrok, and M. Jamin. 2007. Unphosphorylated rhabdoviridae phosphoproteins form elongated dimers in solution. Biochemistry 46:10328-10338. [DOI] [PubMed] [Google Scholar]
- 16.Gerard, F. C., A. Ribeiro Ede, Jr., C. Leyrat, I. Ivanov, D. Blondel, S. Longhi, R. W. Ruigrok, and M. Jamin. 2009. Modular organization of rabies virus phosphoprotein. J. Mol. Biol. 388:978-996. [DOI] [PubMed] [Google Scholar]
- 17.Graham, S. C., R. Assenberg, O. Delmas, A. Verma, A. Gholami, C. Talbi, R. J. Owens, D. I. Stuart, J. M. Grimes, and H. Bourhy. 2008. Rhabdovirus matrix protein structures reveal a novel mode of self-association. PLoS Pathog. 4:e1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green, T. J., and M. Luo. 2009. Structure of the vesicular stomatitis virus nucleocapsid in complex with the nucleocapsid-binding domain of the small polymerase cofactor, P. Proc. Natl. Acad. Sci. U. S. A. 106:11713-11718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green, T. J., X. Zhang, G. W. Wertz, and M. Luo. 2006. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science 313:357-360. [DOI] [PubMed] [Google Scholar]
- 20.Gupta, A. K., D. Blondel, S. Choudhary, and A. K. Banerjee. 2000. The phosphoprotein of rabies virus is phosphorylated by a unique cellular protein kinase and specific isomers of protein kinase C. J. Virol. 74:91-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houben, K., D. Marion, N. Tarbouriech, R. W. Ruigrok, and L. Blanchard. 2007. Interaction of the C-terminal domains of Sendai virus N and P proteins: comparison of polymerase-nucleocapsid interactions within the paramyxovirus family. J. Virol. 81:6807-6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iseni, F., A. Barge, F. Baudin, D. Blondel, and R. W. Ruigrok. 1998. Characterization of rabies virus nucleocapsids and recombinant nucleocapsid-like structures. J. Gen. Virol. 79:2909-2919. [DOI] [PubMed] [Google Scholar]
- 23.Jacob, Y., E. Real, and N. Tordo. 2001. Functional interaction map of lyssavirus phosphoprotein: identification of the minimal transcription domains. J. Virol. 75:9613-9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansson, K., J. M. Bourhis, V. Campanacci, C. Cambillau, B. Canard, and S. Longhi. 2003. Crystal structure of the measles virus phosphoprotein domain responsible for the induced folding of the C-terminal domain of the nucleoprotein. J. Biol. Chem. 278:44567-44573. [DOI] [PubMed] [Google Scholar]
- 25.Kingston, R. L., D. J. Hamel, L. S. Gay, F. W. Dahlquist, and B. W. Matthews. 2004. Structural basis for the attachment of a paramyxoviral polymerase to its template. Proc. Natl. Acad. Sci. U. S. A. 101:8301-8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knobel, D. L., S. Cleaveland, P. G. Coleman, E. M. Fevre, M. I. Meltzer, M. E. Miranda, A. Shaw, J. Zinsstag, and F. X. Meslin. 2005. Re-evaluating the burden of rabies in Africa and Asia. Bull. World Health Organ. 83:360-368. [PMC free article] [PubMed] [Google Scholar]
- 27.Krissinel, E., and K. Henrick. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774-797. [DOI] [PubMed] [Google Scholar]
- 28.Laskowski, R. A., D. S. Moss, and J. M. Thornton. 1993. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231:1049-1067. [DOI] [PubMed] [Google Scholar]
- 29.Liu, P., J. Yang, X. Wu, and Z. F. Fu. 2004. Interactions amongst rabies virus nucleoprotein, phosphoprotein and genomic RNA in virus-infected and transfected cells. J. Gen. Virol. 85:3725-3734. [DOI] [PubMed] [Google Scholar]
- 30.Mavrakis, M., A. A. McCarthy, S. Roche, D. Blondel, and R. W. Ruigrok. 2004. Structure and function of the C-terminal domain of the polymerase cofactor of rabies virus. J. Mol. Biol. 343:819-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mavrakis, M., S. Mehouas, E. Real, F. Iseni, D. Blondel, N. Tordo, and R. W. Ruigrok. 2006. Rabies virus chaperone: identification of the phosphoprotein peptide that keeps nucleoprotein soluble and free from non-specific RNA. Virology 349:422-429. [DOI] [PubMed] [Google Scholar]
- 32.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 33.Ogino, T., and A. K. Banerjee. 2008. Formation of guanosine(5′)tetra- phospho(5′)adenosine cap structure by an unconventional mRNA capping enzyme of vesicular stomatitis virus. J. Virol. 82:7729-7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pasdeloup, D., N. Poisson, H. Raux, Y. Gaudin, R. W. Ruigrok, and D. Blondel. 2005. Nucleocytoplasmic shuttling of the rabies virus P protein requires a nuclear localization signal and a CRM1-dependent nuclear export signal. Virology 334:284-293. [DOI] [PubMed] [Google Scholar]
- 35.Ren, J., S. Sainsbury, N. S. Berrow, D. Alderton, J. E. Nettleship, D. K. Stammers, N. J. Saunders, and R. J. Owens. 2005. Crystal structure of nitrogen regulatory protein IIANtr from Neisseria meningitidis. BMC Struct. Biol. 5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ribeiro, E. A., Jr., A. Favier, F. C. Gerard, C. Leyrat, B. Brutscher, D. Blondel, R. W. Ruigrok, M. Blackledge, and M. Jamin. 2008. Solution structure of the C-terminal nucleoprotein-RNA binding domain of the vesicular stomatitis virus phosphoprotein. J. Mol. Biol. 382:525-538. [DOI] [PubMed] [Google Scholar]
- 37.Ribeiro, E. D., Jr., C. Leyrat, F. C. Gerard, A. A. Albertini, C. Falk, R. W. Ruigrok, and M. Jamin. 2009. Binding of rabies virus polymerase cofactor to recombinant circular nucleoprotein-RNA complexes. J. Mol. Biol. 394:558-575. [DOI] [PubMed] [Google Scholar]
- 38.Schoehn, G., F. Iseni, M. Mavrakis, D. Blondel, and R. W. Ruigrok. 2001. Structure of recombinant rabies virus nucleoprotein-RNA complex and identification of the phosphoprotein binding site. J. Virol. 75:490-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stewart, M. 2007. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 8:195-208. [DOI] [PubMed] [Google Scholar]
- 40.Stuart, D. I., M. Levine, H. Muirhead, and D. K. Stammers. 1979. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 A. J. Mol. Biol. 134:109-142. [DOI] [PubMed] [Google Scholar]
- 41.Takacs, A. M., and A. K. Banerjee. 1995. Efficient interaction of the vesicular stomatitis virus P protein with the L protein or the N protein in cells expressing the recombinant proteins. Virology 208:821-826. [DOI] [PubMed] [Google Scholar]
- 42.Takacs, A. M., T. Das, and A. K. Banerjee. 1993. Mapping of interacting domains between the nucleocapsid protein and the phosphoprotein of vesicular stomatitis virus by using a two-hybrid system. Proc. Natl. Acad. Sci. U. S. A. 90:10375-10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tordo, N., H. Bourhy, S. Sather, and R. Ollo. 1993. Structure and expression in baculovirus of the Mokola virus glycoprotein: an efficient recombinant vaccine. Virology 194:59-69. [DOI] [PubMed] [Google Scholar]
- 45.Toriumi, H., and A. Kawai. 2004. Association of rabies virus nominal phosphoprotein (P) with viral nucleocapsid (NC) is enhanced by phosphorylation of the viral nucleoprotein (N). Microbiol. Immunol. 48:399-409. [DOI] [PubMed] [Google Scholar]
- 46.Vagin, A., and A. Teplyakov. 1997. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30:1022-1025. [Google Scholar]
- 47.Walhout, A. J., and M. Vidal. 2001. High-throughput yeast two-hybrid assays for large-scale protein interaction mapping. Methods 24:297-306. [DOI] [PubMed] [Google Scholar]
- 48.Walter, T. S., J. M. Diprose, C. J. Mayo, C. Siebold, M. G. Pickford, L. Carter, G. C. Sutton, N. S. Berrow, J. Brown, I. M. Berry, G. B. Stewart-Jones, J. M. Grimes, D. K. Stammers, R. M. Esnouf, E. Y. Jones, R. J. Owens, D. I. Stuart, and K. Harlos. 2005. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Crystallogr. D Biol. Crystallogr. 61:651-657. [DOI] [PMC free article] [PubMed] [Google Scholar]