Abstract
Heliothis zea nudivirus 1 (HzNV-1), previously known as Hz-1 virus, is an insect virus able to establish both productive and latent infections in several lepidopteran insect cells. Here, we have cloned and characterized one of the HzNV-1 early genes, hhi1, which maps to the HindIII-I fragment of the viral genome. During the productive viral infection, a 6.2-kb hhi1 transcript was detectable as early as 0.5 h postinfection (hpi). The level of transcript reached a maximum at 2 hpi and gradually decreased after 4 hpi. The transcript was not detectable during the latent phase of viral infection. Upon cycloheximide treatment, much higher levels of hhi1 transcript were detected throughout the productive viral infection cycle, suggesting that newly synthesized proteins are not needed for the expression of hhi1. Nevertheless, viral coinfection can further stimulate the expression of transfected hhi1 promoter in a plasmid. Transient hhi1 expression in latently infected cells resulted in a significant increase in virus titer and viral DNA propagation, suggesting that hhi1 plays a critical role in viral reactivation. Additional experiments showed that six early genes, which possibly function in transcription or DNA replication, were activated in the latent cells upon hhi1 transfection. Among these six genes, orf90 and orf121 expression could be induced by hhi1 alone without the need for other viral genes. Our discovery should be useful for future mechanistic study of the switches of latent/productive HzNV-1 viral infections.
Virus infection of host cells usually results in productive infection in which the viruses express most, if not all, of their genes, replicate their genomes, and finally release progeny viruses. In most cases, host cells are killed by the productive viral infection (3, 14). In latent infections, such as those caused by herpes viruses in nerve tissues, the virus is quiescent, and no infectious virus particles can be detected. Periodic reactivation results in the production of viral progeny and transmission to new hosts (6).
Latent viral infections are difficult to observe and study; consequently, these phenomena are usually neglected and rarely reported (3, 18, 30). In addition, due to the difficulty in establishing latent viral infections in laboratory stocks of culture cells or insects, latent viral infections are usually identified only after an unexpected viral reactivation from previously healthy-looking insects or insect cells (24). So far, we still know very little about genes and mechanisms governing the switch of latent/productive viral infections in mammals and insects.
Heliothis zea nudivirus (HzNV-1), originally named Hz-1 virus, was first found to be a latent virus in the IMC-Hz-l cell line isolated from ovarian tissue of H. zea (8, 10, 14, 16, 21). This virus has been reported to infect many insect cell lines including Trichoplusia ni (TN-368), Spodoptera frugiperda (IPLB-SF-21), H. zea (IPLB-1075), Mamestra brassicae, and Porthetria dispar (IPLB-65Z) (1, 8, 22, 25) and to establish latent infections in most of these insect cell lines, which are permissive to this virus. HzNV-1 virus is an enveloped, rod-shaped, nonoccluded virus with a circular double-stranded DNA genome of 228 kb, which is encased in a 414 ± 30 nm nucleocapsid (1, 3, 4, 11). The HzNV-1 genome has been completely sequenced and is estimated to contain 154 open reading frames (ORFs) (5). Previously, it was classified as a member of the baculovirus family, but due to its lack of an occlusion body and low sequence homology to a typical baculovirus, it has now been relegated to a new nonoccluded genus, the Nudivirus (27, 28).
HzNV-1 virus has two infection cycles, latent and productive, in its life cycle. It can infect insect cells and stay latent for many passages (3, 19, 30). Studies of TN-368 and Sf-21 cells have shown that during the productive infection stage, most cells are killed, and high titers of virus progeny are produced; however, often a small percentage of the cells, usually less than 5%, become latently infected, and virus persists for a prolonged period of time within these cells (3, 30). During latency, virions were not detectable in most of these latently infected cells, and viruses existed either as episomes or inserted into the host genome (19). Sometimes virus particles can be released from very small proportions (usually less than 0.2%) of latently infected cells, resulting in the death of these cells. This small quantity of continuously released virions results in the presence of low viral titers (around 103 PFU/ml) in the culture medium of latently infected cells (4, 19).
The temporal gene expression profiles of the HzNV-1 virus during latent and productive infection have been analyzed. During latent infection, persistency-associated transcript 1 (PAT1), which is expressed by persistency-associated gene 1 (pag1), was found to be involved in the establishment of latent infection of HzNV-1 virus and was the only detectable transcript during latent infection (3, 4). During productive viral infection, an abundant 6.2-kb transcript was detected from the HindIII-I fragment of the genome of HzNV-1. The gene was identified and named hhi1 as it is the first gene identified in the HzNV-1 virus HindIII-I fragment (3, 4, 31). Wu et al. (31) found that hhi1 promoter can be activated in Sf-21 cells by the baculovirus, Autographa californica multiple nucleopolyhedrovirus (AcMNPV). In this study, we further characterized the temporal expression of hhi1 and its function. We found that de novo protein synthesis is not required for hhi1 gene expression upon viral infection. The hhi1 gene is shown to be capable of reactivating viruses from latently infected cells. This discovery highlights hhi1 as a key gene useful for further mechanistic studies of viral gene regulation and switching during latent and productive viral infections.
MATERIALS AND METHODS
Cells and virus.
Spodopter frugiperda IPLB-Sf-21 was incubated in TC-100 insect cell culture medium, which contained 10% fetal bovine serum (FBS) at 26°C (Gibco BRL, Gaithersburg, MD) (23, 31). Standard HzNV-l virus was derived by a serial dilution of the stock viral solution and isolated by plaque purification. SFP4 cells were derived from latently infected Sf-21 cells (3, 4). The titers of the virus clones were estimated by both quantitative PCR (qPCR) (20) and 50% tissue culture infective dose (TCID50) (23).
Computer-assisted sequence analysis.
The contiguous 10,878-bp HindIII-I fragment was sequenced and assembled using Phrep/Phrap and Sequencer software, version 4.1.1 (Sequencher, Ann Arbor, MI). The ORF prediction was performed using GenScan (http://genome.dkfz-heiderlberg/de/cgi-bin/GENSCAN/) and the National Center for Biotechnology Information (NCBI) ORF Finder program (http://www.ncbi.nlm.nih.gov/). The nucleotide and amino acid sequences were analyzed using the NCBI BLAST program and further confirmed by the sequencing results of Cheng et al. (5).
RNA isolation and Northern blot analysis.
Sf-21 cells were inoculated at a multiplicity of infection (MOI) of 10 with HzNV-l virus and incubated at 26°C for 2 h with gentle rocking. After adsorption, total cellular RNA samples were extracted from productively infected cells at 0.5, 1, 2, 4, 6, 8, 10, and 12 h postinfection (hpi) and from latently infected SFP4 cells using Ultraspect RNA (Bioteck, Houston, TX). In some experiments, cycloheximide (50 μg/ml; Sigma, St. Louis, MO) or actinomycin D (10 μg/ml; Sigma) was added to the cells 1 h prior to viral infection or at the indicated times (see Fig. 2) and maintained at the same concentration during the course of experiments. Briefly, approximately 15 μg of total RNA (Qiagen) extracted from virus-infected SF-21 or SFP4 was loaded, blotted onto Hybind-N+ nylon transfer membrane (GE Healthcare), and hybridized with hhi1- or pag1-specific probe, which was labeled with digoxigenin (DIG) by PCR amplification of template HzNV-1 viral DNA and using a PCR DIG Probe Synthesis Kit (Roche Applied Sciences, Burgess Hill, United Kingdom) according to the manufacturer's instructions.
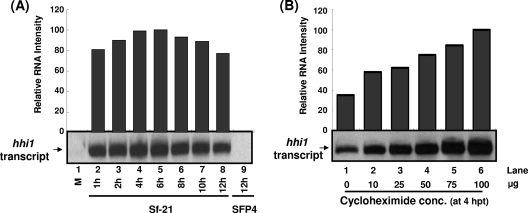
FIG. 2.
Effect of cycloheximide on hhi1 expression in HzNV-1-infected cells. (A) Temporal expression pattern of hhi1 transcripts in the presence of cycloheximide. Cells were treated with cycloheximide (50 μg/ml) 1 h prior to infection with HzNV-1 virus, and total RNA was extracted at different time points postinfection. These RNA samples were analyzed by Northern blotting using DIG-labeled probe specifically for hhi1 transcript. (B) hhi1 transcript accumulation observed by the treatment of cycloheximide at different concentrations.
Plasmid construction.
The 10.8-kb HindIII-I fragment of HzNV-l viral genomic DNA was cloned into the HindIII site of the low-copy-number vector pWSK29 (26). A DNA fragment of 4,518 nucleotides from bp 222659 to 227176 of HzNV-1 virus was PCR amplified using the following primers: HHI1 F, 5′-ATTCCCGGGCTCTCCTCTACAATCATGTCTACCGTG-3′; HHI1 R, 5′-ATTCCCGGGCTCAGATTCACAGTATGGTTCACG-3′ (restriction sites are underlined). The amplified fragment was cloned into T-vector and subsequently released by SmaI cleavage and inserted downstream from the Drosophila heat shock promoter (15) in plasmid pBluescript (Stratagene, La Jolla, CA). The resulting plasmid was pKShH1. We obtained egfp, orf60, orf75, orf90, orf101, orf121, and orf131 coding regions by PCR with SmaI and inserted them downstream of the hsp70 promoter (p-hsp) of plasmid pKShsp70 to give the expression plasmids.
Different HzNV-1 viral early promoter regions, hhi1 (+35 to −688) (31), pag1 (+29 to −727), and orf60, orf75, orf90, orf101, orf121, and orf131 promoters (+1 to −500) were obtained by PCR using HzNV-1 viral DNA as a template. Promoter regions were ligated into pGL3-basic vector (Promega, Madison, WI) to obtain plasmids pGL3-hhi1 and pGL3-pag1 for activity assay with or without HzNV-1 infections and to obtain plasmids pGL3-orf60, pGL3-orf75, pGL3-orf90, pGL3-orf101, pGL3-orf121, and pGL3-orf131, respectively, for activity assays upon hhi1 cotransfection.
DNA transfection into cells.
A total of 2 × 105 cells were seeded in a 24-well culture plate (Corning, Acton, MA) and then transfected with 0.5 μg of appropriate plasmid DNAs using Cellfectin (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol (Gibco BRL). The promoter activities, cellular morphology, or the virus titers of the transfected cells were studied at the indicated time points (see Fig. 4) in different experiments.
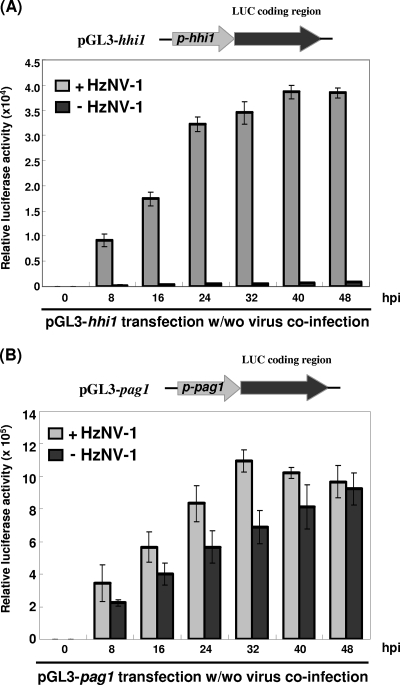
FIG. 4.
Transient expression of hhi1 and pag1 promoters in Sf-21 cells. Luciferase activities driven by hhi1 (A) or pag1 (B) promoters with or without (w/wo) HzNV-1 virus infections were assayed at 8, 16, 24, 32, 40, and 48 hpi.
Luciferase activity assay.
Luciferase assays were conducted as described previously (31, 32). Luciferase activity was measured with a luminometer (Lumat LB 9501; Berthold) by injecting 50 μl of 0.2 mM luciferin (Promega) into each well. The results were plotted as average luciferase activity versus time of infection from triplicate assays of three independent experiments.
Quantitation of viral early gene expression.
Virus RNA was extracted using an RNeasy Mini Kit (Qiagen, Hilden, Germany). The RNA pellet was dissolved in 30 μl of diethyl pyrocarbonate (DEPC) and used for cDNA synthesis. cDNA synthesis was performed using SuperScript III first-strand synthesis system for reverse transcription-PCR (RT-PCR), following the protocol from the manufacturer (Invitrogen). HzNV-1 early genes were RT-PCR amplified using the following primers: p60 R, 5′-ATTAGATCTATGGCTGTATACCACTAGTGT-3′; p60 F, 5′-ATTGGTACCACTGGGCATCACGTTGTTAC-3′; p75 R, 5′-ATTAGATCTATTTAGAAAGATGATT-3′; p75 F, 5′-ATTGGTACCACCGGTCAAGAGTACATTGG-3′; p90 R, 5′-ATTAGATCTATGTTTGCTGCTGAACGCTA-3′; p90 F, 5′-ATTGGTACCTGAGTGATTTGTCATACTTG-3′; p101 R, 5′-ATTAGATCTCTTATCAGCAAATGTTGAT-3′; p101 F, 5′-ATTGGTACCTTGTACCACTGTCGAGTCCA-3′; p121 R, 5′-ATTAGATCTATGTTAAGAGTCTAAAA-3′; p121 F, 5′-ATTGGTACCGAACTTTAAGACTTGAGAAT-3′; p131 R, 5′-ATTAGATCTTCGTGACTACTAAACCACAA-3′; and p131 F, 5′-ATTGGTACCCCTCAAAACACGTATAAATC-3′.
Briefly, amplification was carried out by adding 1 μl of cDNA in the Master Mix Taq polymerase (MBI Fermentas, Vilnius, Lithuania). The resulting DNA products (Amplicon, Brighton, United Kingdom) were analyzed on agarose gel (1.5%) after electrophoresis at 100 V for 30 min.
RNA interference.
For hhi1 knockdown experiments, all small interfering RNAs (siRNAs) were predicted and synthesized by MDbio Inc. (MDbio, Taiwan). The siRNAs used in this study were hhi1 siRNA (5′-CGUUCUGAAAACGACGUAAGAUUAGA-3′) and egfp siRNA (5′-GGCGAUGCCACCUACGGCAAG-3′). Sf-21 cells (4 × 104) in 96-well plates were transfected with 50 nM siRNA using a Silencer siRNA Transfection Kit II (Applied Biosystems). Cells were transfected with designed siRNAs, and then at 4 h posttransfection (hpt), the cells were infected with HzNV-1 virus (MOI of 1), and the number of latently infected colonies was again calculated at 12 days postinfection (4). Several primers were designed for the RT-PCR amplification of hhi1, pag1, and actin as follows: hhi1-1F, 5′-CGATATGAACATTAACGATGACGATC-3′; hhi1-1R, 5′-AAACGGATGCAAAATGGACTCAA-3′; pag1-F, 5′-ACGGGAATTCAGTGTCGAGGACTT-3′; pag1-R, 5′-CATGTCTAGAACCCTACCTACCT-3′; actin-F, 5′-CGTGATGGTGGGCATGGGTCAG-3′; and actin-R, 5′-CTAATGTCACGCACGTATTCC-3′.
Nucleotide sequence accession number.
The nucleotide sequence of the hhi1 gene was submitted to the GenBank database under accession number AF264019.
RESULTS
The hhi1 gene maps to a 10.8-kb HindIII-I fragment of the HzNV-1 viral genome.
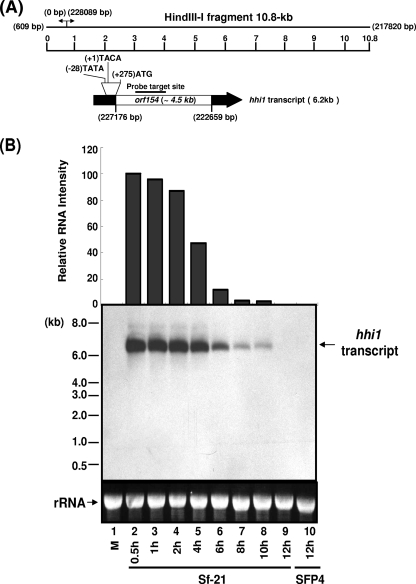
Previously, we reported a 6.2-kb early transcript mapped to the HindIII-I region (10.8 kb) of the HzNV-1 viral genome (3); the gene encoding this 6.2-kb transcript was named hhi1 (Fig. 1A). The temporal expression pattern and the orientation of hhi1 transcript were studied. Sf-21 cells were infected with HzNV-1 virus at an MOI of 10. Total cellular RNA was extracted from these productively infected cells at the indicated time points (Fig. 1B). Total cellular RNA was also isolated from uninfected Sf-21 cells to serve as a negative control. Northern blot analysis showed that a transcript was detected very early at 0.5 hpi. The levels of the transcript remained high from 0.5 hpi to 4 hpi and then decreased gradually. The gene was almost undetectable after 10 hpi (Fig. 1B). Due to its early appearance, it is regarded as an early transcript. This result was further confirmed by RT-qPCR with primers specific for the hhi1 gene, which showed a similar pattern of transcript expression and disappearance (data not shown).
FIG. 1.
Mapping of the hhi1 transcript of HzNV-1 virus. (A) Mapping of hhi1 transcript. The first line shows the corresponding sequence numbers, 609 bp and 217820 bp on the complementary strand, of the nucleotides of HindIII-I fragment (5). The locations of a TATA box-like sequence, initiation site of transcription (+1), and translational start codon (ATG) on the HindIII-I fragment are indicated. (B) Temporal expression profile of the hhi1 gene. Sf-21 cells were infected with HzNV-1 virus at an MOI of 10. Total RNA was extracted and resolved on agarose gel and analyzed by Northern blot analysis using an hhi1-specific probe generated from a DIG labeling reaction. Lane 1, total RNA extracted from mock-infected cells; lanes 2 to 9, total RNA extracted from infected Sf-21 cells at 0.5 to 12 hpi; lane 10, total RNA extracted from latently infected SFP4 cells.
Analysis of hhi1 expression in the presence of cycloheximide.
To study whether hhi1 expression requires viral protein synthesis, cycloheximide, an antibiotic which blocks translation, was added to the cell culture medium. After viral infection, total cellular RNA was extracted from cells and analyzed by Northern hybridizations. The hhi1 transcript was detected throughout the infection cycle in the presence of cycloheximide (50 μg/ml), with a maximal accumulation of hhi1 transcript at 4 to 6 hpi, which then fell slightly at later stages of infection (Fig. 2A). Concentration-dependent experiments showed that HHI1 expression increased proportionally to the increasing concentrations of cycloheximide, suggesting that protein synthesis had a negative effect on hhi1 gene expression (Fig. 2B). These data indicate either that hhi1 expression is blocked or that its transcript is quickly degraded at later stages of viral infection. We also noted that there is no sign of hhi1 expression during the stage of latent viral infection, with or without cycloheximide treatments, suggesting that hhi1 expression is regulated by different mechanisms during latent and productive viral infection stages.
hhi1 and pag1 behave differently in gene expression and RNA stability.
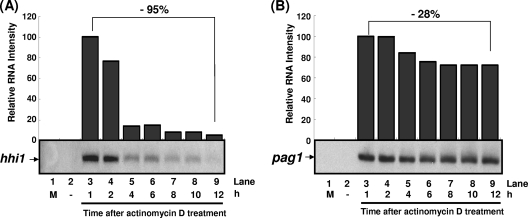
In order to elucidate the stabilities of hhi1 and pag1 transcripts, infected cells were treated with actinomycin D at a concentration of 10 μg/ml, and the transcripts of hhi1 and pag1 were analyzed by Northern hybridizations. Actinomycin D can suppress transcription, enabling measurement of the stability of transcripts already present at the time of drug addition. When actinomycin D was added to the medium 1 h before infection with HzNV-1 virus, neither hhi1 nor pag1 transcripts could be detected in total cellular RNA (Fig. 3, lanes 2). Since both hhi1 and pag1 were early genes and their transcripts were expressed right after viral infection, the stability of their transcripts could therefore be evaluated by adding actinomycin D at 0.5 hpi to the culture medium. Northern blot analysis showed that the level of hhi1 transcripts had decreased by up to 95% by 12 hpi (Fig. 3A), while the level of pag1 transcripts had decreased by only ∼28% over the same period of time (Fig. 3B). These data suggest that pag1 transcript is more stable than hhi1 transcript.
FIG. 3.
Stability analysis of hhi1 and pag1 transcripts upon actinomycin D treatment. The stabilities of hhi1 (A) and pag1 (B) transcripts were analyzed in infected cells treated with actinomycin D. Lane 1, mock-infected cells. In lanes 2 cells were infected with virus 1 h after actinomycin D treatment, and total RNA was harvested at 1 hpi. In lanes 3 to 9, cells were infected with virus 0.5 h prior to actinomycin D treatment, and total RNA was then extracted at the indicated time after actinomycin D addition for the detection of hhi1 and pag1 transcripts.
To investigate whether host factors are sufficient to turn on hhi1 expression, the plasmid pGL3-hhi1 was constructed in which the coding region of luciferase gene was placed under the control of the hhi1 promoter (31). pGL3-hhi1 was transfected into Sf-21 cells with or without HzNV-1 viral infection. No luciferase activity could be detected in cells transfected with pGL3-hhi1, suggesting that the hhi1 promoter is not active or functions weakly in the absence of viral infection. In this experiment, hhi1 promoter was stimulated upon HzNV-1 virus infection (Fig. 4A), indicating that the hhi1 promoter could be activated by HzNV-1 virus infection or transactivated by viral gene products. Since it is known that host factors are sufficient to drive the expression of the early gene pag1 (4), a similar experiment was conducted to compare the promoter activity of pag1 with that of hhi1. Plasmid pGL3-pag1 containing the full-length pag1 promoter region (−727 to +29) to drive the expression of luciferase was constructed previously (4). The pag1 promoter activity was tested in the presence and absence of HzNV-1 virus infection in Sf-21 cells. The pag1 promoter could be activated with or without the coinfection of HzNV-1 virus (Fig. 4B), while viral factor(s) were clearly needed for the optimal expression of hhi1 promoter.
hhi1 stimulates viral reactivation from latent cells.
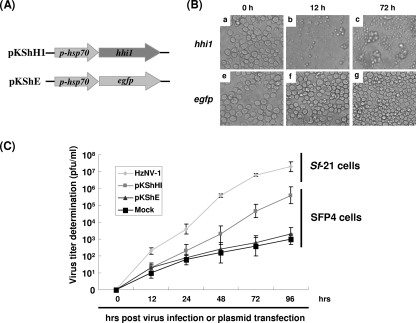
The hhi1 gene was expressed at the very early stage during productive HzNV-1 virus infection but was not detectable during latent HzNV-1 virus infection (3). Thus, it is interesting to study whether hhi1 has any function during latent HzNV-1 virus infection. Two plasmids, pKShE and pKShH1 (Fig. 5A), which express enhanced green fluorescent protein (GFP) and the hhi1 gene product, respectively, under the control of heat shock 70 promoter, were transfected into latently infected SFP4 cells. At 12 hpi with the transfection of plasmid pKShH1, most of the latently infected Sf-21 cells were lysed (Fig. 5B). Supernatants of the transfected cells were harvested, and virus titers were determined. A significantly higher titer of the virus was detected in the hhi1-transfected cells, for which the titer was 127 times greater than that in the pKShE-transfected cells. Furthermore, similar, low titers of viruses were detected in the medium of the latently infected Sf-21 cells with or without the transfection of control plasmid pKShE. These results suggest that the expression of HHI1 can switch HzNV-1 virus infection from a latent to productive viral infection cycle (Fig. 5C).
FIG. 5.
Induction of viral reactivation by the hhi1 gene in latent cells. (A) The map of plasmids pKShH1 and pKShE. In these two plasmids, hhi1 and egfp coding regions were driven by the heat shock promoter. (B) Change of cell morphologies over time after plasmid transfection. Latently infected SFP4 cells were transfected with plasmids pKShH1 (a to c) and pKShE (e to g), and cell images were taken at different time points posttransfection. (C) Viral titers estimated from productively and latently infected cells. Latently infected cells were transfected with plasmids pKShH1 and pKShE separately, and the titers of the released viruses were determined at different time points. Sf-21 cells were infected with wild-type virus (HzNV-1) as a control.
hhi1 transfection can activate many early genes in latently infected cells.
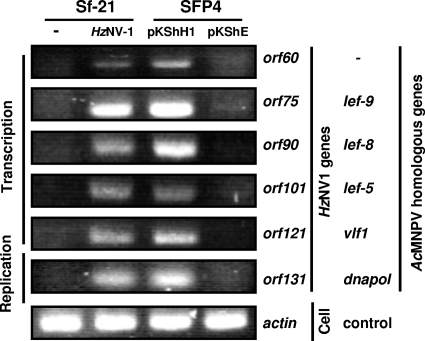
We have shown that hhi1 expression can activate HzNV-1 virus in latently infected Sf-21 cells. Next, HzNV-1 virus early genes that may function in viral DNA transcription (orf60, orf75, orf90, orf101, and orf121) or viral gene replication (orf131), based on their sequence homology to cognate AcMNPV early genes known to function for transcription or DNA replication (28), were tested to see whether they are turned on as a result of entering into the productive cycle of viral infection. The coding region of hhi1 was transfected into latently infected SFP4 cells, and expression of early genes was monitored by RT-PCR. Signals were detected for all the tested genes upon viral reactivation, suggesting that the transient expression of hhi1 turned on the expression of all these early HzNV-1 viral genes either directly or indirectly, and they may eventually contribute to viral reactivation from latency (Fig. 6).
FIG. 6.
Analysis of HzNV-1 viral early transcripts which are induced by hhi1 transfection. Sf-21 and latently infected cells (SFP4) were transfected with plasmid pKShH1 (expresses HHI1) or pKShE (expresses enhanced GFP), and the expression of tested early HzNV-1 transcripts was analyzed by RT-PCR using primers. The early transcripts derived from productive HzNV-1 virus infection of the Sf-21 cells were used as controls; the expression of actin transcript served as another control. These early genes were categorized into two groups based on their predicted functions in either viral DNA replication or viral gene transcription (28).
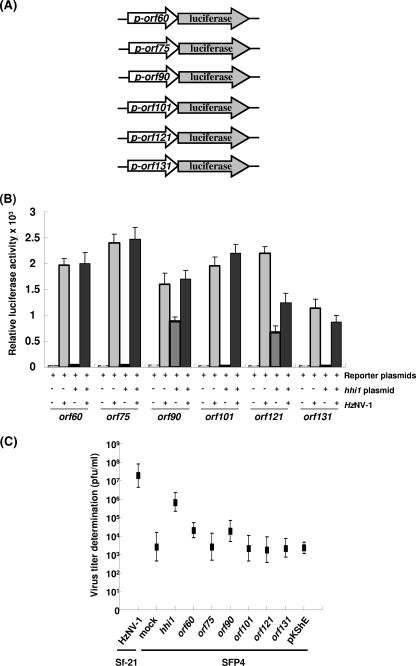
To further confirm the RT-PCR and qPCR results and to determine whether HHI1 activates the expression of these genes directly, the promoter regions of these genes were cloned into plasmid pGL3. Promoter activities were then determined with or without cotransfection of the hhi1 construct (Fig. 7A). Expression of hhi1 could directly stimulate the promoter of both orf90 and orf121, whereas the promoter activities of the other early genes tested could not be stimulated by expression of hhi1 alone. The other early genes became active only upon virus infection, suggesting that their activation requires the assistance of viral gene products (Fig. 7B).
FIG. 7.
Activation analysis of viral early promoters by the hhi1 gene. (A) A list of early promoters for activation assays using luciferase as reporters. (B) The analysis of hhi1 and HzNV-1 in the activation of different viral early promoters. Luciferase activities driven by different promoter constructs were assayed by coinfection of HzNV-1 virus or cotransfected with hhi1 construct (Fig. 6A) in Sf-21 cells. (C) Virus titers estimated from latently infected cells. Latently infected cells were transfected with plasmids expressing several viral early genes, and virus titers were determined at 72 hpt. Sf-21 cells were also infected with HzNV-1 as a control.
To investigate whether hhi1 is solely responsible for switching the virus from latent infection to productive infection, we constructed expression vectors for several of the aforementioned early genes of HzNV-1. Latently infected cells were transfected with these expression vectors, and the supernatants of the transfected cells were harvested at 48 hpt for virus titer analysis. Our results revealed that virus titer was increased by a magnitude of 2 to 3 logs when latently infected cells were transfected with the hhi1-expressing plasmid (Fig. 7C). This result indicates that hhi1 plays a key role in virus reactivation from latency although both orf60 and orf90 also gave low levels of viral reactivation.
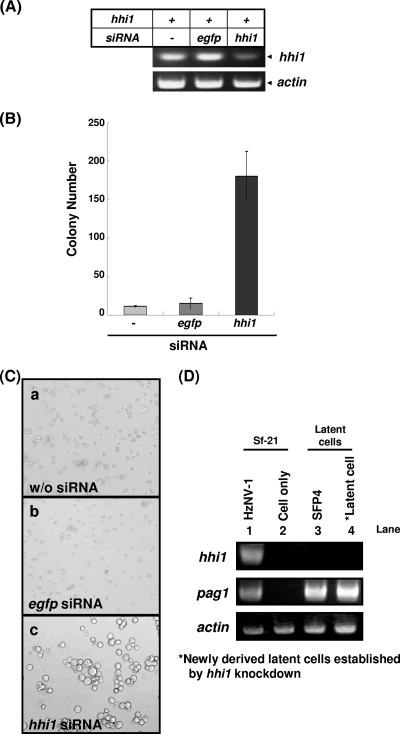
Establishment of latent cells by hhi1 knockdown.
In this report, we found that overexpression of hhi1 in latent cells could stimulate viral DNA accumulation and reactivate cells from the latent to the productive infection stages. We therefore further investigated whether latent infection could be promoted when hhi1 expression was suppressed during productive infection. Sf-21 cells were first transfected with siRNA targeting the hhi1 transcripts, followed by HzNV-1 infection at an MOI of 1. RT-PCR confirmed that suppression of hhi1 expression by siRNA was successful (Fig. 8A). The number of colonies was then recorded at 12 days postinfection. Most of the Sf-21 cells died when they were infected with HzNV-1 virus alone, and only a small percentage of cells became latently infected (Fig. 8B). The percentage of latently infected cells did not increase in cells transfected with siRNA targeting the egfp transcript, which served as a control. However, the number of latently infected cell clones increased drastically when hhi1 expression was suppressed by siRNA (Fig. 8C). We also analyzed gene expression of pag1 and hhi1 in HzNV-1-infected cells at 12 days postinfection with or without hhi1 knockdown (Fig. 8D). hhi1 expression could be detected only in cells productively infected with HzNV-1 (Fig. 8D, lane 1), whereas pag 1 expression could be detected in productive, SFP4, and hhi1 knockdown-induced latent cells (Fig. 8D, lanes 1, 3 and 4, respectively). These results indicate that latent viral infection correlates with the absence of hhi1 expression.
FIG. 8.
Establishment of latent viral infection by hhi1 knockdown. (A) RT-PCR showed that siRNA can efficiently suppress hhi1 expression in HzNV-1-infected cells at 12 hpi. (B) Generation of latently infected colonies by HzNV-1 virus infection in siRNA-transfected cells. Sf-21 cells (4 × 104) were transiently transfected with siRNA (against hhi1 or egfp) followed by HzNV-1 infection. The numbers of surviving cell colonies were calculated at 12 days postinfection. Data (means ± standard deviations) were collected from triplicate assays. (C) Sf-21 cells were infected with HzNV-1, and the images were taken at 12 days postinfection: frame a, Sf-21 cells infected with HzNV-1 only; frame b, Sf-21 cells transfected with egfp siRNA and subsequently infected with HzNV-1; frame c, Sf-21 cells transfected with hhi1 siRNA and subsequently infected with HzNV-1. (D) Confirmation of hhi1 and pag1 expression in cells by RT-PCR. Cells in lane 1 were infected with HzNV-1, and RT-PCR was performed at 2 hpi. Lane 2, cells only; lane 3, latent SFP4 cells; lane 4, hhi1 knockdown-induced latently infected cells (for lane 4, RT-PCR was performed at 20 days postinfection).
DISCUSSION
Previously, we found that during latent viral infection, PAT1 is the only detectable viral transcript while the hhi1 transcript, although not detectable during latent viral infection, was one of the very early transcripts expressed during productive viral infection (3). During productive infection, hhi1 transcript was found to initiate early and then diminish at late viral infection stages. This reduction in hhi1 transcript might be the result of feedback inhibition from hhi1's own gene products or of the functions of other viral gene products which either block the transcription of hhi1 or degrade hhi1 transcript. Since blocking cellular translation with cycloheximide increases the accumulation of hhi1 transcript, it is possible that the presence of certain host or viral factors may negatively regulate hhi1 expression.
Although during productive viral infection several late transcripts other than hhi1 were also expressed by using a probe covering the HindIII-I fragment (data not shown), these late transcripts were no longer detectable after treatment with a protein synthesis inhibitor, cycloheximide. In this work, cycloheximide had a positive effect on the accumulation of hhi1 transcript, indicating that de novo proteins synthesis is not required for hhi1 gene expression. It should be noted, however, that in latently infected cells, no hhi1 expression was detected with or without cycloheximide treatment. This implies that the expression of hhi1 is regulated by different mechanisms during the latent and productive phases of infection.
In our transient expression assay, we found that without viral coinfection, the expression level of hhi1 promoter is relatively low. To study whether hhi1 expression had any effect on latently infected cells, heat shock promoter (15) was used (by plasmid pKShH1) to drive the coding region of the hhi1 gene. In this experiment, we found that transfection of this plasmid into latent cells resulted in viral reactivation and increased viral DNA replication (data not shown), suggesting that hhi1 is the gene responsible, or at least one of the genes responsible, for viral reactivation. Also, although hhi1 functions in viral reactivation, some other genes may be required to further assist the reactivation of the virus from the latent state. Certainly, further studies are needed to investigate all of these possibilities.
Previously, we showed that during latent viral infection, spontaneous viral reactivation occurs in some 0.2% of the cells, which results in the lysis of the host cells and contributes to low background viral titers of an order of 103 in the medium (19). It would be interesting to know the factor(s) and mechanism(s) in individual cells that determine whether viruses remain latent or reactivate. It is possible that host factors may block the hhi1 promoter and suppress its expression. In this case, the virus will remain latent; on the other hand, if the suppression of host factors is not strong enough, perhaps due to poor nutritional conditions, aging of the host cells, or other as yet unknown factors, viral reactivation may result. Alternatively, host factors, or even viral factors that can stimulate the expression of hhi1, may appear by an as-yet-unknown mechanism and turn on hhi1 for viral reactivation.
Upon hhi1 stimulation, the latent cells release viruses. Although the virus titer in these cells is significantly higher than that from originally latently infected cells or pKShE-transfected cells, it is not as high as that observed during productive viral infection. This may be because the latent cells contain a very high proportion of concatemeric or defective viral genomes (19). Thus, after reactivation, these viral genomes may be too big to be enclosed into virus particles. Alternatively, even if they can be packed as viral particles, due to the nature of genome deletion, most of them may become defective interference viral particles and interfere with subsequent titer determination.
Latent baculovirus infections are reported frequently; however, they can be observed only by spontaneous activation or by the detection of trace amounts of viral transcripts or genome in cell lines or insect populations (2, 12, 13). So far, the genes responsible for latent establishment and viral reactivation are largely unknown. Herpes simplex viruses (HSV) can undergo productive infection in epithelial cells and latent infection in sensory neurons. During latency, the virus persists until reactivation, leading to recurrent productive infection and transmission to new hosts (17). The viral immediate-early protein ICP0 of HSV-1 is one of the very few gene products known to cause viral reactivation from latency. ICP0 is a multifunctional gene product which is capable of regulating viral and host cellular gene expression at transcriptional, translational, and posttranslational levels (7, 9). Although there is no significant sequence homology between ICP0 and hhi1, our experiments showed that hhi1 is one of these novel genes that functions in viral reactivation.
HSV-1 and HzNV-1 are quite different viruses and belong to distinct virus families without obvious similarity in genomic sequence or morphology of viral particles (27). The hosts of these viruses are also from different phyla. Thus, their mechanisms for reactivation might be quite different. However, HSV-1 has striking similarities in life cycle with HzNV-1 virus. During productive viral infection, HSV-1 also gives rise to more than 100 transcripts, and during latent viral infection, only one gene, the latency-associate gene 1 (LAT1) (29) is expressed. Interestingly, LAT1 may function similarly to PAT1 since both produce stable noncoding RNAs (4). With such differences in origin, the possibility that they may use similar mechanisms for viral reactivation is even more intriguing. So far, very few genes are known to be involved in viral reactivation in mammals, and hhi1 is currently the only gene known to have a function in viral reactivation in insects.
In our experiments, some viral early genes, which may be involved in either viral DNA replication or viral gene transcription or both, were compared in productive, latent, and reactivated HzNV-1-infected cells. Our results showed that all the tested early genes of HzNV-1 were expressed in the reactivated cells. This suggests that hhi1 can directly or indirectly activate the expression of many early genes, and then viral DNA replication and, perhaps, the transcription of many other genes are turned on. Eventually, the expression of hhi1 leads to the reactivation of HzNV-1 virus from latency. Possible functions of these early genes were predicted by their sequence homology to the cogent genes from AcMNPV (5); however, their actual functions are not yet clear and still await further experiments to elucidate their roles in the life cycle of HzNV-1 virus infection in host cells.
The identification of hhi1 as the gene, or one of the genes, responsible for viral reactivation provides a key for further mechanistic study of viral reactivation. It would be interesting to learn how hhi1 is regulated during productive and latent viral infections. Any interactions of virus and host factors required for hhi1 gene functioning or environmental changes which eventually lead to viral reactivation during latent viral infection are important issues for further study of this fascinating viral infection cycle.
Acknowledgments
We thank Huey-Nan Wu and Tzong-Yueh Chen for valuable discussions and suggestions and Miranda Loney and Harry Wilson for revisions.
This research is funded by grants 098-2811-B-001-025, 098-2811-B-001-036, and 98-2321-B-001-031-MY3 from the National Science Council and by grant 94S-1303 from Academia Sinica, Taiwan.
Footnotes
Published ahead of print on 4 November 2009.
REFERENCES
- 1.Burand, J. P., B. Stiles, and H. A. Wood. 1983. Structural and intracellular proteins of the nonoccluded baculovirus Hz-1. J. Virol. 46:137-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burden, J. P., R. D. Possee, S. M. Sait, L. A. King, and R. S. Hails. 2006. Phenotypic and genotypic characterisation of persistent baculovirus infections in populations of the cabbage moth (Mamestra brassicae) within the British Isles. Arch. Virol. 151:635-649. [DOI] [PubMed] [Google Scholar]
- 3.Chao, Y. C., H. A. Wood, C. Y. Chang, H. J. Lee, W. C. Shen, and H. T. Lee. 1992. Differential expression of Hz-1 baculovirus genes during productive and persistent viral infections. J. Virol. 66:1442-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chao, Y. C., S. T. Lee, M. C. Chang, H. H. Chen, S. S. Chen, T. Y. Wu, F. H. Liu, E. L. Hsu, and R. F. Hou. 1998. A 2.9-kilobase noncoding nuclear RNA functions in the establishment of persistent Hz-1 viral infection. J. Virol. 72:2233-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, C. H., S. M. Liu, T. Y. Chow, Y. Y. Hsiao, D. P. Wang, J. J. Huang, and H. H. Chen. 2002. Analysis of the complete genome sequence of the Hz-1 virus suggests that it is related to members of the Baculoviridae. J. Virol. 76:9024-9035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cliffe, A. R., and D. M. Knipe. 2008. Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection. J. Virol. 24:12030-12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Everett, R. D. 2000. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays 22:761-770. [DOI] [PubMed] [Google Scholar]
- 8.Granados, R. R. 1978. Early events in the infection of Heliothis zea midgut cells by a baculovirus. Virology 90:170-174. [DOI] [PubMed] [Google Scholar]
- 9.Hagglund, R., and B. Roizman. 2004. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J. Virol. 78:2169-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hink, W., and C. M. Ignoffo. 1970. Establishment of a new cell line (IMC-HZ-1) from ovaries of cotton bollworm moths, Heliothis zea (Boddie). Exp. Cell Res. 60:307-309. [DOI] [PubMed] [Google Scholar]
- 11.Huang, Y. S., M. Hedberg, and C. Y. Kawanishi. 1982. Characterization of the DNA of a nonoccluded baculovirus, Hz-1V. J. Virol. 43:174-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughes, D. S., R. D. Possee, and L. A. King. 1993. Activation and detection of a latent baculovirus resembling Mamestra brassicae nuclear polyhedrosis virus in M. brassicae insects. Virology 194:608-615. [DOI] [PubMed] [Google Scholar]
- 13.Hughes, D. S., R. D. Possee, and L. A. King. 1997. Evidence for the presence of a low-level, persistent baculovirus infection of Mamestra brassicae insects. J. Gen. Virol. 78:1801-1805. [DOI] [PubMed] [Google Scholar]
- 14.Ignoffo, C. M., M. Shapiro, and W. F. Hink. 1971. Replication and serial passage of infectious Heliothis nucleopolyhedrosis virus in an established line of Heliothis zea cells. J. Invertebr. Pathol. 18:131-134. [DOI] [PubMed] [Google Scholar]
- 15.Ingolia, T. D., E. A. Craig, and B. J. McCarthy. 1980. Sequence of three copies of the gene for the major Drosophila heat shock induced protein and their flanking regions. Cell 21:669-679. [DOI] [PubMed] [Google Scholar]
- 16.Kelly, D. C., T. Lescott, M. D. Ayres, D. Carey, A. Coutts, and K. A. Harrap. 1981. Induction of a nonoccluded baculovirus persistently infecting Heliothis zea cells by Heliothis armigera and Trichoplusia ni nuclear polyhedrosis viruses. Virology 112:174-189. [DOI] [PubMed] [Google Scholar]
- 17.Kent, J. R., and N. W. Fraser. 2005. The cellular response to herpes simplex virus type 1 (HSV-1) during latency and reactivation. J. Neurovirol. 11:376-383. [DOI] [PubMed] [Google Scholar]
- 18.Lee, J. C., H. H. Chen, H. L. Wei, and Y. C. Chao. 1993. Superinfection-induced apoptosis and its correlation with the reduction of viral progeny in cells persistently infected with Hz-1 baculovirus. J. Virol. 67:6989-6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin, C. L., J. C. Lee, S. S. Chen, H. A. Wood, M. L. Li, C. F. Li, and Y. C. Chao. 1999. Persistent Hz-1 virus infection in insect cells: evidence for insertion of viral DNA into host chromosomes and viral infection in a latent status. J. Virol. 73:128-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo, H. R., and Y. C. Chao. 2004. Rapid titer determination of baculovirus by quantitative real-time polymerase chain reaction. Biotechnol. Prog. 20:354-360. [DOI] [PubMed] [Google Scholar]
- 21.McIntosh, A. H., and C. M. Ignoffo. 1981. Establishment of a persistent baculovirus infection in a lepidopteran cell line. J. Invertebr. Pathol. 8:395-403. [Google Scholar]
- 22.McIntosh, A. H., J. J. Grasela, and C. M. Ignoffo. 2007. In vitro host range of the Hz-1 nonoccluded virus in insect cell lines. In Vitro Cell Dev. Biol. Anim. 43:196-201. [DOI] [PubMed] [Google Scholar]
- 23.O'Reilly, D. R., L. K. Miller, and V. A. Luckow. 1994. Baculovirus expression vectors: a laboratory manual. Oxford University Press, New York, NY.
- 24.Podgwaite, J. D., and H. M. Mazzone. 1986. Latency of insect viruses. Adv. Virus Res. 31:293-320. [DOI] [PubMed] [Google Scholar]
- 25.Ralston, A. L., Y. Huang, and C. K. Kawanishi. 1981. Cell culture studies with the IMC-Hz-1 nonoccluded virus. Virology 115:33-44. [DOI] [PubMed] [Google Scholar]
- 26.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 27.Wang, Y., R. G. Kleespies, A. M. Huger, and J. A. Jehle. 2007. The genome of Gryllus bimaculatus nudivirus indicates an ancient diversification of baculovirus-related nonoccluded nudiviruses of insects. J. Virol. 81:5395-5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, Y., M. M. van Oers, A. M. Crawford, J. M. Vlak, and J. A. Jehle. 2007. Genomic analysis of Oryctes rhinoceros virus reveals genetic relatedness to Heliothis zea virus 1. Arch. Virol. 152:519-531. [DOI] [PubMed] [Google Scholar]
- 29.Whitley, R. J. 2001. Herpes simplex viruses, p. 2461-2510. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 30.Wood, H. A., and J. P. Burand. 1986. Persistent and productive infections with the Hz-1 baculovirus. Curr. Top. Microbiol. Immunol. 131:119-133. [DOI] [PubMed] [Google Scholar]
- 31.Wu, Y. L., C. Y. Liu, C. P. Wu, C. H. Wang, S. T. Lee, and Y. C. Chao. 2008. Cooperation of ie1 and p35 genes in the activation of baculovirus AcMNPV and HzNV-1 promoters. Virus Res. 135:247-254. [DOI] [PubMed] [Google Scholar]
- 32.Wu, C. P., J. Y. Wang, T. Y. Huang, H. R. Lo, and Y. C. Chao. 2008. Identification of baculoviral factors required for the activation of enhancer-like polyhedrin upstream (pu) sequence. Virus Res. 138:7-16. [DOI] [PubMed] [Google Scholar]