Abstract
The evolution of envelope mutations by replicating primate immunodeficiency viruses allows these viruses to escape from the immune pressure mediated by neutralizing antibodies. Vaccine-induced anti-envelope antibody responses may accelerate and/or alter the specificity of the antibodies, thus shaping the evolution of envelope mutations in the replicating virus. To explore this possibility, we studied the neutralizing antibody response and the envelope sequences in rhesus monkeys vaccinated with either gag-pol-nef immunogens or gag-pol-nef immunogens in combination with env and then infected with simian immunodeficiency virus (SIV). Using a pseudovirion neutralization assay, we demonstrate that envelope vaccination primed for an accelerated neutralizing antibody response following virus challenge. To monitor viral envelope evolution in these two cohorts of monkeys, full-length envelopes from plasma virus isolated at weeks 37 and 62 postchallenge were sequenced by single genome amplification to identify sites of envelope mutations. We show that env vaccination was associated with a change in the pattern of envelope mutations. Prevalent mutations in sequences from gag-pol-nef vaccinees included deletions in both variable regions 1 and 4 (V1 and V4), whereas deletions in the env vaccinees occurred only in V1. These data show that env vaccination altered the focus of the antibody-mediated selection pressure on the evolution of envelope following SIV challenge.
Immune containment of human immunodeficiency virus (HIV-1) is complicated by the continuous genetic evolution of the virus. The evolution of the HIV-1 envelope is shaped, in part, by selective pressure of neutralizing antibodies (6, 12, 27, 34-36, 40). Changes in envelope sequence and glycosylation patterns following infection can allow the virus to escape neutralization. If the rate and extent of envelope sequence evolution following infection can be decreased, immune containment of HIV-1 may be improved.
One possible strategy for modifying envelope evolution is vaccination prior to infection. A vaccine-elicited memory immune response could focus and potentiate the humoral immune response that develops following infection. The possible consequence of vaccination has not been assessed, however, because of the limited number of human volunteers who have received highly immunogenic envelope immunogens and subsequently became infected with HIV-1.
Simian immunodeficiency virus (SIV) infection of rhesus monkeys provides a powerful model to study the effect of vaccination on envelope evolution. Like HIV-1, SIV employs both the CD4 molecule and the chemokine receptor CCR5 to enter a target cell and cause an AIDS-like disease in macaques (16, 22). Both SIV and HIV-1 envelopes are heavily glycosylated, with approximately 50% of their mass derived from carbohydrates (14, 21). SIV and HIV-1 envelopes share approximately 40% amino acid homology (10, 11) and have overlapping variable and constant regions, although the variable region 3 (V3) of HIV-1 envelope does not align with the homologous region of SIV envelope (7). Following SIV infection in rhesus monkeys, SIV envelope evolves most rapidly in variable regions 1 and 4 (V1 and V4, respectively), leading to nucleotide additions, deletions, and/or mutations that can potentially translate to changes in glycosylation (7, 9, 13, 15, 19, 29, 30).
Studies done to characterize SIV neutralization suggest that it occurs through mechanisms similar to those seen in HIV-1 neutralization. Amino acid mutations in the envelope of both viruses contribute to the evasion of antibody binding directly by changing recognition sequences and/or envelope conformation. In addition, the glycosylation of envelope serves as a further obstacle to antibody recognition (20, 33, 40). Considerable effort has been devoted to defining neutralizing epitopes of the HIV and SIV envelopes. The known neutralizing human monoclonal antibodies elicited during natural infection are directed against HIV-1 envelope target sites on both gp120 and gp41, including the V3 region, the CD4 binding site, oligomannose residues of gp120, and gp41 (17, 31). The neutralizing epitope profile of SIV envelope includes the CD4 binding site and gp41 but not the V3 region. There is conflicting evidence as to whether V1, V2, and/or V4 of SIV are targets for antibody neutralization (15, 18, 19). The present study addresses whether vaccine-induced immune responses accelerate the generation of autologous neutralizing antibodies following SIV challenge in rhesus monkeys and how this humoral immune response can potentially shape viral sequence evolution.
MATERIALS AND METHODS
RNA isolation.
Viral RNA in 50 μl of plasma was extracted with a QIAamp Viral RNA Mini Kit (Qiagen). RNA recovered from spin columns was eluted into a final volume of 50 μl.
cDNA synthesis.
Twenty-five microliters of isolated RNA was reverse transcribed to single-stranded cDNA using the Superscript III protocol, following the manufacturer's instructions (Invitrogen). RNA, deoxynucleotides (0.5 mM each), and 0.24 μM primer OR9608 (5′-CTCATCTGATACATTTACGGGG-3′) were incubated for 5 min at 65°C. The sample was chilled on ice for 1 min, followed by a brief centrifugation. First-strand cDNA synthesis was completed by adding 10 μl of 5× reaction buffer and 2.5 μl each of 0.1 M dithiothreitol (DTT), RNase Out, and Superscript III RT. The mixture was incubated for 60 min at 50°C, 60 min at 55°C, and 15 min at 70°C. Five units of RNase H was then added, and the mixture was incubated for 20 min at 37°C.
Single genome amplification (SGA) of SIV envelope.
Viral cDNA was diluted in 96-well plates so that fewer than 30% of the wells were positive for amplification to ensure that positive amplifications were a result of a single cDNA template 80% of the time. First-round PCR was carried out in a reaction mixture containing 1× buffer (Platinum Taq HF Kit; Invitrogen), 0.2 mM deoxynucleoside triphosphate (dNTP) mix, 2 mM Mg2SO4, 0.2 μM primer OF6207 (5′-GGGTAGTGGAGGTTCTGGAAG-3′), 0.2 μM primer OR9608 (5′-CTCATCTGATACATTTACGGGG-3′), and 0.025 units of Platinum Taq High Fidelity in a total volume of 20 μl. PCR conditions were as follows: 5 min at 94°C and 35 cycles of 15 s at 94°C, 30 s at 52°C, and 4 min 15 s at 68°C, followed by a final extension time of 10 min at 68°C. For second-round PCR, 2 μl of first-round PCR product was mixed with 1× buffer, 0.2 mM dNTP mix, 2 mM Mg2SO4, 0.3 μM primer IF6428 (5′-CGTGCTATAACACATGCTATTG-3′), 0.3 μM primer IR9351 (5′-CCCTACCAAGTCATCATCTTC-3′), and 0.025 units of Platinum Taq High Fidelity in a total volume of 20 μl. PCR conditions were the following: 5 min at 94°C and 45 cycles of 15 s at 94°C, 30 s at 51°C, and 3 min 30 s at 68°C, followed by a final extension time of 10 min at 68°C. Amplicons were analyzed on precast 1% agarose 96-well E-Gels (Invitrogen). Amplicons from cDNA dilutions resulting in less than 30% positivity were sequenced. A total of 13 to 29 envelope sequences were analyzed for each monkey at each time point. For comparison of potential N-glycosylation (PNG) sites between animals, 13 random env clones were chosen from each animal at each time point to avoid introducing variation due to the different number of clones isolated from each animal.
The reference sequence for SIVmm239 (accession number M33262) was included in the alignment to standardize the numbering for envelope amino acid residues. Each aligned residue was numbered according to the corresponding position in the SIVmm239 sequence.
Construction of SIVmac239 Δenv plasmid.
The two half-genome plasmids of the SIVmac239, molecular clones p239SpSp5′ and p239SpE3′, were obtained through the AIDS Research and Reference Reagent Program. To generate a single full-length infectious clone, the 5′ cellular sequence was replaced with an EcoRI site, and the 3′ cellular sequence was replaced with an XhoI site by PCR-based technology. The full-length SIVmac239 clone was constructed by inserting the ligation product of the 5′ EcoRI-SphI fragment and the 3′ SphI-XhoI fragment into the EcoRI-XhoI site of the pSP73 vector (Promega). env deletion mutants, termed Δenv, were then constructed based on the full-length infectious clone. A PCR-based mutagenesis method was used to generate an 1,156-bp deletion in the wild-type SphI/XhoI fragment beginning at the gp120 start codon. We replaced the wild-type fragment with the mutant Δenv SphI/XhoI fragment to construct the SIVmac239 Δenv full-length clone. All deletion constructs were confirmed by sequencing, and lack of envelope expression was confirmed by Western blotting. The plasmid was propagated in Stbl2 bacterial cells (Invitrogen) at 30°C.
Construction of env-pseudotyped viruses.
Wild-type (WT) and predominant mutant envelope clones were amplified from products of first-round SGA (see above). Five microliters of first-round SGA product was mixed with 0.4 μM F6484 (5′-ACCATTGCCAGTTTTGTTTTCTTA-3′), 0.4 μM R9339 (5′-GTCATCATCTTCCTCATCTACATC-3′), and 25 μl of Easy-A High-Fidelity PCR Master Mix (Invitrogen) in a total volume of 50 μl. PCR conditions were as follows: 5 min at 94°C and 40 cycles of 15 s at 94°C, 30 s at 54°C, and 3 min at 72°C, followed by a final extension time of 7 min at 72°C. Amplicons were analyzed on a 0.7% agarose gel, extracted using a QIAquick gel extraction kit (Qiagen), and inserted into pcDNA3.1/V5-His TOPO TA plasmids following the manufacturer's instructions (Invitrogen). Recombinant plasmids expressing the envelope from the challenge virus or variant viruses after challenge were transformed into TOP10 cells (Invitrogen) and plated overnight at 37°C. A total of 12 to 24 colonies were chosen for miniprep, restriction enzyme digest, and sequencing. Desired clones were grown and purified by a HiSpeed Plasmid Maxi Kit (Qiagen).
Stocks of replication-incompetent SIV Env pseudovirus were produced by cotransfecting 293T/17 cells (2 × 107 cells per T75 flask) with 2 μg of the env expression plasmid and 12 μg of an env-deficient SIVmac239 backbone plasmid using Lipofectamine (Invitrogen). Supernatant was harvested at 24 h following transfection and cleared by centrifugation and 0.45-μm-pore-size filtration. One-milliliter aliquots were stored at −80°C. The 50% tissue culture infectious dose (TCID50) for each pseudovirus was determined by infection of TZM.bl cells as described previously (25).
Pseudovirus neutralization assay.
Neutralizing antibody responses against SIVmac251 were measured using a TZM.bl luciferase-based assay. This assay measures the reduction in luciferase reporter gene expression following a single round of virus infection. Briefly, 3-fold serial dilutions of heat-inactivated plasma or serum samples were performed in duplicate in 10% Dulbecco's modified Eagle's medium (DMEM) growth medium (100 μl/well). Virus was added to each well in a volume of 50 μl, and the plates were incubated for 1 h at 37°C. TZM.bl cells were then added (1 × 104/well in a 100-μl volume) in 10% DMEM growth medium containing DEAE-dextran (Sigma, St. Louis, MO) at a final concentration of 11 μg/ml. Assay controls included replicate wells of TZM.bl cells alone (cell control) and TZM.bl cells with virus (virus control). Following incubation for 48 h at 37°C, 150 μl of assay medium was removed from each well, and 100 μl of Bright-Glo luciferase reagent (Promega, Madison, WI) was added. A total of 150 μl of the cell lysate was then transferred to a 96-well black plate, and luminescence was measured using a Victor 3 luminometer (Perkin Elmer). The 50% inhibitory dose (ID50) titer was calculated as the serum dilution that resulted in 50% reduction in relative luminescence units (RLU) compared to virus control wells after subtraction of cell control RLU.
RESULTS
env vaccination does not result in lower plasma viral load.
Rhesus monkeys from two arms of a previously reported five-arm vaccine study (24) were selected for analysis in the present study. Six monkeys in one arm of the study were immunized with a plasmid DNA construct encoding SIVmac239 gag-pol-nef on a schedule of 0, 4, and 8 weeks at a dose of 4 mg/plasmid/inoculation; at week 40 they were inoculated intramuscularly with a recombinant adenovirus 5 (rAd5) vector encoding gag-pol (23) at a dose of 1011 particles. The six monkeys in the other arm of the study received the same plasmid DNA construct encoding SIVmac239 gag-pol-nef and a plasmid DNA construct encoding SIVmac239 env gp145ΔCFI (8, 24) on the same 0-, 4-, and 8-week schedule at the same dose of 4 mg/plasmid/inoculation; at week 40 they were inoculated with one rAd5 vector encoding SIVmac239 gag-pol and another encoding SIVmac239 env gp140ΔCFI (24), each at a dose of 1011 particles. The monkeys in both arms of the study were challenged intravenously 19 weeks following the rAd immunizations with a highly pathogenic SIVmac251 isolate.
To determine the effect of envelope vaccination on the clinical consequences of an immunodeficiency virus infection, we first evaluated the levels of SIV viremia. We assessed virus replication in these two groups of monkeys by measuring plasma virus RNA levels in each animal through the first 700 days following SIV infection. No differences in levels of viral replication were demonstrable (Fig. 1). Therefore, any divergence that might be present between these two groups of monkeys in env evolution or in the emergence of antibody responses should not be a consequence of differences in the levels of SIV replication in the monkeys. While a sham vaccine arm was included in the original study (24), we selected two vaccinated arms of that study for evaluation in the present analysis, because these two groups were matched for levels of plasma viral RNA at the peak and set point of viral replication. Since higher viral loads can influence the rate of viral evolution independent of the immune response, we chose not to include the sham-vaccinated arm in this study in order to control for these variables.
FIG. 1.
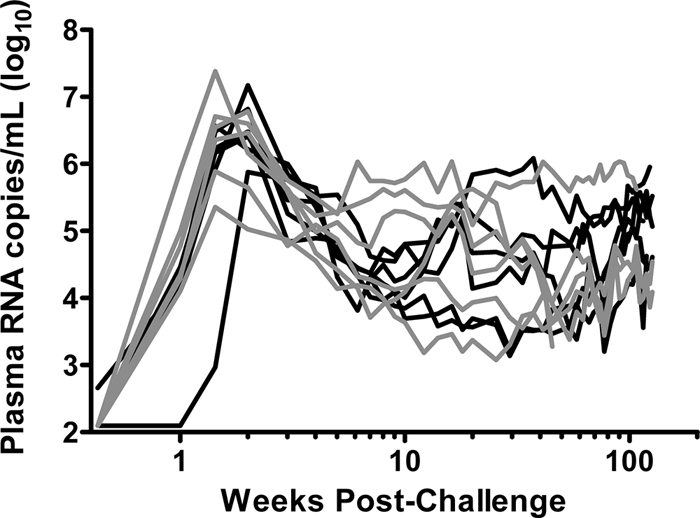
Plasma virus RNA levels following SIVmac251 challenge. Virus loads for monkeys immunized with gag-pol-nef are shown with black lines, and loads for monkeys immunized with gag-pol-nef+env are shown with gray lines. Differences in the virus loads at both the peak and set point in the two groups were nonsignificant by two-way analysis of variance.
The emergence of neutralizing antibody responses.
We first evaluated the envelope gene in the SIV challenge stock to determine the diversity of the challenge stock. Sequencing of the envelope using single genome amplification (SGA) revealed two predominant envelope species at the amino acid level (Fig. 2). Clones 5 and 30 each represent approximately 45% of the total envelope species in the inoculum. An alignment of the two showed that they differ at seven residues, one in the gp120 V1 region and the remaining six in gp41.
FIG. 2.
Amino acid sequence alignment by ClustalW of the two predominant envelope species in the SIVmac251 challenge stock. gp120 variable regions 1 and 4 are highlighted. The V1 (residues 114 to 153) and V4 (residues 403 to 431) regions are in grey. Envelopes 5 and 30 each represent approximately 45% of the total envelope species in the challenge stock. The remaining species are very similar in sequence to these predominant envelopes. The line break preceding residue 23 marks the cleavage site between the signal sequence and gp120, and the line break preceding residue 529 marks the cleavage site between gp120 and gp41.
We then assessed the neutralizing antibody response to this challenge stock envelope in a pseudovirion-based SIV neutralization assay using serum specimens from all animals (26). Clone 30 was randomly selected to be the representative wild-type (WT) env clone since the neutralization profiles of clone 30 and 5 were similar (data not shown). This env plasmid was cotransfected into 293T/17 cells with a plasmid expressing SIVmac239Δenv to generate pseudotyped virions expressing the WT Env. TZM.bl cells expressing both the CD4 and CXCR4 molecules and a Tat-responsive luciferase gene (32, 39) were then infected with the WT pseudovirion in the presence of selected monkey sera.
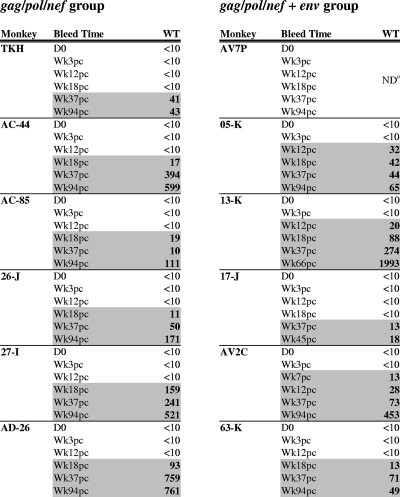
Neutralization activity for the WT Env pseudotyped virions was first detected at week 18 postchallenge in the serum samples from five of the six gag-pol-nef-vaccinated animals (Table 1). The first detectable neutralizing titer in serum of the remaining animal in this group, monkey TKH, was observed by week 37 postinfection (p.i.). The magnitude of these neutralizing titers increased as a function of time and reached titers of >500 in three animals vaccinated without env (AC-44, 27-I, and AD-26). Importantly, sera from animals that were vaccinated with gag-pol-nef and env (gag-pol-nef+env (05-K, 13-K, and AV2C) neutralized the WT Env-expressing pseudovirion with more rapid kinetics than serum from the gag-pol-nef vaccinees, emerging by week 12 or earlier following challenge. The difference in time to the development of a neutralizing antibody response between the two vaccinated groups did not achieve statistical significance (one-sided Wilcoxon exact test, P = 0.10). However, these data suggest that the addition of an env immunogen to the vaccination regimen can contribute to the more rapid emergence of neutralizing antibody response following challenge.
TABLE 1.
Neutralizing antibody response to a SIVmac251 Env clone after challenge in env-vaccinated monkeysb
a ND, not determined due to toxicity to the reporter cells by serum from this monkey.
Serum neutralization of a virus pseudotyped with WT Env (clone 30 of the SIVmac251 challenge stock) was assayed with monkey serum. Serum sample time points are day of SIVmac251 challenge (D0) and noted as weeks post-viral challenge (Wkpc). Titers that are >10 are highlighted.
env immunization alters the mutational pattern of env.
To determine whether env vaccination affected env evolution after SIVmac251 infection, env sequences were determined from plasma viruses obtained from each animal at two time points following challenge. We chose to sequence week 64, because preliminary data from our laboratory show that env evolution was demonstrable at this time point following natural infection of unvaccinated monkeys with the same SIVmac251 stock. We also chose to sequence plasma virus collected at 37 weeks following infection, reasoning that prior env vaccination may have accelerated the rate of escape mutation.
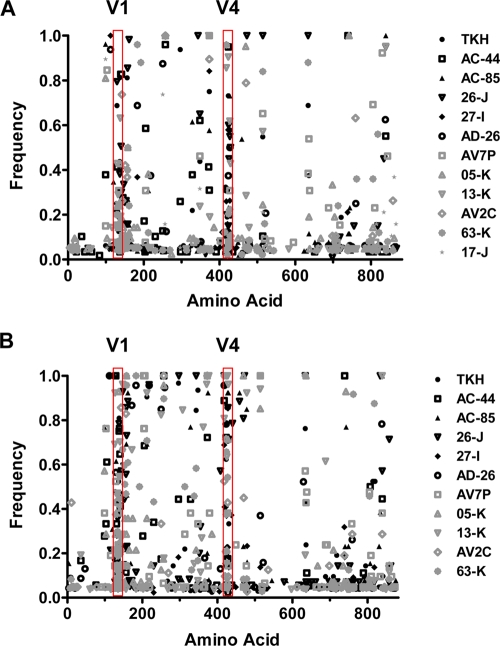
We analyzed the env sequences of the viruses from each animal at 37 and 64 weeks after challenge using Mutation Surveyor (Softgenetics) and determined the frequency of mutation at each amino acid residue of Env. At week 37 (Fig. 3A), most mutations that arose occurred at a frequency of less than 0.4. More mutations accumulated in gp120 than in gp41 (residue 529 marks the N terminus of gp41), and most mutations in gp120 were seen in variable regions 1 and 4. By week 62 (Fig. 3B), further mutations had accumulated in gp120 in the virus of both vaccinated groups while mutations in gp41 had not progressed to a significant extent beyond that observed at week 37 (Fig. 3A).
FIG. 3.
SIV Env evolution in the vaccinated monkeys following SIVmac251 challenge. The frequency of mutations in clones of an animal's circulating viral guasispecies is graphed as a function of the amino acid position at either week 37 (A) or week 62 (B) following challenge. Env mutations in SIVmac251 from challenged monkeys immunized with gag-pol-nef are shown with black symbols, and mutations in monkeys immunized with gag-pol-nef+env are shown with gray symbols. Points indicating zero frequency (no mutation) are not shown. gp120 variable regions 1 and 4 are indicated.
An analysis of these mutations defined only two residues of Env that differed significantly between the two groups of vaccinated monkeys: threonine 417 (T417) and serine 138 (S138). Sequencing of env clones from these monkeys at week 37 following infection showed that T417 of virus from five animals of the gag-pol-nef-vaccinated group evolved to alanine, isoleucine, or asparagine or were deleted, while no clones of viral env from the gag-pol-nef+env group contained a substitution or deletion at this residue. Sequencing of clones of the virus env at week 62 following infection showed that S138 in all gag-pol-nef-vaccinated monkeys evolved to proline, aspartate, leucine, or lysine or were deleted. This mutation was observed in env clones from only one of five gag-pol-nef+env-vaccinated monkeys. These differences in env between the two groups of vaccinated monkeys were statistically significant (two-sided Fisher exact test, P = 0.015, for either the T417 or S138 mutation). However, it is interesting that the T417 and S138 mutations were not persistent since they were not observed in the sequences generated at both time points.
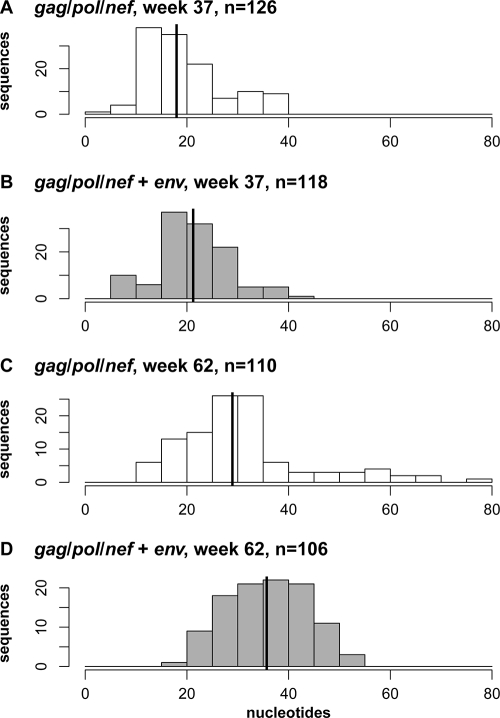
Further analysis of envelope evolution showed that the median distance from the challenge stock to the week 37 env sequences was significantly shorter in the gag-pol-nef-vaccinated group (median of 18.0) than in the gag-pol-nef+env-vaccinated group of monkeys (median of 21.2; P = 0.0032, by one-sided Wilcoxon rank sum test). A similar trend was also observed for env sequences from these monkeys at week 62 following infection with median values of 28.9 and 35.7, respectively (P = 1.5 × 10−6) (Fig. 4). These data suggested that the immune pressure that was generated as a consequence of env vaccination was associated with an increased accumulation of mutations in the replicating viruses.
FIG. 4.
Genetic distances in nucleotides between inoculum consensus SIVmac251 env and env sequences obtained from vaccinated animals following SIVmac251 challenge. Distances are summarized as histograms. Thick vertical lines show the median of each distribution. The upper two panels represent sequences from all clones from week 37, and the lower two panels represent sequences from all clones from week 62. Immunization regimens are indicated on the figure.
env immunization does not alter the total number of envelope potential N-glycosylation sites.
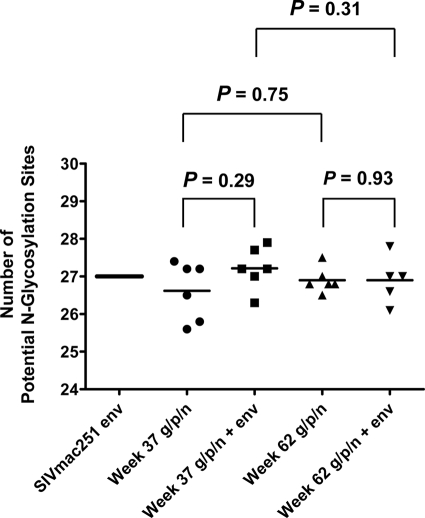
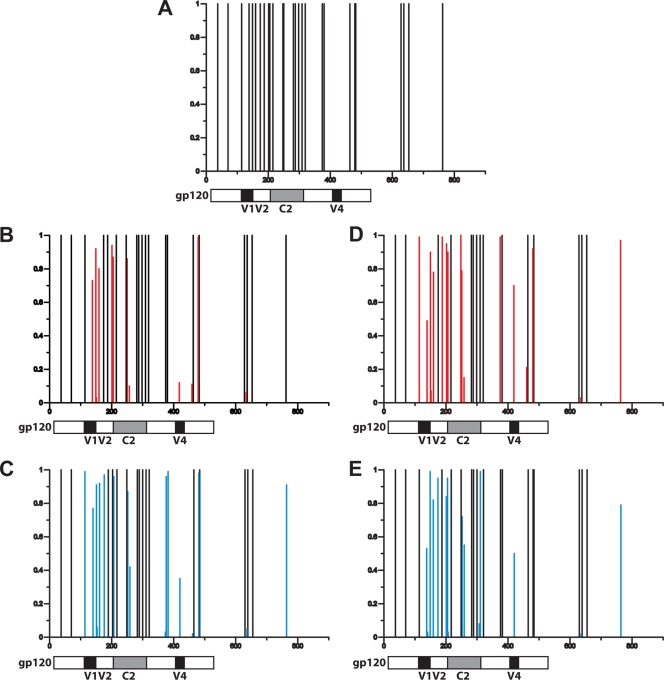
Since the glycan shields of the HIV/SIV envelopes can block antibody-mediated neutralization (33), we reasoned that the divergence between the env-vaccinated group and the other group of monkeys in their evolution of SIVmac251 neutralizing antibody responses may be a consequence of divergent evolution of Env glycosylation. To evaluate this possibility, we first determined the number of PNG sites in each animal's envelope sequences by counting Asn-X-Ser/Thr motifs (where X is not proline) using N-GlycoSite (www.hiv.lanl.gov). Twenty-seven PNG sites were present in clones 5 and 30 of the SIVmac251 challenge inoculum (Fig. 5). Comparison of the clones derived from the infected monkeys revealed that env vaccination was not associated with changes in the total number of PNG sites at weeks 37 or 62.
FIG. 5.
env vaccination did not change the number of SIV Env potential N-glycosylation sites in the circulating virus in the challenged monkeys. Potential N-glycosylation sites were similar in number between the two groups at week 37 and 62 postchallenge as well as within each group over time, as quantified by N-Glycosite. Each value corresponds to the average number of potential N-glycosylation sites from all clones sequenced from a particular animal of that group. The number of potential N-glycosylation sites in the challenge stock of SIVmac251 is presented as a straight line corresponding to 27 potential N-glycosylation sites. P values were determined by a two-sided Wilcoxon exact test. g/p/n, gag-pol-nef.
Although the number of Env PNG sites did not differ in the viruses obtained from the two groups of vaccinated monkeys, the distribution of the glycans may have evolved differently. An analysis of the distribution of the PNG sites in env from the virus challenge stock and the plasma samples from the two groups of monkeys showed that the replicating virus shifted glycosylation sites (Fig. 6). In comparison with the challenge stock virus, PNG sites in the env clones at week 37 from the gag-pol-nef-vaccinated monkeys were present at a lower frequency in variable region 1 (V1), V2, and constant region 2 (C2) and at a higher frequency in V4. Similar changes in the distribution of Env PNG sites occurred from week 37 to week 62 in animals that were vaccinated with env, with fewer sites observed in V1, V2, and C2 and more in V4. Since the PNG site changes were observed in virus from both groups of vaccinated monkeys, these findings suggest that the addition of the env immunogen may have affected the glycan shield in very subtle ways that are not statistically significant.
FIG. 6.
Distribution of potential N-glycosylation sites in the circulating virus in the challenged monkeys. The distribution and prevalence of potential N-glycosylation sites in each vaccinated group at week 37 and 62 postchallenge are represented using N-Glycosite (www.hiv.lanl.gov). The amino acid number of each potential N-glycosylation site is shown on the x axis. The y axis shows the prevalence of each potential N-glycosylation site graphed as a fraction of all sequenced env clones. The glycosylation patterns of clones are shown, as follows: clone 30 of the SIVmac251 challenge stock (A), clones from gag-pol-nef-vaccinated animals at week 37 (B), clones from gag-pol-nef+env vaccinees at week 37 (C), clones from gag-pol-nef vaccinees at week 62 (D), and clones from gag-pol-nef+env vaccinees at week 62 (E). Sequences from gp120 variable regions 1, 2, and 4 and constant region 2 are indicated in schematics below the graphs. The red lines indicate the changes in glycosylation sites in animals that were vaccinated with gag-pol-nef compared to the challenge stock while the blue lines depict changes in glycosylation sites in animals that were vaccinated with gag-pol-nef+env.
Amino acid deletions as a result of env vaccination.
Since a significant fraction of the env mutations occurred in the V1 and V4 regions, we explored these regions in greater depth. Alignment of the sequences of these variable regions with the sequences of the challenge virus revealed the development of in-frame deletions (Fig. 7). Deletions in the V1 region were heterogeneous in length and in location, and a number of these deletions altered an N-glycosylation site. Deletions were more homogeneous in length and location in the V4 region and often involved an addition of a new glycosylation site (Fig. 6).
FIG. 7.
Representative V1 and V4 deletions. A collection of the V1 deletions (A) and V4 deletions (B) observed in these monkeys is shown. The sequences of the gp120 variable regions 1 and 4 are in grey.
An analysis of deletions in the V1 and V4 regions in both vaccinated groups of monkeys is shown in Table 2. At week 37 following infection, env clones from five of the six gag-pol-nef-vaccinated animals had deletions in either V1 or V4 regions. Of the animals vaccinated with gag-pol-nef+env, only clones from animals 05-K, 13-K, and 63-K had deletions in V1, and no viruses from these animals in this group had V4 deletions. The differences in the viruses from the two groups of monkeys with regard to the presence or absence of a V4 deletion reached statistical significance using a two-sided Fisher exact test (P = 0.015). By week 62 following infection, all clones from the gag-pol-nef-vaccinated animals had deletions in V1, and three monkeys had clones that also had V4 deletions. In contrast, env clones at week 62 following infection from all monkeys in the gag-pol-nef+env-vaccinated group had deletions in V1 but not in V4. Differences between the two vaccinated groups of monkeys in the accumulation of either V1 or V4 deletions did not achieve statistical significance at this later time point.
TABLE 2.
A V4 deletion mutation in SIV env does not develop in env-vaccinated rhesus monkeys after virus challengea
| Vaccine | Monkey | % Sequenced clones with a deletion in indicated region at: |
|||
|---|---|---|---|---|---|
| Week 37 |
Week 62 |
||||
| V1 | V4 | V1 | V4 | ||
| gag-pol-nef | TKH | 71 | 24 | 32 | 95 |
| AC-44 | 83 | 3 | 68 | 0 | |
| AC-85 | 0 | 0 | 14 | 0 | |
| 26-J | 48 | 1 | 46 | 85 | |
| 27-I | 14 | 57 | 21 | 63 | |
| AD-26 | 0 | 44 | 17 | 0 | |
| gag-pol-nef +env | AV7P | 0 | 0 | 27 | 0 |
| 05-K | 19 | 0 | 53 | 0 | |
| 13-K | 38 | 0 | 100 | 0 | |
| 17-J | 0 | 0 | —b | —b | |
| AV2C | 0 | 0 | 23 | 0 | |
| 63-K | 35 | 0 | 40 | 0 | |
env clones amplified by SGA from plasma viral cDNA were sequenced and these sequences were aligned to SIVmac251 env of the challenge stock. The predominant env sequence changes that evolved over time were deletions that arose in the V1 and V4 regions.
Monkey 17-J was euthanized at week 45.
Env deletions confer viral escape from neutralizing antibodies.
We hypothesized the V1 and V4 deletions in the replicating virus in the gag-pol-nef-vaccinated group of monkeys might arise as a consequence of escape from neutralizing antibodies directed at these regions of the envelope. If this is the case, serum from the monkeys that were sampled early after infection would neutralize the wild-type Env-expressing pseudovirions but would not neutralize pseudovirions expressing Envs with either a V1 or V4 deletion. The evolution of only V1 deletions in the virus replicating in the gag-pol-nef+env-vaccinated group of monkeys may be a consequence of neutralization directed at the V1 but not the V4 region of Env. If this is true, serum sampled early after infection from these monkeys should not neutralize pseudovirions expressing Env with a V1 deletion.
In fact, the pattern of the specificity and time course of emerging neutralizing antibodies in these two groups of vaccinated monkeys were consistent with these predictions (Table 3). In the gag-pol-nef-vaccinated monkeys, WT virus-specific neutralizing antibodies were first detected in five of six monkeys at week 18 following challenge and in the sixth monkey at week 37 following challenge. Moreover, in four of these six animals, antibodies that neutralized the ΔV1 pseudovirions and antibodies that neutralized the ΔV4 pseudovirions emerged in synchrony. In the other two monkeys, they emerged staggered in time by only a single sampling time. In contrast to these findings, WT virus-specific neutralizing antibodies were first detected in gag-pol-nef+env-vaccinated monkeys 12 weeks or earlier following challenge in three of five monkeys and in the other two monkeys at 18 weeks following infection. Importantly, in four of the five animals in this group of vaccinated monkeys, detectable neutralizing antibodies specific for the ΔV1 Env-expressing pseudovirion were delayed by at least two sampling times in their emergence in comparison with neutralizing antibodies specific for the WT and ΔV4 mutant viruses. In the fifth animal (monkey 63-K), a single sampling time delay was seen in the emergence of the ΔV1-specific neutralizing antibody response in comparison with the WT-specific neutralizing antibodies. Eventually, virus variants containing V1 deletions were neutralized by secondary neutralizing antibodies that emerged at later time points. These findings suggest that the differences in virus sequence evolution between these two groups of vaccinated monkeys are a consequence of different selective pressures related to anti-Env antibody responses elicited by vaccination.
TABLE 3.
A V1 deletion mutation in SIV env confers escape from the emerging neutralizing antibodies in SIV-challenged rhesus monkeysb
aND, not determined due to toxicity to the reporter cells by serum from this monkey.
Serum neutralization of viruses pseudotyped with either the WT Env (clone 30 of the SIVmac251 challenge stock), Env containing a representative V1 deletion, or Env containing a representative V4 deletion. Serum sample time points are day of SIVmac251 challenge (D0) and noted weeks post-viral challenge (Wkpc). Titers that are >10 are highlighted.
DISCUSSION
Infection of rhesus monkeys with SIV provides an ideal model system to study the consequences of a vaccine-elicited immune response for the evolution of the AIDS virus following infection. Our study aimed to examine the role of env vaccination in shaping the generation of neutralizing antibodies and the kinetics of envelope evolution. The inclusion of env immunogens in the regimen with the gag-pol-nef immunogens did not result in better virologic control than was achieved with gag-pol-nef immunization alone, although it is likely that the present study was underpowered for making that comparison. Nevertheless, the comparable levels of viral replication in both groups of animals support our hypothesis that prior env vaccination, and not the level of viral replication, was responsible for the differences in immune responses and viral evolution in these two cohorts of monkeys.
We first examined the effect of env vaccination on the generation of a neutralizing antibody response to the challenge virus (Table 1). In the absence of env vaccination, neutralization of the WT SIV pseudovirion was most often detected in week 18 sera. Although the env immunogen in this study (24) did not induce an appreciable anti-SIV Env neutralizing response after vaccination prior to infection, the env immunization primed for an accelerated anti-SIV pseudovirion neutralizing antibody response that was detected as early as 7 weeks postchallenge in monkey AV2C. This early response is in agreement with other SIV/simian-human immunodeficiency virus (SHIV) studies showing that the env vaccination accelerates the development of a neutralizing antibody response after infection (2-5, 28, 37).
We employed SGA to sequence SIV envelope from the plasma of the two groups of animals to monitor the animals for divergent evolution of envelope sequence diversity. Since SIV infection leads to the generation of genetically diverse viral quasispecies in vivo, an accurate picture of the envelope diversity in each individual monkey can only be obtained using a sensitive PCR-based method that amplifies minority species in the presence of majority species. Although traditional PCR methods will yield envelope clones from the quasispecies, majority species will be preferentially amplified. Moreover, viral sequences generated using bulk amplification may not represent true viruses present in each monkey since template switching may occur during PCR amplification. We therefore used nested PCR amplification and limiting dilution of cDNA (SGA) to generate clones that reflect the in vivo sequence diversity of the viral envelopes (21).
Viral envelope sequences from week 37 and week 62 plasma samples represented chronic virus quasispecies that evolved under antibody selective pressure. By week 37 after infection, we found that env evolution had occurred in all animals, with a majority of mutations developing in variable regions 1 and 4 (Fig. 3A). Mutations in these regions of SIV env have been observed in other SIV envelope studies, and, in agreement with these studies, our data suggest that V1 and V4 regions are hotspots for SIV evolution (1, 7, 9, 13, 15, 17, 19, 29, 30, 38). Compared to env clones isolated at week 37 p.i., env clones from week 62 p.i. revealed a greater accumulation of amino acid mutations throughout envelope, as well as a more focused evolution in the V1 and V4 regions. Since envelope evolution is a mechanism employed by the virus to evade antibody neutralization (34, 40), the greater degree of env evolution observed in the env-vaccinated monkeys suggests that env vaccination, indeed, generated additional immune pressure on the replicating virus.
This immune pressure, however, did not drive dramatic changes in the glycan network. Compared to clones sequenced from monkeys vaccinated with only gag-pol-nef immunogens, clones isolated from gag-pol-nef+env vaccinees displayed a similar number of total PNG sites (Fig. 5). However, we did observe changes in glycosylation sites in variable loops 1 and 2 and constant region 2 as well as an expanded number of PNG sites in variable region 4 in viruses that appeared later in infection than with the SIVmac251 challenge stock. It is interesting that a new PNG site arose in the V4 region of envelope in both vaccinated groups (Fig. 6). Although a new PNG site in the V4 region may have influenced the sensitivity of individual SIV clones to antibody neutralization, it appears that env vaccination did not exert additional immune pressure in this region since the addition of PNG in the V4 region occurred in both groups at the same rate.
Our neutralization data verify earlier reports in the literature that these regions of SIV envelope may be sites for escape from antibody neutralization. In the present study, we chose a representative V1 deletion mutant envelope clone that had a minimum number of random mutations, insertions, and/or deletions in the gp120 or gp41 sequence; no evolution in the V4 region; and a common V1 deletion. A similar strategy was followed in choosing a representative V4 deletion mutant envelope clone. The V1 deletion mutant envelope was cloned from monkey AC-44 plasma, and the V4 deletion mutant envelope was cloned from monkey 27-I plasma. The mutant V4 envelope we used in the present study did not include the addition on one PNG site since the gain of PNG sites was observed in both groups of animals, and we focused our analysis to residues that were different between the two groups. Therefore, the predominant mutant envelopes and monkey sera tested in the neutralization assays did not represent matched monkey and envelope sets. Testing the neutralization of Env pseudoviruses using sera from unrelated monkeys could complicate the intended analysis. This may have contributed to the absence of a correlation between envelope sequences and neutralization data for animals TKH, AD-26, and AC-85 (Tables 2 and 3). Serum from monkey TKH neutralized the WT and mutant pseudovirions very poorly despite the large number of mutant envelope clones sequenced in this animal. This may have partly been the result of a mismatch between the serum and the founder/mutant envelopes. Sequencing envelope from monkey AD-26 showed a complex switch in V1 and V4 deletions between weeks 37 and 62. This switch complicated drawing conclusions about an association between virus neutralization by AD-26 neutralizing antibodies and virus sequence evolution, since neutralization escape could have been conferred by either the V1 or the V4 deletion. It is possible that we did not amplify the critical envelope variants at both time points. Furthermore, our sequencing data also demonstrated that deletions in the V1 and V4 regions were associated with random point mutations throughout Env. Therefore, we cannot rule out the possibility that the delayed neutralization of one or both pseudovirion-expressing mutant envelopes was attributable to mutations other than these deletions.
While unique envelope sequence diversity was observed in each evaluated monkey, env vaccination was associated with the absence of deletions in variable region 4 of viral envelope, suggesting that env vaccination conferred a generalized protection against the evolution of a V4 deletion in Env. If env vaccination had also primed for potent neutralizing antibodies against both the V1 and V4 regions, we would have expected antibody-directed V4 evolution in addition to the V1 evolution observed following infection. This absence of envelope V4 deletions in the gag-pol-nef+env-vaccinated group of monkeys could not be explained by a lower level of viral replication relative to the gag-pol-nef-vaccinated monkeys since env vaccination did not affect viral load in this cohort. This finding therefore suggests that env vaccination may have elicited an anti-SIV envelope antibody response directed against the V1 region only. In the gag-pol-nef+env-vaccinated group, the V4 region may not have been a target for neutralization, or antibodies targeting this region of envelope were not generated by vaccination. It is also possible that the V4 region deletion tracked with other changes in envelope that did not arise in this vaccinated group. A comparison of V1 deletions observed in the two groups of monkeys showed no statistically significant differences at the time points chosen for sequencing (data not shown). This finding suggests that the envelope vaccine construct did not confer protection against, or direct further, V1 evolution. While the mechanism underlying this phenomenon remains unclear, the present data suggest that env immunization may have little beneficial role with regard to antibody-mediated virus neutralization, since vaccination directed evolution to a variable region from which the virus readily mutated to escape neutralization. Hence, what is needed is an envelope immunogen that primes for antibodies directed to a more conserved region of envelope from which the virus cannot easily escape.
Acknowledgments
TZM.bl cells were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from John C. Kappes, Xiaoyun Wu, and Tranzyme, Inc. (4, 23, 26). We are grateful to Yair Benita for helpful discussions regarding sequencing results, Alida Ault and John Paul Todd for coordinating animal procedure logistics, Alan Dodson and Tammy Jenkins at BioQual, Inc., for housing and care of the nonhuman primates used in the study, Rebecca Gelman for help with statistical analyses, Richard M. White for help with Mutation Surveyor, and Joseph Sodroski for manuscript preparation.
This work was supported in part by funds from the intramural research program of the Vaccine Research Center, NIAID, NIH; NIH grant N01-AI30033; K08-AI06995 (W.W.Y.); and the Bill and Melinda Gates Collaboration for AIDS Vaccine Discovery Vaccine Immune Monitoring Consortium grant 38619 to M.S.S.
Footnotes
Published ahead of print on 11 November 2009.
REFERENCES
- 1.Almond, N., A. Jenkins, A. B. Heath, and P. Kitchin. 1993. Sequence variation in the env gene of simian immunodeficiency virus recovered from immunized macaques is predominantly in the V1 region. J. Gen. Virol. 74:865-871. [DOI] [PubMed] [Google Scholar]
- 2.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 3.Barouch, D. H., S. Santra, M. J. Kuroda, J. E. Schmitz, R. Plishka, A. Buckler-White, A. E. Gaitan, R. Zin, J. H. Nam, L. S. Wyatt, M. A. Lifton, C. E. Nickerson, B. Moss, D. C. Montefiori, V. M. Hirsch, and N. L. Letvin. 2001. Reduction of simian-human immunodeficiency virus 89.6P viremia in rhesus monkeys by recombinant modified vaccinia virus Ankara vaccination. J. Virol. 75:5151-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T. M. Fu, W. Wagner, M. Bilska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M. E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, M. G. Lewis, E. A. Emini, J. W. Shiver, and N. L. Letvin. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486-492. [DOI] [PubMed] [Google Scholar]
- 5.Buckner, C., L. G. Gines, C. J. Saunders, L. Vojtech, I. Srivastava, A. Gettie, R. Bohm, J. Blanchard, S. W. Barnett, J. T. Safrit, and L. Stamatatos. 2004. Priming B cell-mediated anti-HIV envelope responses by vaccination allows for the long-term control of infection in macaques exposed to a R5-tropic SHIV. Virology 320:167-180. [DOI] [PubMed] [Google Scholar]
- 6.Bunnik, E. M., L. Pisas, A. C. van Nuenen, and H. Schuitemaker. 2008. Autologous neutralizing humoral immunity and evolution of the viral envelope in the course of subtype B human immunodeficiency virus type 1 infection. J. Virol. 82:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns, D. P., and R. C. Desrosiers. 1991. Selection of genetic variants of simian immunodeficiency virus in persistently infected rhesus monkeys. J. Virol. 65:1843-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakrabarti, B. K., W. P. Kong, B. Y. Wu, Z. Y. Yang, J. Friborg, X. Ling, S. R. King, D. C. Montefiori, and G. J. Nabel. 2002. Modifications of the human immunodeficiency virus envelope glycoprotein enhance immunogenicity for genetic immunization. J. Virol. 76:5357-5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, W. S., C. Collignon, C. Thiriart, D. P. Burns, E. J. Stott, K. A. Kent, and R. C. Desrosiers. 1994. Effects of natural sequence variation on recognition by monoclonal antibodies neutralize simian immunodeficiency virus infectivity. J. Virol. 68:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desrosiers, R. C. 1988. Simian immunodeficiency viruses. Annu. Rev. Microbiol. 42:607-625. [DOI] [PubMed] [Google Scholar]
- 11.Franchini, G., C. Gurgo, H. G. Guo, R. C. Gallo, E. Collalti, K. A. Fargnoli, L. F. Hall, F. Wong-Staal, and M. S. Reitz, Jr. 1987. Sequence of simian immunodeficiency virus and its relationship to the human immunodeficiency viruses. Nature 328:539-543. [DOI] [PubMed] [Google Scholar]
- 12.Gray, E. S., P. L. Moore, I. A. Choge, J. M. Decker, F. Bibollet-Ruche, H. Li, N. Leseka, F. Treurnicht, K. Mlisana, G. M. Shaw, S. S. Karim, C. Williamson, and L. Morris. 2007. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J. Virol. 81:6187-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrington, P. R., J. A. Nelson, K. M. Kitrinos, and R. Swanstrom. 2007. Independent evolution of human immunodeficiency virus type 1 env V1/V2 and V4/V5 hypervariable regions during chronic infection. J. Virol. 81:5413-5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Julenius, K., A. Molgaard, R. Gupta, and S. Brunak. 2005. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology 15:153-164. [DOI] [PubMed] [Google Scholar]
- 15.Jurkiewicz, E., G. Hunsmann, J. Schaffner, T. Nisslein, W. Luke, and H. Petry. 1997. Identification of the V1 region as a linear neutralizing epitope of the simian immunodeficiency virus SIVmac envelope glycoprotein. J. Virol. 71:9475-9481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kannagi, M., J. M. Yetz, and N. L. Letvin. 1985. In vitro growth characteristics of simian T-lymphotropic virus type III. Proc. Natl. Acad. Sci. U. S. A. 82:7053-7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karlsson Hedestam, G. B., R. A. Fouchier, S. Phogat, D. R. Burton, J. Sodroski, and R. T. Wyatt. 2008. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat. Rev. Microbiol. 6:143-155. [DOI] [PubMed] [Google Scholar]
- 18.Kent, K. A. 1995. Neutralising epitopes of simian immunodeficiency virus envelope glycoprotein. J. Med. Primatol. 24:145-149. [DOI] [PubMed] [Google Scholar]
- 19.Kinsey, N. E., M. G. Anderson, T. J. Unangst, S. V. Joag, O. Narayan, M. C. Zink, and J. E. Clements. 1996. Antigenic variation of SIV: mutations in V4 alter the neutralization profile. Virology 221:14-21. [DOI] [PubMed] [Google Scholar]
- 20.Kwong, P. D., M. L. Doyle, D. J. Casper, C. Cicala, S. A. Leavitt, S. Majeed, T. D. Steenbeke, M. Venturi, I. Chaiken, M. Fung, H. Katinger, P. W. Parren, J. Robinson, D. Van Ryk, L. Wang, D. R. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678-682. [DOI] [PubMed] [Google Scholar]
- 21.Leonard, C. K., M. W. Spellman, L. Riddle, R. J. Harris, J. N. Thomas, and T. J. Gregory. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 265:10373-10382. [PubMed] [Google Scholar]
- 22.Letvin, N. L., M. D. Daniel, P. K. Sehgal, R. C. Desrosiers, R. D. Hunt, L. M. Waldron, J. J. MacKey, D. K. Schmidt, L. V. Chalifoux, and N. W. King. 1985. Induction of AIDS-like disease in macaque monkeys with T-cell tropic retrovirus STLV-III. Science 230:71-73. [DOI] [PubMed] [Google Scholar]
- 23.Letvin, N. L., Y. Huang, B. K. Chakrabarti, L. Xu, M. S. Seaman, K. Beaudry, B. Korioth-Schmitz, F. Yu, D. Rohne, K. L. Martin, A. Miura, W. P. Kong, Z. Y. Yang, R. S. Gelman, O. G. Golubeva, D. C. Montefiori, J. R. Mascola, and G. J. Nabel. 2004. Heterologous envelope immunogens contribute to AIDS vaccine protection in rhesus monkeys. J. Virol. 78:7490-7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Letvin, N. L., J. R. Mascola, Y. Sun, D. A. Gorgone, A. P. Buzby, L. Xu, Z. Y. Yang, B. Chakrabarti, S. S. Rao, J. E. Schmitz, D. C. Montefiori, B. R. Barker, F. L. Bookstein, and G. J. Nabel. 2006. Preserved CD4+ central memory T cells and survival in vaccinated SIV-challenged monkeys. Science 312:1530-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, M., F. Gao, J. R. Mascola, L. Stamatatos, V. R. Polonis, M. Koutsoukos, G. Voss, P. Goepfert, P. Gilbert, K. M. Greene, M. Bilska, D. L. Kothe, J. F. Salazar-Gonzalez, X. Wei, J. M. Decker, B. H. Hahn, and D. C. Montefiori. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108-10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montefiori, D. C. 2005. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr. Protoc Immunol. Unit 12.11. doi: 10.1002/0471142735.im1211s64. [DOI] [PubMed]
- 27.Moore, P. L., E. S. Gray, I. A. Choge, N. Ranchobe, K. Mlisana, S. S. Abdool Karim, C. Williamson, and L. Morris. 2008. The c3-v4 region is a major target of autologous neutralizing antibodies in human immunodeficiency virus type 1 subtype C infection. J. Virol. 82:1860-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ourmanov, I., M. Bilska, V. M. Hirsch, and D. C. Montefiori. 2000. Recombinant modified vaccinia virus Ankara expressing the surface gp120 of simian immunodeficiency virus (SIV) primes for a rapid neutralizing antibody response to SIV infection in macaques. J. Virol. 74:2960-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Overbaugh, J., and L. M. Rudensey. 1992. Alterations in potential sites for glycosylation predominate during evolution of the simian immunodeficiency virus envelope gene in macaques. J. Virol. 66:5937-5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Overbaugh, J., L. M. Rudensey, M. D. Papenhausen, R. E. Benveniste, and W. R. Morton. 1991. Variation in simian immunodeficiency virus env is confined to V1 and V4 during progression to simian AIDS. J. Virol. 65:7025-7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pantophlet, R., and D. R. Burton. 2006. GP120: target for neutralizing HIV-1 antibodies. Annu. Rev. Immunol. 24:739-769. [DOI] [PubMed] [Google Scholar]
- 32.Platt, E. J., K. Wehrly, S. E. Kuhmann, B. Chesebro, and D. Kabat. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reitter, J. N., R. E. Means, and R. C. Desrosiers. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679-684. [DOI] [PubMed] [Google Scholar]
- 34.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 100:4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rong, R., F. Bibollet-Ruche, J. Mulenga, S. Allen, J. L. Blackwell, and C. A. Derdeyn. 2007. Role of V1V2 and other human immunodeficiency virus type 1 envelope domains in resistance to autologous neutralization during clade C infection. J. Virol. 81:1350-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rong, R., S. Gnanakaran, J. M. Decker, F. Bibollet-Ruche, J. Taylor, J. N. Sfakianos, J. L. Mokili, M. Muldoon, J. Mulenga, S. Allen, B. H. Hahn, G. M. Shaw, J. L. Blackwell, B. T. Korber, E. Hunter, and C. A. Derdeyn. 2007. Unique mutational patterns in the envelope α2 amphipathic helix and acquisition of length in gp120 hypervariable domains are associated with resistance to autologous neutralization of subtype C human immunodeficiency virus type 1. J. Virol. 81:5658-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rose, N. F., P. A. Marx, A. Luckay, D. F. Nixon, W. J. Moretto, S. M. Donahoe, D. Montefiori, A. Roberts, L. Buonocore, and J. K. Rose. 2001. An effective AIDS vaccine based on live attenuated vesicular stomatitis virus recombinants. Cell 106:539-549. [DOI] [PubMed] [Google Scholar]
- 38.Sato, S., E. Yuste, W. A. Lauer, E. H. Chang, J. S. Morgan, J. G. Bixby, J. D. Lifson, R. C. Desrosiers, and W. E. Johnson. 2008. Potent antibody-mediated neutralization and evolution of antigenic escape variants of simian immunodeficiency virus strain SIVmac239 in vivo. J. Virol. 82:9739-9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]