Abstract
The gp350 glycoprotein encoded by BLLF1 is crucial for efficient Epstein-Barr virus (EBV) infection of resting B cells. Gp350 binds to CD21, but whether this interaction sums up its functions remains unknown. We generated gp350-null EBVs that display CD19-, CD21-, or CD22-specific antibodies at their surface (designated as ΔBLLF1-Ab). Gp350-complemented (ΔBLLF1-C) and ΔBLLF1-Ab were found to bind equally well to B cells. Surprisingly, ΔBLLF1 binding was reduced only 1.7-fold relative to its complemented counterparts. Furthermore, B cells exposed to ΔBLLF1-Ab or ΔBLLF1 viruses presented structural antigens with comparable efficiency and achieved 25 to 80% of the T-cell activation elicited by ΔBLLF1-C. These findings show that the gp350-CD21 interaction pair plays only a modest role during virus transfer to the endosomal compartment. However, primary B cells or Raji B cells infected with ΔBLLF1-C viruses displayed a 35- to 70-fold higher infection rates than those exposed to ΔBLLF1, ΔBLLF1-CD22Ab, or ΔBLLF1-CD19Ab viruses. Complementation of the gp350 knockout phenotype with CD21Ab substantially enhanced infection rates relative to ΔBLLF1 but remained sevenfold (Raji B-cell line) to sixfold (primary B cells) less efficient than with gp350. We therefore infer that gp350 mainly exerts its functions after the internalization step, presumably during release of the viral capsid from the endosomal compartment, and that CD21-dependent but also CD21-independent molecular mechanisms are involved in this process. The latter appear to be characteristic of B-cell infection since transfection of CD21 in 293 cells improved the infection rates with both ΔBLLF1-CD21Ab and ΔBLLF1-C to a similar extent.
The Epstein-Barr virus (EBV) is a B lymphotropic virus that infects its target cells through multiple interactions between envelope glycoproteins and their cognate cellular receptors (for a review, see reference 18). gp350 is the most abundant viral protein in the viral envelope. This large protein is encoded by the BLLF1 gene, is heavily glycosylated and localizes to various subcellular compartments (cytoplasm, endoplasmic reticulum, Golgi, and plasma membrane) of replicating cells (11, 17, 38, 44). Multiple reports provided solid evidence that EBV binds to primary B cells through its interaction with CD21, the complement receptor 2 (CR2) (23, 30-32, 47, 48). Several gp350 domains appear to be involved in the formation of a stable complex with CD21, one of which has been identified as the receptor-binding site (amino acids [aa] 142 to 161) (46). This glycan-free domain is also recognized by the neutralizing gp350-specific antibody 72A1 (15, 46). The introduction of specific mutations has allowed identification of further residues that contribute to gp350-CR2 binding (54). Virions treated with antibodies against gp350 have an enhanced ability to infect epithelial cells (50). Binding of gp350 to CD21 activates several pathways, in particular NF-κB, which results in interleukin-6 (IL-6) upregulation (6, 45). The addition of free gp350 to primary B cells leads to CD19 tyrosine phosphorylation and the subsequent recruitment of the phosphatidylinositol 3-kinase (41). Both of these kinase activities are necessary for efficient EBNA-LP and EBNA2 production shortly after infection, which suggests that gp350 contributes to this process. More generally, preincubation of resting B cells with gp350 markedly enhances the expression of transfected expression plasmids (42). Deletion of gp350 reduces B-cell infection efficiency by 2 orders of magnitude (19). Reciprocally, introduction of CR2 in nonlymphoid cells massively enhances infection rates in nonlymphoid cells (22). However, gp350-null mutants retain a residual infectious potential (19). This contrasts with other viral proteins such as gp110, gp85, or gp42 that are absolutely required for infection (18). This might reflect the fact that these glycoproteins are mainly involved in fusion with the target cell membrane, whereas gp350 appears to be required solely for target cell binding (13, 34, 52). However, it is unclear whether gp350 serves additional functions during viral infection in addition to its clear implication in virus binding. We also wanted to learn whether the gp350-CD21 binding sums up gp350's interactions with cellular proteins. We posited that if this were the case, a CD21-specific antibody should be able to fully complement the gp350-null phenotype. We therefore exchanged gp350 against antibodies directed against CD19, CD21, or CD22 within the viral envelope and investigated the ability of these proteins to confer full infectious potential to mature virions.
MATERIALS AND METHODS
Primary cells and cell lines.
HEK293 is a neuroendocrine cell line obtained by transformation of embryonic epithelial kidney cells with adenovirus (12). Raji is a human EBV-positive Burkitt's lymphoma cell line (36). Elijah 5E5 is a human EBV-negative Burkitt's lymphoma cell line (kindly provided by A. Rickinson). THB-5 is a hybridoma cell line that produces a monoclonal antibody against CD21 (10, 49). WI38 cells are primary human lung embryonic fibroblasts (14). Mononuclear cells were purified from fresh blood buffy coats by density gradient centrifugation. CD19-positive B cells were isolated from total lymphocytes by using M-450 CD19 (PanB) magnetic beads (Dynal). Cells were grown in RPMI 1640 medium supplemented with 10% fetal calf serum.
Plasmids.
A BZLF1 expression plasmid (p509) was used to initiate the viral lytic cycle (37). A cytomegalovirus (CMV)-driven expression plasmid for BLLF1 based on the pcDNA3.1(+) vector (p2385) was used for transcomplementation experiments of the ΔBLLF1 phenotype (19).
cDNA was prepared from primary B cells and used as a template for PCR-mediated amplification of CD19, CD21, and CD22 by using specific primers designed according to sequences deposited in the NCBI database GenBank (NM_001770, NM_001877, and NM_001771). The PCR products were placed under the control of a strong CMV promoter by cloning onto the pRK5 expression plasmid.
Cloning of the THB-5 antibody.
To clone the anti-CD21 heavy- and light-chain expression plasmids, mRNA from THB-5 was reverse transcribed by using a Stratagene cDNA synthesis kit. The thus-obtained cDNA was used as a template for PCR-mediated amplification of both immunoglobulin chains. The following primers specific for the light chain were used: 5′Mk, 5′-GGGAGCTCGAYATTGTGMTSACMCARWCTMCA-3′; 5-5, 5′-TAGCAAAGTGATCCTTTGC-3′; 10-5, 5′-GTCAGCACATTAGGGAGCT-3′; and L3C, 5′-CTAACACTCATTCCTGTTGAAGCTCTTGAC-3′. The degenerated primer 5′Mk is specific for a wide range of kappa chains (53). Primers 5-5 and 10-5 are specific for aberrant kappa chains found in the THB-5 hybridoma. L3C recognizes the murine immunoglobulin kappa chain constant region (accession number NG_005612).
The primers amplifying the heavy chain were VH1BACK (5′-AGGTSMARCTGCAGSAGTCWGG-3′) and H3C (5′-TACATGGTACCAGGCTTTGCTGGATGCTTT-3′). VH1BACK is a degenerated primer that amplifies the immunoglobulin heavy-chain variable regions (35). H3C binds to the immunoglobulin G2a (IgG2a) constant region (accession number J00470). After successful amplification and sequencing of the CD21 antibody variable regions, sequence of the respective leader peptides were obtained from the NCBI GenBank (accession number XM_001472961). This analysis allowed the design of primers that include the following leader peptides. HC US (5′-TAGACAGAATTCATGGCTGTCCTGGTGCTGTTC-3′) primes in the native leader peptide of the anti-CD21 heavy chain. The primer aCD21CH2 DS (5′-CTCGATAGCGCTTTTGGGTTTTGAGATGGTTCT-3′) primes at the 3′ region of CH2 of IgG2a. The CH3 domain of the CD21 antibody heavy chain is not included in these constructs.
All 5′ primers included an EcoRI restriction site, L3C included a HindIII restriction site, and IgVH3 included an Eco47III restriction site that facilitated cloning onto the pRK5 expression plasmid.
Construction of the gp350 antibody heavy-chain fusion proteins.
A fragment of BLLF1 comprising the gp350 transmembrane and intraviral domain (nucleotide positions 2568 to 2721 of BLLF1 [46, 48]) was PCR amplified by using the primers gp350US (5′-GTTGGAATTCAGCGCTTTCTCAAACCTCTCCATGCTA-3′) and gp350DS (5′-AAGCAAGCTTTTATACATAGGTCTCGGCGTCATC-3′) and the plasmid p2385 as a template. The resulting fragment was cloned in pRK5 via its EcoRI and HindIII sites. The additional Eco47III site of gp350US was used for cloning of the immunoglobulin heavy chain.
To generate similar fusion proteins between gp350 and the antibodies directed against CD19 and CD22, the variable regions of the anti-CD21 antibody were swapped with those of pHOG-scFv18-CD19 and pHOG21-CD22scFV-MJ7, respectively, by overlap PCR (16). All plasmids were entirely sequenced, and only clones free of adventitious mutations were selected for further studies.
EBV mutants.
The wild-type EBV recombinant plasmid used in the present study consists of a B95.8 genome cloned onto the prokaryotic F factor origin of replication. This recombinant virus carries the green fluorescent protein (GFP) and gene cassettes that confer resistance against chloramphenicol and hygromycin (8). The EBV BLLF1-negative mutant was constructed by replacing the complete BLLF1 gene (B95.8 coordinates 89433 to 92153; accession number NC_007605) with the kanamycin resistance gene by homologous recombination (33). To do this, composite primers were used whose internal parts (24 bp; underlined in the sequences below) are specific for the kanamycin resistance gene and whose external parts (40 bp) are specific for the BLLF1 gene. These primers (L1-Kan1 [5′-AAAGTGCAAATTTCTGATTAATAAATTTTTATTGACTTTAACCAGTCACGACGTTGTAAAACGAC-3′] and L1-Kan2 [5′-GGCGTAATCATGGTCATAGCTGTTTGTCTCGGCACCGATTTCTAG GCAGCATCCTCTTTAATA-3′]) allowed PCR-mediated amplification of the kanamycin resistance gene through their internal sequences and then homologous recombination of the amplified PCR product with the EBV wild-type genome via their external sequences. PCR amplification products were incubated with the restriction enzyme DpnI to remove traces of the parental plasmid and introduced by electroporation (1,000 V, 25 μF, 100 Ω) into Escherichia coli DH10B cells carrying the EBV wild type and the temperature-sensitive pKD46 helper plasmid that encodes the phage lambda red recombinase to foster homologous recombination and the ampicillin resistance gene (7). Cells were grown in Luria broth (LB) with chloramphenicol (15 μg/ml) at 37°C for an hour and then plated onto LB agar plates containing chloramphenicol (15 μg/ml) and kanamycin (10 μg/ml). Incubation at 42°C led to a progressive loss of the helper plasmid, which restored sensitivity to ampicillin. After selection, DNA of positive clones was purified and analyzed with the XhoI restriction enzyme to ensure that recombination had taken place correctly. Homologous recombination via the flanking regions resulted in the replacement of the BLLF1 gene with the kanamycin resistance cassette and loss of a XhoI restriction site.
Stable clone selection and virus production.
293 cells were transfected with the properly recombined ΔBLLF1 viral DNA by using Lipofectamine (Invitrogen) as described previously (19). At day 1 posttransfection, the cells were transferred to a 150-mm cell culture dish, and hygromycin (100 μg/ml) was added to the culture medium to induce selection of 293 clones stably carrying the EBV recombinant plasmid. Outgrowing GFP-positive colonies were expanded for further investigation. 293 clones carrying the BLLF1-negative EBV mutant, designated 293/ΔBLLF1, were transfected with a BZLF1 expression plasmid to induce the lytic cycle by using lipid micelles (Metafectene; Biontex) as described previously (34). In transcomplementation assays, 293/ΔBLLF1 were cotransfected with a BLLF1 expression plasmid or with expression plasmids for one of the CD19-, CD21-, or CD22-specific antibodies. Virus supernatants were harvested 4 days posttransfection and filtered through 0.45-μm-pore-size filter. All experiments were performed with supernatants purified by centrifugation (3,000 × g, 30 min) using a 300-kDa cutoff centrifugal device (Microsep; Pall Corp.). This procedure also led to a twofold concentration of virus supernatants.
Quantitative real-time PCR.
Structural virus titers were determined by quantitative real-time PCR (qPCR). Supernatants were first digested with DNase I (5 U/50 μl of supernatant) at 37°C for 1 h. After DNase I heat inactivation (10 min at 70°C), supernatants were mixed (1:1 [vol/vol]) with lysis buffer (0.1 mg of proteinase K/ml in water) and incubated for 60 min at 50°C, after which the enzyme was heat inactivated (75°C for 20 min). Water-diluted supernatants (1:10) or genomic DNAs were subjected to qPCR using primers and a probe specific to the BALF5 DNA polymerase (Pol) gene. Amplification reactions were performed in 25-μl volumes containing 12.5 μl of TaqMan Universal master mix (Applied Biosystems), 2.5 μl of forward and reverse Pol primers (2 μM), 1 μl of 5 μM FAM-labeled Pol probe, 1.5 μl of water, and 5 μl of virus supernatant or DNA. After activation of the AmpliTaq Gold DNA Pol (10 min at 95°C), the reaction mixtures were amplified for 40 cycles (15 s at 95°C and 60 s at 60°C), and the fluorescent signals were monitored by using an ABI 7300 sequence detection system (Applied Biosystems). The primer and probe sequences used were as follows: reverse Pol primer, 5′-AGTCCTTCTTGGCTAGTCTGTTGAC-3′; forward Pol primer, 5′-CTTTGGCGCGGATCCTC-3′; and EBV Pol probe, 5′-FAM-CATCAAGAAGCTGCTGGCGGCC-TAMRA-3′. The DNA content was calculated by using a serial dilution of Namalwa DNA, a human Burkitt's lymphoma cell line containing two EBV genome copies per cell, as a reference for the standard curve.
Infection, immortalization, and binding assays.
Functional virus titers were determined by infecting 104 Raji cells with increasing dilutions of supernatants. At 3 days postinfection, GFP-positive cells were counted.
Five thousand 293 cells transfected with either CD19, CD21, or CD22 were infected at a multiplicity of infection (MOI) of 100. At 3 days postinfection, GFP-positive cells were counted in a counting chamber under UV light. Infection experiments were conducted in the presence or absence of chloroquine (50 μM final concentration).
For immortalization experiments, primary B cells were infected at an MOI of 10 genome equivalents (geq) per cell (as defined by qPCR) with infectious supernatants and seeded into U-bottom 96-well microtiter plates coated with 103 gamma-irradiated WI38 feeder cells at a concentration of 2.5 × 104 cells per well. Wells containing outgrowing LCL clones were visually counted.
Elijah 5E5 were incubated with viral particles at an MOI of 10 at 4°C under agitation to measure the efficiency of binding. After 3 h, the cells were washed with phosphate-buffered saline (PBS), and the genomic DNA was extracted by using a DNeasy tissue kit (Qiagen). A total of 350 pg of genomic DNA was used as input DNA for the qPCR as described above.
Western blot.
Western blot analyses were conducted to detect immunoglobulin heavy and light chains. To do this, the cells were resuspended in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 0.5% Nonidet P-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 5 mM EDTA, 20 mM Tris-HCl), lysed by sonication, and denatured in Laemmli buffer for 10 min at 95°C. gp350 immunoblots were also performed, in which case the cells were resuspended in RIPA buffer including glass powder and Laemmli buffer exempt of mercaptoethanol. After vigorous vortexing for 2 min, the tested samples were denatured for 10 min at 95°C.
A similar method was applied to detect proteins in mature virions. Supernatants from induced cells were ultracentrifuged at 20,000 × g for 2 h. Virus pellets were then resuspended in RIPA buffer and Laemmli buffer, and debris was discarded after centrifugation at 16,000 × g for 5 min. Here again, mercaptoethanol-induced protein denaturation was used to facilitate the visualization of immunoglobulin-specific signals.
The samples were separated on a sodium dodecyl sulfate-polyacrylamide gel and electroblotted onto a Hybond ECL membrane (Amersham). After preincubation for 30 min in 5% milk powder in PBS, blots were incubated with a goat anti-mouse IgG conjugated to horseradish peroxidase (Promega) or a mouse monoclonal antibody against gp350 (clone 72A1 [15]) for 1 h at room temperature. The gp350 blot was further incubated for 1 h with a goat anti-mouse antibody coupled with horseradish peroxidase (Promega). Antibody binding was revealed by using an enhanced chemiluminescence detection reagent (Amersham).
Immunostaining.
Cells were washed three times in PBS, dried on glass slides, and treated with pure acetone for 20 min at room temperature. Slides were then incubated for 30 min with a purified mouse monoclonal antibody directed against gp350 (clone 72A1) (15) or with supernatant from cells transfected with the previously described expression plasmids for immunoglobulin heavy and light chains against CD21, CD19, or CD22, respectively. Slides were then washed three times in PBS and incubated for 30 min with a secondary goat anti-mouse antibody conjugated with the Cy3 fluorochrome (Dianova). After several washes in PBS, slides were embedded with 90% glycerol, and the immunofluorescence was evaluated by using a fluorescence microscope (Leica).
T-cell antigen recognition assays.
T-cell antigen recognition assays were used to identify virus internalization in the endosomal-lysosomal compartment as previously described (2). Mini-LCLs were obtained by transformation of primary B cells with the mini-EBV, a recombinant plasmid that contains the EBV latent genes but not all lytic genes and therefore cannot enter the replicative cycle (27). Mini-LCL cells (105/well) were incubated with three increasing dilutions of viral supernatant containing the same MOI for 48 h. T cells (105) were then incubated with the infected B cells and placed in culture for 24 h. Gamma interferon (IFN-γ) released in the culture medium after antigen-specific B-cell/T-cell interactions was quantified with an enzyme-linked immunosorbent assay system (R&D Systems). The CD4+ T-cell clones have been isolated from an EBV-seropositive donor and cultivated as described previously (1).
RESULTS
Construction of a gp350 knockout devoid of the entire BLLF1 open reading frame.
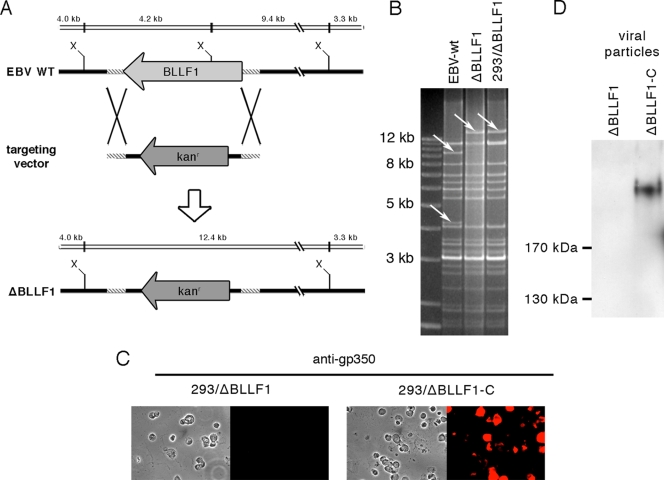
We have previously reported the phenotypic traits of a recombinant EBV devoid of the BLLF1 gene (19). However, only part of the BLLF1 open reading frame in this mutant virus was deleted. Although we have no reason to think that this mutant kept any functional gp350-mediated residual activity, we were concerned that truncated gp350 proteins might interfere with efficient insertion of foreign proteins into the viral envelope. We therefore generated a recombinant virus that is devoid of the complete BLLF1 reading frame. The followed strategy is depicted in Fig. 1A; the kanamycin resistance cassette was exchanged against the entire BLLF1 open reading frame through homologous recombination. Insertion of the targeting vector into the EBV recombinant plasmid led to an alteration of the restriction enzyme pattern that allowed unequivocal identification of properly rearranged clones. Figure 1B illustrates that this strategy led to the exchange of the 4.2- and 9.4-kb XhoI DNA segments against a unique 12.4-kb fragment; this result is in accord with the restriction maps of the BLLF1 locus in the wild-type genome or after BLLF1 deletion as presented in Fig. 1A. Correctly recombined clones were then stably introduced into 293 cells to yield virus producer cell lines, designated 293/ΔBLLF1. These cells were lytically induced, and gp350 expression was eventually visualized by immunostains (Fig. 1C). This test was negative in 293/ΔBLLF1 cells, whereas the positive control (gp350-complemented 293/ΔBLLF1 cells, designated 293/ΔBLLF1-C) exhibited unequivocal signals (Fig. 1C). These results were validated by a Western blot analysis conducted on purified virions; only the complemented ΔBLLF1 displayed gp350-specific signals (Fig. 1D). As a further control, viral episomes were shuttled back from 293/ΔBLLF1 cells into E. coli to allow restriction analysis. This test confirmed both the identity and the integrity of the ΔBLLF1 recombinant virus in the 293 producer cell line (Fig. 1B).
FIG. 1.
Construction and characterization of a ΔBLLF1 recombinant virus. (A) Schematic map of the EBV-wt and the ΔBLLF1 genomes generated by exchange of the BLLF1 gene against the kanamycin resistance cassette (kanr). The regions of homology between the targeting vector and the wild-type genome are indicated (hatched line). The figure includes the cleavage sites for XhoI (X) and the expected fragment sizes after restriction analysis of EBV-wt and ΔBLLF1 genomes. (B) XhoI restriction fragment analysis of EBV-wt (lane 1) and ΔBLLF1 mutant genomes after construction in E. coli (lane 2) and rescue from 293/ΔBLLF1 cells (lane 3). The result is consistent with the predicted restriction pattern (see panel A). (C) Gp350 protein production was examined by immunostaining of induced 293/ΔBLLF1 and 293/ΔBLLF1-C cells with a gp350-specific antibody. (D) Detection of gp350 within purified viral particles produced by 293/ΔBLLF1 (left) and 293/ΔBLLF1-C (right) by Western blot analysis with a gp350-specific antibody.
Construction of gp350-antibody fusion proteins.
Having excised the main binding protein from the EBV genome, we investigated the ability of various antibodies directed against B-cell-specific epitopes to mediate virus-cell binding and infection. To this aim, we generated chimeric proteins that consisted of the gp350 intracytoplasmic and transmembrane domains fused in frame with parts of the antibody heavy chains (Fig. 2A). cDNAs that encode the variable regions of murine antibodies directed against CD19 and CD22 are available (4, 21). In contrast, we needed first to clone the CD21-specific antibody from the THB-5 hybridoma by using a PCR-mediated approach with degenerated primers that cover the majority of the murine immunoglobulin variable-region gene repertoire (35, 53). This approach swiftly led to the isolation of the THB-5 heavy chain that could be introduced into an expression plasmid. Cloning of the corresponding THB-5 light chain proved more difficult since our approach led to the preferential amplification of a previously characterized aberrant gene encoded by the myeloma cell line used to generate the THB-5 hybridoma (5). We therefore screened PCR amplificates cloned on the pGEM-T vector with primers that specifically identifies this aberrant kappa chain (51). Only clones amplified from THB-5 that were not recognized by this set of primers were further examined. Sequencing of these selected clones revealed that they either encoded an additional aberrant light chain that appeared to be nonfunctional as a result of an out-of-frame mutation or an as-yet-unpublished functional kappa chain that we presumed to be THB-5 light-chain gene. Support for this assumption came from a partial protein sequence of the purified THB-5 antibody that included regions from the light-chain variable domain; both DNA and partial protein sequences were concordant (data not shown). Comparison of the identified THB-5 heavy- and light-chain clones with the mouse genome allowed identification of the corresponding signal peptides and generation of complete clones that encompass the signal peptides, the variable and constant immunoglobulin regions. Sequences of both light and heavy chains are available from the GenBank database (accession numbers GQ850526 and GQ850527). THB-5 genes were cloned onto an expression plasmid and transiently transfected into 293 cells. Supernatants from these cells were found to stain 293 cells transfected with a CD21 expression plasmid but not untransfected control cells (Fig. 2B). This provided definitive evidence that the previously identified clones genuinely encode the THB-5 antibody. Similar experiments were performed to confirm the specificity of the CD19 and CD22 antibodies (Fig. 2B). Fusion proteins containing the gp350 transmembrane and intracytoplasmic domains were fused with parts of CD21 antibody heavy chain, including the leader peptide, the variable regions, and some of the constant domain. Preliminary experiments showed that constructs comprising the CH1 and CH2 constant regions yielded the best results in terms of complementation and were selected for further experiments (data not shown). We then cotransfected 293/ΔBLLF1 cells with BZLF1, the THB-5 light chain, and the THB-5 heavy-chain gp350 fusion protein. Expression of the modified CD21 antibody in cells undergoing the EBV lytic cycle should lead to its incorporation in the viral envelope. We therefore carried out a Western blot analysis of both the induced producer cells and the thus-produced viruses with antibodies specific for murine heavy or light chains (Fig. 2C and data not shown). These experiments demonstrated clear expression of the THB-5 light chain and of the THB-5 heavy-chain gp350 fusion protein in both types of samples. The same strategy was applied to the construction of CD19- and CD22-gp350 fusion proteins (data not shown).
FIG. 2.
Construction of gp350-Ab fusion proteins. (A) Schematic representation of immunoglobulin heavy chain-gp350 chimera (top) and of the immunoglobulin light-chain genes (bottom). The organization of leader peptide (lp), variable, and constant region, and in case of the heavy chain, the gp350 intracytoplasmic and trans-membrane domains are depicted. CH1/CH2 correspond to the heavy chain constant domain 1 and 2. (B) Immunostains were carried out on 293 cells (left panel) or 293 cells transfected with CD19, CD21, or CD22 (right panel) with supernatants from 293 cells transfected with CD21, CD19, and CD22 antibodies. (C) Western blot analysis of purified viral particles obtained from induced 293/ΔBLLF1 (lane 1), 293/ΔBLLF1-C (lane 2) or 293/ΔBLLF1-CD21Ab cells (lane 3). Cell extracts from the THB-5 hybridoma were used as a positive control (lane 4). Please note that the THB-5 cell line produces heavy chains, including the heavy-chain constant domain three which accounts for the difference in size. An antibody specific to mouse immunoglobulin heavy and light chains was used to attest incorporation of the gp350-CD21 antibody construct into the viral particles.
CD21-EBV exhibits normal binding and endosome entry but is less infectious than complemented counterparts.
We initiated the study of the 293/ΔBLLF1-CD21Ab phenotypic traits by gauging structural virus titers using a qPCR-based approach (Fig. 3A). 293/ΔBLLF1 mutant cells produced on average 107 virus equivalents per ml of supernatants, a value perfectly in consonance with those pointed out in positive control cells that carry wild-type virus (9) (Fig. 3A). Complementation with a gp350 expression plasmid (293/ΔBLLF1-C) did not significantly alter virus titers, as previously observed (19). This confirms that gp350 is not required for virus maturation. In contrast, 293/ΔBLLF1-Ab viruses suffered a threefold titer reduction, suggesting that incorporation of the Ab-gp350 chimeras had an adverse effect on virus production (Fig. 3A). However, this mild titer reduction did not significantly impede further analysis of 293/ΔBLLF1-Ab's properties.
FIG. 3.
Phenotypic traits of the ΔBLLF1-Ab recombinant viruses. Each chart represents the mean value of at least three experiments. Error bars correspond to the standard deviation. (A) Virus titers (genome equivalents, geq) in supernatants from lytically induced 293/ΔBLLF1, 293/ΔBLLF1-C, 293/ΔBLLF1-CD21Ab, 293/ΔBLLF1-CD19Ab, and 293/ΔBLLF1-CD22Ab were determined by qPCR using primers specific for the BALF5 gene. (B) Raji B cells were infected with decreasing amounts of supernatants from lytically induced 293/ΔBLLF1, 293/ΔBLLF1-C, and 293/ΔBLLF1-Ab cells. This assay delivered functional titers expressed as GFP-positive Raji cells per ml of supernatant. (C) Functional titers in 293/ΔBLLF1, 293/ΔBLLF1-C, and 293/ΔBLLF1-Ab supernatants were determined by immortalization assays in cluster plates at an MOI of 10. The results are given as a percentage of ΔBLLF1-C viruses immortalizing capacity. (D) The numbers of bound EBV genomes per cell after infection of EBV-negative Elijah B cells with 293/ΔBLLF1, 293/ΔBLLF1-C, and 293/ΔBLLF1-Ab at an MOI of 10 was quantitated by qPCR. The results are given as percentages of ΔBLLF1-C virus binding efficiency. (E, F, and G) Mini-LCL cells were pulsed with 0.1, 0.3, or 1.0 geq/cell. Subsequently, T cells specific for gp350, BNRF1, or BcLF1 were added, and IFN-γ release was measured by enzyme-linked immunosorbent assay 44 h later. The results of a representative experiment are presented.
We then carried out infection experiments on the Raji B-cell line and on primary B cells using column-purified virions. Determination of the number of GFP-positive Raji cells or of EBV-transformed B-cell clones provided a readout with which to quantify functional virus titers. The results of three independent experiments are presented in Fig. 3B; they document that gp350 complementation improved virus titers in Raji cells by 70-fold and in primary B cells by 35-fold. Nevertheless, gp350-null viruses retained a residual infectivity, as previously noted (19). Exposure of Raji cells to 293/ΔBLLF1-CD21 yielded intermediate results; titers witnessed a 10-fold increase but were still seven times lower than complemented virions. Concordant results were obtained with B-cell transformation assays; incorporation of gp350-CD21 into virions resulted in a sixfold-increased titer that nevertheless remained sixfold lower than those measured after genuine complementation with gp350 (Fig. 3C). Complementation with CD22Ab had a modest effect (1.8-fold) on primary B-cell infection that could not be reproduced with Raji cells, whereas infection with 293/ΔBLLF1-CD19Ab and gp350-negative viruses yielded indistinguishable results.
Next, we conducted a series of experiments that aimed at investigating the various steps of infection. We first evaluated the efficiency of binding by carrying out three independent binding assays during which viruses were allowed to bind to their target cells under conditions that preclude virus internalization (4°C) (29). These experiments therefore exclusively detect EBV bound at the cell surface. After elimination of free viruses by extensive washings, the average number of virions bound per cell was determined by using qPCR. The results of this approach, summarized in Fig. 3D, demonstrate that ΔBLLF1's binding ability is impaired by 1.7-fold relative to complemented counterparts and is therefore far from being abolished. ΔBLLF1-CD21Ab and ΔBLLF1-CD19Ab viruses bound to their target cells as efficiently as ΔBLLF1-C, whereas ΔBLLF1-CD22Ab viruses reached 80% of the complemented level. Therefore, the CD19Ab- and CD21Ab-gp350 chimeras can efficiently compensate for the loss of gp350 with regard to B-cell binding.
A thorough analysis of postbinding events is difficult in EBV-infected cells, not least because the low EBV virus titers impede direct observation of the infectious process by electron microscopy. We therefore used an indirect approach to gauge virus transfer to the endosome-lysosome compartment. Previous work has established that B cells exposed to virions that are able to access the endosome efficiently present viral antigens to major histocompatibility complex (MHC) class II-restricted T cells (2). We assessed here the B-cell presentation of three virus epitopes derived from gp350, the tegument protein BNRF1, and the capsid protein BcLF1 (Fig. 3E, F, and G).
T cells specific for gp350 recognized solely B cells infected with ΔBLLF1-C virions, indirectly demonstrating that ΔBLLF1 viruses lack this glycoprotein (Fig. 3E). All tested samples elicited a BNRF1- and BcLF1-specific T-cell response, although only ΔBLLF1-C reached cytokine release levels seen after infection with wild-type virions (Fig. 3F and G). ΔBLLF1-Ab and ΔBLLF1 triggered similar levels of cytokine release that amounted to between 40 and 80% of the control levels (293/ΔBLLF1-C).
Enhanced infection of 293 cells that express lymphoid markers with pseudotyped virions.
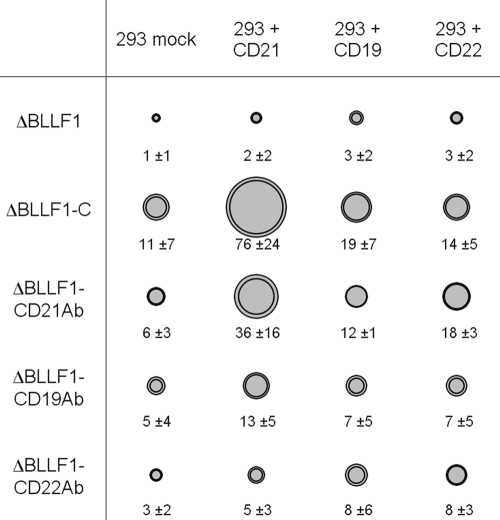
It has previously been reported that transfection of CD21 into epithelial cells markedly enhances EBV infection rates (22). We pursued a similar strategy to assess the ability of the antibody-complemented virions to infect 293 cells transfected with CD19, CD21, or CD22 (Fig. 4). It emerged from three independent experiments that 293 cell transfection with CD21 or CD22, but not CD19, increased infection rates with virions expressing the respective specific antibody, although the effect observed after CD22 transfection was marginal. In contrast, the improvement in the mean value of infection rates compared to mock-transfected cells reached 6.0- and 6.9-fold after CD21 transfection for ΔBLLF1-CD21Ab and ΔBLLF1-C, respectively. It is interesting that transfection of CD19 or CD22 in 293 cells also led to a 1.3- to 3.0-fold increase in infection rates with these viruses relative to mock transfectants. However, it remains difficult to ascertain that this represents a genuine effect, rather than a statistical fluctuation (Fig. 4). We then repeated these experiments in the presence of chloroquine, which prevents the acidification of endosomal vesicles (29). We found that chloroquine largely blocked infection, thereby demonstrating that EBV infected 293 cells along an endosomal route in that case (data not shown).
FIG. 4.
293 cell infection rates after transfection with CD19, CD21, or CD22 and infection with ΔBLLF1, ΔBLLF1-C, and the ΔBLLF1-Ab viruses. Infection rates are given as surface graph. The outer circle represents standard deviation. The number of positive cells per 5,000 infected cells is indicated.
DISCUSSION
The marked tropism for primary B cells that epitomizes EBV infection is mirrored at the molecular level by the interaction between CD21, a characteristic if not specific B-cell marker and gp350, the most abundant viral envelope protein. Binding between these two molecules has been seen as evidence to characterize gp350 as the main mediator of virus binding to B cells (31). We have taken these issues further by exchanging gp350 against antibodies specific for CD19, CD21, and CD22 at the viral surface. Fusion of these immunoglobulins with gp350's transmembrane domain led to their efficient incorporation into the viral envelope, thereby adding to the evidence that herpesvirus surface glycoproteins can be replaced by various cellular proteins, such as cellular receptor ligands or antibodies, as recently demonstrated for herpes simplex virus type 1 (HSV-1) (20, 25, 55).
The central observation of the present study is that CD21Ab only partly complemented the gp350-null phenotype during B-cell infection. Although we favor the view that this finding implies that gp350's functions extend beyond mere binding to CD21, it is important to exclude that the differences noticed between gp350 and CD21Ab were not a reflection of a lower binding capacity of the CD21Ab relative to gp350 when inserted in the viral envelope. In contrast, the CD21-specific antibody might bind so tightly to its target that it hinders virus release. Although exclusion of these alternative scenarios would require exact determination of the respective affinity of CD21Ab and gp350 to CD21, the finding that ΔBLLF1-C and ΔBLLF1-CD21Ab bind equally well to B cells suggests that this was not the case. A significant difference between the CD21-specific antibody and gp350 in terms of their respective affinity for CD21 would be expected to lead to different infection rates in CD21-positive 293 cells. However, transfection of CD21 in 293 cells equally boosted infection rates after exposure to ΔBLLF1-CD21Ab or to ΔBLLF1-C, relative to mock-transfected cells.
Interestingly, BLLF1-null viruses retained a substantial binding capacity; while deletion of gp350 reduced infection rates by up to 70-fold, it only halved the binding efficiency. From these findings we infer that viral molecules other than gp350 are equally important for EBV binding and that gp350 serves functions that extend beyond cell surface binding. Indeed, there is one report that glycoproteins involved in virus fusion can contribute to epithelial cell binding (26). Double mutants that lack both gp350 and one of the remaining viral glycoproteins should help clarifying this issue.
For many viruses, including HSV-1, binding is a very transitory event immediately followed by fusion with various cell membranes to ensure access to intracellular organelles (28, 43). In contrast, EBV must first enter the primary B-cell endosomal compartment, where the actual fusion takes place (18). The fusion itself has been found to require at least the three viral glycoproteins gp110, gp42, and gp85 (13, 34, 52). How EBV accesses the endosome has not been studied in detail at the molecular level, but the common perception is that it uses the CD21-coupled vesicle cargo system to reach the endosome. This suggests that the interactions between CD21 and gp350 extend from the initial binding step at the cellular surface until incorporation into the endosome and fusion. We have previously found that only a minority of viruses bound to the B-cell surface ultimately reach the cell nucleus (40). This shows that binding is to some extent independent from internalization. Therefore, no reliable conclusions about internalization can be drawn from recorded levels of binding. In an attempt to understand the role of the gp350-CD21 pair at this stage of the infection, we monitored access to the endosomal compartment through assessment of MHC class II-restricted viral epitopes presentation efficiency. This experiment indicated very efficient presentation of gp350, BNRF1, and BcRF1 after infection with ΔBLLF1-C, at levels indistinguishable from those observed with wild-type virus. Infection with ΔBLLF1-Ab and ΔBLLF1 elicited two- to threefold weaker T-cell responses against BcLF1 and BNRF1. As expected, infection with all of these samples failed to induce cytokine release with gp350-specific T cells.
From these data we conclude that gp350 is dispensable for transfer to the endosome and antigen presentation. It is also likely that gp350 induces endosome transfer more efficiently than CD21Ab but that the effect remains modest, as already noticed for surface binding. Whether the BLLF1-null viruses access the endosomal compartment through the same route as their wild-type homologues is unclear at present. However, this is probably the case since it is unlikely that the cellular proteins involved in gp350-independent virus entry would segregate from CD21 during internalization; this would have deleterious consequences for the efficiency of the infection process.
The mild effects of gp350 on cell surface binding and transfer to the endosome cannot account for the 70-fold drop in infection that follows its deletion. gp350's main functions must therefore take place downstream of the endosome transfer. Enveloped viruses that invade their target cells through the endosome characteristically go through a process of de-envelopment by fusion with the endosomal membranes that allows the release of envelope-free capsids into the cytoplasm, although there is only sparse evidence that this applies to EBV (29). If one posits that this model is correct, we must conclude that gp350 plays a crucial role at some stage during this de-envelopment. The results of infections carried out with ΔBLLF1-CD21Ab also suggest that binding to CD21 did not entirely account for gp350's functions. Comparison of gp350 and CD21Ab's effects needs to take into account ΔBLLF1-CD21Ab's reduced incorporation into the endosome relative to ΔBLLF1-C. However, we deem it unlikely that this effect could explain the six- to sevenfold difference in B-cell infection rates observed after exposure to either ΔBLLF1-C or ΔBLLF1-CD21Ab.
Pseudotyping or alteration of virus tropism is a recurring theme. It can be achieved by incorporation of a protein from one viral species into a virus from a different species (e.g., G protein from VSV and HSV [3]). At the other end of the spectrum, restriction of viral tropism can be sought by incorporation of cellular proteins that specifically recognize cells that belong to a particular lineage (e.g., retargeting HSV-1 to HER2 [24]). Our results suggest that the suitability of EBV for such a strategy might be rather limited. Although incorporation of CD19Ab and CD22Ab efficiently mediated binding to B cells, it did not translate into an increased infection rate. Transfection of CD22 into 293 cells weakly enhanced infection with viruses expressing the CD22Ab, suggesting that a CD21-independent EBV infection pathway might exist. However, CD22 is also linked to the endosome (39). Furthermore, it remains important to note that BLLF1-null viruses retained a substantial residual infectious potential that might impede specific targeting of non-B cells. Defining the viral and cellular proteins involved in the CD21-independent binding might help in defining a strategy to circumvent this problem.
Acknowledgments
This study was funded by grant 106168 from the Deutsche Krebshilfe to H.-J.D. and by the Deutsche Forschungsgemeinschaft (SFB455 [J.M.]). C.B. received a Ph.D. stipend from Genzyme Virotech GmbH.
Footnotes
Published ahead of print on 4 November 2009.
REFERENCES
- 1.Adhikary, D., U. Behrends, H. Boerschmann, A. Pfunder, S. Burdach, A. Moosmann, K. Witter, G. W. Bornkamm, and J. Mautner. 2007. Immunodominance of lytic cycle antigens in Epstein-Barr virus-specific CD4+ T-cell preparations for therapy. PLoS ONE 2:e583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adhikary, D., U. Behrends, A. Moosmann, K. Witter, G. W. Bornkamm, and J. Mautner. 2006. Control of Epstein-Barr virus infection in vitro by T helper cells specific for virion glycoproteins. J. Exp. Med. 203:995-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, D. B., S. Laquerre, K. Ghosh, H. P. Ghosh, W. F. Goins, J. B. Cohen, and J. C. Glorioso. 2000. Pseudotyping of glycoprotein D-deficient herpes simplex virus type 1 with vesicular stomatitis virus glycoprotein G enables mutant virus attachment and entry. J. Virol. 74:2481-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arndt, M. A., J. Krauss, R. Schwarzenbacher, B. K. Vu, S. Greene, and S. M. Rybak. 2003. Generation of a highly stable, internalizing anti-CD22 single-chain Fv fragment for targeting non-Hodgkin's lymphoma. Int. J. Cancer 107:822-829. [DOI] [PubMed] [Google Scholar]
- 5.Carroll, W. L., E. Mendel, and S. Levy. 1988. Hybridoma fusion cell lines contain an aberrant kappa transcript. Mol. Immunol. 25:991-995. [DOI] [PubMed] [Google Scholar]
- 6.D'Addario, M., T. A. Libermann, J. Xu, A. Ahmad, and J. Menezes. 2001. Epstein-Barr Virus and its glycoprotein-350 upregulate IL-6 in human B-lymphocytes via CD21, involving activation of NF-κB and different signaling pathways. J. Mol. Biol. 308:501-514. [DOI] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delecluse, H. J., T. Hilsendegen, D. Pich, R. Zeidler, and W. Hammerschmidt. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. USA 95:8245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feederle, R., H. Bannert, H. Lips, N. Muller-Lantzsch, and H. J. Delecluse. 2009. The Epstein-Barr virus alkaline exonuclease BGLF5 serves pleiotropic functions in virus replication. J. Virol. 83:4952-4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fingeroth, J. D., J. J. Weis, T. F. Tedder, J. L. Strominger, P. A. Biro, and D. T. Fearon. 1984. Epstein-Barr virus receptor of human B lymphocytes is the C3d receptor CR2. Proc. Natl. Acad. Sci. USA 81:4510-4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong, M., and E. Kieff. 1990. Intracellular trafficking of two major Epstein-Barr virus glycoproteins, gp350/220 and gp110. J. Virol. 64:1507-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59-74. [DOI] [PubMed] [Google Scholar]
- 13.Haddad, R. S., and L. M. Hutt-Fletcher. 1989. Depletion of glycoprotein gp85 from virosomes made with Epstein-Barr virus proteins abolishes their ability to fuse with virus receptor-bearing cells. J. Virol. 63:4998-5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayflick, L. 1965. The limited in vitro lifetime of human diploid cell strains. Exp. Cell Res. 37:614-636. [DOI] [PubMed] [Google Scholar]
- 15.Hoffman, G. J., S. G. Lazarowitz, and S. D. Hayward. 1980. Monoclonal antibody against a 250,000-dalton glycoprotein of Epstein-Barr virus identifies a membrane antigen and a neutralizing antigen. Proc. Natl. Acad. Sci. USA 77:2979-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 17.Hummel, M., D. Thorley-Lawson, and E. Kieff. 1984. An Epstein-Barr virus DNA fragment encodes messages for the two major envelope glycoproteins (gp350/300 and gp220/200). J. Virol. 49:413-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutt-Fletcher, L. M. 2007. Epstein-Barr virus entry. J. Virol. 81:7825-7832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janz, A., M. Oezel, C. Kurzeder, J. Mautner, D. Pich, M. Kost, W. Hammerschmidt, and H. J. Delecluse. 2000. Infectious Epstein-Barr virus lacking major glycoprotein BLLF1 (gp350/220) demonstrates the existence of additional viral ligands. J. Virol. 74:10142-10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamiyama, H., G. Zhou, and B. Roizman. 2005. Herpes simplex virus 1 recombinant virions exhibiting the amino-terminal fragment of urokinase-type plasminogen activator can enter cells via the cognate receptor. Gene Ther. 13:621-629. [DOI] [PubMed] [Google Scholar]
- 21.Kipriyanov, S. M., G. Moldenhauer, G. Strauss, and M. Little. 1998. Bispecific CD3 × CD19 diabody for T cell-mediated lysis of malignant human B cells. Int. J. Cancer 77:763-772. [DOI] [PubMed] [Google Scholar]
- 22.Li, Q. X., L. S. Young, G. Niedobitek, C. W. Dawson, M. Birkenbach, F. Wang, and A. B. Rickinson. 1992. Epstein-Barr virus infection and replication in a human epithelial cell system. Nature 356:347-350. [DOI] [PubMed] [Google Scholar]
- 23.Lowell, C. A., L. B. Klickstein, R. H. Carter, J. A. Mitchell, D. T. Fearon, and J. M. Ahearn. 1989. Mapping of the Epstein-Barr virus and C3dg binding sites to a common domain on complement receptor type 2. J. Exp. Med. 170:1931-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menotti, L., A. Cerretani, H. Hengel, and G. Campadelli-Fiume. 2008. Construction of a fully retargeted herpes simplex virus 1 recombinant capable of entering cells solely via human epidermal growth factor receptor 2. J. Virol. 82:10153-10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menotti, L., G. Nicoletti, V. Gatta, S. Croci, L. Landuzzi, C. De Giovanni, P. Nanni, P. L. Lollini, and G. Campadelli-Fiume. 2009. Inhibition of human tumor growth in mice by an oncolytic herpes simplex virus designed to target solely HER-2-positive cells. Proc. Natl. Acad. Sci. USA 106:9039-9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molesworth, S. J., C. M. Lake, C. M. Borza, S. M. Turk, and L. M. Hutt-Fletcher. 2000. Epstein-Barr virus gH is essential for penetration of B cells but also plays a role in attachment of virus to epithelial cells. J. Virol. 74:6324-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moosmann, A., N. Khan, M. Cobbold, C. Zentz, H. J. Delecluse, G. Hollweck, A. D. Hislop, N. W. Blake, D. Croom-Carter, B. Wollenberg, P. A. Moss, R. Zeidler, A. B. Rickinson, and W. Hammerschmidt. 2002. B cells immortalized by a mini-Epstein-Barr virus encoding a foreign antigen efficiently reactivate specific cytotoxic T cells. Blood 100:1755-1764. [PubMed] [Google Scholar]
- 28.Morgan, C., H. M. Rose, and B. Mednis. 1968. Electron microscopy of herpes simplex virus. I. Entry. J. Virol. 2:507-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nemerow, G. R., and N. R. Cooper. 1984. Early events in the infection of human B lymphocytes by Epstein-Barr virus: the internalization process. Virology 132:186-198. [DOI] [PubMed] [Google Scholar]
- 30.Nemerow, G. R., R. A. Houghten, M. D. Moore, and N. R. Cooper. 1989. Identification of an epitope in the major envelope protein of Epstein-Barr virus that mediates viral binding to the B lymphocyte EBV receptor (CR2). Cell 56:369-377. [DOI] [PubMed] [Google Scholar]
- 31.Nemerow, G. R., C. Mold, V. K. Schwend, V. Tollefson, and N. R. Cooper. 1987. Identification of gp350 as the viral glycoprotein mediating attachment of Epstein-Barr virus (EBV) to the EBV/C3d receptor of B cells: sequence homology of gp350 and C3 complement fragment C3d. J. Virol. 61:1416-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nemerow, G. R., J. J. Mullen III, P. W. Dickson, and N. R. Cooper. 1990. Soluble recombinant CR2 (CD21) inhibits Epstein-Barr virus infection. J. Virol. 64:1348-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neuhierl, B., and H. J. Delecluse. 2005. Molecular genetics of DNA viruses: recombinant virus technology. Methods Mol. Biol. 292:353-370. [DOI] [PubMed] [Google Scholar]
- 34.Neuhierl, B., R. Feederle, D. Adhikary, B. Hub, K. Geletneky, J. Mautner, and H.-J. Delecluse. 2009. Primary B-cell infection with a ΔBALF4 Epstein-Barr virus comes to a halt in the endosomal compartment yet still elicits a potent CD4-positive cytotoxic T-cell response. J. Virol. 83:4616-4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orlandi, R., D. H. Gussow, P. T. Jones, and G. Winter. 1989. Cloning immunoglobulin variable domains for expression by the polymerase chain reaction. Proc. Natl. Acad. Sci. USA 86:3833-3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pulvertaft, R. J. V. 1964. Cytology of Burkitt's tumour (African lymphoma). Lancet 283:238-240. [DOI] [PubMed] [Google Scholar]
- 37.Rooney, C. M., D. T. Rowe, T. Ragot, and P. J. Farrell. 1989. The spliced BZLF1 gene of Epstein-Barr virus (EBV) transactivates an early EBV promoter and induces the virus productive cycle. J. Virol. 63:3109-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Serafini-Cessi, F., N. Malagolini, M. Nanni, F. Dall'Olio, G. Campadelli-Fiume, J. Tanner, and E. Kieff. 1989. Characterization of N- and O-linked oligosaccharides of glycoprotein 350 from Epstein-Barr virus. Virology 170:1-10. [DOI] [PubMed] [Google Scholar]
- 39.Shan, D., and O. W. Press. 1995. Constitutive endocytosis and degradation of CD22 by human B cells. J. Immunol. 154:4466-4475. [PubMed] [Google Scholar]
- 40.Shannon-Lowe, C., G. Baldwin, R. Feederle, A. Bell, A. Rickinson, and H. J. Delecluse. 2005. Epstein-Barr virus-induced B-cell transformation: quantitating events from virus binding to cell outgrowth. J. Gen. Virol. 86:3009-3019. [DOI] [PubMed] [Google Scholar]
- 41.Sinclair, A. J., and P. J. Farrell. 1995. Host cell requirements for efficient infection of quiescent primary B lymphocytes by Epstein-Barr virus. J. Virol. 69:5461-5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sinclair, A. J., I. Palmero, G. Peters, and P. J. Farrell. 1994. EBNA-2 and EBNA-LP cooperate to cause G0 to G1 transition during immortalization of resting human B lymphocytes by Epstein-Barr virus. EMBO J. 13:3321-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spear, P. G., and R. Longnecker. 2003. Herpesvirus entry: an update. J. Virol. 77:10179-10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strnad, B. C., M. R. Adams, and H. Rabin. 1983. Glycosylation pathways of two major Epstein-Barr virus membrane antigens. Virology 127:168-176. [DOI] [PubMed] [Google Scholar]
- 45.Sugano, N., W. Chen, M. L. Roberts, and N. R. Cooper. 1997. Epstein-Barr virus binding to CD21 activates the initial viral promoter via NF-κB induction. J. Exp. Med. 186:731-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szakonyi, G., M. G. Klein, J. P. Hannan, K. A. Young, R. Z. Ma, R. Asokan, V. M. Holers, and X. S. Chen. 2006. Structure of the Epstein-Barr virus major envelope glycoprotein. Nat. Struct. Mol. Biol. 13:996-1001. [DOI] [PubMed] [Google Scholar]
- 47.Tanner, J., J. Weis, D. Fearon, Y. Whang, and E. Kieff. 1987. Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping, and endocytosis. Cell 50:203-213. [DOI] [PubMed] [Google Scholar]
- 48.Tanner, J., Y. Whang, J. Sample, A. Sears, and E. Kieff. 1988. Soluble gp350/220 and deletion mutant glycoproteins block Epstein-Barr virus adsorption to lymphocytes. J. Virol. 62:4452-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tedder, T. F., L. T. Clement, and M. D. Cooper. 1984. Expression of C3d receptors during human B-cell differentiation: immunofluorescence analysis with the HB-5 monoclonal antibody. J. Immunol. 133:678-683. [PubMed] [Google Scholar]
- 50.Turk, S. M., R. Jiang, L. S. Chesnokova, and L. M. Hutt-Fletcher. 2006. Antibodies to gp350/220 enhance the ability of Epstein-Barr virus to infect epithelial cells. J. Virol. 80:9628-9633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vidarsson, G., J. G. van de Winkel, and M. A. van Dijk. 2001. Multiplex screening for functionally rearranged immunoglobulin variable regions reveals expression of hybridoma-specific aberrant V-genes. J. Immunol. Methods 249:245-252. [DOI] [PubMed] [Google Scholar]
- 52.Wang, X., and L. M. Hutt-Fletcher. 1998. Epstein-Barr virus lacking glycoprotein gp42 can bind to B cells but is not able to infect. J. Virol. 72:158-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, Z., M. Raifu, M. Howard, L. Smith, D. Hansen, R. Goldsby, and D. Ratner. 2000. Universal PCR amplification of mouse immunoglobulin gene variable regions: the design of degenerate primers and an assessment of the effect of DNA polymerase 3′ to 5′ exonuclease activity. J. Immunol. Methods 233:167-177. [DOI] [PubMed] [Google Scholar]
- 54.Young, K. A., A. P. Herbert, P. N. Barlow, V. M. Holers, and J. P. Hannan. 2008. Molecular basis of the interaction between complement receptor type 2 (CR2/CD21) and Epstein-Barr virus glycoprotein gp350. J. Virol. 82:11217-11227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou, G., and B. Roizman. 2005. Characterization of a recombinant herpes simplex virus 1 designed to enter cells via the IL13Rα2 receptor of malignant glioma cells. J. Virol. 79:5272-5277. [DOI] [PMC free article] [PubMed] [Google Scholar]