Abstract
In infected cells, hepatitis C virus (HCV) induces the formation of membrane alterations referred to as membranous webs, which are sites of RNA replication. In addition, HCV RNA replication also occurs in smaller membrane structures that are associated with the endoplasmic reticulum. However, cellular mechanisms involved in the formation of HCV replication complexes remain largely unknown. Here, we used brefeldin A (BFA) to investigate cellular mechanisms involved in HCV infection. BFA acts on cell membranes by interfering with the activation of several members of the family of ADP-ribosylation factors (ARF), which can lead to a wide range of inhibitory actions on membrane-associated mechanisms of the secretory and endocytic pathways. Our data show that HCV RNA replication is highly sensitive to BFA. Individual knockdown of the cellular targets of BFA using RNA interference and the use of a specific pharmacological inhibitor identified GBF1, a guanine nucleotide exchange factor for small GTPases of the ARF family, as a host factor critically involved in HCV replication. Furthermore, overexpression of a BFA-resistant GBF1 mutant rescued HCV replication in BFA-treated cells, indicating that GBF1 is the BFA-sensitive factor required for HCV replication. Finally, immunofluorescence and electron microscopy analyses indicated that BFA does not block the formation of membranous web-like structures induced by expression of HCV proteins in a nonreplicative context, suggesting that GBF1 is probably involved not in the formation of HCV replication complexes but, rather, in their activity. Altogether, our results highlight a functional connection between the early secretory pathway and HCV RNA replication.
Hepatitis C virus (HCV) is an important human pathogen. It mainly infects human hepatocytes, and this often leads to chronic hepatitis, cirrhosis, or hepatocarcinoma. HCV studies have been hampered for many years by the difficulty in propagating this virus in vitro. Things have recently changed with the development of a cell culture model referred to as HCVcc (34, 60, 65), which allows the study of the HCV life cycle in cell culture and facilitates studies of the interactions between HCV and the host cell.
HCV is an enveloped positive-strand RNA virus belonging to the family Flaviviridae (35). The viral genome contains a single open reading frame, which is flanked by two noncoding regions that are required for translation and replication. All viral proteins that are produced after proteolytic processing of the initially synthesized polyprotein are membrane associated (15, 43). This reflects the fact that virtually all steps of the viral life cycle occur in close association with cellular membranes.
Interactions of HCV with cell membranes begin during entry. Several receptors, coreceptors, and other entry factors have been discovered over the years, which link HCV entry to specialized domains of the plasma membrane, such as tetraspanin-enriched microdomains and tight junctions (8, 16, 59). The internalization of the viral particle occurs by clathrin-mediated endocytosis (5, 40). The fusion of the viral envelope with the membrane of an acidic endosome likely mediates the transfer of the viral genome to the cytosol of the cell (5, 40, 57). However, little is known regarding the pre- and postfusion intracellular transport steps of entering viruses in the endocytic pathway.
HCV RNA replication is also associated with cellular membranes. Replication begins with the translation of the genomic RNA of an incoming virus. This leads to the production of viral proteins, which in turn initiate the actual replication of the viral RNA. Mechanisms regulating the transition from the translation of the genomic RNA to its replication are not yet known. All viral proteins are not involved in RNA replication. Studies performed with subgenomic replicons demonstrated that proteins NS3-4A, NS4B, NS5A, and NS5B are necessary and sufficient for replication (6, 27, 37). RNA replication proceeds through the synthesis of a cRNA strand (negative strand), catalyzed by the RNA-dependent RNA polymerase activity of NS5B, which is then used as a template for the synthesis of new positive strands.
Electron microscopy studies using a subgenomic replicon model suggested that replication takes place in membrane structures made of small vesicles, referred to as “membranous webs,” which are induced by the virus (26). Membranous webs are detectable not only in cells carrying subgenomic replicons but also in infected cells (50). They appear to be associated with the endoplasmic reticulum (ER) (26). In addition to the membranous webs, a second type of ER-associated replicase that is smaller and more mobile has recently been described (63). Cellular mechanisms leading to these membrane alterations are still poorly understood. In cells replicating and secreting infectious viruses effectively, the situation appears to be even more complex, since replicase components appear to be, at least in part, associated with cytoplasmic lipid droplets (41, 50, 56). This association depends on the capsid protein (41) and may reflect a coupling between replication and assembly. Indeed, HCV assembly and secretion show some similarities with very-low-density lipoprotein (VLDL) maturation and secretion (24, 64).
Our knowledge of the cellular membrane mechanisms involved in the HCV life cycle is still limited. The expression of NS4B alone induces membrane alterations that are reminiscent of membranous webs (19). However, cellular factors that participate in this process are still unknown. On the other hand, several cellular proteins potentially involved in the HCV life cycle have been identified through their interactions with viral proteins. For some of these proteins, a functional role in infection was recently confirmed using RNA interference (48). It is very likely that other cellular factors critical to HCV infection have yet to be identified.
To gain more insight into cellular mechanisms underlying HCV infection, we made use of brefeldin A (BFA), a macrocyclic lactone of fungal origin that exhibits a wide range of inhibitory actions on membrane-associated mechanisms of the secretory and endocytic pathways (30). BFA acts on cell membranes by interfering with the activation of several members of the family of ADP-ribosylation factors (ARFs). ARFs are small GTP-binding proteins of the Ras superfamily. They function as regulators of vesicular traffic, actin remodeling, and phospholipid metabolism by recruiting effectors to membranes. BFA does not actually interfere directly with ARF GTPases but rather interferes with their activation by regulators known as guanine nucleotide exchange factors (GEFs) (14, 25). We now report the identification of an ARF GEF as a cellular BFA-sensitive factor that is required for HCV replication.
MATERIALS AND METHODS
Chemicals.
Dulbecco's modified Eagle's medium (DMEM), phosphate-buffered saline (PBS), OptiMEM, Oligofectamine, Geneticin, goat serum, horse serum, and fetal calf serum (FCS) were purchased from Invitrogen. 4′,6-Diamidino-2-phenylindole (DAPI) was from Molecular Probes. Golgicide A and Mowiol 3-88 were from Calbiochem. Fugene-6 was from Roche. ExGen500 was purchased from Euromedex. Dharmafect-1 was from ThermoFischer Scientific. Other chemicals were from Sigma.
Antibodies.
Rat anti-E2 monoclonal antibody (MAb) 3/11 (20), mouse anti-E1 MAb A4 (17), and mouse anti-bovine viral diarrhea virus (anti-BVDV) NS3 MAb Osc-23 (7) were produced in vitro by using a MiniPerm apparatus (Heraeus) as recommended by the manufacturer. Anti-BVDV NS3 MAb was purified by affinity chromatography using protein A-Sepharose and conjugated to Alexa 488 using a kit from Invitrogen. Rabbit antiserum to HCV NS4 (both NS4A and NS4B of strain H) was obtained by immunization against a bacterially expressed TrpE fusion protein. Mouse anti-CD81 MAb 5A6 (47) was kindly provided by S. Levy (Stanford University). Mouse anti-E2 MAb AP33 (11) was kindly provided by A. H. Patel (Institute of Virology, Glasgow). Sheep anti-NS5A antiserum (38) was kindly provided by M. Harris (University of Leeds). Mouse anti-NS5A MAb was purchased from Austral Biologicals. Mouse anti-HCV NS3 MAb (1848) was from Virostat. Mouse anti-GM130, anti-Rab5, and anti-GBF1 MAbs were from Transduction Laboratories. Mouse anti-BIG1 and anti-BIG2 MAbs were from Bethyl Laboratories. Rabbit anticalnexin polyclonal antibody was from Stressgen. Biotinylated anti-CD63 MAb was from Ancell. Mouse antitubulin MAb (clone TUB 2.1) was from Sigma. Goat antiactin polyclonal antibody (I-19) and peroxidase-conjugated donkey anti-goat immunoglobulin G (IgG) were from Santa Cruz Biotechnology. Alexa 594-conjugated and Alexa 555-conjugated goat anti-mouse IgG, Alexa 488-conjugated donkey anti-mouse IgG, and Alexa 555-conjugated donkey anti-sheep IgG antibodies were from Molecular Probes. Peroxidase-conjugated goat anti-mouse, anti-rabbit, and anti-rat IgG were from Jackson Immunoresearch. Peroxidase-conjugated neutravidin was from Pierce.
Cell culture.
HEK 293T cells and Huh-7 cells were grown in DMEM supplemented with glutamax-I and 10% fetal bovine serum. UHCV-11 cells were kindly provided by D. Moradpour (University of Lausanne, Switzerland). They were grown in DMEM supplemented with glutamax-1, 10% fetal bovine serum, 1 μg/ml puromycin, 0.4 mg/ml Geneticin, and 1 μg/ml tetracycline (42). MDBK cells were grown in DMEM supplemented with glutamax-I and 10% horse serum.
HCVcc.
The virus used in this study was based on JFH1 (60), kindly provided by T. Wakita (National Institute of Infectious Diseases, Tokyo, Japan), and contained cell culture-adaptive mutations CS and N6 (13). To facilitate the detection of E1 in infected cells, the substitutions T197S, S199G, S200L, and M202H were introduced by overlapping PCR using primers 5′-CAGGTGAA GAATAGCAGTGGCCTCTACCATGTGACCAATGACTGC-3′, 5′-GCAGTCATTGGTCACATGGTAGAGGCCACTGCTATTCTTCACCTG-3′, 5′-GAGAGCCATAGTGGTCTGCGG-3′, and 5′-CCCGCTAACGATGTCTATGATGACCTCG-3′, followed by ligation into pJFH1/CS-N6 at the AgeI and BsiWI restriction sites. These substitutions reconstituted an epitope that is recognized by anti-E1 MAb A4 (17). The resulting plasmid was named pJFH1/CS-N6-A4. HCVcc expressing Renilla luciferase was as previously described (49). An in-frame deletion introduced in the E1E2 region of constructs with a Renilla luciferase reporter or puromycin acetyltransferase selection marker was as previously described (60). Nonreplicative controls contained a GND mutation in the NS5B active site, as previously reported (60).
To generate genomic HCV RNA, plasmids were linearized at the 3′ end of the HCV cDNA by XbaI digestion. Following treatment with mung bean nuclease, the linearized DNA was then used as a template for in vitro transcription with the MEGAscript kit from Ambion. In vitro-transcribed RNA was delivered to Huh-7 cells by electroporation as previously described (29). Viral stocks were obtained as previously reported (13).
For the HCVcc infection assay, Huh-7 cells grown in 24-well plates were infected for 2 h at 37°C. For experiments using small interfering RNA (siRNA), equal numbers of cells treated with each siRNA were infected. For experiments with BFA or golgicide A, cells were preincubated for 30 min before infection, and the drug was added to the infection and culture media up to 6 h postinfection (for dose-response experiments), or up to the end of the experiment (22 or 24 h postinfection), except where otherwise stated. In some experiments, BFA was added at various time points postinfection. Infections were scored by measuring luciferase activity in cell lysates using a Renilla luciferase assay system from Promega or by indirect immunofluorescence microscopy at 24 h postinfection.
Subgenomic replicon.
The subgenomic replicon used in this work was kindly provided by Stanley M. Lemon (University of Texas Medical Branch, Galveston) (27). To generate subgenomic replicon RNA, the plasmid was linearized by XbaI digestion and then used as a template for in vitro transcription with the MEGAscript kit from Ambion. In vitro-transcribed RNA was delivered to Huh-7 cells by electroporation. Cells containing the subgenomic replicon were selected in culture medium supplemented with 0.5 mg/ml Geneticin, and individual clones were isolated using cloning cylinders. The presence of the subgenomic replicon was confirmed by immunoblot and immunofluorescence analyses.
HCVpp.
Pseudotyped particles were produced as described previously (1). Briefly, 293T cells were cotransfected with a murine leukemia virus (MLV)-based transfer vector encoding luciferase (46), a murine leukemia virus Gag-Pol packaging construct, and an envelope glycoprotein-expressing vector, phCMV-E1E2 (1), using Exgen 500 as recommended by the manufacturer. The phCMV-G, and phCMV-RD114 expression vectors, encoding the vesicular stomatitis virus G protein (VSV G), and the feline endogenous virus RD114 glycoprotein, respectively, were used to produce control pseudotyped particles harboring VSV G or RD114 envelope glycoproteins on murine leukemia virus cores (VSVpp and RD114pp, respectively). The luciferase-based HCVpp infection assay was as previously described (46).
Other viruses.
Bovine viral diarrhea virus (BVDV) strain NADL and human adenovirus 5 were used as controls. BVDV was produced as previously described (33). MDBK cells were plated in six-well plates and infected 4 h later. Cells were preincubated for 30 min, infected for 1 h at 37°C with BVDV at a multiplicity of infection (MOI) of about 1, and cultured for 15 h in the presence of increasing doses of BFA. Infected and control cells were trypsinized, rinsed in PBS, fixed with 3% paraformaldehyde, incubated for 1 h at 4°C with Alexa 488-conjugated MAb to NS3 in the presence of 0.05% saponin to detect intracellular signals, and rinsed twice with PBS. NS3-positive cells were counted by flow cytometry using a FACS Beckman EPICS-XL MCL.
A recombinant adenovirus expressing the green fluorescent protein (GFP) was kindly provided by Didier Monté (Institut de Biologie de Lille). The stock of adenovirus was produced as previously described (2). Huh-7 cells were plated in six-well plates and infected the day after. Cells were preincubated for 30 min, infected for 1 h at 37°C, and cultured in the presence of BFA or golgicide A. The drug was removed at 7 h postinfection, and the cells were cultured for 16 h with no BFA. Cells were trypsinized, rinsed with PBS, and fixed with 3% paraformaldehyde. GFP-expressing cells were counted by flow cytometry.
RNA interference.
RNA interference experiments were carried out with pools of four different synthetic double-stranded siRNAs to the same target (on-target plus smart pool reagents from Dharmacon). For HCVcc experiments, subconfluent cultures of Huh-7 cells in six-well plates were transfected with 80 pmol of siRNA complexed with 4 μl of Oligofectamine in a total volume of 1 ml of OptiMEM for 6 h. Cells were trypsinized 24 h after siRNA transfection, plated in 24-well plates, and infected 24 h after trypsinization. Just before infection, extra wells of cells treated with each siRNA were counted to ensure that equal numbers of cells were infected. Relative levels of targeted proteins were analyzed by immunoblotting equal amounts of cell lysates.
For experiments with the subgenomic replicon, subconfluent cultures of replicon-containing Huh-7 cells in six-well plates were transfected with 100 pmol of synthetic double-stranded siRNA complexed with 3 μl of Dharmafect-1 (Dharmacon), in a total volume of 1 ml serum-free DMEM, and 2 ml of DMEM containing 10% FCS was added 6 h later. Cells were trypsinized 24 h after siRNA transfection and plated in 24-well plates. Relative levels of NS5A were analyzed by immunoblotting equal amounts of cell lysates at 6 days after siRNA transfection.
GBF1 complementation.
Huh-7 cells were grown on glass coverslips and transfected with expression vectors for yellow fluorescent protein (YFP)-tagged wild-type, inactive mutant (E794K), or BFA-resistant mutant (M832L) GBF1 (45), kindly provided by C. L. Jackson (Laboratoire d'Enzymologie et Biochimie Structurales, CNRS, Gif-sur-Yvette, France) or with pEYFP-C1 (Clontech). Endotoxin-free plasmid DNA (0.25 μg) was incubated for 20 min with 1 μl of FuGene-6 in 50 μl of serum-free medium. This mix was used to transfect 2 subconfluent wells of a 24-well plate. Transfected and control untransfected cells were rinsed three times, and the medium was changed at 24 h posttransfection. Cells were infected with JFH1-CS-N6-A4 virus at 48 h posttransfection. Similar transfection efficiencies of the different constructs were verified before infection by observation of YFP fluorescence. Cells were infected for 2 h and cultured for 24 h in the presence of 100 ng/ml BFA or 0.002% ethanol (BFA stock solvent). Cells were fixed and processed for immunofluorescence detection of E1 using MAb A4 and Alexa 594-conjugated secondary antibody. Nuclei were labeled with DAPI. About 20 independent fields of each coverslip were imaged using identical settings. E1-positive cells and nuclei were counted. Infections were scored as the ratio of E1-positive cells to the total number of cells (nuclei). About 8,000 cells were counted by coverslip.
Quantitative RT-PCR.
Cellular RNA was isolated by using the RNeasy mini kit (Qiagen) according to the manufacturer's protocol. HCV RNA positive- and negative-strand-specific quantification was carried out by real-time PCR using TaqMan, as previously reported (31). The primer pair and the probe were located in the 5′ HCV noncoding region (9). Briefly, 0.5 μg of total RNA was used for cDNA synthesis in a 20-μl reaction mixture containing 7.5 U of Thermoscript (Invitrogen) and 10 pmol of reverse transcription (RT) primer. The HCV-AS2 primer (5′-TCC AAG AAG GAC CCR GT-3′) and the tag-HCV-S1 primer (5′-ggc cgt cat ggt ggc gaa taa TCC CGG GAG AGC CAT AGT G-3′) (31) were used for the positive- and negative-strand reverse transcription assays, respectively (lowercase letters correspond to the non-HCV sequence, i.e., the tag sequence, and uppercase letters correspond to the HCV genomic sequence). cDNA was synthesized at 60°C for 1 h and then treated with 20 U of RNase H (Invitrogen) for 20 min at 37°C.
Real-time PCR was performed with 2 μl of cDNA in a 25-μl reaction mixture containing 12.5 μl of TaqMan universal PCR master mix (Applied Biosystems), 300 nM of HCV-S1 primer (5′-TCC CGG GAG AGC CAT AGT G-3′), 300 nM of HCV-AS2 primer, and 300 nM TaqMan minor-groove binding (MGB) probe labeled with 6-carboxyfluorescein (5′-FAM-TCT GCG GAA CCG GTG-MGB-3′) for positive-strand amplification. A 300 nM concentration of tag (5′-ggc cgt cat ggt ggc gaa taa-3′) instead of HCV-S1 was used for negative-strand amplification. Samples were placed in an ABI 7900 instrument (Applied Biosystems) at 50°C for 2 min and then at 95°C for 10 min with cycling parameters set to 95°C for 15 s and 60°C for 60 s for 50 cycles.
The positive- and negative-strand copy numbers in each reaction were determined by linear regression analysis based on the slope and intercept generated with an external standard curve (9). The results for positive- and negative-strand HCV RNA were expressed in copy number per reaction.
Translation assay.
HCV internal ribosome entry site (IRES) activity was monitored with the bicistronic reporter pIRF1b, kindly provided by Annie Cahour (Hôpital de la Pitié-Salpétrière, Paris, France) (32). The pIRF1b reporter is composed of firefly luciferase (F-Luc) followed by the HCV 1b 5′ untranslated region (UTR) sequence and then by Renilla luciferase (R-Luc). The plasmid was linearized by NotI restriction, and the linearized DNA was then used as a template for in vitro transcription with the mMESSAGE mMACHINE T7 kit from Ambion, as recommended by the manufacturer. In vitro-transcribed capped RNA was delivered to Huh-7 cells by electroporation. Cells were cultured for 8 h in the presence of increasing concentrations of BFA. Firefly and Renilla luciferase activities were measured with a dual-luciferase reporter assay system from Promega. IRES-dependent translation and cap-dependent translation are expressed as R-Luc activity and F-Luc activity, respectively.
UHCV-11 cell induction.
UHCV-11 cells were cultured for 3 days in a medium containing 0.1 μg/ml tetracycline, rinsed three times with PBS to remove tetracycline, and cultured for 8 h in tetracycline-free medium containing increasing concentrations of BFA. Control noninduced cells were cultured for 8 h in the presence of 1 μg/ml tetracycline.
Indirect immunofluorescence microscopy.
Infected cells were processed for immunofluorescent detection of viral proteins as previously described (50). Nuclei were stained by a 5-min incubation in PBS containing 1 μg/ml DAPI. Coverslips were mounted on glass slides using a Mowiol-based medium and observed with a Zeiss Axiophot equipped with a 20× magnification, 0.5 numerical aperture objective. Fluorescent signals were collected with a Coolsnap ES camera (Photometrix) using specific fluorescence excitation and emission filters. Images were processed using Adobe Photoshop software. For quantification, images of randomly picked areas from each coverslip were recorded. Cells labeled with anti-E1 MAb A4 were counted as infected cells. The total number of cells was obtained from DAPI-labeled nuclei. The infections were scored as the ratio of infected cells to total cells. For colocalization experiments, confocal microscopy was performed with an LSM710 confocal microscope (Zeiss) using a 63×/1.4 numerical aperture oil immersion objective. Signals were sequentially collected by using single fluorescence excitation and acquisition settings to avoid crossover. Images were assembled by using Adobe Photoshop software.
Immunoblotting.
Cells were lysed in 50 mM Tris-Cl buffer (pH 7.5) containing 100 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium dodecyl sulfate (SDS), and protease inhibitors for 30 min on ice. Cells were collected, and the nuclei were pelleted. The protein concentration in the postnuclear supernatants was determined by the bicinchoninic acid method as recommended by the manufacturer (Sigma), using bovine serum albumin as the standard. Proteins were separated by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (Hybond-ECL; Amersham) by using a Trans-Blot apparatus (Bio-Rad). The proteins of interest were revealed with specific primary antibodies, followed by species-specific secondary antibodies conjugated to peroxidase, and enhanced chemiluminescence detection (Amersham) as recommended by the manufacturer. CD63 was revealed using a biotinylated primary antibody and peroxidase-conjugated neutravidin. For quantification, the films were scanned and quantified with the NIH Image software.
Viability assay.
Subconfluent cell cultures grown in 96-well plates were incubated with BFA. An MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium]-based viability assay (CellTiter 96 aqueous nonradioactive cell proliferation assay from Promega) was conducted as recommended by the manufacturer.
Electron microscopy.
For ultrastructural analysis, cells were fixed in 4% paraformaldehyde and 1% glutaraldehyde in 0.1 M phosphate buffer (pH 7.2) for 48 h. Cells were then washed in phosphate buffer, harvested, and postfixed with 1% osmium tetroxide for 1 h. They were then dehydrated in a graded series of ethanol solutions, and cell pellets were embedded in Epon resin, which was allowed to polymerize for 48 h at 60°C. Ultrathin sections were cut on an ultramicrotome (Reichert, Heidelberg, Germany), collected on copper grids, and stained with 5% uranyl acetate-5% lead citrate. The grids were then observed with a 1010 XC electron microscope (JEOL, Tokyo, Japan).
RESULTS
BFA inhibits HCV infection.
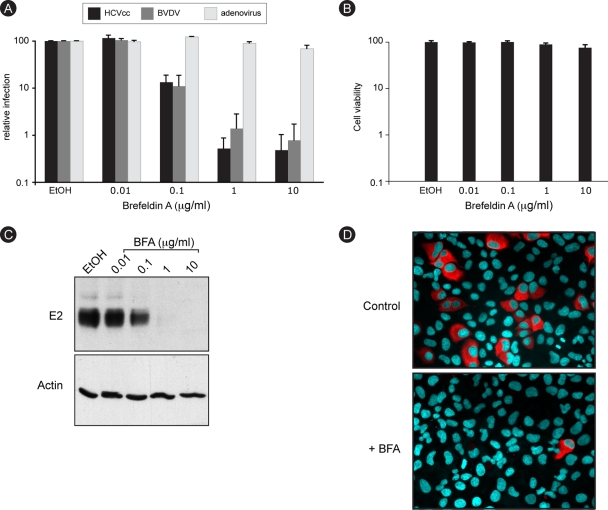
To investigate the function of cellular membranes during HCV infection, we made use of BFA, a drug that blocks different intracellular transport steps. BFA-treated Huh-7 cells were infected with a recombinant virus expressing Renilla luciferase (HCV-RLuc). BFA was removed at 6 h postinfection to minimize the toxicity of the drug, and luciferase activity in cell lysates was measured 18 h later. A dose-dependent reduction in luciferase activity was observed (Fig. 1A), suggesting an inhibitory effect of BFA on HCV infection. These experimental conditions did not induce any cell toxicity, as measured by an MTS assay (Fig. 1B). Adenovirus infection was very weakly affected by BFA treatment (99.5% inhibition for HCV and 10% inhibition for adenovirus at 1 μg/ml), thereby confirming that BFA action on HCV did not result from cell toxicity. Much like with HCV, the infection of BVDV, a close relative of HCV, was also strongly inhibited in BFA-treated MDBK cells in a dose-dependent manner (98.5% inhibition at 1 μg/ml), indicating that the effect of BFA was not restricted to HCV or to Huh-7 cells.
FIG. 1.
HCV infection is sensitive to BFA. (A) Huh-7 cells were infected with HCV or GFP-expressing adenovirus, and MDBK cells were infected with BVDV in the presence of 0.2% ethanol (EtOH) or increasing concentrations of BFA. BFA was present for 8 h for Huh-7 cells (HCVcc and adenovirus) or throughout the experiment for MDBK cells (BVDV). At 24 h postinfection, cells were harvested for luciferase assays (HCVcc) or fluorescence-activated cell sorter analysis (BVDV and adenovirus). The luciferase activity or number of infected cells for ethanol-treated cells is expressed as 100%. Error bars indicate standard errors of the means for 4 (HCV), 6 (BVDV), or 3 (adenovirus) experiments. (B) Huh-7 cells were incubated for 8 h in the presence of 0.2% EtOH or increasing concentrations of BFA and then cultured for 18 h without drug. Viability was assessed using an MTS assay. The absorbance of the ethanol-treated sample is expressed as 100%. (C) Huh-7 cells were infected with HCVcc as for panel A. At 30 h postinfection, E2 and actin expression levels were analyzed by immunoblotting. (D) Huh-7 cells were infected for 2 h with HCVcc in the presence of 0.2% EtOH or 1 μg/ml BFA. BFA was removed at 6 h postinfection. Cells were fixed at 30 h postinfection and processed for immunofluorescence detection of E2.
The inhibition of HCV infection in BFA-treated cells was confirmed with a nonrecombinant, cell culture-adapted virus. A dose-dependent decrease in envelope glycoprotein E2 expression was observed in BFA-treated cells (Fig. 1C). Immunofluorescence microscopy indicated that this reduced expression resulted from a reduced number of infected cells but apparently not from a reduced expression of the protein in individual infected cells (Fig. 1D). Altogether, these results indicate that BFA inhibits HCV infection.
BFA inhibits a postentry step of infection.
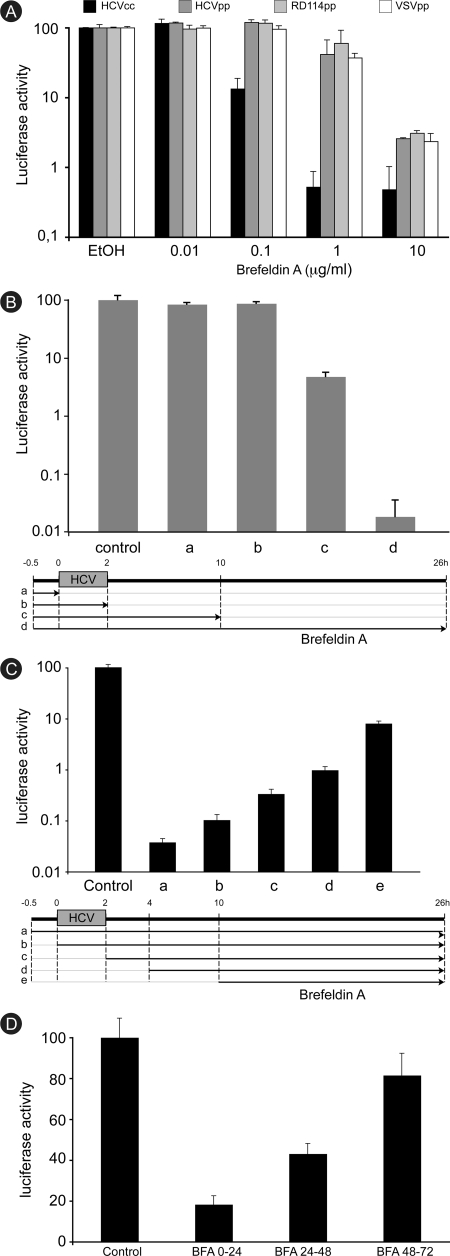
BFA is known to inhibit intracellular transport in the endocytic pathway (30). Therefore, we sought to determine whether it interferes with HCV entry by using retroviral particles pseudotyped with HCV envelope glycoproteins (HCVpp). Particles pseudotyped with the glycoprotein G of vesicular stomatitis virus (VSVpp) or with the envelope protein of the retrovirus RD114 (RD114pp) were used as controls. A very similar dose-dependent decrease in luciferase activity was measured for all pseudotypes (Fig. 2A), indicating an envelope-independent inhibition of pseudoparticle infection. Furthermore, this nonspecific inhibition was not as strong as the one observed with HCVcc. At a concentration of 1 μg/ml of BFA, the infection of HCVpp and control pseudoparticles was reduced by about 50%, whereas HCV-RLuc infection was reduced by more than 99%. These different sensitivities to BFA suggest that different BFA targets are likely involved in HCVcc and retrovirus infections.
FIG. 2.
Brefeldin A inhibits a postentry step of HCV infection. (A) Huh-7 cells were infected with HCVcc, HCVpp, RD114pp, or VSVpp in the presence of 0.2% ethanol (EtOH) or increasing concentrations of BFA. BFA was removed at 6 h postinfection. Luciferase activity was measured at 24 h or 48 h postinfection for HCVcc and pseudoparticles, respectively. The luciferase activity from ethanol-treated cells is expressed as 100%. Error bars indicate standard deviations. (B) Huh-7 cells were pretreated for 30 min with 50 ng/ml BFA and infected for 2 h. BFA was removed either before infection (a), just after infection (b), or at 8 h postinfection (c) or was present throughout the experiment (d). Luciferase activity was measured at 24 h postinfection. The luciferase activity from untreated cells (control) is expressed as 100%. (C) BFA was added to HCVcc-infected cells either 30 min before infection (a), at the beginning of the infection (b), just after infection (c), 2 h postinfection (d), or 8 h postinfection (e). Luciferase activity was measured at 24 h postinfection. The luciferase activity from untreated cells (control) is expressed as 100%. (D) HCVcc-infected cells were treated with 50 ng/ml BFA from 0 to 24 h, from 24 to 48 h, or from 48 to 72 h. Luciferase activity was measured at 72 h postinfection for all the samples. The luciferase activity from untreated cells (control) is expressed as 100%.
To further investigate the action of BFA during entry, we tested its inhibitory effect on HCVcc when it was removed at various time points postinfection. Cells were pretreated with BFA for 30 min, infected for 2 h, and lysed 24 h later. BFA was removed either before infection, just after virus inoculation, or 8 h postinfection, or it was present during the whole experiment (Fig. 2B). In these experiments, BFA was used at a concentration of 50 ng/ml. These experimental conditions were chosen because they showed a strong inhibitory effect on HCV-RLuc infection with no toxicity to the cells (data not shown). When the drug was removed at 8 h postinfection, a strong inhibition (95%) of the infection was observed. However, HCV infection was not inhibited when BFA was omitted during postinfection steps, whether it was present or not during virus inoculation (Fig. 2B). These results suggested that BFA interferes with a postentry step of HCV infection.
The postentry inhibition of HCV infection was confirmed with experiments during which BFA was added at various time points after virus inoculation. A very strong inhibition was recorded when the drug was added after virus entry. HCV infection was inhibited by 99% or more when BFA was added to infected cells at 2 h or 4 h after virus withdrawal (Fig. 2C and data not shown). When added at 8 h postinfection, BFA still inhibited infection by more than 90%, even though it was in contact with the cells for only 16 h instead of 26 h in the control condition (Fig. 2C).
We also tested the inhibitory potency of BFA later during infection. Cells were infected for 2 h and lysed 72 h later. BFA was added to infected cells for 24 h, either from the beginning of the infection or 24 or 48 h later. The results show that the inhibition of the infection was stronger when the drug was added earlier (Fig. 2D). A modest reduction (about 20%) was obtained when the drug was added at 48 h postinfection, whereas the inhibition observed with the drug present during the first 24 h was much stronger (80%).
Altogether, these results indicate that BFA inhibits a postentry step of HCV infection, that this inhibition appears more efficient early during infection, and that BFA has a weaker effect on an established infection.
BFA inhibits RNA replication.
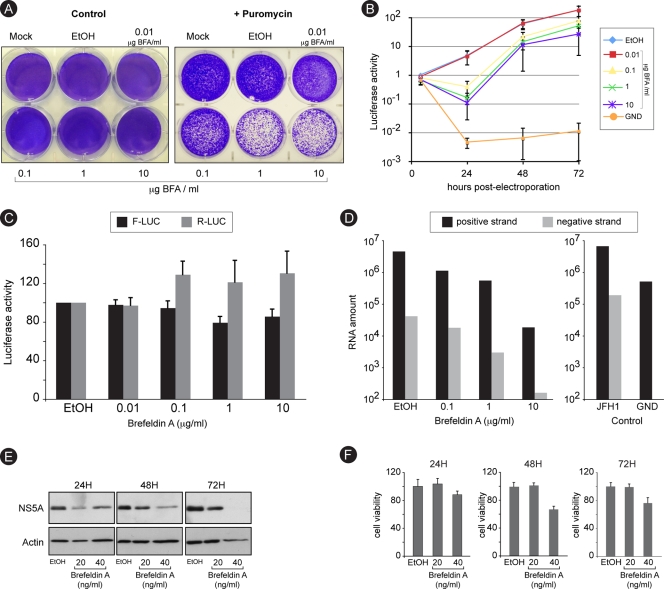
We next sought to determine which postentry step of the intracellular viral cycle was actually inhibited by BFA. We used a colony formation assay to assess the impact of BFA on replication. An in vitro-transcribed recombinant JFH1 genomic RNA expressing a puromycin acetyltransferase as a selection marker was electroporated into Huh-7 cells. To avoid reinfection, we used a construct with an in-frame deletion in the E1E2-coding sequence (60). Following 8 h of BFA treatment and 6 days of puromycin selection, we observed a dose-dependent decrease in the number of resistant colonies (Fig. 3A), suggesting that replication was sensitive to BFA. To quantify this effect in the absence of puromycin, we used a similar construct expressing a Renilla luciferase reporter instead of the puromycin acetyltransferase selection marker. Replication was assessed by measuring luciferase activity over a 72-h time course, after an initial 8-h treatment with increasing doses of BFA. Again replication was inhibited in a dose-dependent manner (Fig. 3B). An initial decrease in luciferase activity was measured at 24 h posttransfection in BFA-treated cells, and then luciferase activity increased in parallel with the control, suggesting that the initial inhibition of replication imposed by BFA was reversible.
FIG. 3.
Brefeldin A inhibits RNA replication. (A) Huh-7 cells were electroporated with a recombinant HCV genome containing a deletion in E1E2 and expressing a puromycin acetyltransferase selection marker. Cells were cultured in the presence of increasing concentrations of BFA for 8 h and then in the presence or the absence of 1 μg/ml puromycin for 6 days. Cells were stained with crystal violet. (B) Huh-7 cells were electroporated with a recombinant HCV genome containing a deletion in E1E2 and expressing Renilla luciferase and were cultured in the presence of BFA for 8 h and then in the absence of the drug. For comparison, Huh-7 cells were electroporated with a nonreplicative (GND) HCV genome and cultured in the absence of BFA. Samples were harvested for luciferase assay at 4, 24, 48, and 72 h postelectroporation. The luciferase activity from ethanol-treated cells at 4 h postelectroporation is expressed as 1. Error bars indicate standard errors of the means for 3 experiments. (C) Huh-7 cells were electroporated with in vitro-transcribed and capped RNA constructs expressing firefly luciferase (F-Luc) under cap control and Renilla luciferase (R-Luc) under HCV IRES control. Cells were cultured in the presence of increasing concentrations of BFA and harvested for dual-luciferase assay at 8 h postelectroporation. (D) Huh-7 cells were infected with HCVcc, treated with increasing concentrations of BFA for 8 h, and harvested at 24 h postinfection for quantitative RT-PCR quantification of HCV plus and minus strands. To confirm the specificity of negative-strand amplification, cells electroporated with in vitro-transcribed HCV replicative (JFH1) or nonreplicative (GND) genomes were processed in parallel (control). (E and F) Huh-7 cells harboring a subgenomic replicon were cultured in the presence of the indicated concentrations of BFA. Samples were harvested after 24, 48, or 72 h of treatment for immunoblot detection of NS5A and actin (E) or for analysis of cell viability (F).
An initial step in replication is thought to be the IRES-dependent translation of the incoming viral RNA. To determine whether BFA affects IRES-dependent translation, we used a bicistronic construct expressing the Renilla luciferase under the control of HCV IRES and the firefly luciferase under the control of the cap of the RNA (32). To avoid a potential promoter-like function of the HCV IRES in plasmid DNA (18), we transfected in vitro-transcribed capped RNA. After 8 h of BFA treatment, transfected cells expressed slightly increased levels of Renilla luciferase and slightly reduced levels of firefly luciferase, indicating that BFA did not inhibit IRES-dependent translation (Fig. 3C).
To confirm the action of BFA on replication, we quantified HCV RNA. Huh-7 cells were infected for 2 h in the presence of increasing concentrations of BFA. BFA was removed at 6 h postinfection. Cells were harvested 16 h after BFA removal, and viral RNA was quantified by quantitative RT-PCR. A dose-dependent decrease of the amounts of both plus- and minus-strand RNA was observed (Fig. 3D). This result is consistent with an inhibitory action of BFA on replication.
To further confirm the inhibition of replication, we treated with BFA Huh-7 cells containing a subgenomic replicon. Cells were treated over a 72-h time course with 20 or 40 ng/ml BFA. Cell viability was monitored with an MTS-based assay, and NS5A expression was analyzed by immunoblotting. Lower NS5A expression levels were observed at each time point for both concentrations of BFA (Fig. 3E). NS5A expression was reduced by 26% and 63% after 48 h of incubation with 20 and 40 ng/ml, respectively. Under these experimental conditions, cell toxicity was undetectable (20 ng/ml) or limited (40 ng/ml), as measured by the MTS assay (Fig. 3F), suggesting that the decrease in NS5A expression reflected an inhibition of replication. Collectively, these results indicate that BFA inhibits the RNA replication step of the HCV life cycle.
GBF1 is the BFA-sensitive factor required for HCV replication.
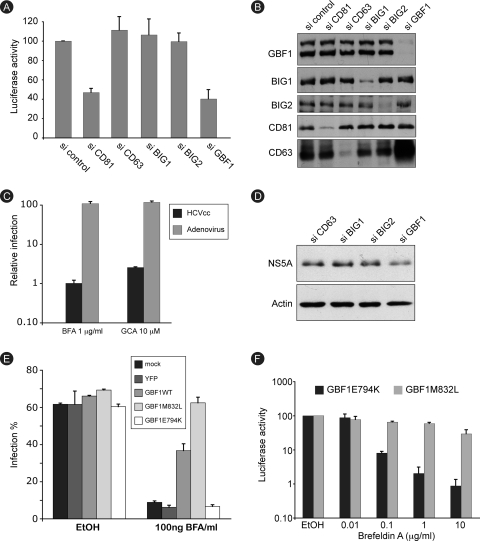
BFA is known to inhibit various cellular pathways. However, we could not also exclude a direct action of the drug on a viral factor. The best-documented cellular targets of BFA are members of the ARF family. Only three ARF GEFs (out of 15 identified) are known to be sensitive to BFA (14, 25). Therefore, we assessed their function in HCV infection, using siRNA technology. Huh-7 cells were transfected with siRNA pools targeting each of the BFA-sensitive ARF GEFs: brefeldin A-inhibited guanine nucleotide exchange factor 1 (BIG1), BIG2, or Golgi BFA resistance factor 1 (GBF1). The tetraspanin CD81, an HCV coreceptor, was targeted as a control of RNA interference efficiency, and negative controls were a nontargeting siRNA pool and an siRNA pool targeting CD63, another tetraspanin not known to be involved in HCV infection. Transfected cells were infected with HCV-RLuc, and luciferase expression was measured at 24 h postinfection. As shown in Fig. 4A, luciferase activity was reduced by about 60% in cells transfected with siRNA to GBF1 or to CD81, whereas it was unaffected in other conditions. The efficiency and specificity of siRNA-mediated downregulation were verified by immunoblot analysis of the targeted proteins. For each protein, the downregulation was estimated to be 80% or more, and no impact on the expression of nontargeted proteins was observed, with the notable exception of CD63, which was dramatically upregulated in GBF1-depleted cells (Fig. 4B). Consistent with the impact of GBF1 depletion, BFA treatment resulted in a similar upregulation of CD63. However, CD63 overexpression is likely not implicated in the inhibition of HCV infection, since no difference was observed regarding BFA inhibition of HCV infection in CD63-depleted and control cells (data not shown). The simultaneous depletion of BIG1 and BIG2 had no impact on HCV infection (data not shown). These results indicate that GBF1 is a cellular factor required for HCV infection.
FIG. 4.
GBF1 is the brefeldin A-sensitive factor required for HCV replication. (A) Huh-7 cells were transfected with the indicated siRNAs and infected with HCV. Luciferase activity was measured at 24 h postinfection. The luciferase activity from control siRNA-transfected cells (si control) is expressed as 100%. Error bars indicate standard errors of the means for 6 experiments. (B) siRNA-mediated depletion of target proteins was verified by immunoblot analysis. (C) Huh-7 cells were infected with HCV-RLuc or GFP-expressing adenovirus in the presence of 1 μg/ml BFA, 0.02% ethanol (BFA stock solvent), 10 μM golgicide A (GCA), or 0.02% dimethyl sulfoxide (DMSO) (golgicide A stock solvent). Both drugs were present for 8 h. At 24 h postinfection, cells were harvested for luciferase assay (HCVcc) or fluorescence-activated cell sorter analysis (adenovirus). The luciferase activity and number of adenovirus-infected cells from ethanol- or DMSO-treated samples are expressed as 100%. (D) Huh-7 cells harboring a subgenomic replicon were transfected with the indicated siRNAs. Cells were lysed, and cell lysates were analyzed by immunoblotting with antibodies to NS5A and actin. (E) Huh-7 cells were transfected with expression plasmids for GBF1, BFA-resistant mutant GBF1-M832L, GBF1 inactive mutant E794K, or YFP. Transfected cells were infected with HCVcc and cultured in the presence or absence of BFA. Cells were fixed at 24 h postinfection and processed for immunofluorescence detection of E1. Results are presented as percentage of infected cells. (F) Huh-7 cells were transfected with expression plasmids for GBF1-M832L or GBF1-E794K, infected with HCV-RLuc in the presence of 0.2% ethanol or the indicated concentration of BFA, and cultured in the presence of BFA for 8 h and then in the absence of the drug. Luciferase activity was measured at 24 h postinfection. The luciferase activity from ethanol-treated cells is expressed as 100%.
The requirement for GBF1 in HCV infection was confirmed with the use of golgicide A, a recently described specific GBF1 inhibitor with no action on BIG1 or BIG2 (52). Huh-7 cells were infected with HCV-RLuc or with adenovirus as a control and treated for 8 h with 10 μM golgicide A or 1 μg/ml BFA, and the infection was scored 18 h later. Luciferase activity was similarly reduced in samples treated with golgicide A or BFA (97% and 99% inhibition, respectively). Adenovirus infection was not significantly affected in either condition (Fig. 4C).
Since BFA inhibits the replication step of the HCV life cycle, we asked whether the BFA-sensitive factor GBF1 was required for replication. GBF1 siRNA was transfected in cells harboring a subgenomic replicon, and replication was assessed by monitoring NS5A expression by immunoblot analysis. As controls, we also transfected siRNAs to BIG1, BIG2, and CD63. A 45% decrease in NS5A expression was observed in GBF1-depleted cells but not in controls (Fig. 4D), confirming that GBF1 is indeed required for replication.
To further confirm that GBF1 is the BFA-sensitive factor required for HCV infection, we undertook a complementation approach. It has been shown that GBF1 overexpression can rescue BFA-induced disruption of the Golgi apparatus (10). Therefore, we sought to determine whether GBF1 overexpression could reverse BFA inhibition of HCV infection. Huh-7 cells were transfected with expression plasmids for wild-type GBF1 or GBF1-M832L, a BFA-resistant mutant of GBF1 (45). Cells were infected in the presence of BFA, fixed at 24 h postinfection, and processed for immunofluorescence analysis of E1 expression. As controls, we transfected cells with expression plasmids for YFP or GBF1-E794K, an inactive mutant of GBF1 (28). In the absence of BFA treatment, 60 to 69% of the cells were infected, whether they were transfected with either construct or left untransfected, indicating that GBF1 overexpression had no major effect on HCV infection. The number of infected cells was down to about 6 to 9% in the presence of BFA for the controls, whereas it was 35 to 40% in wild-type GBF1-overexpressing cells and 60 to 65% in cells overexpressing the BFA-resistant mutant GBF1-M832L (Fig. 4E), indicating a protective effect of GBF1 overexpression over BFA-induced inhibition of HCV infection.
The protective effect of GBF1-M832L was confirmed using higher BFA concentrations. Huh-7 cells expressing GBF1-M832L, or the inactive mutant GBF1-E794K as a control, were infected with HCV-RLuc in the presence of BFA. BFA was removed at 6 h postinfection, and luciferase activity in cell lysates was measured 18 h later. A dose-dependent reduction in luciferase activity was observed for both GBF1 constructs (Fig. 4F). However, the inhibition was dramatically reduced in cells expressing the BFA-resistant construct M832L (40% versus 99% inhibition in cells expressing GBF1-E794K, at a concentration of 1 μg/ml of BFA). These results strongly suggest that GBF1 is the cellular BFA-sensitive factor required for HCV infection.
GBF1 is not a component of HCV replication complexes.
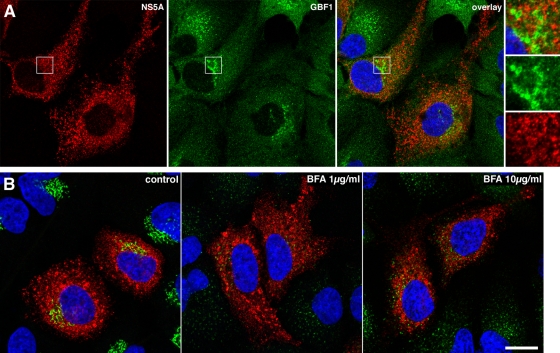
We analyzed GBF1 intracellular localization in HCV-infected Huh-7 cells using immunofluorescence confocal microscopy. GBF1 staining was observed in Golgi-like perinuclear structures and in cytoplasmic small dot-like structures, as previously documented with other cells (10, 23, 45). Similar intracellular GBF1 distributions were observed in infected and noninfected cells (Fig. 5A). GBF1 staining did not overlap with NS5A, a marker of replication complexes, indicating that GBF1 is not recruited to HCV replication complexes.
FIG. 5.
Immunofluorescence analysis of GBF1 localization and BFA effect in HCV-infected cells. (A) HCV-infected cells were fixed and processed for double-label immunofluorescence for NS5A (red) and GBF1 (green) at 24 h postinfection. Representative confocal images are shown together with the merge image and an enlargement of the indicated area. (B) HCV-infected cells were treated for 1 h with the indicated concentration of BFA, fixed, and processed for double-label immunofluorescence for NS5A (red) and the Golgi marker GM130 (green). Representative confocal images are shown.
To further examine GBF1 function in HCV replication, we analyzed the morphology of replication complexes in infected cells treated with BFA. NS5A staining was unchanged in cells treated for 1 h with 1 or 10 μg/ml of BFA (Fig. 5B). Similar observations were made for cells treated for up to 8 h with BFA or with 10 μM golgicide A (data not shown). In contrast, the morphology of the Golgi apparatus was dramatically altered in cells treated with BFA or golgicide A, as indicated by the scattered labeling of the cis-Golgi marker GM130 (Fig. 5B and data not shown). This effect on cis-Golgi morphology specifically results from GBF1 inhibition (10, 39). These results indicate that, in contrast to the case for the Golgi complex, the morphology of HCV replication complexes is not disrupted when GBF1 function is inhibited.
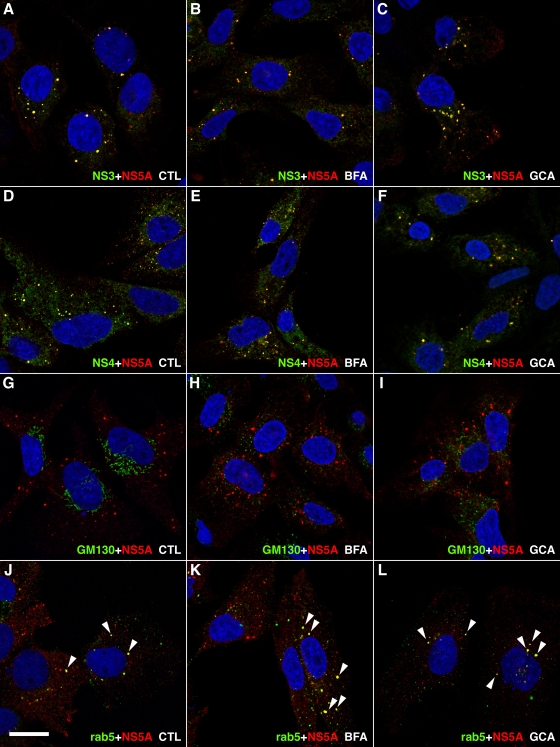
We investigated whether we could detect differences in the composition of HCV replication complexes when GBF1 function is inhibited. To facilitate this analysis, we used Huh-7 cells harboring a subgenomic replicon, in which replication complexes form more discrete structures than in JFH1-infected cells. Replicon cells were treated for 8 h with 1 μg/ml BFA or 10 μM golgicide A and analyzed by immunofluorescence confocal microscopy. As previously reported (19, 26, 63), replication complexes appeared as dot-like structures of various sizes in Huh-7 cells harboring a subgenomic replicon (Fig. 6A to L). These structures could be labeled with antibodies to NS3, with antibodies to NS5A, and with an antiserum to both NS4A and NS4B (NS4A-B) and were not affected by BFA- or golgicide A-mediated GBF1 inhibition (Fig. 6A to F). In contrast, the morphology of the Golgi apparatus was dramatically altered, as indicated by scattered GM130 staining in cells treated with BFA or golgicide A (Fig. 6G to I).
FIG. 6.
Immunofluorescence analysis of GBF1 inhibition in subgenomic replicon-harboring cells. Huh-7 cells harboring a subgenomic replicon were treated for 8 h in the presence of ethanol 0.2% (A, D, G, and J), 1 μg/ml BFA (B, E, H, and K), or 10 μM golgicide A (C, F, I, and L) and processed for double-label immunofluorescence using anti-NS5A antibody together with antibodies to NS3 (A to C), to NS4A-B (D to F), to cis-Golgi marker GM130 (G to I), or to Rab5 (J to L). Representative confocal images are shown, with NS5A in red and other markers in green. Colocalization of both markers in dot-like structures appears in yellow and is marked by white arrowheads in panels J to L. Bar, 20 μm.
It has been reported previously that the small GTPase Rab5 is recruited to HCV replication complexes (54). Therefore, we investigated whether Rab5 recruitment is GBF1 dependent. Rab5 immunostaining appeared as a cytoplasmic punctuated compartment in cells harboring a subgenomic replicon (Fig. 6J), as well as in naïve Huh-7 cells (data not shown), which represents early endosomes. In addition to this endosomal staining, anti-Rab5 antibody also decorated some NS5A positive structures in about 10 to 20% of replicon-harboring cells. In contrast to NS3 and NS4A-B, for which the colocalization with NS5A was observed in all dot-like structures (Fig. 6A to F), NS5A and Rab5 colocalized only in larger, NS5A-positive dot-like structures (Fig. 6J to L). This partial colocalization was insensitive to BFA or golgicide A treatments (Fig. 6J to L).
Taken together, these results suggest that GBF1 is not a component of HCV replication complexes and that its inhibition does not alter their composition or their morphology at the resolution of light microscopy.
BFA alters the membranous web ultrastructure.
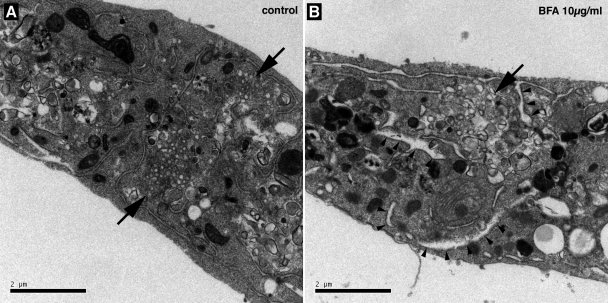
We next examined the ultrastructure of BFA-treated, HCV-infected cells using electron microscopy. As previously reported (50), membrane alterations composed of clusters of small membrane-bound vesicles were observed in infected cells (Fig. 7). These structures were reminiscent of the membranous webs previously identified in U-2 OS human osteosarcoma-derived cell lines inducibly expressing HCV polyprotein and in Huh-7 cells harboring a subgenomic replicon (19). When infected cells were treated for 8 h in the presence of 10 μg/ml BFA, membranous web-like structures could still be observed, but they appeared less organized, with vesicles more variable in shape and size than in control cells (Fig. 7). As previously reported (36), BFA treatment also led to the swelling of ER cisternae (Fig. 7B). Similar observations were made with cells that had been treated for 8 h with 1 μg/ml BFA (data not shown). These results suggest that GBF1 function is required for the ultrastructural organization of HCV membranous webs.
FIG. 7.
Membrane alterations in BFA-treated cells visualized by electron microscopy. HCVcc-infected Huh-7 cells were incubated for 8 h in the presence of 0.2% ethanol (A) or 10 μg/ml BFA (B) and processed for electron microscopy. Membranous webs are indicated by arrows. Arrowheads indicate examples of swollen ER cisternae in BFA-treated cells. Bars, 2 μm.
BFA does not inhibit membranous web formation.
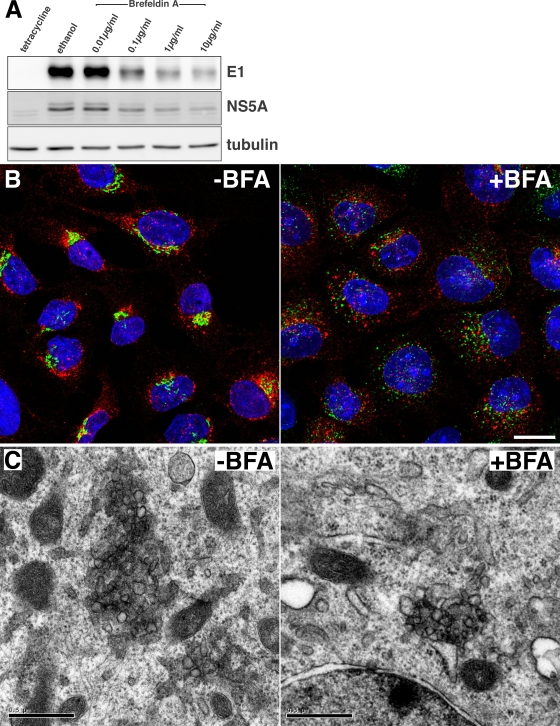
To further investigate the function of GBF1 during HCV infection, we sought to determine whether it is required for the remodeling of cellular membranes that leads to the formation of membranous webs. To test this hypothesis, we used UHCV-11 cells, a U-2 OS-derived cell line expressing the entire HCV polyprotein in an inducible manner (44). Upon tetracycline withdrawal, HCV polyprotein is expressed and processed, and nonstructural proteins induce membranous web-like structures (19) very similar to those observed in replicon-containing cells (26) or in JFH1-infected cells (50). When UHCV-11 cells were induced for 8 h in the presence of BFA, a dose-dependent decrease in E1 and NS5A expression was observed (Fig. 8A). A very similar dose-dependent decrease in E2 expression was also observed (data not shown), indicating that BFA had a negative effect on HCV polyprotein expression in UHCV-11 cells. The formation of membranous webs was assessed by immunofluorescence analysis of NS5A. Typically, over 90% of the cells were positive for NS5A staining after 8 h of induction, with some heterogeneity in the intensity of the staining in individual cells, as previously reported (44). NS5A staining was found in cytoplasmic perinuclear dot-like structures and was very similar in BFA-treated and control cells, although the dot-like structures appeared more scattered in some cells after BFA treatment (Fig. 8B). In contrast, the morphology of the Golgi apparatus was dramatically altered in BFA-treated cells, indicating that UHCV-11 cells were indeed sensitive to BFA (Fig. 8B). NS3 colocalized with NS5A in the dot-like structures, in both BFA-treated and control cells (data not shown), confirming that they were membranous web-like structures.
FIG. 8.
Effect of brefeldin A on membranous web-like structure formation in UHCV-11 cells. (A) UHCV-11 cells were induced by tetracycline withdrawal and incubated for 8 h in the presence of different concentrations of BFA or ethanol (0.2%). Control cells were incubated for 8 h without induction (tetracycline). HCV protein expression was assessed by immunoblot analysis of E1, NS5A, and tubulin. (B) Cells induced for 8 h in the presence or absence of BFA (1 μg/ml) were fixed and processed for immunofluorescence detection of NS5A (red) and the Golgi marker GM130 (green) Nuclei were stained with DAPI (blue). Bar, 20 μm. (C) Cells induced for 8 h in the presence or absence of BFA (10 μg/ml) were fixed and processed for electron microscopy detection of membranous web-like membrane alterations. Bars, 0.5 μm.
The formation of membranous webs in the presence of BFA was further confirmed by electron microscopy. Both in BFA-treated and in control cells, clusters of vesicles that were absent in noninduced cells were observed (Fig. 8C). These membrane structures were similar to the membranous web-like structures previously described (19); however, they were reduced in size, which was probably due to the limited expression time used in these experiments in order to avoid BFA-induced toxicity. Again, BFA sensitivity of the cells could be confirmed by the observation of swollen ER cisternae by electron microscopy, as previously described (36). These results indicate that the morphological changes associated with the formation of membranous web-like structures are not sensitive to BFA.
DISCUSSION
Many positive-strand RNA viruses assemble cytoplasmic replication complexes by modifying cellular membranes. HCV-induced membranous webs and smaller, recently described HCV replicases are thought to be derived from the ER, with which they appear to be in close association (26, 63). Accordingly, cellular factors derived from the early secretory pathway which are involved in HCV replication have been identified, such as VAP-A (22), Rab1, and the Rab1 GTPase-activating protein TBC1D20 (53). In this paper, we show that HCV replication strongly depends on GBF1, a regulator of membrane traffic in the early secretory pathway between the ER and Golgi apparatus (14). We demonstrate that BFA inhibits HCV replication and that GBF1 is the BFA-sensitive cellular factor required for HCV replication.
BFA could potentially inhibit multiple steps of the HCV life cycle. BFA is well known for its inhibitory effect on secretion, and it has been reported that BFA indeed blocks the egress of mature HCV particles (24). BFA also interferes with VLDL lipidation (51) and might therefore also disrupt late steps of HCV assembly, which shares common features with VLDL lipidation, although this potential action of BFA on HCV assembly has not been reported so far. The assays we used in this study did not allow us to assess the impact of BFA on HCV assembly and secretion, but only that on earlier steps of the viral life cycle, such as entry and replication. BFA is known to inhibit transport steps in the endocytic pathway (30), and it could have inhibited HCV entry, which occurs by endocytosis (5, 40, 57). However, we show that BFA inhibition of HCV entry is very unlikely. Although we cannot completely rule out an action of BFA on postfusion late entry events, our results clearly show an inhibitory action of BFA on replication.
GBF1, the BFA-sensitive factor involved in HCV replication, has recently been shown to be required for the replication of two other plus-strand RNA viruses: poliovirus and murine hepatitis virus (MHV) (4, 58). Like HCV, both of them induce membranous replication complexes, and MHV replication complexes are associated with ER membranes (12). In contrast, Sindbis virus, another plus-strand RNA virus, which assembles replication complexes on membranes derived from lysosomes (21), is much less sensitive to BFA (reference 58 and our unpublished observation), suggesting that GBF1 is not required for its replication. These observations suggest that GBF1 might be a cellular factor required for the replication of positive-strand RNA viruses, provided that they assemble replication complexes derived from membranes of the early secretory pathway. In light of this, it would be interesting to determine whether GBF1 is also required for the replication of other viruses of the family Flaviviridae, such as Kunjin virus and dengue virus, which also induce ER-associated membrane alterations (61, 62), or BVDV, the infection of which is inhibited by BFA (Fig. 1).
Regarding GBF1 function in HCV replication, two mechanisms could be considered. GBF1 could promote membrane alterations that are to harbor replication complexes. BFA would then inhibit the formation of the membrane alterations, and this would result in reduced replication and hence in reduced expression of viral proteins and luciferase reporter. Alternatively, GBF1 could be involved in the activity or the maturation of HCV replication complexes rather than their formation. BFA would then block HCV replication with no direct effect on membrane mechanisms associated with the formation of the replication complexes. For poliovirus, BFA did not block membrane alterations, suggesting that GBF1 is not involved in the formation of poliovirus replication complexes (4). For HCV, these alternative hypotheses are quite difficult to distinguish, because membrane alterations and RNA replication likely occur concomitantly during HCV infection. Our results with UHCV-11 cells suggest that in a nonreplicative context, BFA does not inhibit the formation of membranous web-like structures. If we consider that this observation holds true in a replicative context, this suggests that GBF1 is required for the activity of HCV replication complexes rather than for their assembly. This is consistent with our observation that BFA does not alter their morphology, once they have formed, but only their ultrastructural organization. This is also consistent with the observation that BFA inhibits HCV replication throughout the entire time course of the infection, even though the inhibitory effect appeared to be weaker when the infection was already established. The stronger BFA inhibition of luciferase expression at earlier time points may reflect at least in part the fact that new copies of viral RNA already produced in replication complexes before BFA addition are probably still translationally active after BFA addition, since BFA does not inhibit IRES-dependent translation. Therefore, we conclude that GBF1 is likely involved in the maturation or the activity of HCV replication complexes, rather than in their formation.
In contrast to what was found for poliovirus (3), GBF1 did not appear to be recruited to HCV replication complexes. BFA treatment, which is known to stabilize GBF1 on membranes, did not allow us to observe any redistribution of GBF1 into NS5A-positive structures in infected Huh-7 cells or in induced UHCV-11 cells (data not shown), confirming the absence of transient GBF1 recruitment to replication complexes. Therefore, it is unlikely that GBF1 directly interacts with replication complexes. Consequently, we propose that GBF1 functions to activate effectors involved in HCV replication. This is consistent with the recent finding that COP-I, a downstream effector of GBF1, is involved in HCV replication (55). COP-I is a coat complex involved in vesicular transport between the cis-Golgi apparatus and the ER. We hypothesize that GBF1-associated mechanisms function to deliver proteins or lipids to HCV replication complexes. Whether other GBF1 effectors also participate in HCV replication must await further study.
In conclusion, our results highlight a functional connection between HCV RNA replication and the early secretory pathway of the host cell. Identifying more precisely the function of GBF1 effectors and other regulators of membrane traffic in the early secretory pathway should provide clues about the cellular mechanisms underlying HCV RNA replication.
Acknowledgments
We thank Sophana Ung for help with preparing the figures. We are grateful to S. Levy, A. Patel, J. McKeating, and M. Harris for providing us with antibodies; to T. Wakita for the plasmid pJFH1; to C. L. Jackson for GBF1 expression vectors; to D. Moradpour for UHCV-11 cells; to D. Monté for the recombinant adenovirus; to A. Cahour for the bicistronic IRES reporter; to S. M. Lemon for the subgenomic replicon; and to F.-L. Cosset for plasmids. Some data were generated with the help of the Imaging Core Facility of the Calmette campus (MICPAL) and the RIO/IBiSA Electron Microscopy Facility of the François Rabelais University (Tours).
This work was supported in part by grants from the “Agence Nationale de Recherche sur le Sida et les Hépatites Virales” (ANRS) to Y.R. and from the “Agence Nationale de la Recherche” (ANR) to J.D. L.G. was supported by predoctoral fellowships from the Institut Pasteur de Lille, the Région Nord-Pas-de-Calais, and ANRS. J.D. is an international scholar of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 11 November 2009.
REFERENCES
- 1.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belouzard, S., D. Delcroix, and Y. Rouillé. 2004. Low levels of expression of leptin receptor at the cell surface result from constitutive endocytosis and intracellular retention in the biosynthetic pathway. J. Biol. Chem. 279:28499-28508. [DOI] [PubMed] [Google Scholar]
- 3.Belov, G. A., N. Altan-Bonnet, G. Kovtunovych, C. L. Jackson, J. Lippincott-Schwartz, and E. Ehrenfeld. 2007. Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. J. Virol. 81:558-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belov, G. A., Q. Feng, K. Nikovics, C. L. Jackson, and E. Ehrenfeld. 2008. A critical role of a cellular membrane traffic protein in poliovirus RNA replication. PLoS Pathog 4:e1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanchard, E., S. Belouzard, L. Goueslain, T. Wakita, J. Dubuisson, C. Wychowski, and Y. Rouillé. 2006. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J. Virol. 80:6964-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 7.Boulanger, D., S. Waxweiler, L. Karelle, M. Loncar, B. Mignon, J. Dubuisson, E. Thiry, and P. P. Pastoret. 1991. Characterization of monoclonal antibodies to bovine viral diarrhoea virus: evidence of a neutralizing activity against gp48 in the presence of goat anti-mouse immunoglobulin serum. J. Gen. Virol. 7:1195-1198. [DOI] [PubMed] [Google Scholar]
- 8.Burlone, M. E., and A. Budkowska. 2009. Hepatitis C virus cell entry: role of lipoproteins and cellular receptors. J. Gen. Virol. 90:1055-1070. [DOI] [PubMed] [Google Scholar]
- 9.Castelain, S., V. Descamps, V. Thibault, C. Francois, D. Bonte, V. Morel, J. Izopet, D. Capron, P. Zawadzki, and G. Duverlie. 2004. TaqMan amplification system with an internal positive control for HCV RNA quantitation. J. Clin. Virol. 31:227-234. [DOI] [PubMed] [Google Scholar]
- 10.Claude, A., B. P. Zhao, C. E. Kuziemsky, S. Dahan, S. J. Berger, J. P. Yan, A. D. Armold, E. M. Sullivan, and P. Melançon. 1999. GBF1: A novel Golgi-associated BFA-resistant guanine nucleotide exchange factor that displays specificity for ADP-ribosylation factor 5. J. Cell Biol. 146:71-84. [PMC free article] [PubMed] [Google Scholar]
- 11.Clayton, R. F., A. Owsianka, J. Aitken, S. Graham, D. Bhella, and A. H. Patel. 2002. Analysis of antigenicity and topology of E2 glycoprotein present on recombinant hepatitis C virus-like particles. J. Virol. 76:7672-7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.David-Ferreira, J. F., and R. A. Manaker. 1965. An electron microscope study of the development of a mouse hepatitis virus in tissue culture cells. J. Cell Biol. 24:57-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delgrange, D., A. Pillez, S. Castelain, L. Cocquerel, Y. Rouillé, J. Dubuisson, T. Wakita, G. Duverlie, and C. Wychowski. 2007. Robust production of infectious viral particles in Huh-7 cells by introducing mutations in hepatitis C virus structural proteins. J. Gen. Virol. 88:2495-2503. [DOI] [PubMed] [Google Scholar]
- 14.D'Souza-Schorey, C., and P. Chavrier. 2006. ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7:347-358. [DOI] [PubMed] [Google Scholar]
- 15.Dubuisson, J. 2007. Hepatitis C virus proteins. World J. Gastroenterol. 13:2406-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubuisson, J., F. Helle, and L. Cocquerel. 2008. Early steps of the hepatitis C virus life cycle. Cell. Microbiol. 10:821-827. [DOI] [PubMed] [Google Scholar]
- 17.Dubuisson, J., H. H. Hsu, R. C. Cheung, H. B. Greenberg, D. G. Russell, and C. M. Rice. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J. Virol. 68:6147-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dumas, E., C. Staedel, M. Colombat, S. Reigadas, S. Chabas, T. Astier-Gin, A. Cahour, S. Litvak, and M. Ventura. 2003. A promoter activity is present in the DNA sequence corresponding to the hepatitis C virus 5′ UTR. Nucleic Acids Res. 31:1275-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egger, D., B. Wölk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flint, M., C. Maidens, L. D. Loomis-Price, C. Shotton, J. Dubuisson, P. Monk, A. Higginbottom, S. Levy, and J. A. McKeating. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Froshauer, S., J. Kartenbeck, and A. Helenius. 1988. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 107:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao, L., H. Aizaki, J. W. He, and M. M. Lai. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 78:3480-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.García-Mata, R., and E. Sztul. 2003. The membrane-tethering protein p115 interacts with GBF1, an ARF guanine-nucleotide-exchange factor. EMBO Rep. 4:320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gastaminza, P., G. Cheng, S. Wieland, J. Zhong, W. Liao, and F. V. Chisari. 2008. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J. Virol. 82:2120-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gillingham, A. K., and S. Munro. 2007. The small G proteins of the Arf family and their regulators. Annu. Rev. Cell Dev. Biol. 23:579-611. [DOI] [PubMed] [Google Scholar]
- 26.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikeda, M., M. Yi, K. Li, and S. M. Lemon. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson, C. L., and J. E. Casanova. 2000. Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 10:60-67. [DOI] [PubMed] [Google Scholar]
- 29.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808-1817. [DOI] [PubMed] [Google Scholar]
- 30.Klausner, R. D., J. G. Donaldson, and J. Lippincott-Schwartz. 1992. Brefeldin A: insights into the control of membrane traffic and organelle structure. J. Cell Biol. 116:1071-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komurian-Pradel, F., M. Perret, B. Deiman, M. Sodoyer, V. Lotteau, G. Paranhos-Baccalà, and P. André. 2004. Strand specific quantitative real-time PCR to study replication of hepatitis C virus genome. J. Virol. Methods 116:103-106. [DOI] [PubMed] [Google Scholar]
- 32.Laporte, J., I. Malet, T. Andrieu, V. Thibault, J. J. Toulme, C. Wychowski, J. M. Pawlotsky, J. M. Huraux, H. Agut, and A. Cahour. 2000. Comparative analysis of translation efficiencies of hepatitis C virus 5′ untranslated regions among intraindividual quasispecies present in chronic infection: opposite behaviors depending on cell type. J. Virol. 74:10827-10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lecot, S., S. Belouzard, J. Dubuisson, and Y. Rouillé. 2005. Bovine viral diarrhea virus entry is dependent on clathrin-mediated endocytosis. J. Virol. 79:10826-10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wölk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 35.Lindenbach, B. D., and C. M. Rice. 2003. Molecular biology of flaviviruses. Adv. Virus Res. 59:23-61. [DOI] [PubMed] [Google Scholar]
- 36.Lippincott-Schwartz, J., L. C. Yuan, J. S. Bonifacino, and R. D. Klausner. 1989. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell 56:801-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lohmann, V., F. Körner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 38.Macdonald, A., K. Crowder, A. Street, C. McCormick, K. Saksela, and M. Harris. 2003. The hepatitis C virus non-structural NS5A protein inhibits activating protein-1 function by perturbing ras-ERK pathway signaling. J. Biol. Chem. 278:17775-17784. [DOI] [PubMed] [Google Scholar]
- 39.Manolea, F., A. Claude, J. Chun, J. Rosas, and P. Melançon. 2008. Distinct functions for Arf guanine nucleotide exchange factors at the Golgi complex: GBF1 and BIGs are required for assembly and maintenance of the Golgi stack and trans-Golgi network, respectively. Mol. Biol. Cell 19:523-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meertens, L., C. Bertaux, and T. Dragic. 2006. Hepatitis C virus entry requires a critical postinternalization step and delivery to early endosomes via clathrin-coated vesicles. J. Virol. 80:11571-11578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyanari, Y., K. Atsuzawa, N. Usuda, K. Watashi, T. Hishiki, M. Zayas, R. Bartenschlager, T. Wakita, M. Hijikata, and K. Shimotohno. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089-1097. [DOI] [PubMed] [Google Scholar]
- 42.Moradpour, D., P. Kary, C. M. Rice, and H. E. Blum. 1998. Continuous human cell lines inducibly expressing hepatitis C virus structural and nonstructural proteins. Hepatology 28:192-201. [DOI] [PubMed] [Google Scholar]
- 43.Moradpour, D., F. Penin, and C. M. Rice. 2007. Replication of hepatitis C virus. Nat. Rev. Microbiol. 5:453-463. [DOI] [PubMed] [Google Scholar]
- 44.Moradpour, D., T. Wakita, J. R. Wands, and H. E. Blum. 1998. Tightly regulated expression of the entire hepatitis C virus structural region in continuous human cell lines. Biochem. Biophys. Res. Commun. 246:920-924. [DOI] [PubMed] [Google Scholar]
- 45.Niu, T. K., A. C. Pfeifer, J. Lippincott-Schwartz, and C. L. Jackson. 2005. Dynamics of GBF1, a brefeldin A-sensitive Arf1 exchange factor at the Golgi. Mol. Biol. Cell 16:1213-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Op De Beeck, A., C. Voisset, B. Bartosch, Y. Ciczora, L. Cocquerel, Z. Keck, S. Foung, F. L. Cosset, and J. Dubuisson. 2004. Characterization of functional hepatitis C virus envelope glycoproteins. J. Virol. 78:2994-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oren, R., S. Takahashi, C. Doss, R. Levy, and S. Levy. 1990. TAPA-1, the target of an antiproliferative antibody, defines a new family of transmembrane proteins. Mol. Cell. Biol. 10:4007-4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Randall, G., M. Panis, J. D. Cooper, T. L. Tellinghuisen, K. E. Sukhodolets, S. Pfeffer, M. Landthaler, P. Landgraf, S. Kan, B. D. Lindenbach, M. Chien, D. B. Weir, J. J. Russo, J. Ju, M. J. Brownstein, R. Sheridan, C. Sander, M. Zavolan, T. Tuschl, and C. M. Rice. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. U. S. A. 104:12884-12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rocha-Perugini, V., C. Montpellier, D. Delgrange, C. Wychowski, F. Helle, A. Pillez, H. Drobecq, F. Le Naour, S. Charrin, S. Levy, E. Rubinstein, J. Dubuisson, and L. Cocquerel. 2008. The CD81 partner EWI-2wint inhibits hepatitis C virus entry. PLoS One 3:e1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rouillé, Y., F. Helle, D. Delgrange, P. Roingeard, C. Voisset, E. Blanchard, S. Belouzard, J. McKeating, A. H. Patel, G. Maertens, T. Wakita, C. Wychowski, and J. Dubuisson. 2006. Subcellular localization of hepatitis C virus structural proteins in a cell culture system that efficiently replicates the virus. J. Virol. 80:2832-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rustaeus, S., K. Lindberg, J. Borén, and S. O. Olofsson. 1995. Brefeldin A reversibly inhibits the assembly of apoB containing lipoproteins in McA-RH7777 cells. J. Biol. Chem. 270:28879-28886. [DOI] [PubMed] [Google Scholar]
- 52.Sáenz, J. B., W. J. Sun, J. W. Chang, J. Li, B. Bursulaya, N. S. Gray, and D. B. Haslam. 2009. Golgicide A reveals essential roles for GBF1 in Golgi assembly and function. Nat. Chem. Biol. 5:157-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sklan, E. H., R. L. Serrano, S. Einav, S. R. Pfeffer, D. G. Lambright, and J. S. Glenn. 2007. TBC1D20 is a Rab1 GTPase-activating protein that mediates hepatitis C virus replication. J. Biol. Chem. 282:36354-36361. [DOI] [PubMed] [Google Scholar]
- 54.Stone, M., S. Jia, W. D. Heo, T. Meyer, and K. V. Konan. 2007. Participation of rab5, an early endosome protein, in hepatitis C virus RNA replication machinery. J. Virol. 81:4551-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tai, A. W., Y. Benita, L. F. Peng, S. S. Kim, N. Sakamoto, R. J. Xavier, and R. T. Chung. 2009. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe 5:298-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Targett-Adams, P., S. Boulant, and J. McLauchlan. 2008. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication. J. Virol. 82:2182-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tscherne, D. M., C. T. Jones, M. J. Evans, B. D. Lindenbach, J. A. McKeating, and C. M. Rice. 2006. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J. Virol. 80:1734-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Verheije, M. H., M. Raaben, M. Mari, E. G. Te Lintelo, F. Reggiori, F. J. van Kuppeveld, P. J. Rottier, and C. A. de Haan. 2008. Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLoS Pathog. 4:e1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.von Hahn, T., and C. M. Rice. 2008. Hepatitis C virus entry. J. Biol. Chem. 283:3689-3693. [DOI] [PubMed] [Google Scholar]
- 60.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Welsch, S., S. Miller, I. Romero-Brey, A. Merz, C. K. Bleck, P. Walther, S. D. Fuller, C. Antony, J. Krijnse-Locker, and R. Bartenschlager. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Westaway, E. G., J. M. Mackenzie, M. T. Kenney, M. K. Jones, and A. A. Khromykh. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 71:6650-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wölk, B., B. Buchele, D. Moradpour, and C. M. Rice. 2008. A dynamic view of hepatitis C virus replication complexes. J. Virol. 82:10519-10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ye, J. 2007. Reliance of host cholesterol metabolic pathways for the life cycle of hepatitis C virus. PLoS Pathog. 3:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]