Abstract
Only a few archaeal viruses have been subjected to detailed structural analyses. Major obstacles have been the extreme conditions such as high salinity or temperature needed for the propagation of these viruses. In addition, unusual morphotypes of many archaeal viruses have made it difficult to obtain further information on virion architectures. We used controlled virion dissociation to reveal the structural organization of Halorubrum pleomorphic virus 1 (HRPV-1) infecting an extremely halophilic archaeal host. The single-stranded DNA genome is enclosed in a pleomorphic membrane vesicle without detected nucleoproteins. VP4, the larger major structural protein of HRPV-1, forms glycosylated spikes on the virion surface and VP3, the smaller major structural protein, resides on the inner surface of the membrane vesicle. Together, these proteins organize the structure of the membrane vesicle. Quantitative lipid comparison of HRPV-1 and its host Halorubrum sp. revealed that HRPV-1 acquires lipids nonselectively from the host cell membrane, which is typical of pleomorphic enveloped viruses.
In recent years there has been growing interest in viruses infecting hosts in the domain Archaea (43). Archaeal viruses were discovered 35 years ago (52), and today about 50 such viruses are known (43). They represent highly diverse virion morphotypes in contrast to the vast majority (96%) of head-tail virions among the over 5,000 described bacterial viruses (1). Although archaea are widespread in both moderate and extreme environments (13), viruses have been isolated only for halophiles and anaerobic methanogenes of the kingdom Euryarchaeota and hyperthermophiles of the kingdom Crenarchaeota (43).
In addition to soil and marine environments, high viral abundance has also been detected in hypersaline habitats such as salterns (i.e., a multipond system where seawater is evaporated for the production of salt) (19, 37, 50). Archaea are dominant organisms at extreme salinities (36), and about 20 haloarchaeal viruses have been isolated to date (43). The majority of these are head-tail viruses, whereas electron microscopic (EM) studies of highly saline environments indicate that the two other described morphotypes, spindle-shaped and round particles, are the most abundant ones (19, 37, 43). Thus far, the morphological diversity of the isolated haloarchaeal viruses is restricted compared to viruses infecting hyperthermophilic archaea, which are classified into seven viral families (43).
All of the previously described archaeal viruses have a double-stranded DNA (dsDNA) genome (44). However, a newly characterized haloarchaeal virus, Halorubrum pleomorphic virus 1 (HRPV-1), has a single-stranded DNA (ssDNA) genome (39). HRPV-1 and its host Halorubrum sp. were isolated from an Italian (Trapani, Sicily) solar saltern. Most of the studied haloarchaeal viruses lyse their host cells, but persistent infections are also typical (40, 44). HRPV-1 is a nonlytic virus that persists in the host cells. In liquid propagation, nonsynchronous infection cycles of HRPV-1 lead to continuous virus production until the growth of the host ceases, resulting in high virus titers in the growth medium (39).
The pleomorphic virion of HRPV-1 represents a novel archaeal virus morphotype constituted of lipids and two major structural proteins VP3 (11 kDa) and VP4 (65 kDa). The genome of HRPV-1 is a circular ssDNA molecule (7,048 nucleotides [nt]) containing nine putative open reading frames (ORFs). Three of them are confirmed to encode structural proteins VP3, VP4, and VP8, which is a putative ATPase (39). The ORFs of the HRPV-1 genome show significant similarity, at the amino acid level, to the minimal replicon of plasmid pHK2 of Haloferax sp. (20, 39). Furthermore, an ∼4-kb region, encoding VP4- and VP8-like proteins, is found in the genomes of two haloarchaea, Haloarcula marismortui and Natronomonas pharaonis, and in the linear dsDNA genome (16 kb) of spindle-shaped haloarchaeal virus His2 (39). The possible relationship between ssDNA virus HRPV-1 and dsDNA virus His2 challenges the classification of viruses, which is based on the genome type among other criteria (15, 39).
HRPV-1 is proposed to represent a new lineage of pleomorphic enveloped viruses (39). A putative representative of this lineage among bacterial viruses might be L172 of Acholeplasma laidlawii (14). The enveloped virion of L172 is pleomorphic, and the virus has a circular ssDNA genome (14 kb). In addition, the structural protein pattern of L172 with two major structural proteins, of 15 and 53 kDa, resembles that of HRPV-1.
The structural approach has made it possible to reveal relationships between viruses where no sequence similarity can be detected. It has been realized that several icosahedral viruses infecting hosts in different domains of life share common virion architectures and folds of their major capsid proteins. These findings have consequences for the concept of the origin of viruses. A viral lineage hypothesis predicts that viruses within the same lineage may have a common ancestor that existed before the separation of the cellular domains of life (3, 5, 8, 26). Currently, limited information is available on the detailed structures of viruses infecting archaea. For example, the virion structures of nontailed icosahedral Sulfolobus turreted icosahedral virus (STIV) and SH1 have been determined (21, 23, 46). However, most archaeal viruses represent unusual, sometimes nonregular, morphotypes (43), which makes it difficult to apply structural methods that are based on averaging techniques.
A biochemical approach, i.e., controlled virion dissociation, gives information on the localization and interaction of virion components. In the present study, controlled dissociation was used to address the virion architecture of HRPV-1. A comparative lipid analysis of HRPV-1 and its host was also carried out. Our results show that the unique virion type is composed of a flexible membrane decorated with the glycosylated spikes of VP4 and internal membrane protein VP3. The circular ssDNA genome resides inside the viral membrane vesicle without detected association to any nucleoproteins.
MATERIALS AND METHODS
Purification of HRPV-1.
Halorubrum sp. strain PV6 was used for the propagation of HRPV-1. The culture conditions, as well as the virus production and purification methods, have been previously described (39). In brief, cells were removed from an infected overnight culture of Halorubrum sp. strain PV6, and impurities were precipitated from the supernatant with 6% (wt/vol) polyethylene glycol (PEG) 6000. Virus particles were then precipitated with 11% (wt/vol) PEG and resuspended in HRPV-1-buffer (1.5 M NaCl, 100 mM MgCl2, 2 mM CaCl2, 20 mM Tris-HCl [pH 7.5]). After the removal of aggregates from the concentrate, viruses were purified in a linear 5 to 20% (wt/vol) sucrose gradient by rate zonal centrifugation (Sorvall AH629, 116,000 × g, 4 h, 15°C) yielding “1× purified” virus. The “1× virus” was further purified by equilibrium centrifugation in a cesium chloride gradient with a mean density of 1.3 g/ml (Sorvall AH629, 82,000 × g, 20 h, 15°C), yielding “2× purified” virus. The “2× virus” was concentrated by differential centrifugation (Sorvall T647.5, 136,000 × g, 3 h, 15°C). HRPV-1-buffer was used throughout the purification procedure.
Inactivation of HRPV-1.
HRPV-1 virus stocks contained 23% (wt/vol) salt water containing 3.15 M NaCl, 113 mM MgCl2, 119 mM MgSO4, 3.8 mM CaCl2, 72 mM KCl, and 61 mM Tris-HCl (pH 7.2). The salt water stock was prepared as described elsewhere (HaloHandbook [http://www.haloarchaea.com/resources/halohandbook/Halohandbook_2008_v7.pdf]). The sensitivity of HRPV-1 to lowered ionic strength was determined by diluting the virus stock in sterile deionized water in volume ratios 1:1, 1:4, and 1:9 (stock to water). Different NaCl concentrations were tested by diluting the virus stock in 23% (wt/vol) salt water devoid of NaCl in appropriate volume ratios. For MgCl2, CaCl2, and KCl determinations, a buffer containing 1.5 M NaCl, 222 mM MgCl2, 3.8 mM CaCl2, 72 mM KCl, and 61 mM Tris-HCl (pH 7.2) was prepared. The concentrations of MgCl2, CaCl2, or KCl in the buffer were decreased one at a time, and the virus stock was diluted 1,000-fold in the buffer. The diluted samples were incubated 18 h at 4°C. The infectivity of the treated viruses was determined by plaque assay after the dilution and incubation.
Sensitivity to pH changes was determined by diluting the HRPV-1 stock 1,000-fold in the appropriate pH buffer. The buffer contained 20 mM potassium phosphate buffer (pH 5.0 to 7.0) or Tris-HCl (pH 7.0 to 9.0), 1.5 M NaCl, 100 mM MgCl2, and 2 mM CaCl2 (modified HRPV-1-buffer). The diluted samples were incubated 30 min at 22°C. The thermal stability of HRPV-1 was tested by incubation of the virus stocks at different temperatures for 30 min. After each treatment, the infectivity in the samples was determined.
Virion dissociation.
All of the experiments were carried out with freshly made “2× purified” virus material. For protease treatments at high salinity, purified HRPV-1 particles (210 μg of protein/ml) were incubated with proteinase K (0.1 mg/ml; Finnzymes), trypsin (0.1 mg/ml; Sigma), or bromelain (1.0 mg/ml; ICN Biomedicals, Inc.) in HRPV-1-buffer for 3 h at 37°C. After incubation, virus titer was determined by plaque assay and dissociation products were analyzed in a linear 5 to 20% (wt/vol) sucrose gradient in HRPV-1-buffer by rate zonal centrifugation (Sorvall TH641, 210,000 × g, 3 h, 15°C). Gradients were fractionated and protein, DNA, and lipid (when appropriate) compositions as well as virus titer (when appropriate) of the fractions were analyzed. DNA was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) having a stacking gel and 16% (wt/vol) acrylamide concentration in the separation gel (35). After electrophoresis, gels were stained with ethidium bromide (EtBr) and visualized by UV. Proteins were separated by Tricine-SDS-PAGE (see Protein analysis) and lipids were analyzed by thin-layer chromatography (TLC) or by lipid staining (see below).
For further experiments, proteinase K-treated particles (the treatment as described above) were purified in a linear 5 to 20% (wt/vol) sucrose gradient in HRPV-1-buffer (Sorvall AH629, 116,000 × g, 5 h, 15°C), and the light-scattering zone was collected. The collected particles were concentrated and washed with HRPV-1-buffer using ultrafiltration (Amicon Ultra Centrifugal Filter Devices, Millipore, 50,000 nominal molecular weight limit; Eppendorf Centrifuge 5810R, A-4-62 rotor, 3,220 × g, 15°C). For further dissociation, proteinase K-treated particles (50 μg of protein/ml) were incubated in 0.03% (vol/vol) Triton X-100 (TX-100) in a buffer containing 375 mM NaCl, 25 mM MgCl2, 0.5 mM CaCl2, and 20 mM Tris-HCl (pH 7.5) for 15 min at 22°C. After incubation, dissociation products were analyzed as described for the protease treatments. TX-100 dissociation was performed with or without a protease inhibitor cocktail (Complete EDTA-free; Roche).
For protease treatments at low salinity, purified HRPV-1 particles (100 μg of protein/ml) were incubated with proteinase K (0.2 mg/ml; Finnzymes) in a buffer containing 75 mM NaCl, 5 mM MgCl2, 0.1 mM CaCl2, and 20 mM Tris-HCl (pH 7.5) for 1 h at 37°C. As a control, HRPV-1 particles were incubated in the same buffer without proteinase K for 1 h at 37°C. For virion dissociation by Nonidet P40 (NP-40), HRPV-1 virions (210 μg of protein/ml) were incubated in HRPV-1-buffer containing 0.1% (vol/vol) NP-40 for 1 h at 22°C. After the incubations, the infectivity was determined, and dissociation products were analyzed as described for the protease treatments at high salinity.
For low-salinity dissociation, HRPV-1 virions (95 μg of protein/ml) were incubated in a buffer containing 75 mM NaCl, 5 mM MgCl2, 0.1 mM CaCl2, and 20 mM Tris-HCl (pH 7.5) for 1 h at 4 or 60°C. After incubation, the virus infectivity was determined, and dissociation products were analyzed in a linear 5 to 20% (wt/vol) sucrose gradient in a buffer containing 50 mM NaCl and 20 mM Tris-HCl (pH 7.5) by rate zonal centrifugation (Sorvall TH641, 210,000 × g, 3 h 45 min, 15°C). After centrifugation, gradients were analyzed as described for the protease treatments.
Soluble proteins were collected from the top of the sucrose gradients for further analyses (see below). The collected proteins were concentrated and washed by using ultrafiltration (Amicon Ultra Centrifugal Filter Devices, Millipore, 10,000 nominal molecular weight limit; Eppendorf Centrifuge 5810R, A-4-62 rotor, 3,220 × g, 15°C). For washing, either HRPV-1-buffer or a buffer containing 50 mM NaCl and 20 mM Tris-HCl (pH 7.5) was used.
Extraction and analysis of lipids.
Cellular lipids and lipids from “2× purified” virus preparations were extracted according to the method of Folch et al. (16) modified for halophiles (22). Lipid extracts were first analyzed by TLC as described previously (4), and the phosphorus content of lipid spots was determined as described by Bartlett et al. (6). For identification of individual lipids, the lipid-containing areas were scraped from a preparative TLC plate, re-extracted, and analyzed by mass spectrometry (MS) essentially as previously described (4). Lipids were identified based on (i) their m/z value, (ii) product ion analysis, and (iii) precursor ion scans. Neutral lipids were analyzed by TLC as described previously (29). For lipid staining, samples were separated in Tricine-SDS-polyacrylamide gels. After electrophoresis, gels were stained with Coomassie blue to detect proteins and then with Sudan Black B (Sigma) to detect lipids according to the manufacturer's instructions.
Protein analysis.
Protein concentrations were measured by the Coomassie blue method (10) using bovine serum albumin as a standard. Proteins were separated by using either Tricine-SDS-PAGE having a stacking gel and 14% or 17% (wt/vol) acrylamide concentration in the separation gel (48) or SDS-PAGE having a stacking gel and 16% (wt/vol) acrylamide concentration in the separation gel (35).
For deglycosylation, soluble proteins from the low-salinity dissociation at 60°C (see above) were lyophilized. Lyophilized proteins were deglycosylated by using a GlycoProfile IV chemical deglycosylation kit (Sigma) according to the manufacturer's instructions. After deglycosylation, samples were concentrated and washed with 50 mM NaCl and 20 mM Tris-HCl (pH 7.5) buffer using ultrafiltration (see above for the concentrations of the soluble proteins).
For glycoprotein staining, proteins were separated in Tricine-SDS-polyacrylamide gels. After electrophoresis, the gels were stained with a Pro-Q Emerald 300 glycoprotein gel stain kit (Invitrogen) according to the manufacturer's instructions and visualized by UV transillumination at 302 nm. After glycoprotein staining, gels were stained with Coomassie blue.
For N-terminal protein sequencing, proteins were electroblotted from Tricine-SDS-polyacrylamide gels onto a polyvinylidene difluoride membrane, followed by Coomassie brilliant blue staining (32). Sequencing was performed by using a Procise 494A HT sequencer (Perkin-Elmer, Applied Biosystems Division). Liquid chromatography electrospray ionization tandem mass spectrometry (LC-ESI MS/MS) was performed by using a Q-TOF instrument (Micromass, Ltd.) connected to an Ultimate nano-chromatograph (Dionex Corp.). For LC-ESI MS/MS, proteins in the SDS-PAGE bands were digested “in gel” essentially as previously described (49). The recovered peptides were then analyzed as described earlier (42). The analyses were performed in the Protein Chemistry Core Facility of the Institute of Biotechnology, University of Helsinki.
Gel filtration and sedimentation analyses.
Soluble proteins from the NP-40 or low-salinity at 60°C dissociations were analyzed by gel filtration and sedimentation. In gel filtration, the column (Tricorn Superdex 200 10/300 GL; Amersham Biosciences) was equilibrated with either HRPV-1-buffer or a buffer containing 50 mM NaCl and 20 mM Tris-HCl (pH 7.5). Apoferritin (443 kDa), β-amylase (200 kDa), PRD1 P3 trimer (120 kDa), bovine serum albumin (66 kDa), carbonic anhydrase (29 kDa), and RNase A (13.7 kDa) were used as molecular mass standards. Fractions collected from the filtration were studied in SDS-polyacrylamide gels. Sedimentation analyses (Sorvall TH660, 217,000 × g, 28 h, 15°C) were performed in linear 10 to 40% (wt/vol) sucrose gradients in either HRPV-1-buffer or a buffer containing 50 mM NaCl and 20 mM Tris-HCl (pH 7.5). The same standards as in the gel filtration were used. After centrifugation, gradients were fractionated and analyzed as in the gel filtration.
Electron microscopy.
EM samples from the dissociation experiments were fixed with 2.5% (vol/vol) glutaraldehyde for 1.5 h at 22°C. For fixation, Tris-HCl-buffer (pH 7.5) in samples was replaced with 2-(N-morpholino)-ethanesulfonic acid buffer (pH 6.7). After fixation, samples were negatively stained with 1% (wt/vol) ammonium molybdate (pH 7.0). Staining-times varied from 10 to 20 s depending on the sample. The micrographs were taken with JEOL 1200EX electron microscope operating at 60 kV at the Electron Microscopy Unit of the Institute of Biotechnology, University of Helsinki.
RESULTS
HRPV-1 infectivity depends on salinity, pH, and temperature.
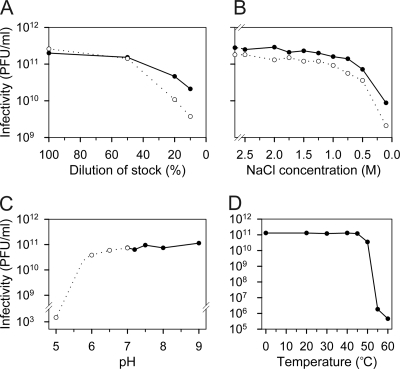
Stocks of HRPV-1 reached titers of ∼2 × 1011 PFU/ml, and the virus was stable for several months when stored at 4°C. Stability of the HRPV-1 infectivity in various conditions was studied, and the results are summarized in Fig. 1. The virus stock contains various salts (see Materials and Methods), and the effects of the total ionic strength (Fig. 1A) and different salts at variable concentrations were separately examined in order to determine the most relevant buffer constituents affecting the infectivity. NaCl was the most critical one (Fig. 1B). A decrease in the MgCl2 or CaCl2 concentration below the concentrations in the virus stock caused only a slight inactivation of HRPV-1, and changes in the KCl concentration had no detectable effect (data not shown). Based on these results, the HRPV-1-buffer containing 1.5 M NaCl, 100 mM MgCl2, 2 mM CaCl2, and 20 mM Tris-HCl (pH 7.5) was designed. The tolerance of HRPV-1 to changes in pH and temperature is represented in Fig. 1C and D, respectively.
FIG. 1.
Inactivation of haloarchaeal virus HRPV-1. (A) Effect of total ionic strength. The virus stock was diluted in deionized water. The x axis indicates the proportion of the virus stock in the dilution. (B) Effect of NaCl concentration. The virus stock was diluted to decrease NaCl concentration (see Materials and Methods). In panels A and B the number of the infective virus particles was measured immediately after dilution (•) and after an 18-h incubation period at 4°C (○). The first point on the y axis represents the infectivity of the undiluted virus stock. (C) HRPV-1 sensitivity to pH changes. The virus stock was diluted in modified HRPV-1-buffer containing either potassium phosphate buffer (○) or Tris-HCl-buffer (•) and incubated for 30 min at 22°C. (D) HRPV-1 sensitivity to temperature changes. The undiluted virus stock was incubated at different temperatures for 30 min.
Inactivation experiments suggested conditions that could be utilized in the dissociation of the HRPV-1 virion. With this information, we did a systematic screening of conditions potentially favorable for controlled virion dissociation. Treatments included freezing and thawing, lowered ionic strength, lowered pH, elevated temperature, proteases, urea, guanidine hydrochloride, chloroform, dithiothreitol, mercaptoethanol, and several detergents. Both anionic detergents (sodium dodecyl sulfate and sodium deoxycholate) and nonionic detergents (octyl-β-d-glucopyranoside, TX-100, TX-114, and NP-40) were used. In addition, many of the treatments were carried out in high- and low-salinity conditions.
After each treatment, the infectivity was determined and dissociation products were analyzed by rate zonal centrifugation. Gradients were fractionated, and the protein pattern in each fraction was determined by Tricine-SDS-PAGE. Most of the tested conditions inactivated the virus but did not dissociate any of the viral proteins or, alternatively, all of the viral proteins were solubilized. Interestingly, the HRPV-1 virion remained intact even in 3 M urea (data not shown). A number of treatments resulted in a close-to-quantitative dissociation of the virion to its components and revealed important aspects about the virion structure. The most successful conditions are summarized in Table 1 and were chosen for more detailed analyses. Also, a stepwise dissociation of HRPV-1 combining proteinase K and TX-100 treatment was developed (see below).
TABLE 1.
Effect of virion dissociation on infectivity
| Condition(s) | Titer (PFU/ml) |
|---|---|
| “2× purified” virus in HRPV-1-buffer at 4°C (starting material)a | 1.5 × 1013 |
| Proteinase K (0.1 mg/ml) incubation in HRPV-1-buffer for 3 h at 37°C | 7.6 × 106 |
| Trypsin (0.1 mg/ml) incubation in HRPV-1-buffer for 3 h at 37°C | 1.1 × 106 |
| Bromelain (1.0 mg/ml) incubation in HRPV-1-buffer for 3 h at 37°C | 3.9 × 109 |
| 0.1% (vol/vol) NP-40 incubation in HRPV-1-buffer for 1 h at 22°C | 1.6 × 104 |
| Incubation at low salinityb for 1 h at: | |
| 4°C | 7.8 × 1012 |
| 37°C | 5.9 × 1011 |
| 60°C | 2.4 × 103 |
Specific infectivity, ∼2.8×1013 PFU/mg of protein.
Low salinity = 75 mM NaCl, 5 mM MgCl2, 0.1 mM CaCl2, and 20 mM Tris-HCl (pH 7.5).
VP4 forms spikes on the virion surface.
To determine protease sensitivity of the HRPV-1 virion, virus particles were treated with proteinase K, trypsin, or bromelain in HRPV-1-buffer (Table 1). Protease concentrations were chosen based on those described previously for another haloarchaeal virus, SH1 (24). The only virion protein observed to be sensitive to proteases was the major structural protein VP4. With an increasing proteinase K treatment time, it was observed that practically all of the VP4 was digested (Fig. 2A and B). Bromelain digestion was not as efficient as that of proteinase K, and trypsin produced several digestion products cosedimenting with the treated particles (data not shown). Thus, proteinase K treatment was chosen for further analyses.
FIG. 2.
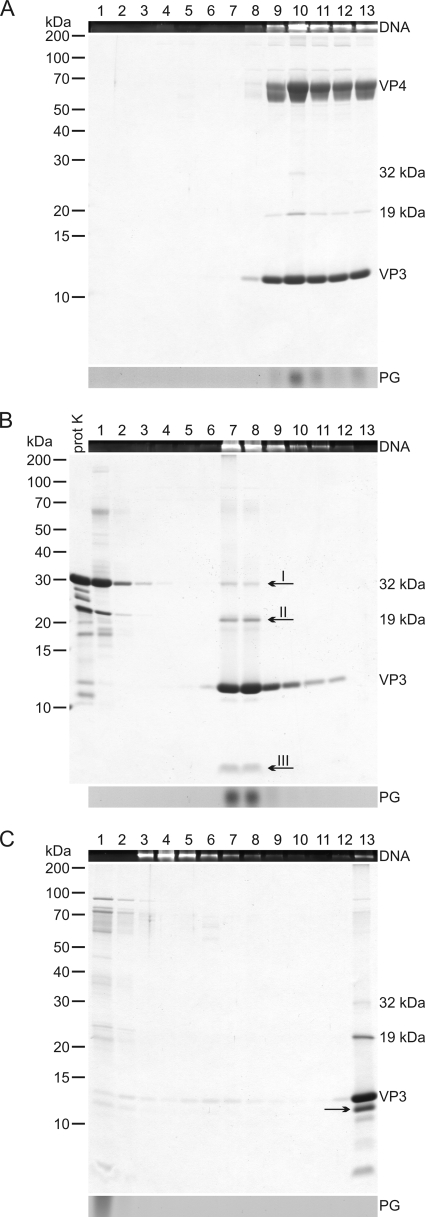
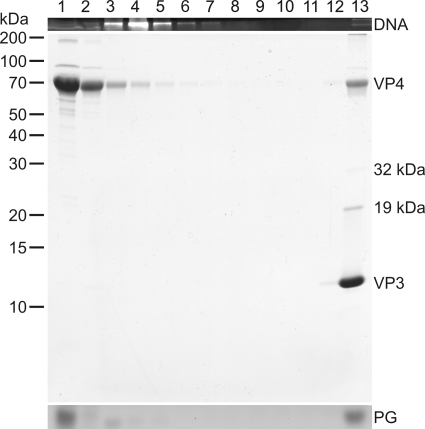
Proteinase K treatment of HRPV-1 virions and further dissociation with TX-100. (A) Untreated “2× purified” virus particles analyzed by rate zonal centrifugation in HRPV-1-buffer. (B) Particles treated with proteinase K in HRPV-1-buffer and analyzed in the same conditions as in panel A. (C) Proteinase K-treated particles further dissociated with TX-100 without a protease inhibitor and analyzed as in panel A. (A to C) After centrifugation, gradients were fractionated, and numbers at the top indicate fractions 1 to 13 from the top to the bottom (fraction 13 is the resuspended pellet). The upper panels show the presence of DNA in EtBr-stained SDS-polyacrylamide gels. The middle panels show protein compositions in Coomassie blue-stained 14% Tricine-SDS-polyacrylamide gels. Numbers on the left indicate the molecular masses (in kilodaltons) of the markers. In the lower panels lipids were analyzed by TLC, but only the PG band of the viral lipids (see Fig. 7) is indicated. In panel A, the infectivity was also determined, and the infectivity peak was in fractions 9 to 13. In panel B, a sample of proteinase K (prot K) was also included in the Tricine-SDS-PAGE, and the arrows I to III indicate the bands analyzed by N-terminal sequencing or mass spectrometry. In panel C, the PG band is distorted due to TX-100 interference, and the arrow indicates a likely digestion product of VP3.
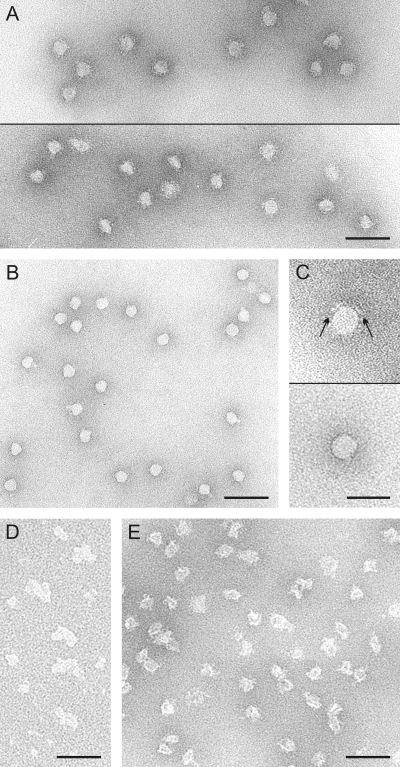
Proteinase K treatment resulted in DNA-containing lipid vesicles sedimenting more slowly than the untreated virions (Fig. 2A and B). Based on negative-stain transmission electron microscopy (TEM), the untreated HRPV-1 virions had a rough surface with extensions (Fig. 3A and C), and the proteinase K treated particles had a smooth one (Fig. 3B and C). The average dimensions of the proteinase K-treated virions (of 50 particles) were 31 by 34 nm. The respective dimensions of the untreated virions are 44 by 55 nm (39).
FIG. 3.
Negative-stain electron micrographs of dissociation products of HRPV-1. (A) Untreated “2× purified” virions. Two micrographs are shown. (B) Proteinase K-treated particles. (C) A single particle of the untreated “2× purified” virions (upper panel) and of the proteinase K-treated particles (lower panel). Arrows in the upper panel indicate surface extensions missing in the treated particles. (D) Aggregates in the pellet fraction of the TX-100 dissociation of the proteinase K-treated particles (see Fig. 2C). (E) Disrupted particles obtained after dissociation of the virions at low salinity at 60°C. Bars, 100 nm in panels A, B, D, and E and 50 nm in panel C.
A specific digestion product was observed in the proteinase K-treated particles (Fig. 2B, arrow III). Edman degradation of this ∼4-kDa protein band gave the N-terminal sequences FGSGSTDTML, GSGSTDTML, and GSTDTM, indicating that the respective band contained C-terminal portions of VP4. The sequence FGSGSTDTML corresponds to a C-terminal fragment of 32 amino acids. The 19- and 32-kDa protein species (Fig. 2B, arrows I and II) were not digestion products since they were also observed in untreated virion samples (Fig. 2A). Their identity could not be determined as the tryptic peptide LC-ESI MS/MS analysis revealed only peptides matching to major virion proteins VP3 and VP4 in both protein bands (Fig. 2B, arrows I and II).
In order to release the genome from the lipid vesicles, the proteinase K-treated particles were exposed to nonionic detergent TX-100. Concentrations from 0.025 to 0.1% (vol/vol) were tested. In 0.03% TX-100, the genome was released, and the rest of the proteins (VP3, the C-terminal portion of VP4, and 19-kDa and 32-kDa protein species) were found in the pellet (Fig. 2C). In negative-stain TEM the pellet fraction revealed aggregative material (Fig. 3D). None of the proteins cosedimented with DNA, and the viral lipids as well as TX-100 were detected at the top of the gradient (Fig. 2C). When 0.03% TX-100 dissociation was carried out with a protease inhibitor, the additional protein band (Fig. 2C, arrow), most likely a digestion product of VP3, was not observed. If lower TX-100 concentrations were used, only a partial genome release was achieved, and in higher concentrations VP3 was detected throughout the gradient (data not shown).
Spike protein VP4 can be removed by low salinity and elevated temperature or by NP-40.
Low salinity decreased the infectivity of the purified HRPV-1 virions, and the effect was greatly enhanced at an elevated temperature (Table 1). Rate zonal centrifugation followed by Tricine-SDS-PAGE revealed that spike protein VP4 was partly removed. At 4°C only a minor dissociation of VP4 was detected, but at 37°C and 60°C the majority of VP4 became soluble (Fig. 4A and see Fig. S1 in the supplemental material). The spike dissociation was the most quantitative at 60°C since all particles lost most of the spikes (see Fig. S1B in the supplemental material). At 37 and 60°C, the genome was released without associated proteins (Fig. 4A and see Fig. S1B in the supplemental material). In addition, the released genome was completely degraded by RQ1 DNase I, indicating that there is no protein-caused protection (data not shown). TEM revealed that subviral particles produced at 60°C were empty (Fig. 3E). In gel filtration, VP4 dissociated at 60°C had an apparent mass of 270 kDa (see Fig. S2A in the supplemental material), and in sedimentation only low-molecular-mass particles (40 kDa) were observed (see Fig. S2B in the supplemental material). An elevated temperature alone was not enough to dissociate VP4. Virions incubated in HRPV-1-buffer at 60°C did not lose VP4, but the genome was released from about half of the virions (data not shown).
FIG. 4.
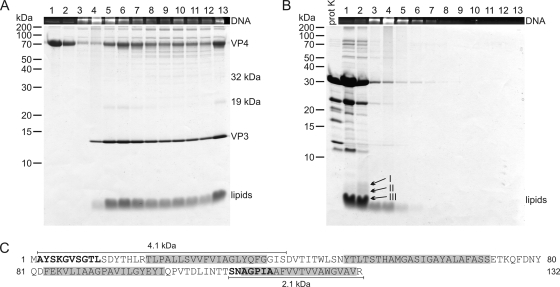
Proteinase K treatment of HRPV-1 virions at low salinity. (A) HRPV-1 virions dissociated at low salinity at 37°C. (B) HRPV-1 virions treated with proteinase K at low salinity at 37°C. (A and B) Particles were analyzed by rate zonal centrifugation in HRPV-1-buffer as in Fig. 2. The upper panels show the presence of DNA in EtBr-stained SDS-polyacrylamide gels. The lower panels show protein and lipid compositions in Coomassie blue- and Sudan Black B-stained 17% Tricine-SDS-polyacrylamide gels. Numbers on the left indicate the molecular mass markers (in kilodaltons). In panel B, a sample of proteinase K (prot K) was included, and the arrows I to III indicate the bands analyzed by N-terminal sequencing. (C) Amino acid sequence of VP3 indicating the sequences derived from N-terminal sequencing of the peptides (in boldface) and the predicted TM domains (gray shading). The 2.1-kDa fragment illustrates the peptide detected in the band indicated by arrow III, and the 4.1-kDa fragment illustrates the peptide in the band indicated by arrow I.
HRPV-1 virions were treated with several detergents, but controlled dissociation was obtained only with nonionic NP-40. The treatment of the virions with 0.1% (vol/vol) NP-40 (Table 1) led to the release of the genome without associated proteins (Fig. 5). The vast majority of VP4 with about half of the total viral lipids along with the detergent were detected at the top of the gradient. The other half of the total viral lipids and the rest of the viral proteins were found in the pellet (Fig. 5). In gel filtration, dissociated VP4 eluted mainly as a large particle, which could not be resolved by the filtration column used (separation range for globular proteins, 10 to 600 kDa), and in sedimentation analysis both small (90 kDa) and large (400 kDa) particles were observed (data not shown).
FIG. 5.
NP-40 dissociation of HRPV-1 virions. Virions were treated with NP-40, and the dissociation products were analyzed by rate zonal centrifugation in HRPV-1-buffer as in Fig. 2. The data of the fractions are presented as in Fig. 2.
Spike protein VP4 is glycosylated.
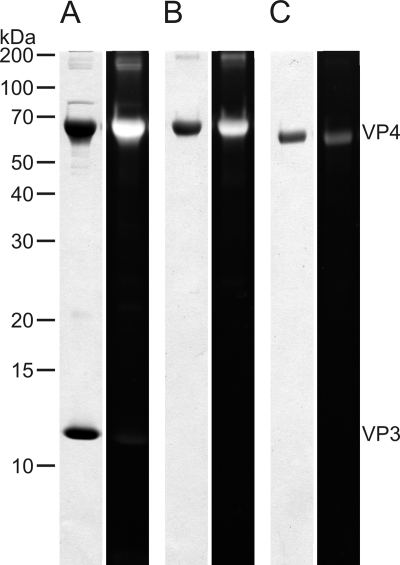
Previously, it has been observed that the apparent (65 kDa) and calculated (53 kDa) molecular masses of VP4 differ (39). To study possible glycosylation, Tricine-SDS-PAGE-separated proteins from highly purified virions were stained with a carbohydrate-specific stain. A positive signal was observed with VP4 (Fig. 6A). To further confirm this result, dissociated VP4 was deglycosylated using nonselective trifluoromethanesulfonic acid that removes both N-linked and O-linked oligosaccharide motifs. After deglycosylation treatment, VP4 gave a strongly reduced glycosylation signal in glycoprotein staining compared to the untreated form. Furthermore, deglycosylated VP4 migrated faster in Tricine-SDS-PAGE, and its apparent molecular mass was ∼55 kDa (Fig. 6B and C).
FIG. 6.
Glycoprotein analysis of HRPV-1. (A) Proteins of “2× purified” HRPV-1. (B) VP4. (C) Deglycosylated VP4. (A to C) Protein samples were separated by 14% Tricine-SDS-PAGE, and the gel was stained with Pro-Q Emerald 300 glycoprotein gel stain to detect the presence of glycosylated proteins. After UV illumination, the gel was stained with Coomassie blue to detect the proteins. The first lane represents the Coomassie blue, and the second lane represents the glycoprotein staining. Numbers on the left indicate the molecular mass markers (in kilodaltons). In panels B and C, VP4 was obtained from the dissociation of HRPV-1 at a low salinity at 60°C.
VP3 is an internal membrane protein.
Protease treatments of the virions in HRPV-1-buffer did not digest VP3 (Fig. 2B), and even in an excessive proteinase K concentration (0.5 mg/ml) VP3 remained intact (data not shown). When HRPV-1 virions were treated with proteinase K at low salinity at 37°C, the genome was released, both VP3 and VP4 were digested, and the empty lipid vesicles formed a band close to the top of the gradient (Fig. 4B). The infectivity was completely lost (<102 PFU/ml). Specific digestion products were obtained (Fig. 4B) and Edman degradation of the ∼5-kDa band (Fig. 4B, arrow I) gave an N-terminal sequence AYSKGVSGTL matching to the N terminus of VP3 (Fig. 4C). In the ∼4-kDa band (Fig. 4B, arrow II) no peptides were detected. In addition, the gel band containing the lipids (Fig. 4B, arrow III), which corresponds to ∼3 kDa, was analyzed, and the N-terminal sequence SNAGPIA was obtained, indicating that the respective band contained a C-terminal fragment of VP3 (Fig. 4C).
Comparison of lipids extracted from HRPV-1 and its host.
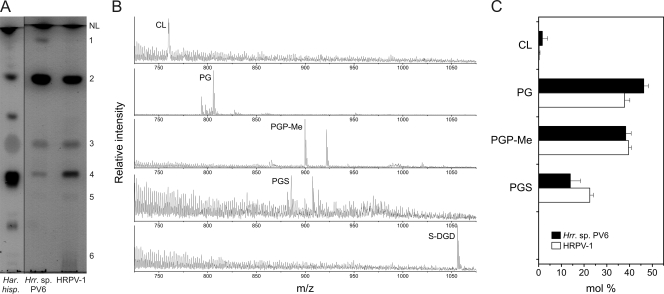
The total lipid extracts of highly purified HRPV-1 and its host Halorubrum sp. strain PV6 were examined by thin-layer chromatography. As seen from Fig. 7A, Halorubrum sp. strain PV6 and HRPV-1 contained three major polar lipids (spots 2 to 4). In addition, there were minor lipids present in both extracts (spots 1, 5, and 6). Most of the lipids from HRPV-1 and Halorubrum sp. strain PV6 extracts coeluted with those from halophilic archaeon Haloarcula hispanica (Fig. 7A) (4). This indicated that the two halophilic microorganisms and HRPV-1 share a similar polar lipid composition. TLC analysis also showed that neutral lipid compositions in HRPV-1 and its host were similar. However, the amounts of the neutral lipids seemed to be somewhat lower than those in Haloarcula hispanica (data not shown).
FIG. 7.
Lipid analysis of HRPV-1 and its Halorubrum host. (A) TLC image of the lipids extracted from Halorubrum sp. strain PV6 host (middle lane) and HRPV-1 (right lane). The pattern of Haloarcula hispanica lipids (left lane) from the same TLC analysis is included for comparison. The total amounts of phospholipids in the middle and right lanes are similar. Spots of polar lipids are designated by numbers 1 to 6 according to their migration. (B) Negative ion mass spectra of the extracts from the lipid-containing TLC spots of Halorubrum sp. strain PV6 (spots 1 to 5, CL to S-DGD). The prevalent peaks of doubly charged PGP-Me and PGS are not shown for clarity. Spot 6 could not be identified. (C) Phospholipid compositions of HRPV-1 and Halorubrum sp. strain PV6. Proportions are expressed as the mol% of the total phospholipids. Abbreviations: NL, neutral lipids; CL, cardiolipin; PG, phosphatidylglycerol; PGP-Me, phosphatidylglycerophosphate methyl ester; PGS, phosphatidylglycerosulfate; S-DGD, sulfated diglycosylglycerol diether.
To specifically identify the polar lipids of HRPV-1 and its host, the separated lipids were subjected to mass spectrometric analysis. Based on the m/z values of the intact molecules, as well as the profiles of the lipid fragments, the lipids in spots 1 to 4 were identified as cardiolipin (CL; doubly charged peak at m/z 759.7), phosphatidylglycerol (PG; major peak at m/z 805.8), phosphatidylglycerophosphate methyl ester (PGP-Me; m/z 899.8), and phosphatidylglycerosulfate (PGS; m/z 885.8), respectively (Fig. 7B), which are negatively charged. These are also the major phospholipids of Haloarcula hispanica (4). Quantitative analysis of the phospholipids revealed that HRPV-1 and its host contained similar proportions of all phospholipids, except that PG was slightly more prevalent in the host, while PGS was more prevalent in HRPV-1 (Fig. 7C).
Staining of the TLC plates with specific reagents revealed that spot 5 contained glycolipids. This glycolipid did not, however, coelute with the triglycosylglycerodiether or with any other of the lipids from Haloarcula hispanica (Fig. 7A). The mass of the lipid from spot 5 was 1,055.9 Da. This corresponds to the mass of sulfodiglycosyldiglyceride (S-DGD), a negatively charged glycolipid typical of various Halorubrum strains and of certain other halophilic microorganisms (30, 33, 45). These data strongly suggest the presence of nonphosphorus S-DGD in HRPV-1 and its host.
DISCUSSION
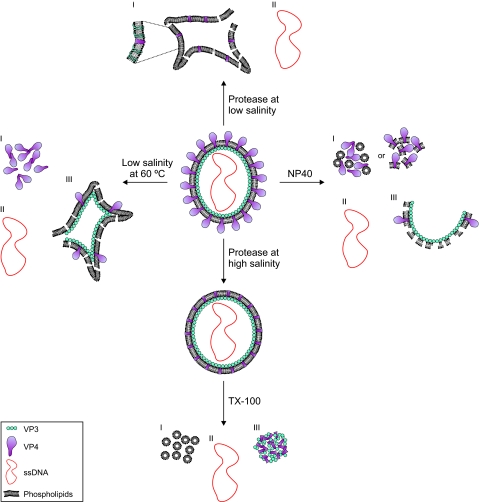
The initial description of the HRPV-1 virion suggested a pleomorphic membrane vesicle enclosing the ssDNA genome (39). Since structural biology methods are less advantageous for pleomorphic enveloped virus particles, we tackled the HRPV-1 structure by biochemical means. The dissociation pathways described here revealed the main structural features of the virion (Fig. 8).
FIG. 8.
Schematic presentation of structural organization of HRPV-1 virion. The main dissociation conditions are indicated (see Results). The left panel (low salinity at 60°C) illustrates the dissociation of spike protein VP4 (I), the release of the genome (II), and the empty particles after the partial VP4 release (III). The right panel (NP-40) illustrates the two lipid fractions (I and III) and the released genome (II) obtained by NP-40 dissociation. The lower panel (protease at high salinity and TX-100) illustrates protease digestion of VP4 at high salinity and further dissociation of the lipid vesicles with TX-100, resulting in solubilized lipids (I), the released genome (II), and aggregated VP3 and the hydrophobic domain of VP4 (III). The upper panel (protease at low salinity) illustrates the lipid vesicles (I) and the released genome (II). The inset shows the membrane associated domains of VP3, which are not indicated in the other graphs.
VP4, the larger major structural protein of HRPV-1, was shown to form spikelike protrusions on the virion surface. Mature VP4 has one putative transmembrane (TM) helix at the C terminus (39) and, after protease digestion, only the C-terminal fragment of VP4 containing this region remained associated with the viral membrane (Fig. 2B). Without the spikes, the lipid vesicles were close to spherical (Fig. 3) and clearly more uniform than the native virus particles (compare the sedimentation in Fig. 2A and B) implying that the spikes contribute to the pleomorphism of the HRPV-1 virion.
HRPV-1 originates from a solar saltern (39) and, as expected, a high sodium chloride concentration is necessary for infectivity (Fig. 1B). Most haloarchaeal viruses, e.g., icosahedral SH1 (41) and spindle-shaped His2 (7), require a high salt concentration for infectivity, but the head-tail haloarchaeal virus ΦN is an exception since it remains infective even in distilled water (54). Temperature is also critical for the infectivity of HRPV-1 (Fig. 1D). The infectivity was lost in the same temperature range, as is the case with SH1 and His2 (7, 41). The combination of lowered ionic strength and elevated temperature led to the dissociation of VP4 (Fig. 4A and see Fig. S1B in the supplemental material). Spike protein VP4 seems to be strongly asymmetric since gel filtration of dissociated VP4 indicated a multimeric protein and sedimentation smaller than a monomer (see Fig. S2 in the supplemental material).
NP-40 treatment also released VP4 (Fig. 5). The treatment dissected the two major structural proteins into soluble (mainly VP4) and aggregated (mainly VP3) fractions. The intriguing result was that the viral lipids were detected approximately in the same ratio in these fractions. This was in contrast to the TX-100 dissociation of the protease-treated particles, where all of the viral lipids were solubilized (Fig. 8).
Glycosylation of spike protein VP4 was elucidated by both carbohydrate staining and deglycosylation (Fig. 6). The apparent molecular mass of deglycosylated VP4 (55 kDa) was close to the calculated molecular mass of VP4 (53 kDa). Among archaeal viruses, glycosylation has been previously detected on the major capsid protein of STIV infecting hyperthermophilic crenarchaeon Sulfolobus solfataricus. In addition, STIV encodes a glycosyltransferase, and it is proposed that the major capsid protein might have novel glycosylation chemistry (27, 31). Rod-shaped virus ARV1 infecting hyperthermophilic crenarchaea of the genus Acidianus also has a glycosylated major coat protein (53).
VP3, the smaller major structural protein of HRPV-1, is predicted to contain four TM helices (39). In the intact virion, VP3 was completely resistant to proteases (Fig. 2B). However, when the virion vesicle was opened leading to the genome release, VP3 was quantitatively digested by proteinase K, leaving two membrane-associated peptides, one from the N terminus and other from the C terminus (Fig. 4). Both of these fragments are predicted to contain one TM domain. We conclude that VP3 is a membrane protein facing the inner membrane surface of the virion (Fig. 8).
Lipid analyses showed that HRPV-1 and its host contained almost the same proportions of all phospholipids (Fig. 7). Such similarity between the viral and host membranes indicates that the lipids are nonselectively incorporated into the viral membrane. It has been previously reported that pleomorphic mycoplasmavirus L172 and its host also have similar phospholipid compositions (2). Unlike pleomorphic viruses, nontailed icosahedral lipid-containing prokaryotic viruses commonly display selective lipid acquisition from the host cell membrane, as seen in bacteriophages with either an inner membrane (PM2, PRD1, and Bam35) or with an outer one (Φ6) (11, 12, 28, 29). Also, the archaeal viruses STIV (31) and SH1 (4) having the lipid membrane underneath the icosahedral protein capsid selectively acquire lipids from the host lipid pool. The geometrical constraints within the icosahedral protein capsid most likely drive this selectivity (4, 11, 12, 28, 29, 31).
Budding is a typical exit mechanism for enveloped viruses (18, 55). Both the nonselective lipid acquisition and the structural organization of the HRPV-1 virion support the presence of such a maturation mechanism. The nature of the infection cycle is also in accordance with the presence of a budding mechanism since HRPV-1 does not lyse the host cells, but progeny viruses are released while the host growth continues (39). Most likely, the viral proteins are incorporated into the plasma membrane of the host cell, after which these proteins interact with the genome to bud through the membrane. The size of the vesicle should then be determined by the genome length. If budding occurs, it remains to be determined how HRPV-1 passes the proteinaceous S layer (25) of the host cell. However, we were previously unable to detect budding viruses using transmission or scanning EM (unpublished data).
Our results strongly indicated that the genome of HRPV-1 does not form a nucleoprotein complex. The common nominator for all four main dissociation pathways determined was the release of the genome without associated proteins and the presence of separate lipid and protein fractions (Fig. 8). This is in contrast to other enveloped viruses that usually have a nucleoprotein associated with the genome (9, 17, 34, 47, 51). However, several enveloped viruses, such as orthomyxoviruses, paramyxoviruses, and retroviruses also have a matrix protein underneath the membrane facilitating the interaction of the envelope and genome (17, 34, 51). The genome of HRPV-1 may have interactions with VP3 inside the vesicle, but these interactions cannot be particularly strong since the genome is released with relatively gentle treatments. In addition, the domain of VP3 facing the naked genome has only a few positively charged amino acid residues. Since all phospholipids and the glycolipid of HRPV-1 are negatively charged, strong ionic interactions between the genome and membrane are unlikely.
Electron cryotomography of Uukuniemi virus (family Bunyaviridae) gives important insights into the architecture of pleomorphic enveloped viruses (38). The structure of Uukuniemi virus was determined at 5.9-nm resolution and shows that the glycoprotein spikes in the envelope are arranged in the most regular particles on an icosahedral lattice with T=12 triangulation. Because ribonucleoprotein complexes do not seem to be particularly organized inside the envelope and because bunyaviruses have no matrix protein, lateral interactions between the two glycoproteins most likely determine the virion architecture (38). Our dissociation studies imply similar architecture for HRPV-1, but membrane protein VP3 is most likely a crucial structural determinant in addition to glycoprotein VP4. Electron cryotomography seems attractive to gain further structural information on the HRPV-1 particle, although the high salinity needed for the virus infectivity is a challenge for this approach.
We showed here that HRPV-1 is structurally novel among the studied archaeal viruses. It may resemble pleomorphic bacterial viruses infecting mycoplasmas (14), and it is closely related to another pleomorphic haloarchaeal virus, Haloarcula hispanica pleomorphic virus 1, recently isolated from a different Italian solar saltern (E. Roine, P. Kukkaro, L. Paulin, S. Laurinavičius, A. Domanska, P. Somerharju, and D. H. Bamford, unpublished data). Accordingly, HRPV-1-type viruses may form a structure-based lineage or clade of viruses, possibly with a common origin and infecting hosts in different domains of life. To validate this lineage, it is necessary to perform similar structural analyses for other pleomorphic enveloped viruses. Structure-based viral lineages have thus far been proposed for icosahedral viruses only (3, 5, 8, 26). A lineage is proposed now for pleomorphic enveloped viruses, which have no detectable nucleoprotein.
Supplementary Material
Acknowledgments
This research was supported by Academy of Finland Centre of Excellence Programme in Virus Research grant 1129684 (2006-2011) (D.H.B.).
We thank Sari Korhonen for excellent technical assistance. We thank Nisse Kalkkinen and Gunilla Rönnholm for the protein sequencing and mass spectrometry. We thank Pentti Somerharju for providing facilities and materials for the lipid analyses. M.K.P. is a fellow of Viikki Graduate School in Molecular Biosciences.
Footnotes
Published ahead of print on 28 October 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ackermann, H. W. 2007. 5,500 phages examined in the electron microscope. Arch. Virol. 152:227-243. [DOI] [PubMed] [Google Scholar]
- 2.Al-Shammari, A. J. N., and P. F. Smith. 1981. Lipid composition of two mycoplasmaviruses, MV-Lg-L172 and MVL2. J. Gen. Virol. 54:455-458. [Google Scholar]
- 3.Bamford, D. H. 2003. Do viruses form lineages across different domains of life? Res. Microbiol. 154:231-236. [DOI] [PubMed] [Google Scholar]
- 4.Bamford, D. H., J. J. Ravantti, G. Rönnholm, S. Laurinavičius, P. Kukkaro, M. Dyall-Smith, P. Somerharju, N. Kalkkinen, and J. K. H. Bamford. 2005. Constituents of SH1, a novel lipid-containing virus infecting the halophilic euryarchaeon Haloarcula hispanica. J. Virol. 79:9097-9107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bamford, D. H., J. M. Grimes, and D. I. Stuart. 2005. What does structure tell us about virus evolution? Curr. Opin. Struct. Biol. 15:655-663. [DOI] [PubMed] [Google Scholar]
- 6.Bartlett, E. M., and D. H. Lewis. 1970. Spectrophotometric determination of phosphate esters in the presence or absence of orthophosphate. Anal. Biochem. 36:159-167. [DOI] [PubMed] [Google Scholar]
- 7.Bath, C., T. Cukalac, K. Porter, and M. L. Dyall-Smith. 2006. His1 and His2 are distantly related, spindle-shaped haloviruses belonging to the novel virus group, Salterprovirus. Virology 350:228-239. [DOI] [PubMed] [Google Scholar]
- 8.Benson, S. D., J. K. H. Bamford, D. H. Bamford, and R. M. Burnett. 2004. Does common architecture reveal a viral lineage spanning all three domains of life? Mol. Cell 16:673-685. [DOI] [PubMed] [Google Scholar]
- 9.Bishop, D. H. L. 1990. Bunyaviridae, p. 1155-1173. In B. N. Fields, D. M. Knipe, R. M. Chanock, M. S. Hirsch, J. L. Melnick, T. P. Monath, and B. Roizman (ed.), Fields virology, 2nd ed., vol. 1. Raven Press, Ltd., New York, NY. [Google Scholar]
- 10.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 11.Braunstein, S. N., and R. M. Franklin. 1971. Structure and synthesis of a lipid-containing bacteriophage. V. Phospholipids of the host BAL-31 and of the bacteriophage PM2. Virology 43:685-695. [DOI] [PubMed] [Google Scholar]
- 12.Brewer, G. J., and R. M. Goto. 1983. Accessibility of phosphatidylethanolamine in bacteriophage PM2 and in its gram-negative host. J. Virol. 48:774-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLong, E. F., and N. R. Pace. 2001. Environmental diversity of bacteria and archaea. Syst. Biol. 50:470-478. [PubMed] [Google Scholar]
- 14.Dybvig, K., J. A. Nowak, T. L. Sladek, and J. Maniloff. 1985. Identification of an enveloped phage, mycoplasma virus L172, that contains a 14-kilobase single-stranded DNA genome. J. Virol. 53:384-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fauquet, C. M., M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.). 2005. Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, London, United Kingdom.
- 16.Folch, J., M. Lees, and G. H. Sloane Stanley. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226:497-509. [PubMed] [Google Scholar]
- 17.Ganser-Pornillos, B. K., M. Yeager, and W. I. Sundquist. 2008. The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 18:203-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garoff, H., R. Hewson, and D. J. E. Opstelten. 1998. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 62:1171-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guixa-Boixareu, N., J. I. Calderón-Paz, M. Heldal, G. Bratbak, and C. Pedrós-Alió. 1996. Viral lysis and bacterivory as prokaryotic loss factors along a salinity gradient. Aquat. Microb. Ecol. 11:215-227. [Google Scholar]
- 20.Holmes, M. L., F. Pfeifer, and M. L. Dyall-Smith. 1995. Analysis of the halobacterial plasmid pHK2 minimal replicon. Gene 153:117-121. [DOI] [PubMed] [Google Scholar]
- 21.Jäälinoja, H. T., E. Roine, P. Laurinmäki, H. M. Kivelä, D. H. Bamford, and S. J. Butcher. 2008. Structure and host-cell interaction of SH1, a membrane-containing, halophilic euryarchaeal virus. Proc. Natl. Acad. Sci. USA 105:8008-8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kates, M. 1972. Techniques of lipidology: isolation, analysis and identification of lipids. North-Holland Publishing Co., Amsterdam, The Netherlands.
- 23.Khayat, R., L. Tang, E. T. Larson, C. M. Lawrence, M. Young, and J. E. Johnson. 2005. Structure of an archaeal virus capsid protein reveals a common ancestry to eukaryotic and bacterial viruses. Proc. Natl. Acad. Sci. USA 102:18944-18949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kivelä, H. M., E. Roine, P. Kukkaro, S. Laurinavičius, P. Somerharju, and D. H. Bamford. 2006. Quantitative dissociation of archaeal virus SH1 reveals distinct capsid proteins and a lipid core. Virology 356:4-11. [DOI] [PubMed] [Google Scholar]
- 25.König, H., R. Rachel, and H. Claus. 2007. Proteinaceous surface layers of Archaea: ultrastructure and biochemistry, p. 315-340. In R. Cavicchioli (ed.), Archaea: molecular and cellular biology. ASM Press, Washington, DC.
- 26.Krupovič, M., and D. H. Bamford. 2008. Virus evolution: how far does the double beta-barrel viral lineage extend? Nat. Rev. Microbiol. 6:941-948. [DOI] [PubMed] [Google Scholar]
- 27.Larson, E. T., D. Reiter, M. Young, and C. M. Lawrence. 2006. Structure of A197 from Sulfolobus turreted icosahedral virus: a crenarchaeal viral glycosyltransferase exhibiting the GT-A fold. J. Virol. 80:7636-7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laurinavičius, S., R. Käkelä, D. H. Bamford, and P. Somerharju. 2004. The origin of phospholipids of the enveloped bacteriophage phi6. Virology 326:182-190. [DOI] [PubMed] [Google Scholar]
- 29.Laurinavičius, S., R. Käkelä, P. Somerharju, and D. H. Bamford. 2004. Phospholipid molecular species profiles of tectiviruses infecting gram-negative and gram-positive hosts. Virology 322:328-336. [DOI] [PubMed] [Google Scholar]
- 30.Lizama, C., M. Monteoliva-Sánchez, A. Suárez-García, R. Rosello-Mora, M. Aguilera, V. Campos, and A. Ramos-Cormenzana. 2002. Halorubrum tebenquichense sp. nov., a novel halophilic archaeon isolated from the Atacama Saltern, Chile. Int. J. Syst. Evol. Microbiol. 52:149-155. [DOI] [PubMed] [Google Scholar]
- 31.Maaty, W. S. A., A. C. Ortmann, M. Dlakić, K. Schulstad, J. K. Hilmer, L. Liepold, B. Weidenheft, R. Khayat, T. Douglas, M. J. Young, and B. Bothner. 2006. Characterization of the archaeal thermophile Sulfolobus turreted icosahedral virus validates an evolutionary link among double-stranded DNA viruses from all domains of life. J. Virol. 80:7625-7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsudaira, P. 1987. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 262:10035-10038. [PubMed] [Google Scholar]
- 33.McGenity, T. J., and W. D. Grant. 1995. Transfer of Halobacterium saccharovorum, Halobacterium sodomense, Halobacterium trapanicum NRC 34021 and Halobacterium lacusprofundi to the genus Halorubrum gen. nov., as Halorubrum saccharovorum comb. nov., Halorubrum sodomense comb. nov., Halorubrum trapanicum comb. nov., and Halorubrum lacusprofundi comb. nov. Syst. Appl. Microbiol. 18:237-243. [Google Scholar]
- 34.Nayak, D. P., E. K. Hui, and S. Barman. 2004. Assembly and budding of influenza virus. Virus Res. 106:147-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olkkonen, V. M., and D. H. Bamford. 1989. Quantitation of the adsorption and penetration stages of bacteriophage phi 6 infection. Virology 171:229-238. [DOI] [PubMed] [Google Scholar]
- 36.Oren, A. 2002. Molecular ecology of extremely halophilic Archaea and Bacteria. FEMS Microbiol. Ecol. 39:1-7. [DOI] [PubMed] [Google Scholar]
- 37.Oren, A., G. Bratbak, and M. Heldal. 1997. Occurrence of virus-like particles in the Dead Sea. Extremophiles 1:143-149. [DOI] [PubMed] [Google Scholar]
- 38.Överby, A. K., R. F. Pettersson, K. Grünewald, and J. T. Huiskonen. 2008. Insights into bunyavirus architecture from electron cryotomography of Uukuniemi virus. Proc. Natl. Acad. Sci. USA 105:2375-2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pietilä, M. K., E. Roine, L. Paulin, N. Kalkkinen, and D. H. Bamford. 2009. An ssDNA virus infecting archaea: a new lineage of viruses with a membrane envelope. Mol. Microbiol. 72:307-319. [DOI] [PubMed] [Google Scholar]
- 40.Porter, K., B. E. Russ, and M. L. Dyall-Smith. 2007. Virus-host interactions in salt lakes. Curr. Opin. Microbiol. 10:418-424. [DOI] [PubMed] [Google Scholar]
- 41.Porter, K., P. Kukkaro, J. K. H. Bamford, C. Bath, H. M. Kivelä, M. L. Dyall-Smith, and D. H. Bamford. 2005. SH1: a novel, spherical halovirus isolated from an Australian hypersaline lake. Virology 335:22-33. [DOI] [PubMed] [Google Scholar]
- 42.Poutanen, M., L. Salusjärvi, L. Ruohonen, M. Penttilä, and N. Kalkkinen. 2001. Use of matrix-assisted laser desorption/ionization time-of-flight mass mapping and nanospray liquid chromatography/electrospray ionization tandem mass spectrometry sequence tag analysis for high sensitivity identification of yeast proteins separated by two-dimensional gel electrophoresis. Rapid Commun. Mass Spectrom. 15:1685-1692. [DOI] [PubMed] [Google Scholar]
- 43.Prangishvili, D., P. Forterre, and R. A. Garrett. 2006. Viruses of the Archaea: a unifying view. Nat. Rev. Microbiol. 4:837-848. [DOI] [PubMed] [Google Scholar]
- 44.Prangishvili, D., R. A. Garrett, and E. V. Koonin. 2006. Evolutionary genomics of archaeal viruses: unique viral genomes in the third domain of life. Virus Res. 117:52-67. [DOI] [PubMed] [Google Scholar]
- 45.Qiu, D. F., M. P. L. Games, X. Y. Xiao, D. E. Games, and T. J. Walton. 2000. Characterisation of membrane phospholipids and glycolipids from a halophilic archaebacterium by high-performance liquid chromatography/electrospray mass spectrometry. Rapid Commun. Mass Spectrom. 14:1586-1591. [DOI] [PubMed] [Google Scholar]
- 46.Rice, G., L. Tang, K. Stedman, F. Roberto, J. Spuhler, E. Gillitzer, J. E. Johnson, T. Douglas, and M. Young. 2004. The structure of a thermophilic archaeal virus shows a double-stranded DNA viral capsid type that spans all domains of life. Proc. Natl. Acad. Sci. USA 101:7716-7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rojek, J. M., and S. Kunz. 2008. Cell entry by human pathogenic arenaviruses. Cell. Microbiol. 10:828-835. [DOI] [PubMed] [Google Scholar]
- 48.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 49.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 50.Srinivasiah, S., J. Bhavsar, K. Thapar, M. Liles, T. Schoenfeld, and K. E. Wommack. 2008. Phages across the biosphere: contrasts of viruses in soil and aquatic environments. Res. Microbiol. 159:349-357. [DOI] [PubMed] [Google Scholar]
- 51.Takimoto, T., and A. Portner. 2004. Molecular mechanism of paramyxovirus budding. Virus Res. 106:133-145. [DOI] [PubMed] [Google Scholar]
- 52.Torsvik, T., and I. D. Dundas. 1974. Bacteriophage of Halobacterium salinarium. Nature 248:680-681. [DOI] [PubMed] [Google Scholar]
- 53.Vestergaard, G., M. Häring, X. Peng, R. Rachel, R. A. Garrett, and D. Prangishvili. 2005. A novel rudivirus, ARV1, of the hyperthermophilic archaeal genus Acidianus. Virology 336:83-92. [DOI] [PubMed] [Google Scholar]
- 54.Vogelsang-Wenke, H., and D. Oesterhelt. 1988. Isolation of a halobacterial phage with a fully cytosine-methylated genome. Mol. Gen. Genet. 211:407-414. [Google Scholar]
- 55.Welsch, S., B. Müller, and H. G. Kräusslich. 2007. More than one door: budding of enveloped viruses through cellular membranes. FEBS Lett. 581:2089-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.