Abstract
The genome of the severe acute respiratory syndrome-associated coronavirus (SARS-CoV) contains eight open reading frames (ORFs) that encode novel proteins. These accessory proteins are dispensable for in vitro and in vivo replication and thus may be important for other aspects of virus-host interactions. We investigated the functions of the largest of the accessory proteins, the ORF 3a protein, using a 3a-deficient strain of SARS-CoV. Cell death of Vero cells after infection with SARS-CoV was reduced upon deletion of ORF 3a. Electron microscopy of infected cells revealed a role for ORF 3a in SARS-CoV induced vesicle formation, a prominent feature of cells from SARS patients. In addition, we report that ORF 3a is both necessary and sufficient for SARS-CoV-induced Golgi fragmentation and that the 3a protein accumulates and localizes to vesicles containing markers for late endosomes. Finally, overexpression of ADP-ribosylation factor 1 (Arf1), a small GTPase essential for the maintenance of the Golgi apparatus, restored Golgi morphology during infection. These results establish an important role for ORF 3a in SARS-CoV-induced cell death, Golgi fragmentation, and the accumulation of intracellular vesicles.
The severe acute respiratory syndrome-associated coronavirus (SARS-CoV) genome encodes several smaller open reading frames (ORFs) located in the 3′ region of the genome that are predicted to express eight novel proteins termed accessory proteins. The accessory proteins are designated ORFs 3a, 3b, 6, 7a, 7b, 8a, 8b, and 9b and range in size from 39 to 274 amino acids (35, 50). These SARS-CoV-specific ORFs are not present in other coronaviruses and do not display significant homology with any known proteins in the NCBI database. Five of these are predicted to code for polypeptides of greater than 50 amino acids (35, 50). Antibodies reactive against all of the SARS-CoV proteins have been detected in sera isolated from SARS patients, indicating that these proteins are expressed by the virus in vivo (7, 9, 17-19, 45, 59). Expression of three of the ORF proteins has been demonstrated during infection using protein-specific antibodies and include the ORFs 3a, 6, and 7a (12, 37, 41, 60). Six of the eight group-specific ORFs, including ORFs 3a, 3b, 6, 7a, 7b, and 9b, were deleted from recombinant SARS-CoV and shown to be dispensable for in vitro and in vivo replication (66).
Related coronaviruses also encode unique accessory proteins in the 3′ region of the genome, often referred to as group-specific ORFs. Similar to SARS-CoV, several of these proteins are dispensable for viral replication. Murine hepatitis virus (MHV) expresses accessory proteins ORFs 2a, 4, and 5a. A recombinant virus in which ORF 2a was deleted replicated normally in vitro but caused attenuated disease in vivo (55). Deletion of the group-specific ORF 7 in porcine coronavirus TGEV also results in reduced replication and virulence in vivo despite normal replication in vitro (38). Similarly, in feline infectious peritonitis virus (FIPV), group-specific proteins are dispensable for replication in cell culture but contribute to pathogenesis in vivo (20). Thus, while the SARS-CoV group specific proteins are unnecessary for in vitro and in vivo replication, their expression may underlie the devastating pathology associated with SARS disease. Detailed characterization of these novel proteins may contribute to a better understanding of SARS pathogenesis and host-virus interactions.
The ORF 3a protein is expressed from subgenomic RNA3, which contains the 3a and 3b ORFs (35, 50). The 3a protein, which is the largest group-specific SARS-CoV accessory protein at 274 amino acids, has been reported to localize to the Golgi apparatus, the plasma membrane, and intracellular vesicles of unknown origin (67, 68). The protein is efficiently transported to the cell surface and is also internalized during the process of endocytosis (60).
The mechanism of SARS-CoV-induced cell death has been investigated by several groups. Studies to date have used overexpression of individual SARS-CoV ORFs to evaluate their intrinsic cytotoxicity. Using this approach, the following proteins have been reported to cause apoptosis: the 3CL-like protease; spike; ORFs 3a, 3b, and 7a; and the envelope (E), membrane (M), and nucleocapsid (N) proteins (23, 31, 32, 36, 46, 58, 61, 65, 69). However, since all of these reports utilize overexpression of individual proteins, it is unclear whether these effects may be attributable to high, nonphysiological levels of protein and whether they occur during infection. Analysis of recombinant viruses with specific mutations or deletions is necessary to determine the relative contribution of these proteins to the cytotoxicity of SARS-CoV during infection (63). Therefore, the cytotoxic component(s) of SARS-CoV have not been fully defined.
Here, we have investigated the function of the ORF 3a protein in the context of SARS-CoV infection and by overexpression. We confirm that ORF 3a contributes to SARS-CoV cytotoxicity using a recombinant strain deficient for expression of ORF 3a. While characterizing this deficient strain, we observed that SARS-CoV-induced vesicle formation, a feature that has been documented in cells from infected SARS patients, is dependent on ORF 3a. Furthermore, we observed that SARS-CoV infection causes Golgi fragmentation by ORF 3a. Additional characterization of 3a in transfected cells revealed that the protein colocalizes with markers of the trans-Golgi network (TGN) and late endosomal pathways and causes an accumulation of these vesicles. Finally, we report that Arf1 overexpression rescued SARS-CoV or 3a-induced Golgi fragmentation, suggesting that the ORF 3a protein may perturb Arf1-mediated vesicle trafficking.
MATERIALS AND METHODS
Cell lines and viruses.
Vero cells were obtained from the American Type Culture Collection (Rockville, MD). Cells were maintained in Dulbecco modified Eagle medium with 4.5 g of glucose/liter, supplemented with 2 mM l-glutamine, 1% penicillin-streptomycin solution, and 10% fetal bovine serum. The construction and characterization of ΔORF3a SARS-CoV has been described elsewhere (66). Vero cell monolayers were infected with SARS-CoV (Urbani strain), ΔORF3a SARS-CoV at a multiplicity of infection of 5. All work with infectious SARS-CoV virus was performed in a biosafety level 3 facility by personnel wearing powered air purifying respirators (3M HEPA AirMate, Saint Paul, MN).
Plasmids and DNA transfection.
An expression construct for SARS-CoV 3a was generated by using RNA extracted from SARS-CoV-infected Vero cells. Reverse transcription (RT) followed by PCR was carried out by with the following gene specific primers: forward primer, 5′-CCGGAATTCAGATTTTTTACTCTTAGATC-3′, and reverse primer, 5′-TCCCCCCGGGCCAAAGGCACGCTAGTAGTCG-3′.
The resulting PCR products were cloned into the EcoRI-XmaI sites of the p3xFlag-green fluorescent protein (GFP) vector, which is expressed under the cytomegalovirus promoter. The fidelity of the resulting construct was confirmed by sequencing. Expression constructs for Arf1 and Gal have been described elsewhere (33). GFP-LC3 was kindly provided by N. Mizushima and T. Yoshimori and RFP-LC3 was kindly provided by Marja Jaattela. Vero cells (106) were transfected with 1 μg of DNA (single transfections) or 0.5 μg of each plasmid (cotransfections) by Amaxa nucleofection, using V solution, program O-17 (Gaithersburg, MD). HEK293T cells were transfected by the calcium phosphate precipitation method using 2 μg of DNA.
Detection of SARS-CoV nucleic acid.
Detection and quantification of SARS-CoV was determined by using a QuantiTect Probe RT-PCR kit (catalog no. 204443; Qiagen, Valencia, CA) according to the manufacturer's protocol. In brief, RNA samples were quantified with a spectrophotometer (Agilent Technologies, Palo Alto, CA) and an aliquot diluted to a concentration of 100 ng/μl in preparation for real-time RT-PCR analyses. Each 50-μl reaction mixture contained 25 μl of 2× Master Mix; 0.5 μl of RT mix; SARS N3-specific primers and probe (forward, 5′-GAA GTA CCA TCT GGG GCT GAG-3′; reverse, 5′-CCG AAG AGC TAC CCG ACG-3′; probe, 5′-HEX-CTC TTT CAT TTT GCC GTC ACC ACC AC-BHQ1-3′) at final concentrations of 0.4 and 0.2 μM, respectively; 18.5 μl of nuclease-free water; and 1 μl of RNA (100 ng) that was heat denatured at 95°C for 2 min on a PTC-100 Peltier thermal cycler (MJ Research, Waltham, MA) prior to addition. Amplification was carried out in 96-well plates on a Mx4000 multiplex quantitative PCR system (Stratagene, La Jolla, CA). Thermocycling conditions consisted of 30 min at 50°C for RT; 15 min at 95°C for polymerase activation; and 40 cycles of 30 s at 95°C, 1 min at 56°C, and 30 s at 76°C. Each sample was also run under identical conditions with primers and probe specific for the 18S housekeeping gene (forward, 5′-GGT ACA GTG AAA CTG CGA AT-3′; reverse, 5′-CAG TTA TCC AAG TGG GAG AG-3′; probe, 5′-6-FAM-ATT AAA TCA GTT ATG GTT CCT TTG GTC G-BHQ-6-FAM-3′). A 10-fold serial dilution standard curve was generated for each set of primers with the Urbani strain of SARS-CoV (1.55 × 105 50% tissue culture infective doses/μl).
Microscopy.
Transfected cells were subcultured in a four-chamber borosilicate chambered coverglass system (Nunc), fixed with 4% paraformaldehyde in phosphate-buffered saline (pH 7.4), and analyzed under a confocal fluorescence microscope (Leica SP2-AOBS-UV405; Leica Microsystems, Wetzlar, Germany) or imaged live while maintaining cells at 37°C and 5% CO2 with a heated stage. Cells were imaged by using a 63× objective with 3× digital zoom or 63× objective with 1× digital zoom for high and low magnifications, respectively. Anti-GM130 monoclonal antibody was obtained from Becton Dickinson Transduction Laboratories (Franklin Lakes, NJ). For staining for GM130, cells were fixed, permeabilized, and stained by using permeabilization solution and blocking solution from Molecular Probes according to the instructions. Primary and secondary antibodies were diluted 1:1,000. For evaluation of morphology of the Golgi apparatus, cells were imaged at random using a 63× objective lens. Then, using ImageJ software, the numbers of cells per field containing a bright and distinct juxtanuclear Golgi stack or a dispersed Golgi apparatus were determined. Electron microscopy was carried out as described previously (16). Briefly, cells were fixed in 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4), postfixed in 1% osmium tetroxide for 1 h, embedded in Epon-substitute/araldite resin, sectioned, double stained with uranyl acetate and lead citrate, and analyzed using a Philips 410LS transmission electron microscope. For each treatment or control group, at least 100 cells from randomly chosen transmission electron microscopy (TEM) fields were analyzed for quantification of morphological features.
RESULTS
ORF 3a contributes to cell death caused by SARS-CoV.
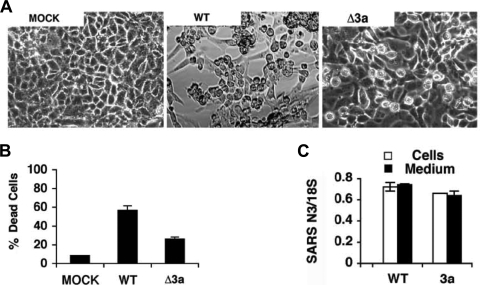
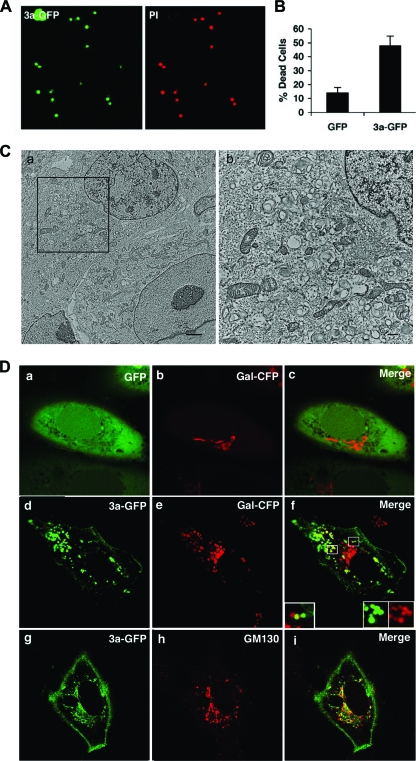
Previous reports have documented cell death as a result of overexpression of ORF 3a (31, 39). To determine the physiological importance of ORF 3a, we infected Vero cells with wild-type (WT) or 3a-deficient (Δ3a) SARS-CoV, which has been described previously (66). At 48 h postinfection (hpi), we observed a reduction in cell death for Δ3a compared to the WT (Fig. 1A and B). To determine whether the observed reduction in cell death was a result of impaired replication of the virus, we assessed the levels of intracellular and extracellular viral RNA by real-time PCR. We observed that RNA levels and titers were equivalent for both strains, indicating that reduced cell death was unlikely a consequence of impaired replication (Fig. 1C). These results are in agreement with a previous report that reported equivalent replication for Δ3a and WT virus in Vero cells, assessed by plaque assay, at 32 hpi (66). Thus, 3a contributes to SARS-CoV-induced cell death during infection.
FIG. 1.
Cell death caused by SARS-CoV is reduced by deletion of 3a. (A) Vero cells were mock infected (MOCK) or infected with wild-type (WT) or 3a-deficient (Δ3a) SARS-CoV. After 48 h, cells were examined by phase-contrast light microscopy. (B) Quantification of cell death in samples in panel A after trypan blue staining. (C) Amplification of viral nucleic acid in medium and cells of samples in panel A determined by quantitative real-time PCR and represented as the mean threshold cycle normalized by 18S rRNA.
ORF 3a is necessary for the formation of intracellular vesicles.
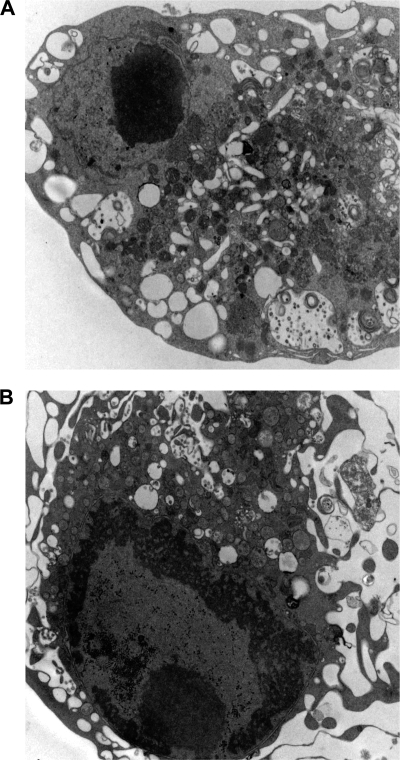
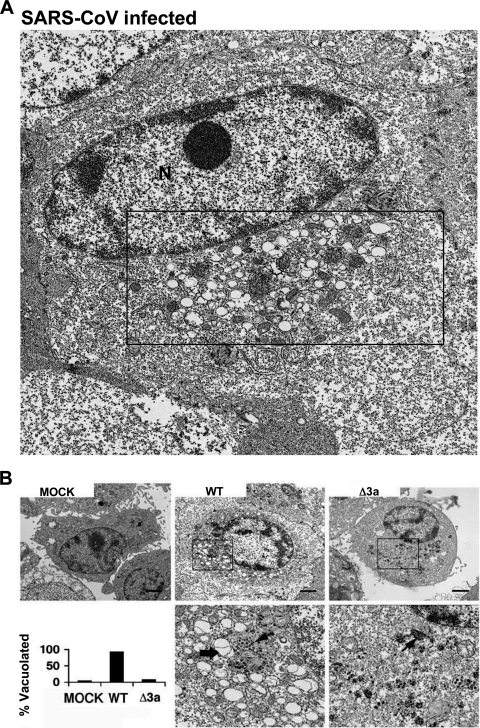
SARS-CoV induces extensive rearrangement of host cellular membranes, which may serve as sites for viral RNA replication and protect replicating virus from host immune responses (15, 53). A reticulovesicular network of modified endoplasmic reticulum (ER) containing vesicles and convoluted membranes has recently been characterized in SARS-CoV-infected cells (27). However, the mechanism of membrane rearrangement and vesicle formation remains poorly understood. By TEM analysis, we observed that Vero cells infected with WT SARS-CoV for 24 and 48 h also exhibit vesicles similar to those found in published images of human cells analyzed ex vivo (Fig. 2 and Fig. 3A) (15). These vesicles are primarily localized in the juxtanuclear region of the cell and are largely devoid of contents, which is likely due to the fixation methodology since others have reported different results using cryofixation and freeze substitution (54). Most vesicles appeared to be surrounded by a single membrane and therefore might not have originated by the cellular process of autophagy. Strikingly, vesicles were absent in cells infected with Δ3a SARS-CoV (Fig. 3B). Intracellular virus particles were visible and present in similar quantities in cells infected by both strains (Fig. 3B, lower panel). Thus, these results indicate that 3a expression is required for vesicle formation.
FIG. 2.
SARS-CoV-infected cells exhibit cytoplamsic vesicles. (A and B) Vero cells were infected for 48 h with WT SARS-CoV and were fixed and analyzed by TEM. Infected cells show numerous vesicles and vacuoles and do not show characteristics of apoptosis such as reduction in cytoplasmic volume or nuclear condensation.
FIG. 3.
Membrane rearrangement and vesicle formation by SARS-CoV is dependent on 3a. (A) Vero cells were infected with WT SARS-CoV and 24 hpi were fixed and analyzed by TEM. Boxed region contains paranuclear vesicles. N, nucleus. (B) TEM of Vero cells either mock infected (MOCK) or infected with wild-type (WT) or Δ3a SARS-CoV (Δ3a) viruses as indicated. Higher magnifications of boxed paranuclear regions of WT and Δ3a SARS-CoV-infected cells are shown in the lower panels. Membrane-bound vesicles (thick arrow) and virus particles (thin arrows) are shown (bar, 1 μm). The lower left panel shows histograms representing the fraction of cells containing vacuoles based on TEM.
SARS-CoV causes fragmentation of the Golgi apparatus by ORF 3a.
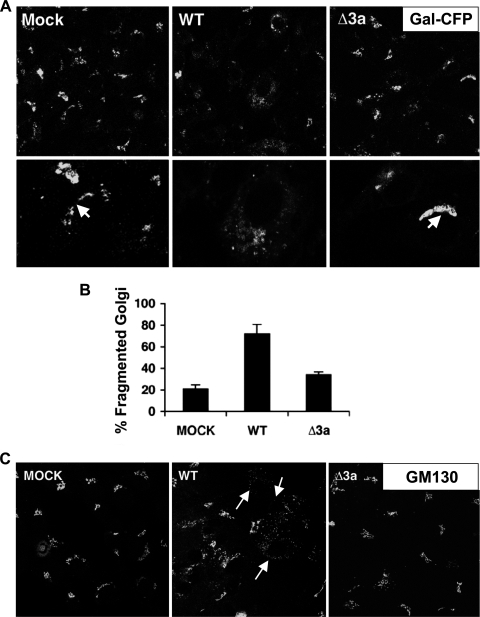
Since we observed that ORF 3a was necessary for vesicle formation and an important contributor to virus-induced cell death, we next investigated the role of vesicle formation and their origin in infected and dying cells. Possible sources of intracellular vesicles include the ER, Golgi apparatus, autophagic vesicles, and the plasma membrane via the endocytic pathway. Other coronaviruses have been shown to interact with the Golgi apparatus or ER-Golgi intermediate compartment (ERGIC) to acquire the viral envelope (26). While investigating the impact of virus infection on organelle morphology, we found that SARS-CoV infection of Vero cells caused fragmentation of the Golgi apparatus. Upon infection with WT SARS-CoV, the Golgi structural proteins galactosyltransferase (Gal) and GM130 redistributed from a paranuclear stack to dispersed vesicles (Fig. 4A and C). Since ORF 3a is important for vesicle formation, we also evaluated the role of ORF 3a in Golgi fragmentation. Infection of Vero cells with WT SARS-CoV induced Golgi fragmentation in 74% of cells compared to a background level of 21% in mock-infected cells. Golgi fragmentation was reduced to 34% when cells were infected with Δ3a SARS-CoV (Fig. 4B). These data show that the ORF 3a glycoprotein contributes to Golgi fragmentation during SARS-CoV infection.
FIG. 4.
Golgi fragmentation caused by SARS-CoV infection depends on 3a. (A) Vero cells were transfected with Golgi marker Gal-CFP and after 24 h were mock infected (MOCK) or infected with SARS-CoV (WT) or Δ3a SARS-CoV (Δ3a) at a multiplicity of infection of 5 and examined by confocal microscopy at 24 hpi. The lower panels show higher magnifications of infected cells and intact juxtanuclear Golgi stacks (white arrows). (B) Histogram showing the fraction of cells that contain fragmented Golgi (percent fragmented Golgi) ± the standard deviation based on confocal microscopy analysis as described in Materials and Methods (n > 100 cells) in three replicates. (C) Cells were infected as in panel A. After 24 h, cells were fixed and stained for GM130 and examined by confocal microscopy. White arrows indicate examples of cells that display what was designated “fragmented Golgi.”
Overexpression of ORF 3a is sufficient to cause cell death, vesicle formation, and Golgi fragmentation.
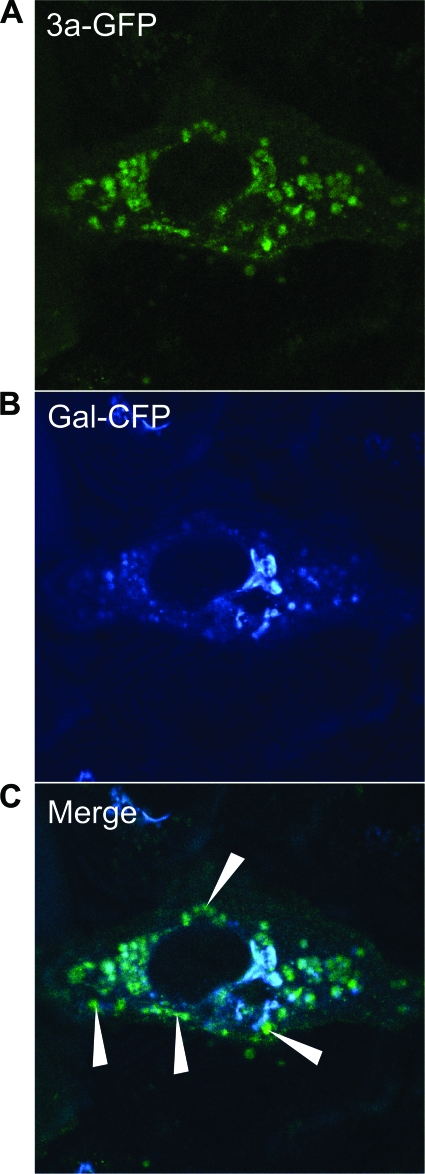
Having documented the importance of 3a in the processes of cell death, vesicle formation, and Golgi fragmentation using the 3a-deficient virus, we next wanted to determine whether the protein was capable of inducing similar effects when overexpressed or might require additional SARS-CoV proteins. Consistent with published reports, we also observed that overexpression of ORF 3a as a fusion protein with the green fluorescent protein (3a-GFP) caused pronounced cell death compared to GFP control (Fig. 5A and B) (31, 39). TEM analysis of transfected cells showed the presence of intracellular vesicles similar to those seen during infection with SARS-CoV (Fig. 5C). Overexpression of ORF 3a also caused Golgi fragmentation, also indicated by the redistribution of Gal and GM130 from a juxtanuclear stack, as seen in control cells, to dispersed vesicles (Fig. 5D and Fig. 6). Golgi disassembly occurs transiently during mitosis and has the effect of enhancing replication in some cases of viral infection (1, 3, 51). Intriguingly, Golgi fragmentation has also been observed during infection of a related coronavirus, MHV, and is associated with syncytium formation (29, 30). During mitotic Golgi fragmentation, the membrane components of the Golgi apparatus sequentially redistributes into isolated fragments that then disassemble further when the membrane components redistribute into the ER or to ER exit sites (2). Rather than inducing distribution to the ER, overexpression of 3a-GFP causes Gal to localize to isolated fragments, and 3a-GFP and Gal-CFP appear to partially colocalize (Fig. 5Df, inset, and Fig. 6). These results demonstrate that SARS-CoV 3a is necessary and sufficient to cause Golgi fragmentation.
FIG. 5.
Expression of SARS-CoV 3a is sufficient to cause cell death, vesicle formation, and Golgi fragmentation. (A) Vero cells transfected with 3a-GFP were examined by confocal microscopy 48 hpt after propidium iodide (PI) staining to identify dead cells. (B) GFP or 3a-GFP transfected cells were harvested at 48 hpt, and cell death was calculated. (C) Vero cells were transfected with 3a-GFP and harvested for TEM analysis 24 h after transfection. Panel b is an enlargement of the boxed region in panel a and depicts juxtanuclear vesicles similar to those seen after infection with SARS-CoV. (Scale bars: a, 2 μm; b, 0.5 μm) (D) Fluorescence photomicrographs of cells cotransfected with GFP alone and the Golgi marker Gal-CFP fusion protein (a to c), pseudocolored in red or the 3a-GFP fusion protein and Gal-CFP (d to f), or 3a-GFP and stai ned for the Golgi protein GM130 (g to i) and examined by confocal microscopy at 24 hpt. The boxed regions in panel f depict areas of colocalized and noncolocalized protein.
FIG. 6.
SARS-CoV 3a-GFP partially colocalizes with the Golgi marker Gal-CFP. (A) Vero cells were cotransfected with 3a-GFP and Gal-CFP and analyzed at 24 hpt by confocal microscopy. 3a-GFP (A) and Gal-CFP (B) can be seen to partially colocalize in the merged micrograph (C). White arrows indicate examples of 3a-GFP punctae that do not colocalize with Gal-CFP.
Vesicles containing 3a-GFP also exhibit markers for late endosomes.
When overexpressed, 3a-GFP can be observed to localize to the plasma membrane, as well as to numerous cytoplasmic vesicles. We therefore sought to characterize these vesicles further and to determine their origin. We first observed the behavior of 3a-GFP in live cells by confocal microscopy. Vesicles containing 3a-GFP are dynamic (see Video S1 in the supplemental material). Although a few vesicles remained stationary during the observation period, most vesicles exhibited saltatory movement within the cytoplasm. It was also possible to observe vesicles moving to and from the plasma membrane, thus suggesting a possible interaction with the exocytic or endocytic pathways.
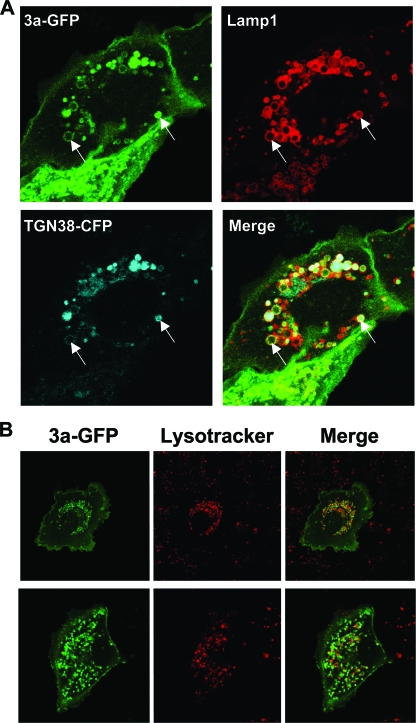
As mentioned above, we observed that the intracellular vesicles containing 3a-GFP only partially colocalized with the Golgi markers Gal-CFP (Fig. 5D, inset, and Fig. 6) and therefore hypothesized that the 3a-protein might localize to the TGN or endocytic vesicles. In order to test this hypothesis, 3a-GFP or GFP only were co-overexpressed with the TGN marker, TGN38-CFP. TGN38 is a type I membrane protein involved in vesicle formation from the trans-Golgi apparatus and cycles between the TGN and the plasma membrane (4, 22, 43). In addition, we sought to determine whether the vesicles also contained Lamp1, a major transmembrane glycoprotein found in late endosomes and lysosomes (8). Vesicles containing 3a-GFP exhibited extensive colocalization with both TGN38 with Lamp1 (Fig. 7A). To test whether the internal environment of these vesicles was also acidic, cells were transfected with 3a-GFP and stained at 24 h posttransfection (hpt) with LysoTracker Red DND-99 (Invitrogen/Molecular Probes), which is a fluorophore in the form of a weakly basic amine that selectively accumulates in acidic compartments and exhibits red fluorescence when used appropriately (13). We observed that many of the 3a-GFP-positive vesicles costained with LysoTracker Red (Fig. 7B). Together, these data indicate that 3a-GFP vesicles exhibit markers of late endosomes and lysosomes.
FIG. 7.
3a vesicles in live cells contain endosomal markers and are acidic. (A) Vero cells were cotransfected with 3a-GFP, TGN38-CFP, and Cherry-Lamp1, and live cells were analyzed by confocal microscopy at 24 hpt. Arrows indicate examples of vesicles that contain 3a, TGN38, and Lamp1. (B) Vero cells were transfected with 3a-GFP and analyzed by confocal microscopy 24 hpi after counterstaining with LysoTracker Red DND 99 according to the manufacturer's instructions. Two examples of transfected cells are shown. Areas of colocalization appear orange in merge image.
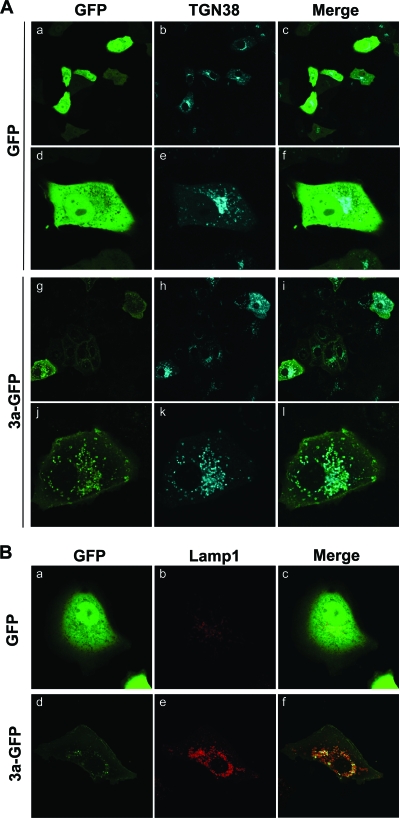
Overexpression of 3a-GFP causes redistribution of Lamp1 and TGN38.
TGN38 is transported on vesicles that bud from the trans-Golgi apparatus to the plasma membrane and may be involved in vesicle formation (4). From the plasma membrane, TGN38 cycles from endocytic vesicles back to the TGN. However, mislocalization of TGN38 to late endosomes or lysosomes has been reported (49). In addition to colocalizing with TGN38 and Lamp1, we also observed that overexpression of 3a-GFP induces relocalization of TGN38. Compared to cells expressing GFP only, cells expressing 3a-GFP exhibit an increase in the number of TGN vesicles and show dispersal of TGN38 from paranuclear Golgi structures (Fig. 8A). We also observed a similar effect on the localization of Lamp1, with a noticeable increase in vesicles containing Lamp1 (Fig. 8B). These results suggest that 3a-GFP induces an accumulation of vesicles containing TGN38 and Lamp1, which could occur as a result of increased endocytosis or by inhibiting the transit of TGN38 from late endosomes to the TGN and therefore causing mislocalization of the protein to acidic compartments.
FIG. 8.
Expression of 3a causes redistribution and accumulation of Lamp1 and TGN38 in 3a-GFP vesicles. (A) Vero cells were cotransfected with GFP (a to f) or 3a-GFP (g to l) and TGN38-CFP and analyzed by confocal microscopy at 24 hpt. Examples of cells imaged at lower (a to c, g to i) and higher (d to f, j to l) magnifications are depicted. (B) Vero cells were cotransfected with GFP (a to c) or 3a-GFP (d to f) and Cherry-Lamp1 and analyzed in live cells by confocal microscopy at 24 hpt.
Golgi fragmentation is reduced by overexpression of Arf1.
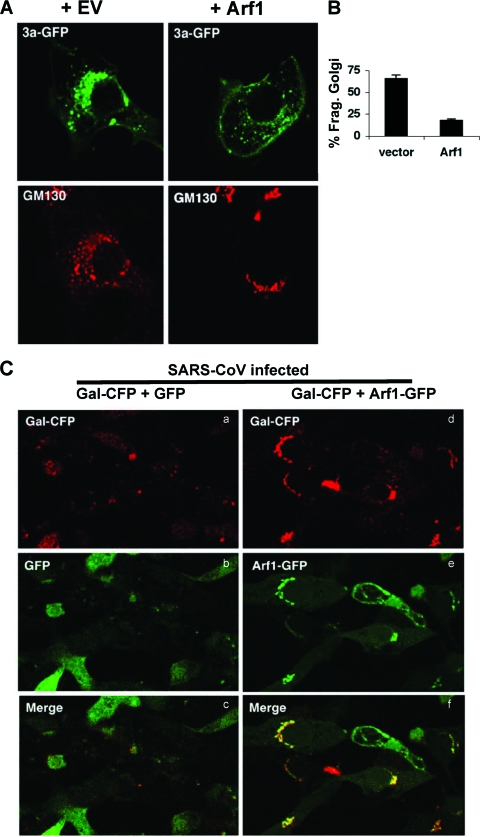
Recently, viruses that cause proliferation of intracellular vesicles but are unrelated to SARS-CoV have been shown to modulate Arf1 activity during infection. Arf1 is a Golgi body-associated small Ras-related GTPase that maintains Golgi structure and function (10). The activity of Arf1 is critical for assembling coat proteins during vesicle formation by recruiting COPI to Golgi membranes and regulating clathrin adaptor proteins at the TGN (11, 56). In addition, Golgi fragmentation can be induced by treating cells with brefeldin A, a fungal metabolite that inhibits Arf1 binding to GTP and thus inactivates the protein (25, 42, 48). We therefore hypothesized that Arf1 might be involved in ORF 3a-induced Golgi fragmentation. To test this hypothesis, we cotransfected expression constructs for 3a and either empty vector or Arf1. Overexpression of Arf1 potently inhibited 3a-induced Golgi fragmentation (Fig. 9A and B). Next, using WT SARS-CoV, we infected cells that had been cotransfected with either Arf1-GFP or control GFP and the Golgi marker Gal-CFP. In cells that showed overexpression of Arf1, Gal-CFP was found to localize to intact paranuclear stacks (Fig. 9C). Therefore, Golgi fragmentation induced by SARS-CoV likely results from inhibition of the Arf1 pathway.
FIG. 9.
Overexpression of Arf1 reduces Golgi fragmentation. (A) Cells were cotransfected with 3a-GFP and empty vector (EV) (left panel) or Arf1-CFP (right panel, +Arf1). After 24 h, cells were stained with an antibody specific for the Golgi marker GM130 (shown in red) and examined by confocal microscopy. (B) The percent fragmented Golgi of cells in panel A was determined by morphological analysis. (C) Cells were cotransfected with the Golgi marker Gal-CFP and GFP (left) or Arf1-GFP (right). Transfected cells were infected with the WT SARS-CoV and examined by confocal microscopy at 24 hpi.
DISCUSSION
SARS-CoV infection results in severe disease with high morbidity and mortality. This is in contrast to other human coronaviruses, which usually cause a mild upper respiratory tract infection. As is the case with other coronaviruses, the novel SARS-CoV accessory proteins that are unique to SARS-CoV likely contribute to the highly pathogenic nature of the virus. Thus, further characterization of these proteins is likely to elucidate mechanisms by which SARS-CoV disrupts cellular functions and causes disease.
In detailed studies of the 3a protein, we discovered that it causes intracellular vesicle formation, which is a prominent feature of cells from SARS patients (15, 53) and is both necessary and sufficient for Golgi fragmentation during virus infection. Vesicles containing ORF 3a protein are acidified compartments that also contain the trans-Golgi protein TGN38 and the marker of late endosomes and lysosomes, Lamp1. These data are consistent with previous findings that ORF 3a interacts with caveolin and thus the endocytic pathway (40). It will be important to discover whether the 3a-mediated processes of Golgi fragmentation and vesicle formation are directly related or whether they are coincidental disruptions that occur during infection. Finally, Golgi fragmentation was reduced by overexpression of the Golgi regulator protein, Arf1, suggesting that the functioning of this cellular protein may be disrupted by ORF 3a.
Positive-strand RNA viruses utilize intracellular membranes on which to replicate their RNA (28). It is thought that the membrane acts as a nucleation site for replicase proteins and polymerases. Many RNA viruses, including the human enterovirus poliovirus and the group II coronavirus, MHV, cause a proliferation in cellular membranes or the formation of intracellular vesicles, which is believed to be advantageous for virus replication (21, 57, 64). Interestingly, poliovirus-induced membrane rearrangement occurs through activation of Arf1 (5, 6). Electron microscopy analysis of MHV-infected cells shows the presence of double-membrane vesicles (DMVs), which are thought to be produced by autophagy. In one study, inhibition of cellular autophagy reduced MHV replication (44). We found that the formation of intracellular vesicles during SARS-CoV infection does not affect replication, since depletion of ORF 3a from SARS-CoV abrogated vesicle formation but did not reduce the levels of viral RNA isolated from the cells and from the media. Although SARS-CoV may use vesicular membranes as sites of RNA replication (54), intracellular vesicles may not be necessary, and perhaps not even beneficial, to the virus.
These observations raise the question, if not for replication, why would SARS-CoV cause such dramatic membrane rearrangement? Several possible explanations exist. Evidence suggests that poliovirus, which also induces intracellular vesicles and DMVs, may utilize the vesicles for nonlytic release of virus particles (24). One study reported a deficiency in SARS-CoV release when 3a expression was reduced by RNA interference (RNAi) (34). However, targeted disruption of group-specific ORFs by RNAi may also reduce genomic RNA and confound this interpretation. Our data suggest that release of genomic RNA is not affected by depletion of ORF 3a, since RNA levels observed in the media were equivalent for Δ3a and WT virus. Alternatively, rearrangement of intracellular membranes can coincide with disruption of the secretory pathway, which could facilitate immune evasion by inhibiting antiviral cytokine secretion or antigen presentation by major histocompatibility complex on the cell surface. Disruption of the secretory pathway by ORF 3a may require additional viral proteins, since expression of SARS-CoV ORF 3a protein by replication-deficient particles of Venezuelan equine encephalitis virus does not inhibit secretion of beta interferon (14). It will also be important to determine the membrane location on which the virus replicates in the absence of 3a. Mitochondrial membranes represent a potential site of coronavirus replication. Several SARS-CoV proteins, including 3b and 9b, localize to mitochondria (our unpublished observations) and have been demonstrated to bind replicase proteins nsp8 and nsp14 (62).
Except for the recent study on the 7a protein (52), studies on death induction by SARS proteins have been limited to transfection/overexpression studies of the individual proteins, most likely due to the difficulty of working with this lethal pathogen. Consistent with previous reports, we have observed that overexpression of 3a is sufficient to cause cell death (31, 39). Importantly, our experiments with SARS-CoV Δ3a clearly demonstrate that this protein contributes to the cytotoxicity of the virus and illustrate its biological significance. Residual cytotoxicity of the SARS-CoV Δ3a virus may be attributable to the 7a protein, which is intact in this mutant strain and also contributes to virus-induced cell death (52). Future studies that incorporate additional deletion mutants will be necessary to discern the mechanism and importance of SARS-CoV-induced cell death. An animal model that recapitulates the characteristics of human SARS does not currently exist, and it therefore remains difficult to determine the relative importance of ORF 3a to SARS-CoV pathogenesis. However, recent adaptation of SARS-CoV in mice has led to a lethal model that can be used to investigate the contribution of group-specific ORFs to SARS disease (47) and thus might provide further insight into the functioning of these novel proteins.
Supplementary Material
Acknowledgments
We thank Elaine Lamirande, Leatrice Vogel, and Anjeanette Roberts for help with SARS-CoV infection experiments; Kunio Nagashima, SAIC Frederick, National Cancer Institute at Frederick, for electron micrograph assistance; Laurie Mueller for expertise with electron microscopy processing; and Richard Siegel and Karla Kirkegaard for critical readings of the manuscript.
E.C.F. is a National Institutes of Health—University of Oxford Biomedical Research Scholar.
This research was supported by the Intramural Research Program of the National Institutes of Allergy and Infectious Diseases, National Institutes of Health, and NIH AID grant AI059136 to R.S.B.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the funding agency.
Footnotes
Published ahead of print on 4 November 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Altan-Bonnet, N., R. D. Phair, R. S. Polishchuk, R. Weigert, and J. Lippincott-Schwartz. 2003. A role for Arf1 in mitotic Golgi disassembly, chromosome segregation, and cytokinesis. Proc. Natl. Acad. Sci. USA 100:13314-13319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altan-Bonnet, N., R. Sougrat, W. Liu, E. L. Snapp, T. Ward, and J. Lippincott-Schwartz. 2006. Golgi inheritance in mammalian cells is mediated through endoplasmic reticulum export activities. Mol. Biol. Cell 17:990-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avitabile, E., P. L. Ward, C. Di Lazzaro, M. R. Torrisi, B. Roizman, and G. Campadelli-Fiume. 1994. The herpes simplex virus UL20 protein compensates for the differential disruption of exocytosis of virions and viral membrane glycoproteins associated with fragmentation of the Golgi apparatus. J. Virol. 68:7397-7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banting, G., and S. Ponnambalam. 1997. TGN38 and its orthologues: roles in post-TGN vesicle formation and maintenance of TGN morphology. Biochim. Biophys. Acta 1355:209-217. [DOI] [PubMed] [Google Scholar]
- 5.Belov, G. A., N. Altan-Bonnet, G. Kovtunovych, C. L. Jackson, J. Lippincott-Schwartz, and E. Ehrenfeld. 2007. Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. J. Virol. 81:558-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belov, G. A., C. Habbersett, D. Franco, and E. Ehrenfeld. 2007. Activation of cellular Arf GTPases by poliovirus protein 3CD correlates with virus replication. J. Virol. 81:9259-9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, W. S., C. Wu, S. C. Chow, T. Cheung, K. F. To, W. K. Leung, P. K. Chan, K. C. Lee, H. K. Ng, D. M. Au, and A. W. Lo. 2005. Coronaviral hypothetical and structural proteins were found in the intestinal surface enterocytes and pneumocytes of severe acute respiratory syndrome (SARS). Mod Pathol. 18:1432-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, J. W., T. L. Murphy, M. C. Willingham, I. Pastan, and J. T. August. 1985. Identification of two lysosomal membrane glycoproteins. J. Cell Biol. 101:85-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow, S. C., C. Y. Ho, T. T. Tam, C. Wu, T. Cheung, P. K. Chan, M. H. Ng, P. K. Hui, H. K. Ng, D. M. Au, and A. W. Lo. 2006. Specific epitopes of the structural and hypothetical proteins elicit variable humoral responses in SARS patients. J. Clin. Pathol. 59:468-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donaldson, J. G., and A. Honda. 2005. Localization and function of Arf family GTPases. Biochem. Soc. Trans. 33:639-642. [DOI] [PubMed] [Google Scholar]
- 11.Donaldson, J. G., and R. D. Klausner. 1994. ARF: a key regulatory switch in membrane traffic and organelle structure. Curr. Opin. Cell Biol. 6:527-532. [DOI] [PubMed] [Google Scholar]
- 12.Fielding, B. C., Y. J. Tan, S. Shuo, T. H. Tan, E. E. Ooi, S. G. Lim, W. Hong, and P. Y. Goh. 2004. Characterization of a unique group-specific protein (U122) of the severe acute respiratory syndrome coronavirus. J. Virol. 78:7311-7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freundt, E. C., M. Czapiga, and M. J. Lenardo. 2007. Photoconversion of LysoTracker Red to a green fluorescent molecule. Cell Res. 17:956-958. [DOI] [PubMed] [Google Scholar]
- 14.Frieman, M., K. Ratia, R. E. Johnston, A. D. Mesecar, and R. S. Baric. 2009. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-κB signaling. J. Virol. 83:6689-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldsmith, C. S., K. M. Tatti, T. G. Ksiazek, P. E. Rollin, J. A. Comer, W. W. Lee, P. A. Rota, B. Bankamp, W. J. Bellini, and S. R. Zaki. 2004. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 10:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldsmith, C. S., T. Whistler, P. E. Rollin, T. G. Ksiazek, P. A. Rota, W. J. Bellini, P. Daszak, K. T. Wong, W. J. Shieh, and S. R. Zaki. 2003. Elucidation of Nipah virus morphogenesis and replication using ultrastructural and molecular approaches. Virus Res. 92:89-98. [DOI] [PubMed] [Google Scholar]
- 17.Guan, M., K. H. Chan, J. S. Peiris, S. W. Kwan, S. Y. Lam, C. M. Pang, K. W. Chu, K. M. Chan, H. Y. Chen, E. B. Phuah, and C. J. Wong. 2004. Evaluation and validation of an enzyme-linked immunosorbent assay and an immunochromatographic test for serological diagnosis of severe acute respiratory syndrome. Clin. Diagn. Lab. Immunol. 11:699-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guan, M., H. Y. Chen, S. Y. Foo, Y. J. Tan, P. Y. Goh, and S. H. Wee. 2004. Recombinant protein-based enzyme-linked immunosorbent assay and immunochromatographic tests for detection of immunoglobulin G antibodies to severe acute respiratory syndrome (SARS) coronavirus in SARS patients. Clin. Diagn. Lab. Immunol. 11:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo, J. P., M. Petric, W. Campbell, and P. L. McGeer. 2004. SARS corona virus peptides recognized by antibodies in the sera of convalescent cases. Virology 324:251-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haijema, B. J., H. Volders, and P. J. Rottier. 2004. Live, attenuated coronavirus vaccines through the directed deletion of group-specific genes provide protection against feline infectious peritonitis. J. Virol. 78:3863-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson, W. T., T. H. Giddings, Jr., M. P. Taylor, S. Mulinyawe, M. Rabinovitch, R. R. Kopito, and K. Kirkegaard. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3:e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, S. M., J. R. Crosby, J. Salamero, and K. E. Howell. 1993. A cytosolic complex of p62 and rab6 associates with TGN38/41 and is involved in budding of exocytic vesicles from the trans-Golgi network. J. Cell Biol. 122:775-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan, S., B. C. Fielding, T. H. Tan, C. F. Chou, S. Shen, S. G. Lim, W. Hong, and Y. J. Tan. 2006. Over-expression of severe acute respiratory syndrome coronavirus 3b protein induces both apoptosis and necrosis in Vero E6 cells. Virus Res. 122:20-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirkegaard, K., and W. T. Jackson. 2005. Topology of double-membraned vesicles and the opportunity for non-lytic release of cytoplasm. Autophagy 1:182-184. [DOI] [PubMed] [Google Scholar]
- 25.Klausner, R. D., J. G. Donaldson, and J. Lippincott-Schwartz. 1992. Brefeldin A: insights into the control of membrane traffic and organelle structure. J. Cell Biol. 116:1071-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klumperman, J., J. K. Locker, A. Meijer, M. C. Horzinek, H. J. Geuze, and P. J. Rottier. 1994. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 68:6523-6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knoops, K., M. Kikkert, S. H. E. van den Worm, J. C. Zevenhoven-Dobbe, Y. van der Meer, A. J. Koster, A. M. Mommaas, and E. J. Snijder. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6:1957-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krogerus, C., D. Egger, O. Samuilova, T. Hyypia, and K. Bienz. 2003. Replication complex of human parechovirus 1. J. Virol. 77:8512-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lavi, E., Q. Wang, A. Stieber, Y. Chen, S. Weiss, and N. K. Gonatas. 1995. Fragmentation and rearrangement of the Golgi apparatus during MHV infection of L-2 cells. Adv. Exp. Med. Biol. 380:103-104. [DOI] [PubMed] [Google Scholar]
- 30.Lavi, E., Q. Wang, S. R. Weiss, and N. K. Gonatas. 1996. Syncytium formation induced by coronavirus infection is associated with fragmentation and rearrangement of the Golgi apparatus. Virology 221:325-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Law, P. T., C. H. Wong, T. C. Au, C. P. Chuck, S. K. Kong, P. K. Chan, K. F. To, A. W. Lo, J. Y. Chan, Y. K. Suen, H. Y. Chan, K. P. Fung, M. M. Waye, J. J. Sung, Y. M. Lo, and S. K. Tsui. 2005. The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J. Gen. Virol. 86:1921-1930. [DOI] [PubMed] [Google Scholar]
- 32.Lin, C. W., K. H. Lin, T. H. Hsieh, S. Y. Shiu, and J. Y. Li. 2006. Severe acute respiratory syndrome coronavirus 3C-like protease-induced apoptosis. FEMS Immunol. Med. Microbiol. 46:375-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, W., R. Duden, R. D. Phair, and J. Lippincott-Schwartz. 2005. ArfGAP1 dynamics and its role in COPI coat assembly on Golgi membranes of living cells. J. Cell Biol. 168:1053-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu, W., B. J. Zheng, K. Xu, W. Schwarz, L. Du, C. K. Wong, J. Chen, S. Duan, V. Deubel, and B. Sun. 2006. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc. Natl. Acad. Sci. USA 103:12540-12545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 36.Mizutani, T., S. Fukushi, M. Saijo, I. Kurane, and S. Morikawa. 2004. Importance of Akt signaling pathway for apoptosis in SARS-CoV-infected Vero E6 cells. Virology 327:169-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson, C. A., A. Pekosz, C. A. Lee, M. S. Diamond, and D. H. Fremont. 2005. Structure and intracellular targeting of the SARS-coronavirus Orf7a accessory protein. Structure 13:75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ortego, J., I. Sola, F. Almazan, J. E. Ceriani, C. Riquelme, M. Balasch, J. Plana, and L. Enjuanes. 2003. Transmissible gastroenteritis coronavirus gene 7 is not essential but influences in vivo virus replication and virulence. Virology 308:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Padhan, K., R. Minakshi, M. A. Bin Towheed, and S. Jameel. 2008. Severe acute respiratory syndrome coronavirus 3a protein activates the mitochondrial death pathway through p38 MAP kinase activation. J. Gen. Virol. 89:1960-1969. [DOI] [PubMed] [Google Scholar]
- 40.Padhan, K., C. Tamar, A. Hussain, P. Y. Hui, M. Y. Lee, C. Y. Cheung, J. S. M. Peiris, and S. Jameel. 2007. Severe acute respiratory syndrome coronavirus Orf3a protein interacts with caveolin. J. Gen. Virol. 88:3067-3077. [DOI] [PubMed] [Google Scholar]
- 41.Pewe, L., H. Zhou, J. Netland, C. Tangudu, H. Olivares, L. Shi, D. Look, T. Gallagher, and S. Perlman. 2005. A severe acute respiratory syndrome-associated coronavirus-specific protein enhances virulence of an attenuated murine coronavirus. J. Virol. 79:11335-11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peyroche, A., B. Antonny, S. Robineau, J. Acker, J. Cherfils, and C. L. Jackson. 1999. Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol. Cell 3:275-285. [DOI] [PubMed] [Google Scholar]
- 43.Ponnambalam, S., M. Girotti, M. L. Yaspo, C. E. Owen, A. C. Perry, T. Suganuma, T. Nilsson, M. Fried, G. Banting, and G. Warren. 1996. Primate homologues of rat TGN38: primary structure, expression and functional implications. J. Cell Sci. 109(Pt. 3):675-685. [DOI] [PubMed] [Google Scholar]
- 44.Prentice, E., W. G. Jerome, T. Yoshimori, N. Mizushima, and M. R. Denison. 2004. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 279:10136-10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qiu, M., Y. Shi, Z. Guo, Z. Chen, R. He, R. Chen, D. Zhou, E. Dai, X. Wang, B. Si, Y. Song, J. Li, L. Yang, J. Wang, H. Wang, X. Pang, J. Zhai, Z. Du, Y. Liu, Y. Zhang, L. Li, J. Wang, B. Sun, and R. Yang. 2005. Antibody responses to individual proteins of SARS coronavirus and their neutralization activities. Microbes Infect. 7:882-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren, L., R. Yang, L. Guo, J. Qu, J. Wang, and T. Hung. 2005. Apoptosis induced by the SARS-associated coronavirus in Vero cells is replication-dependent and involves caspase. DNA Cell Biol. 24:496-502. [DOI] [PubMed] [Google Scholar]
- 47.Roberts, A., D. Deming, C. D. Paddock, A. Cheng, B. Yount, L. Vogel, B. D. Herman, T. Sheahan, M. Heise, G. L. Genrich, S. R. Zaki, R. Baric, and K. Subbarao. 2007. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 3:23-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robineau, S., M. Chabre, and B. Antonny. 2000. Binding site of brefeldin A at the interface between the small G protein ADP-ribosylation factor 1 (ARF1) and the nucleotide-exchange factor Sec7 domain. Proc. Natl. Acad. Sci. USA 97:9913-9918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roquemore, E. P., and G. Banting. 1998. Efficient trafficking of TGN38 from the endosome to the trans-Golgi network requires a free hydroxyl group at position 331 in the cytosolic domain. Mol. Biol. Cell 9:2125-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 51.Sandoval, I. V., and L. Carrasco. 1997. Poliovirus infection and expression of the poliovirus protein 2B provoke the disassembly of the Golgi complex, the organelle target for the antipoliovirus drug Ro-090179. J. Virol. 71:4679-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schaecher, S. R., E. Touchette, J. Schriewer, R. M. Buller, and A. Pekosz. 2007. Severe acute respiratory syndrome coronavirus gene 7 products contribute to virus-induced apoptosis. J. Virol. 81:11054-11068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shieh, W. J., C. H. Hsiao, C. D. Paddock, J. Guarner, C. S. Goldsmith, K. Tatti, M. Packard, L. Mueller, M. Z. Wu, P. Rollin, I. J. Su, and S. R. Zaki. 2005. Immunohistochemical, in situ hybridization, and ultrastructural localization of SARS-associated coronavirus in lung of a fatal case of severe acute respiratory syndrome in Taiwan. Hum. Pathol. 36:303-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Snijder, E. J., Y. van der Meer, J. Zevenhoven-Dobbe, J. J. Onderwater, J. van der Meulen, H. K. Koerten, and A. M. Mommaas. 2006. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 80:5927-5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sperry, S. M., L. Kazi, R. L. Graham, R. S. Baric, S. R. Weiss, and M. R. Denison. 2005. Single-amino-acid substitutions in open reading frame (ORF) 1b-nsp14 and ORF 2a proteins of the coronavirus mouse hepatitis virus are attenuating in mice. J. Virol. 79:3391-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stearns, T., M. C. Willingham, D. Botstein, and R. A. Kahn. 1990. ADP-ribosylation factor is functionally and physically associated with the Golgi complex. Proc. Natl. Acad. Sci. USA 87:1238-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tan, Y. J., B. C. Fielding, P. Y. Goh, S. Shen, T. H. Tan, S. G. Lim, and W. Hong. 2004. Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J. Virol. 78:14043-14047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tan, Y. J., P. Y. Goh, B. C. Fielding, S. Shen, C. F. Chou, J. L. Fu, H. N. Leong, Y. S. Leo, E. E. Ooi, A. E. Ling, S. G. Lim, and W. Hong. 2004. Profiles of antibody responses against severe acute respiratory syndrome coronavirus recombinant proteins and their potential use as diagnostic markers. Clin. Diagn. Lab. Immunol. 11:362-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tan, Y. J., E. Teng, S. Shen, T. H. Tan, P. Y. Goh, B. C. Fielding, E. E. Ooi, H. C. Tan, S. G. Lim, and W. Hong. 2004. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 78:6723-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan, Y. X., T. H. Tan, M. J. Lee, P. Y. Tham, V. Gunalan, J. Druce, C. Birch, M. Catton, N. Y. Fu, V. C. Yu, and Y. J. Tan. 2007. Induction of apoptosis by the severe acute respiratory syndrome coronavirus 7a protein is dependent on its interaction with the Bcl-XL protein. J. Virol. 81:6346-6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.von Brunn, A., C. Teepe, J. C. Simpson, R. Pepperkok, C. C. Friedel, R. Zimmer, R. Roberts, R. Baric, and J. Haas. 2007. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS ONE 2:e459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weiss, S. R., and S. Navas-Martin. 2005. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 69:635-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wileman, T. 2006. Aggresomes and autophagy generate sites for virus replication. Science 312:875-878. [DOI] [PubMed] [Google Scholar]
- 65.Yan, H., G. Xiao, J. Zhang, Y. Hu, F. Yuan, D. K. Cole, C. Zheng, and G. F. Gao. 2004. SARS coronavirus induces apoptosis in Vero E6 cells. J. Med. Virol. 73:323-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yount, B., R. S. Roberts, A. C. Sims, D. Deming, M. B. Frieman, J. Sparks, M. R. Denison, N. Davis, and R. S. Baric. 2005. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 79:14909-14922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu, C. J., Y. C. Chen, C. H. Hsiao, T. C. Kuo, S. C. Chang, C. Y. Lu, W. C. Wei, C. H. Lee, L. M. Huang, M. F. Chang, H. N. Ho, and F. J. Lee. 2004. Identification of a novel protein 3a from severe acute respiratory syndrome coronavirus. FEBS Lett. 565:111-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yuan, X., J. Li, Y. Shan, Z. Yang, Z. Zhao, B. Chen, Z. Yao, B. Dong, S. Wang, J. Chen, and Y. Cong. 2005. Subcellular localization and membrane association of SARS-CoV 3a protein. Virus Res. 109:191-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yuan, X., Y. Shan, Z. Zhao, J. Chen, and Y. Cong. 2005. G0/G1 arrest and apoptosis induced by SARS-CoV 3b protein in transfected cells. Virol. J. 2:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.