Abstract
Fragments of double-stranded DNA (dsDNA) forming a right-handed helical structure (B-DNA) stimulate cells to produce type I interferons (IFNs). While an adaptor molecule, IFN-β promoter stimulator 1 (IPS-1), mediates dsDNA-induced cellular signaling in human cells, the underlying molecular mechanism is not fully understood. Here, we demonstrate that the extrachromosomal histone H2B mediates innate antiviral immune responses in human cells. H2B physically interacts with IPS-1 through the association with a newly identified adaptor, CIAO (COOH-terminal importin 9-related adaptor organizing histone H2B and IPS-1), to transmit the cellular signaling for dsDNA but not immunostimulatory RNA. Extrachromosomal histone H2B was biologically crucial for cell-autonomous responses to protect against multiplication of DNA viruses but not an RNA virus. Thus, the present findings provide evidence indicating that the extrachromosomal histone H2B is engaged in the signaling pathway initiated by dsDNA to trigger antiviral innate immune responses.
Fragments of nucleic acids derived from either infectious agents or host cells activate cell-autonomous responses to inhibit multiplication of certain viruses by inducing type I interferon (IFN) production (5). Such effects are more evident when double-stranded DNA (dsDNA) is transduced into the intracellular compartment by use of a transfection agent or electroporation method, suggesting that the DNA sensing system recognizes aberrant DNA fragments inside the cell (6, 21, 23). dsDNA forming a right-handed helical structure, i.e., B-DNA, has a greater ability to induce type I IFNs than Z-DNA, which has a left-handed zig-zag structure (6). dsDNA activates type I IFN production in a wide variety of cell types, including immune cells, such as dendritic cells and macrophages, and nonimmune cells, such as fibroblasts, epithelial cells, and thyroid cells (6, 23). Such effects of dsDNA were corroborated by the observation in mice deficient for DNase II, in which intracellular accumulation of undegraded DNA fragments resulted in hyperproduction of IFN-β, dysregulation of erythropoiesis, and symptoms resembling rheumatoid arthritis (12, 28). The loss-of-function mutation of the DNase I gene has been found in patients with systemic lupus erythematosus (SLE) and, in fact, DNase I−/− mice manifest SLE-like symptoms with anti-DNA antibody (Ab) production (18, 27).
The immunostimulatory property of dsDNA is quite similar to that of immunostimulatory RNA (isRNA), such as dsRNA and 5′-triphosphate RNA (2, 6). Indeed, the signaling pathways engaged by dsDNA in part are shared with those for isRNA. Retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5) directly associate with isRNA and trigger signaling, while it has been demonstrated that RIG-I does not directly interact with dsDNA but mediates its signaling in a human hepatoma cell line, Huh7 (2). IFN-β promoter stimulator 1 (IPS-1, also known as mitochondrial antiviral signaling), mediates the downstream signaling induced by dsDNA or isRNA in humans, while IPS-1 solely mediates isRNA but not dsDNA signaling in mice (2, 6, 15, 22). In contrast, TANK-binding kinase 1 (TBK1) and inducible IκB kinase (IKKi) are essential for dsDNA- or isRNA-induced type I IFN production in both humans and mice (2, 6). While examining distinct molecules involved in dsDNA-mediated but not isRNA-mediated upstream signaling, Z-DNA binding protein 1 (ZBP1, also known as DNA-dependent activator of IFN regulatory factors [DAI]) was identified as a candidate cytosolic DNA sensor, at least in a mouse connective tissue cell line, L929, although its in vivo role was dispensable (7, 24, 26). Recently, a PYHIN family member, Absent in melanoma 2 (AIM2) protein, was shown to associate with an inflammasome signaling adaptor, apoptosis-associated speck-like protein containing a CARD (ASC), and to play a critical role for caspase 1 activation and interleukin-1β (IL-1β) secretion in response to dsDNA (1, 3, 4, 20).
In the present study, we show that extrachromosomal histone H2B is responsible for the dsDNA-induced type I IFN production in human cells and for the innate immune response to DNA virus infection.
MATERIALS AND METHODS
Cells and reagents.
HEK293T, HEK293, HeLa, NIH 3T3, Vero, and L929 cells were purchased from the American Type Culture Collection (Manassas, VA) and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and 50 μg/ml penicillin-streptomycin. SCC-4 squamous carcinoma cells (JCRB9118) were obtained from Human Science Research Resources Bank (Osaka, Japan). Human umbilical vein endothelial cells (HUVEC) were obtained from Cambrex BioScience (Waikersville, MD). IPS-1−/− murine embryonic fibroblasts (MEF) were previously described (15). Encephalomyocarditis virus (EMCV) and mouse cytomegalovirus (MCMV) strain MW97.01 were kindly provided by T. Fujita (Kyoto University, Kyoto, Japan) and U. H. Koszinowski (Max von Pettenkofer Institute for Virology, Munich, Germany), respectively. poly(dA·dT)·poly(dA·dT) [designated poly(dA·dT)] and poly(dG·dC)·poly(dG·dC) [designated poly(dC·dG)] were purchased from Amersham Biosciences (Piscataway, NJ). Poly(I:C) and human tumor necrosis factor alpha (TNF-α) were obtained from Invivogen (San Diego, CA) and PeproTech EC (London, United Kingdom), respectively. MVAdE3L was a kind gift from Holger Ludwig (Paul-Ehrlich Institute, Langen, Germany). The human papillomavirus (HPV) genome cloned in pBR322 (pHPV11) was obtained from JCRB GenBank (Osaka, Japan). Each nucleic acid was mixed for 15 min with TransIT-LT1 (Mirus Bio, Madison, WI) or Lipofectamine 2000 reagent (Invitrogen) at a constant weight/volume ratio of 1:3 in serum-free medium before use for stimulation.
Screening of the cDNA expression library.
A cDNA expression library originated from human bone marrow (Invitrogen) was screened. Escherichia coli DH5α cells transformed with a human bone marrow cDNA library (Invitrogen) were subdivided into ∼20 independent clones or 4,800 pools. Each pool was cultured at 30°C for 48 h, and then library plasmid DNA was extracted using the Wizard SV 96 plasmid DNA purification system (Promega, Madison, WI). HEK293T cells stably transfected with pGL3 IFN-β were transiently transfected with each pool of the library plasmids using FuGENE6 transfection reagent. Twenty-four hours after transfection, cells were washed and then stimulated with 0.5 μg/ml of poly(dA·dT). Luciferase assays were performed using the Bright-Glo luciferase assay system 24 h after stimulation. The three pools that showed the highest luciferase activities were transformed into DH5α, and then library plasmid DNA from 50 independent clones was recovered. A second screening was performed using these single plasmids in the same manner. Finally, insert cDNA from selected clones was sequenced using a Genetic Analyzer 310 apparatus (PE Applied Biosystems, Foster City, CA) and then characterized with the BLAST program.
RNA interference.
Interference of histone mRNA was performed as described previously (6). dsRNA was chemically synthesized by Invitrogen (stealth RNAi; Carlsbad, CA) or iGENE (Hokkaido, Japan). Sequences of each RNA were as follows: histone H1 sense, 5′-GCC CAA GAA AGU AGC UAA AAG CCC UAG-3′; histone H1 antisense, 5′-AGG GCU UUU AGC UAC UUU CUU GGG CAU-3′; histone H2A sense, 5′-CGC AAC GAC GAG GAA CUG AAC AAG CAG-3′; histone H2A antisense, 5′-GCU UGU UCA GUU CCU CGU CGU UGC GAU-3′; histone H2Bf116 sense, 5′-CCG UUU ACG UGU ACA AGG UGC UGA A-3′; histone H2Bf116 antisense, 5′-UUC AGC ACC UUG UAC ACG UAA ACG G-3′; histone H2Bf169 sense, 5′-UCC AAG GCC AUG GGC AUC AUG AAC U-3′; histone H2Bf169 antisense, 5′-AGU UCA UGA UGC CCA UGG CCU UGG A-3′; histone H3 sense, 5′-GAG AUC GCU CAG GAC UUU AAG ACC GAG-3′; histone H3 antisense, 5′-CGG UCU UAA AGU CCU GAG CGA UCU CAU-3′; histone H4 sense, 5′-GGG ACA AUA UCC AAG GCA UUA CAA AAG-3′; histone H4 antisense, 5′-UUU GUA AUG CCU UGG AUA UUG UCC CAU-3′; CIAO sense, 5′-AUG GAC AGU AUG AAG GCA AAG UCA GAG-3′; CIAO antisense, 5′-CUG ACU UUG CCU UCA UAC UGU CCA UAU-3′; ZBP1A sense, 5′-AUU UCA UGU GGA UUC UCU GGG CGG C-3′; ZBP1A antisense, 5′-GCC GCC CAG AGA AUC CAC AUG AAA U-3′; ZBP1B sense, 5′-UGU UGC UGU UGC CGA UGG UGG CGU C-3′; ZBP1B antisense, 5′-GAC GCC ACC AUC GGC AAC AGC AAC A-3′; ZBP1C sense, 5′-UUC AUC CAC AUA GUG GCU GCC UUC U-3′; ZBP1C antisense, 5′-AGA AGG CAG CCA CUA UGU GGA UGA A-3′; RIG-IA sense, 5′-UUA GGA UUC UCA UUG CUG GGA UCC C-3′; RIG-IA antisense, 5′-GGG AUC CCA GCA AUG AGA AUC CUA A-3′; RIG-IB sense, 5′-AUG UCU UGU ACU UCA CAU GGA UUC C-3′; RIG-IB antisense, 5′-GGA AUC CAU GUG AAG UAC AAG ACA U-3′; RIG-IC sense, 5′-UGG ACA UGA AUU CUC ACU AAG AUU C-3′; RIG-IC antisense, 5′-GAA UCU UAG UGA GAA UUC AUG UCC A-3′. The cells (6 × 105) were transfected with 120 pmol of each dsRNA by using the Lipofectamine RNAi MAX reagent (Invitrogen) according to the manufacturer's protocol.
Generation of mammalian expression plasmids.
H2B, RIG-I, ZBP1, TBK1, and CIAO cDNA was amplified by PCR using a human or mouse spleen cDNA library or the isolated library plasmid as a template. cDNA fragments were verified by sequencing and then introduced into pFLAG-CMV4 (Sigma), pFLAG-CMV5 (Sigma), or pCIneo-HA (25), or pCAGGS-CFP, pCAGGS-YFP, or pcDNA3-mRFP (10). To obtain full-length and truncated mutants of CIAO, the full-length CIAO open reading frame (ORF; amino acids [aa] 1 to 249), aa 1 to 121, aa 91 to 249, or aa 194 to 249 was fused to the green fluorescent protein (GFP) ORF and introduced into pFLAG-CMV4 (FLAG-CIAO FL-GFP, FLAG-CIAO N′-3a-GFP, FLAG-CIAO C′-4a-GFP, or FLAG-CIAO C′-2a-GFP, respectively). To obtain a truncated mutant of histone H2B, the full-length histone H2B ORF (aa 1 to 126), aa 1 to 103, aa 1 to 86, aa 1 to 53, or aa 1 to 37 (nuclear localization signal), or aa 38 to 126 (α-helical region) was fused to the GFP ORF and introduced into pCIneo-HA [HA-H2B FL (N′-1α2α3α4α)-GFP, HA-H2B N′-1α2α3α-GFP, HA-H2B N′-1α2α-GFP, HA-H2B N′-1α-GFP, HA-H2B N′-tail-GFP, or HA-H2B αH-GFP, respectively]. To obtain full-length and truncated mutants of IPS-1, the full-length IPS-1 ORF (aa 1 to 540), aa 1 to 100, aa 91 to 180, aa 1 to 170, aa 1 to 100 plus aa 170 to 540, or aa 1 to 514 was either left alone or fused to the GFP ORF and then introduced into pCIneo-HA (HA-IPS-1 FL, HA-IPS-1 CARD-GFP, HA-IPS-1 PRD-GFP, HA-IPS-1-CARD-PRD, HA-IPS-1ΔPRD, or HA-IPS-1ΔTMD, respectively).
Yeast two-hybrid screening.
Yeast two-hybrid screening was performed as described previously (25). Briefly, H2B cDNA was introduced in frame into the GAL4 DNA-binding domain of pGBKT7 as a bait plasmid (BD Clontech, Palo Alto, CA). A Saccharomyces cerevisiae strain, AH109, was transformed with a bait plasmid and the human bone marrow Matchmaker cDNA library (BD Clontech). After screening 1 × 106 clones on synthetic dropout selection agar plates (SD/-Leu/-Trp/-Ade/-His/5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside; BD Clontech), positive clones were picked and the pACT2 library plasmids were recovered. The insert cDNA was sequenced and then characterized with the BLAST program.
Transient transfection and reporter gene assay.
The cells (1 × 104) were transfected with 25 ng of pTK-RL (Promega) plus 25 ng of either reporter plasmid encoding an IFN-α4 or IFN-β promoter firefly luciferase (FFL) gene cassette (pGL3 IFN-α4 or pGL3 IFN-β) or pNF-κB-Luc (Stratagene). In some cases, the cells were cotransfected with expression plasmids for H2B, ZBP1, and RIG-I. Twenty-four hours after transfection, cells were stimulated with 0.1, 0.5, or 2.5 μg/ml of dsDNA [poly(dA·dT) or poly(dG·dC)] for 24 h or with 40 ng/ml of TNF-α for 8 h. The Dual-Glo luciferase assay system was used to measure both FFL activity and Renilla luciferase activity in the same sample. FFL activity of each sample was normalized against Renilla luciferase activity to obtain relative luciferase activity.
Pull-down assay.
HEK293 cells were transiently transfected with H2B-FLAG and stimulated with 0.5 μg/ml of biotin-poly(dA·dT) or biotin-poly(dG·dC) for 4 or 16 h. Then, the cells were lysed in lysis buffer (25 mM Tris-HCl [pH 7.5], 1 mM EDTA, 0.1 mM EGTA, 5 mM MgCl2, 100 mM NaCl, 10% glycerol, and 1% Nonidet P-40) containing both proteinase and phosphatase inhibitor cocktails (Sigma) on ice for 30 min. After centrifugation, the supernatants were recovered, mixed with streptavidin-agarose, and rotated at 4°C for 30 min. The complexes were washed with lysis buffer five times and suspended in SDS sample buffer, and then the supernatants were subjected to immunoblot analysis using anti-FLAG M2 Ab.
Viral genome binding assay.
HEK293 cells were transiently transfected with hemagglutinin (HA)-GFP, HA-H2B FL-GFP, HA-H2B N′-tail-GFP, or HA-H2B αH-GFP. Twenty-four hours after the first transfection, the cells were transfected with pHPV18. Forty-eight hours after the second transfection, the cells were lysed in lysis buffer containing both proteinase and phosphatase inhibitor cocktails (Sigma) on ice for 30 min. After centrifugation, the supernatants were recovered and HA-fusion molecules were precipitated using anti-HA Ab. The complexes were washed with lysis buffer five times and then subjected to standard PCR targeting the subtype-specific E6 gene using the following primer set: HPV18 E6, 5′-CCT GCG GTG CCA GAA ACC GT-3′ and 5′-CGT TGG AGT CGT TCC TGT CG-3′. The PCR products were separated on 2% agarose gels and visualized under UV light after ethidium bromide staining.
Immunoprecipitation and immunoblotting analysis.
Immunoprecipitation and immunoblotting analyses were performed using anti-ZBP1 (clone RG7D12; kindly provided by Stefan Rothenburg [National Institutes of Health]), anti-H2B (BioVision, Mountain View, CA), anti-phospho-IRF-3 (Ser396) 4G4D, anti-IRF3 (Cell Signaling, Danvers, MA), anti-RIG-I (AnaSpec, Inc., San Jose, CA), anti-IPS-1 (MAVS) (Bethyl, Montgomery, TX), anti-importin 9, anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) 6C5 (Abcam, Cambridge, United Kingdom), anti-Sp1, anti-STAT1, anti-extracellular signal-regulated kinase (ERK), anti-phospho-STAT1 (Santa Cruz Biotechnology, Santa Cruz, CA), anti-FLAG M2 (Sigma), or anti-HA Ab (Roche Diagnostics, Indianapolis, IN) as described previously (25).
FRET.
For flow cytometric analysis of fluorescence resonance energy transfer (FRET), 293-F cells (Invitrogen) were transfected with cyan fluorescent protein (CFP) and/or yellow fluorescent protein (YFP) fusion proteins encoding histone H2B, IPS-1, or CIAO in 293 expression medium (Invitrogen) for 24 h, and measurements of YFP (excitation, 488 nm; emission, 530 nm), CFP (excitation, 407 nm; emission, 510 nm), FRET (excitation, 407 nm; emission, 535 nm) were performed using FACSAria (Becton Dickinson) and BD FACSDiVa software. FRET is shown as the YFP emission obtained by CFP excitation divided by the CFP emission by CFP excitation (11).
Transient replication of HPV.
SCC-4 cells treated with control siRNA or H2B siRNA were transfected with the pHPV11 or -18 (VG012 and VG013; Human Science Research Resources Bank, Osaka, Japan) using Lipofectamine 2000 transfection reagent (Invitrogen). After incubation at 37°C for 24, 36, 60, or 72 h, the cells were washed with phosphate-buffered saline (PBS) and suspended in Hirt lysis buffer (0.6% SDS and 20 mM EDTA, pH 8.0). After 15 min, NaCl was added to a final concentration of 1 M, and then the mixture was placed at 4°C for 12 h. After centrifugation, the supernatants were treated with phenol-chloroform, and then DNA was precipitated with isopropanol. To distinguish replicated DNA from input DNA (pHPV11 or pHPV18), the samples were treated with DpnI endonuclease to degrade input DNA that had been propagated in E. coli DH5α. Resultant samples were suspended in Tris-EDTA, the concentrations were adjusted by measuring the optical density at 260 nm, and then 200 ng of DNA sample was subjected to standard PCR, targeting the subtype-specific E6 gene using the following primer sets: HPV11 E6, 5′-CTC CAC GTC TGC AAC ATC TA-3′ and 5′-TGA CAC AGG TAA CAA CGA AT-3′; HPV16 E6, 5′-CAC CAA AAG AGA ACT GCA ATG-3′ and 5′-TCA CGT CGC AGT AAC TGT TG-3′. The PCR products were separated on 2% agarose gels and visualized under UV light after ethidium bromide staining.
Virus multiplication study.
Twenty-four hours after HEK293 or NIH 3T3 cells were infected with adenovirus (AdV) type 5 or MCMV strain MW97.01, respectively, total DNA was collected using a DNeasy tissue kit (Qiagen). One hundred nanograms of each sample was subjected to PCR using primers for the AdV hexon 5 gene, 5′-TGA AGC TGC TAC TGC TCT TGA A-3′ and 5′-GCA TTC AAC TGC CAT GCT TGG C-3′ (18 cycles), the human IFN-β gene promoter region (−110 to +20), 5′-CTA AAA TGT AAA TGA CAT AGG-3′ and 5′-AAA GGT TGC AGT TAG AAT GTC-3′ (18 cycles), the MCMV DNA polymerase gene, 5′-ACG CCG AGA AAG AGT ACG TGC TCA A-3′ and 5′-TCG GAC TGC ATC CTC TCG CAG TAG-3′ (22 cycles), or the mouse Cd63 gene, 5′-GGT CTT GGG AAT TAT CTT CTC CTG CTG-3′ and 5′-CAC AGG CGG CAA AAT TCT TAA ACA TTC-3′ (26 cycles). After cell supernatants were recovered, the number of infectious viruses in 1 ml of each sample was determined by viral plaque assay or the 50% tissue culture infective dose (TCID50) as described previously (10).
Statistical analysis.
Student's t test was used for statistical analyses.
RESULTS
Identification of histone H2B as a mediator of dsDNA-induced IFN-β promoter activation.
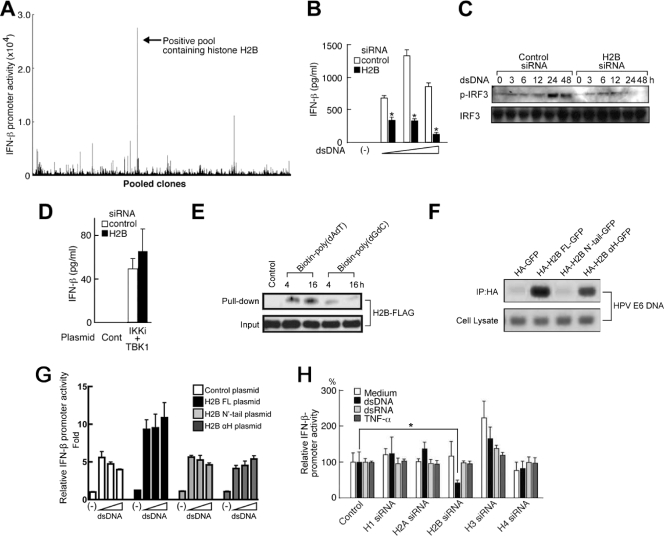
To identify molecules responsible for dsDNA-mediated type I IFN production in human cells, we screened a human bone marrow cDNA expression library using HEK293T cells stably expressing the Luc gene under the control of the IFN-β promoter. Among >960,000 independent clones examined, one of the positive clones encompassed the histone H2B1B ORF and exhibited a striking enhancement of dsDNA-induced IFN-β promoter activation (Fig. 1A). To confirm the role of histone H2B in dsDNA-induced innate immune responses, RNA interference was performed by using a small interfering RNA (siRNA). H2B siRNA treatment significantly suppressed the level of H2B protein expression (see Fig. S1 in the supplemental material). H2B siRNA also suppressed the levels of IFN-β production and IFN regulatory factor 3 (IRF3) phosphorylation induced by dsDNA in HEK293 cells (Fig. 1B and C, respectively), and knockdown of histone H2B inhibited dsDNA-induced activation of various gene promoters regulating IFN-related gene expression (see Fig. S2 in the supplemental material). dsDNA stimulates IRF kinases, such as TBK1 and IKKi (6). Overexpression of TBK1 and IKKi elicited comparable levels of IFN-β production in both control and H2B knockdown HEK293 cells, suggesting that H2B functions upstream of these kinases (Fig. 1D). To confirm the intracellular interaction of histone H2B and dsDNA, we performed pull-down and immunoprecipitation analyses. As a result, histone H2B was coprecipitated with biotinylated poly(dA·dT) or poly(dG·dC) that had been transfected into cells (Fig. 1E). To further identify the dsDNA interaction domain of histone H2B, we performed immunoprecipitation analysis using H2B mutants. It was found that the COOH-terminal α-helical region (αH domain) of histone H2B is sufficient for the interaction with HPV DNA (Fig. 1F). Furthermore, complementation of full-length histone H2B in knockdown cells restored the levels of IFN-β promoter activation induced by dsDNA but not that of the NH2-terminal tail region (N′-tail) or the αH domain alone (Fig. 1G). These results suggest that histone H2B binds to dsDNA through the αH domain, but both the N′-tail and αH domain are required for dsDNA-mediated signaling. When different siRNAs, each targeting histone H1, H2A, H2B, H3, or H4 were tested, only H2B siRNA attenuated IFN-β promoter activation induced by dsDNA (Fig. 1H). Such an effect of H2B siRNA was not observed following dsRNA or TNF-α stimulation (Fig. 1H). These results suggest that histone H2B is involved in the signal activation triggered by dsDNA.
FIG. 1.
Histone H2B mediates dsDNA-induced cellular activation. (A) A human bone marrow expression cDNA library was screened based on the ability to mediate dsDNA-induced IFN-β promoter activation. (B to D) HEK293 cells were transfected with control siRNA or siRNA targeting H2B. (B) The cells were treated with 0, 0.1, 0.5, or 2.5 μg/ml of dsDNA [poly(dA·dT)], and the supernatants were subjected to an enzyme-linked immunosorbent assay (ELISA) for IFN-β (PBL). (C) The cells were treated with 0.5 μg/ml of dsDNA [poly(dA·dT)] for 0, 3, 6, 12, 24, and 48 h, and the levels of phosphorylated IRF3 (p-IRF3) in the nucleus and IRF3 (normalization control) in the whole-cell lysates were examined by immunoblotting analysis. (D) The cells were further transfected with expression plasmids for TBK1 and IKKi, and the supernatants were subjected to ELISA for IFN-β (PBL). (E) HEK293 cells were transfected with histone H2B-FLAG and treated with biotin-poly(dA·dT) or biotin-poly(dG·dC). The cell lysates were collected, and a pull-down assay was performed with streptavidin-agarose. The complex was analyzed by immunoblotting using anti-FLAG Ab. (F) HEK293 cells were transfected with HA-GFP, HA-H2B-GFP, HA-H2B N′-tail-GFP, or HA-H2B αH-GFP. Twenty-four hours after the first transfection, the cells were further transfected with the HPV18 genome. Forty-eight hours after the second transfection, the cell lysates were collected and immunoprecipitation was performed with anti-HA Ab. The standard PCR targeting the HPV18 E6 gene was conducted with each cell lysate and immunoprecipitated complex. (G) HEK293 cells were transfected with H2B siRNA and then further transfected with pGL3 IFN-β and pTK-RL plus control, H2B FL, the H2B N′-tail, or the H2B αH plasmid. After treatment with 0, 0.1, 0.5, or 2.5 μg/ml of dsDNA [poly(dA·dT)] for 24 h, a luciferase assay was performed. (H) HEK293 cells were transfected with control siRNA or siRNA targeting either H2B, H1, H2A, H3, or H4. The cells were further transfected with pGL3 IFN-β and pTK-RL and treated with or without 0.5 μg/ml of dsDNA [poly(dA·dT)], dsRNA [poly(I:C)], or TNF-α (50 ng/ml) for 24 h, and then a luciferase assay was performed. The luciferase activity is depicted as the IFN-β promoter activity relative to samples obtained from the cells treated with control siRNA in each stimulation (relative IFN-β promoter activity). All data except for those in panel represent means ± standard deviations (SD) of six to eight samples. *, P < 0.05.
Histone H2B is involved in the signal activation triggered by dsDNA but not that by dsRNA nor TLR ligands.
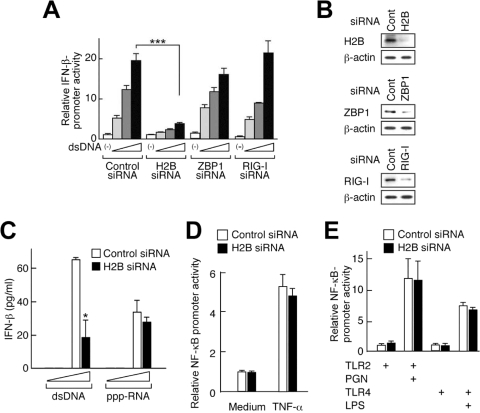
ZBP1 or RIG-I is known as a cytosolic DNA sensor in a mouse connective tissue cell line, L929, and a human hepatoma cell line, Huh7, respectively. dsDNA-induced IFN-β promoter activation was significantly suppressed in the cells treated with histone H2B siRNA, but not with siRNA targeting ZBP1 or RIG-I (Fig. 2A), although each siRNA specifically downregulated the level of target protein expression in HEK293 cells (Fig. 2B). Knockdown of histone H2B resulted in a suppression of IFN-β production in response to dsDNA, but not to 5′-triphosphate RNA stimulation in HUVEC (Fig. 2C). Although histone H2B is an essential component of the chromosome, H2B knockdown cells used in our present study could normally proliferate and respond to TNF-α or Toll-like receptor (TLR) ligands comparably to those treated with control siRNA (Fig. 2D and E), suggesting that chromosomal histone H2B that is essential for cell survival or TNF receptor (TNFR)- or TLR-mediated signaling pathways were not impaired by the use of these siRNAs. These results in toto suggest that histone H2B is involved in the signal activation triggered by dsDNA, but not that by dsRNA or TLR ligands.
FIG. 2.
Histone H2B is involved in the signal activation triggered by dsDNA, but not that of dsRNA or TLR ligands. HEK293 (A, B, D, and E) or HUVEC (C) cells were transfected with control siRNA or siRNA targeting either H2B, ZBP1, or RIG-I. (A and B) The cells were further transfected with expression plasmids for histone H2B, ZBP1, and RIG-I, in the presence of pGL3 IFN-β plus pTK-RL. (A) Twenty-four hours after transfection, cells were treated with 0, 0.1, 0.5, or 2.5 μg/ml of dsDNA [poly(dA·dT)] for 24 h, and then a luciferase assay was performed. (B) Twenty-four hours after transfection, levels of protein expression were examined by immunoblotting analysis using anti-RIG-I, anti-ZBP1, anti-H2B, or anti-β-actin Ab. (C) The cells were treated with 0.1 or 1.0 μg/ml of dsDNA [poly(dA·dT)] or 5′-triphosphate RNA (ppp-RNA) for 24 h, and the supernatants were subjected to an ELISA for IFN-β. (D) The cells were further transfected with pNF-κB-Luc plus pTK-RL and treated with or without TNF-α (50 ng/ml) for 8 h, and then a luciferase assay was performed. (E) The cells were further transfected with pNF-κB-Luc plus pTK-RL in the presence of either expression plasmid for TLR2 or TLR4 plus MD2 and treated with or without peptidoglycan (PGN; 1 μg/ml) or lipopolysaccharide (LPS; 100 ng/ml) for 24 h, and then a luciferase assay was performed. *, P < 0.05; ***, P < 0.001.
Extrachromosomal histone H2B associates with IPS-1 in the cytoplasm.
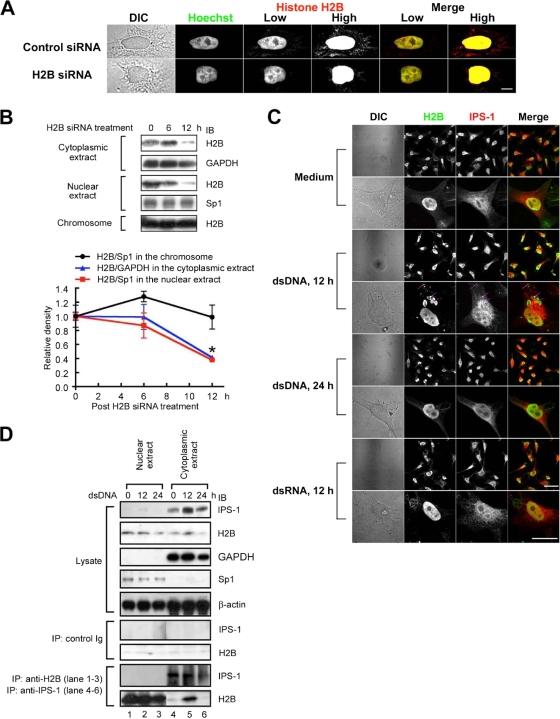
Immunofluorescence analysis revealed that although most histone H2B colocalized with Hoechst staining (chromosomes), there was some H2B staining in the extrachromosomal area (Fig. 3A). The levels of such extrachromosomal H2B were diminished when cells were treated with H2B siRNA, while chromosomal H2B in the same cell was present at a level similar to that in the control cells. Since it is known that histones are also released from the nucleus (14), the levels of histone H2B in each cellular fraction were evaluated by immunoblotting analysis. In accordance with the results of immunofluorescence analysis, H2B siRNA significantly decreased extrachromosomal H2B, but not choromosomal H2B (Fig. 3B). Only when the cells were stimulated with dsDNA for 12 h, but not when they were stimulated with dsRNA, was a punctate signal of H2B fluorescence detected in the cytoplasm (Fig. 3C), suggesting that dsDNA induces aggregation of histone H2B in the cytoplasm. Of interest, the colocalization of H2B with IPS-1 was observed following dsDNA stimulation (Fig. 3C, magenta arrows). To further examine the molecular interaction between histone H2B and IPS-1, immunoprecipitation analysis was performed. The cytoplasmic extracts and postchromosomal nuclear extracts were segregated from HEK293 cells stimulated with or without dsDNA. The purity of each extract was confirmed by immunoblotting analysis with anti-GAPDH (a cytoplasmic enzyme) or anti-Sp1 (a nuclear transcription factor) Ab as shown in Fig. 3D. IPS-1 was predominantly present in the cytoplasmic extracts, while histone H2B was in both extracts. When IPS-1 was immunoprecipitated from the cytoplasmic extracts, coprecipitation of histone H2B was not detected before stimulation but was detected 12 h after dsDNA stimulation. In contrast, when H2B was immunoprecipitated from the nuclear extracts, no detectable IPS-1 coprecipitated even after dsDNA stimulation. Taken together, these results suggest that the interaction between histone H2B and IPS-1 takes place in the cytoplasm following dsDNA stimulation.
FIG. 3.
Extrachromosomal histone H2B interacts with IPS-1. (A) HeLa cells were transfected with control or H2B siRNA. Twelve hours after transfection, the cells were fixed, stained with Hoechst 33258 and anti-H2B Ab followed by Alexa 488-conjugated anti-mouse IgG Ab, and then examined under a confocal microscope. The pictures taken under low and high lighting conditions are shown as low and high, respectively. Bar, 20 μm. (B) The cells were collected 0, 6, or 12 h after H2B siRNA treatment, and the cell lysates were fractionated into the cytoplasmic extract, the nuclear extract, and the chromosome fraction. Each fraction was analyzed by immunoblotting using anti-H2B, anti-GAPDH, or anti-Sp1 Ab. The density of each H2B band was normalized to the density of the corresponding GAPDH band (the cytoplasmic extract) or Sp1 band (the nuclear extract and the chromosome fraction) and is shown on the graph (n = 3). (C) After stimulation with or without dsDNA or dsRNA for 12 or 24 h, HeLa cells were fixed and stained with anti-H2B Ab and anti-IPS-1 Ab followed by Alexa 488-conjugated anti-mouse IgG Ab and Alexa 555-conjugated anti-rabbit IgG Ab. The cells were then examined under a confocal microscope. Bar, 100 μm in low-magnification pictures (upper panels) and 50 μm in high-magnification pictures (lower panels). (D) HEK293 cells were collected 0, 12, or 24 h after dsDNA stimulation, and the cell lysates were fractionated into the cytoplasmic extract and nuclear extract. Each fraction was immunoprecipitated with control IgG, anti-H2B, or anti-IPS-1 Ab. The immune complexes were analyzed by immunobloting using either anti-H2B or anti-IPS-1 Ab. *, P < 0.05.
Identification of CIAO as a species-specific molecule that links histone H2B and IPS-1.
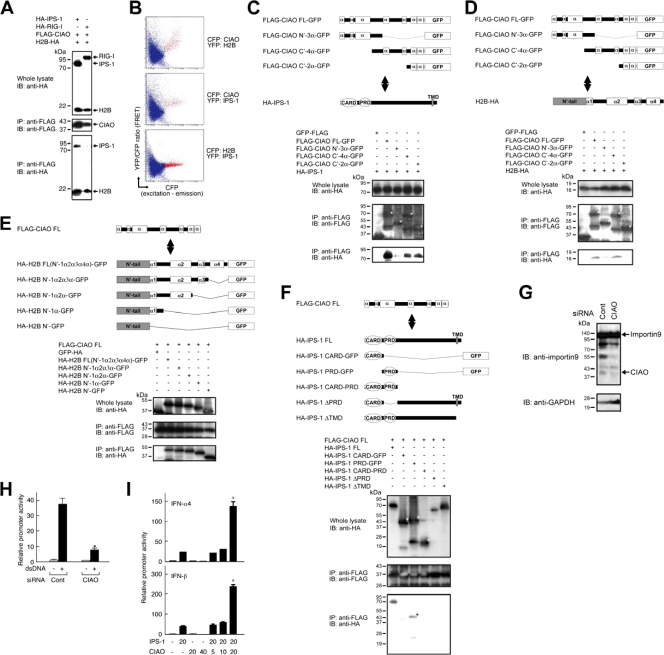
To identify the molecules directly associating with histone H2B, yeast two-hybrid screening was performed. As a result, 3 out of 10 positive clones were identified as KIAA1192, whose function has not yet been characterized but this nucleotide sequence is identical to the COOH-terminal end of importin 9, as a possible alternative splicing transcript of importin 9 (see Table S1 in the supplemental material). We renamed it to CIAO, based on its novel role, described below. Immunoprecipitation analysis clearly revealed that CIAO binds to histone H2B as well as IPS-1 but not to RIG-I (Fig. 4A). Full-length importin 9 interacted with histone H2B but not with IPS-1 (data not shown), suggesting that CIAO, a possible alternative splicing variant of importin 9, gains a peculiar function as an interacting partner of IPS-1. The spatial disposition of H2B, CIAO, and IPS-1 in the cell was further examined by FRET analysis. As shown in Fig. 4B, the higher levels of the specific FRET signal were detected in cells expressing CFP-CIAO and YFP-H2B as well as CFP-CIAO and YFP-IPS-1, compared to those expressing CFP-H2B and YFP-IPS-1, suggesting that CIAO and H2B or CIAO and IPS-1 juxtapose in a close spatiality and that CIAO disposes between H2B and IPS-1 in a molecular complex, thereby acting as an adaptor linking histone H2B to IPS-1. We further characterized the domains of CIAO, histone H2B, and IPS-1 that are responsible for the molecular interaction. Immunoprecipitation analysis demonstrated that the COOH-terminal region of CIAO (aa 194 to 249) is sufficient to interact with IPS-1, while the central region of CIAO (aa 91 to 193) is required for interaction with H2B (Fig. 4C and D). The N′-tail of H2B (aa 1 to 37) is sufficient, while the proline-rich domain (PRD) of IPS-1 (aa 94 to 170) is required for the interaction with CIAO (Fig. 4E and F). CIAO was endogenously expressed in HeLa and HEK293 cells (Fig. 4G and data not shown), and the treatment of siRNA targeting the CIAO ORF significantly suppressed the expression level of CIAO but not importin 9 protein (Fig. 4G). Knockdown of CIAO resulted in a marked reduction in dsDNA-induced activation of the IFN-β promoter (Fig. 4H), while overexpression of CIAO enhanced IPS-1-induced activation of type I IFN promoters (Fig. 4I). Taken together, these results indicate that CIAO links histone H2B to IPS-1, thereby enabling H2B-mediated type I IFN production in human cells.
FIG. 4.
CIAO associates with histone H2B and IPS-1 to constitute a signaling complex, thereby transmitting a signal leading to type I IFN production. (A and C to F) Cell lysates from HEK293 cells transfected with the indicated expression plasmid were immunoprecipitated with anti-FLAG Ab. The immune complexes were analyzed by immunoblotting using either anti-HA or anti-FLAG Ab. (B) 293-F cells transfected with CFP and YFP fusion proteins encoding histone H2B, IPS-1, or CIAO. FRET results are shown as the YFP emission obtained by CFP excitation divided by the CFP emission by CFP excitation. (C to F) Schematic diagrams of CIAO, IPS-1, H2B, and their truncated mutants. α, a putative α-helical region of CIAO and H2B; *, a predicted gene product of the target molecule. (G) HeLa cells were transfected with control siRNA or CIAO siRNA. Immunoblotting analysis was conducted using anti-importin 9 (COOH-terminal end) monoclonal Ab. (H and I) HeLa cells were transfected with or without the indicated reporter plasmid plus the indicated amounts (in ng) of expression plasmid(s). A luciferase assay was performed as described for Fig. 1. Data represent means ± SD of the relative luciferase activity of six to eight samples. *, P < 0.05.
Species-specific involvement of histone H2B and IPS-1 in dsDNA-mediated signaling.
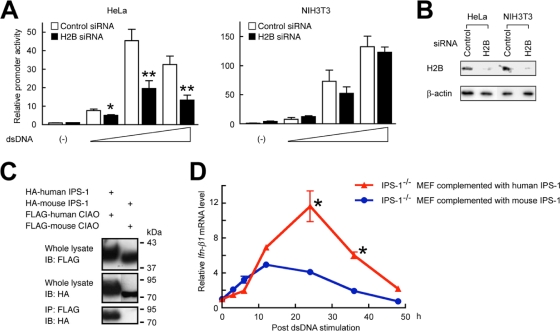
Previous reports have demonstrated that there are mechanistic differences in dsDNA signaling among species, e.g., human IPS-1 but not mouse IPS-1 is involved in dsDNA-mediated cellular signaling (2, 6, 15, 22). To examine a molecular mechanism underlying the species-specific signaling pathway induced by dsDNA, we compared the effects of H2B knockdown in human and mouse cells. Our results showed that knockdown of H2B resulted in a suppression of dsDNA-mediated IFN-β promoter activation in HeLa cells but not in NIH 3T3 cells (Fig. 5A and B), suggesting that there is a difference in the action of histone H2B between these species. Of interest, however, although an interaction was observed between human CIAO and IPS-1, an interaction between mouse CIAO and IPS-1 was not observed (Fig. 5C). While high similarities of amino acid sequences were seen among human and mouse H2B subtypes (human H2B1A, -1B, -1C, -1D, -1H, -1J, -1K, -1L, -1M, -1N, -1O, -2E, -2F, -3B, and -FS and mouse H2B1A, -1B, -1C, -1F, -1H, -1K, -1M, -1P, and -3A) (>70.1%) and between human and mouse CIAO (99.2%), a lower level of identity was seen between human and mouse IPS-1 amino acid sequences (30.3%). Thus, it was suggested that human IPS-1, but not mouse IPS-1, has the potential to interact with histone H2B and transmit signaling. To further examine a human IPS-1-specific mechanism in dsDNA-mediated signaling, we complemented IPS−/− mouse embryonic fibroblasts (MEF) with mouse IPS-1 (mouse IPS-1 MEF) or human IPS-1 (human IPS-1 MEF) and tested their responses to dsDNA. While Ifnb1 mRNA expression was induced within 3 h, its level peaked at 12 h after dsDNA stimulation and thereafter declined in mouse IPS-1 MEF (Fig. 5D). In contrast, the level of Ifnb1 mRNA expression peaked at 24 h after dsDNA stimulation in human IPS-1 MEF, and the overall level was significantly higher in human IPS-1 MEF than in mouse IPS-1 MEF (Fig. 5D). Thus, these results, taken together, suggest that human IPS-1 mediates the later-phase induction of IFN-β compared with that mediated by the dsDNA signaling endogenously present in MEF (Fig. 5D).
FIG. 5.
Species-specific involvement of histone H2B and IPS-1 in dsDNA-mediated signaling. (A and B) HeLa or NIH 3T3 cells were transfected with either control or H2B siRNA. (A) The cells were further transfected with pGL3 IFN-β and pTK-RL and treated with 0, 0.1, 0.5, or 2.5 μg/ml of dsDNA [poly(dA·dT)] for 24 h, and then a luciferase assay was performed. (B) The levels of H2B protein expression were examined by immunoblotting analysis. (C) The immunoprecipitation analysis was performed after HEK293 cells were transfected with the expression plasmids for human or mouse IPS-1 and CIAO. (D) IPS-1−/− MEF stably complemented with mouse or human IPS-1 were stimulated with 0.5 μg/ml of dsDNA [poly(dA·dT)] for 3, 6, 12, 24, 36, and 48 h. The cells were subjected to real-time PCR analysis for Ifnb1 mRNA and 18S rRNA. The levels of Ifnb1 mRNA were normalized with the corresponding levels of 18S rRNA and are shown on the graph (n = 4). *, P < 0.05; **, P < 0.01.
Extrachromosomal histone H2B is involved in suppression of DNA virus multiplication.
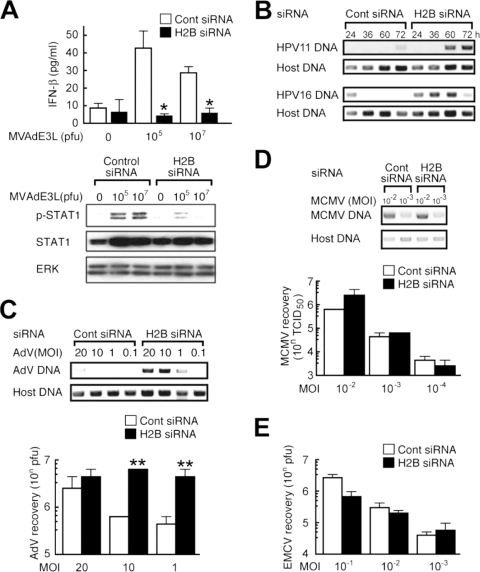
We further evaluated the biological role of histone H2B on cell-autonomous antiviral responses. Knockdown of histone H2B suppressed vaccinia virus-induced IFN-β production as well as STAT1 phosphorylation (Fig. 6A). Multiplication of human papilloma viruses (HPV 11 and HPV 16) and AdV type 5 was significantly enhanced in the H2B knockdown cells (Fig. 6B and C, respectively) but not in RIG-I or ZBP-1-knockdown cells (see Fig. S3 in the supplemental material). H2B knockdown, however, had virtually no effect on the multiplication of MCMV in mouse cells and RNA virus, EMCV, in human cells (Fig. 6D and E). These findings suggest that extrachromosomal histone H2B is involved in a sensing mechanism of DNA virus infection and mediates cell-autonomous antiviral innate immune responses in human cells.
FIG. 6.
Histone H2B is a crucial element for suppression of DNA virus replication. (A to E) HEK293 (A and C), SCC-4 (B), NIH 3T3 (D), or HeLa cells (E) were pretreated with control siRNA or H2B siRNA. (A) The cells were infected with MVAdE3L (1 × 105 or 1 × 107 PFU). Twenty-four hours after infection, cell lysates were subjected to immunoblotting for total STAT1, phosphorylated STAT1 (p-STAT1), or ERK. (B) The cells were transfected with the HPV11 or -16 genome, and the episomal DNA fractions were recovered 24, 36, 60, or 72 h after transfection. Viral multiplication was determined by PCR. (C and D) The cells were infected with AdV type 5 or MCMV strain MW97.01. Twenty-four hours after infection, viral multiplication was determined by PCR amplification of the viral genomic DNA and by a plaque assay (AdV) or by measuring the TCID50 (MCMV). (E) The cells were infected with EMCV. Twenty-four hours after infection, viral multiplication was determined by a plaque assay. *, P < 0.05; **, P < 0.01.
DISCUSSION
Our results provide direct evidence indicating that extrachromosomal histone H2B plays an important role in the signaling pathway triggered by dsDNA. Biologically, dsDNA is released following unusual or “dangerous” situations in cells, e.g., infection, apoptosis, and tissue damage (6, 18, 23). Therefore, the signaling pathway mediated by the signalsome consisting of extrachromosomal histone H2B and IPS-1 may have evolved not just in response to pathogen recognition but also perhaps for auto-recognition of unusual self DNA.
Although histone H2B is usually assembled in the nucleosome as a core histone, histone exchange and deposition often take place in living cells, with ∼3% of total H2B having a t1/2 of ∼6 min, ∼40% with a t1/2 of ∼130 min, and >50% with a t1/2 of ∼8.5 h. Indeed, nucleosome assembly protein 1 mediates chromatin fluidity by exchanging H2A/H2B and assisting nucleosome sliding (13). Nuclear import of the cytoplasmic core histones occurs along multiple redundant pathways mediated by importin family members. Importin 9 mediates one of the most productive pathways in H2B import through the nuclear core complex (8, 17). Interestingly, karyopherin 114p, a yeast homologue of mammalian importin 9, bind to the H2A-H2B complex in the cytoplasm (16). Such evidence strongly supports our present observation that the extrachromosomal H2B that is released from the chromosome plays an important role in the signal transmission induced by dsDNA.
Lines of evidence in recent decades indicate how histones are assembled with the genomic DNA to compose the chromosomes. Most studies have focused on the assumption that histones target only the genomic DNA under physiological conditions. Recent studies, however, have demonstrated that subtypes of histones play distinct roles in extrachromosomal settings. H2A.X is phosphorylated and recruited where DNA double-strand breaks take place (19), and H3.3 accumulates in the condensed chromatin where gene transcription is activated (9). More striking evidence is that H1.2 transmigrates from the nucleus to mitochondria to transmit apoptotic signals arising from DNA damage, and this crucially regulates the Bak-dependent release of cytochrome c from mitochondria (14). Thus, further characterization of extrachromosomal histone H2B as a component of the signaling complex may help in understanding the biological roles of the extrachromosomal histones and the mechanism of cell-autonomous protection against invading pathogens after dsDNA sensing.
CIAO encodes a 750-bp ORF identical to the COOH-terminal end of the importin 9 ORF (aa 793 to 1041) with no apparent domains or regions responsible for signal transduction. Although we have identified that the COOH-terminal region of CIAO is sufficient for the interaction with the IPS-1 PRD and the central region of CIAO is required for the interaction with the NH2-terminal tail region of H2B, the dynamics of such signaling complex formation under physiological conditions or during viral infection remain to be elucidated. To examine such dynamics, we are now generating Ab and agents to discriminate endogenous CIAO from importin 9 or the extrachromosomal histone H2B from the chromosomal histone by targeting specific residues of each molecule that have been modified before or after the complex formation.
It has been demonstrated that RIG-I plays a significant role in dsDNA-induced antiviral responses through IPS-1 in the human hepatoma cell line, Huh7 (2). However, we observed that histone H2B but not RIG-I or ZBP1 is involved in dsDNA-mediated signaling in HEK293 cells, suggesting that there are some cell-type-specific as well as redundant mechanisms in the signaling pathway activated by dsDNA. In fact, although ZBP1, also known as DAI, is involved in dsDNA-mediated signaling in mouse L929 cells, its in vivo role is redundant (7, 24).
The present data indicate that extrachromosomal histone H2B recognizes and interacts with not only “transfected” dsDNA but also viral DNA exposed to the cytoplasm. This mechanism is crucial for cell-autonomous innate responses to infection with vaccinia virus, AdV, and HPV, but not with EMCV. Our ongoing analysis shows that replication of HIV is also enhanced in H2B knockdown Magic5 cells (data not shown), suggesting that proviral dsDNA rather than genomic RNA of HIV is sensed by extrachromosomal histone H2B. Thus, histone H2B seems to discriminate between foreign DNA and RNA upon viral infection to evoke IPS-1-mediated signaling through the association with a novel adaptor protein, CIAO, and it is suggested that human IPS-1 has evolutionarily gained the potential to transmit dsDNA- and histone H2B-mediated signaling to combat against human viruses that produce DNA intermediates within the cell.
In conclusion, our present work demonstrates that extrachromosomal histone H2B physically interacts with IPS-1 through CIAO to form a distinct signaling complex that transmits dsDNA-induced type I IFN production in human cells. Such a molecular platform may act as a sensor of the dsDNA aberrantly present within the cell, alerting cells to urgent hazards, such as pathogens (infection), apoptosis (hormonal stimuli, chemical agents, or irradiation), and necrosis (injury). Thus, this mechanism may also play some roles in autoimmunity, transplantation rejection, gene-mediated vaccines, and other therapeutic applications.
Supplementary Material
Acknowledgments
We declare no conflicts of interest.
This work was supported in part by the Strategic Research Project of Yokohama City University (K18022 to F.T.), Advancement of Medical Sciences from Yokohama Medical Foundation (to F.T.), the National Institute of Biomedical Innovation (to K.O.), Mitsubishi Pharma Research Foundation (to K.S.), and a Grant-in-Aid for Scientific Researches (B and C) and Innovative Areas (18590432, 20590477, and 20200074 to F.T., 18659126 to K.O., and 15390296 to K.S.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and from the Japan Science and Technology Corporation.
Footnotes
Published ahead of print on 11 November 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Burckstummer, T., C. Baumann, S. Bluml, E. Dixit, G. Durnberger, H. Jahn, M. Planyavsky, M. Bilban, J. Colinge, K. L. Bennett, and G. Superti-Furga. 2009. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat. Immunol. 10:266-272. [DOI] [PubMed] [Google Scholar]
- 2.Cheng, G., J. Zhong, J. Chung, and F. V. Chisari. 2007. Double-stranded DNA and double-stranded RNA induce a common antiviral signaling pathway in human cells. Proc. Natl. Acad. Sci. U. S. A. 104:9035-9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandes-Alnemri, T., J. W. Yu, P. Datta, J. Wu, and E. S. Alnemri. 2009. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458:509-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hornung, V., A. Ablasser, M. Charrel-Dennis, F. Bauernfeind, G. Horvath, D. R. Caffrey, E. Latz, and K. A. Fitzgerald. 2009. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458:514-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isaacs, A., R. A. Cox, and Z. Rotem. 1963. Foreign nucleic acids as the stimulus to make interferon. Lancet ii:113-116. [DOI] [PubMed] [Google Scholar]
- 6.Ishii, K. J., C. Coban, H. Kato, K. Takahashi, Y. Torii, F. Takeshita, H. Ludwig, G. Sutter, K. Suzuki, H. Hemmi, S. Sato, M. Yamamoto, S. Uematsu, T. Kawai, O. Takeuchi, and S. Akira. 2006. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat. Immunol. 7:40-48. [DOI] [PubMed] [Google Scholar]
- 7.Ishii, K. J., T. Kawagoe, S. Koyama, K. Matsui, H. Kumar, T. Kawai, S. Uematsu, O. Takeuchi, F. Takeshita, C. Coban, and S. Akira. 2008. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature 451:725-729. [DOI] [PubMed] [Google Scholar]
- 8.Jakel, S., J. M. Mingot, P. Schwarzmaier, E. Hartmann, and D. Gorlich. 2002. Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 21:377-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janicki, S. M., T. Tsukamoto, S. E. Salghetti, W. P. Tansey, R. Sachidanandam, K. V. Prasanth, T. Ried, Y. Shav-Tal, E. Bertrand, R. H. Singer, and D. L. Spector. 2004. From silencing to gene expression: real-time analysis in single cells. Cell 116:683-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jounai, N., F. Takeshita, K. Kobiyama, A. Sawano, A. Miyawaki, K. Q. Xin, K. J. Ishii, T. Kawai, S. Akira, K. Suzuki, and K. Okuda. 2007. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc. Natl. Acad. Sci. U. S. A. 104:14050-14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawai, T., S. Sato, K. J. Ishii, C. Coban, H. Hemmi, M. Yamamoto, K. Terai, M. Matsuda, J. Inoue, S. Uematsu, O. Takeuchi, and S. Akira. 2004. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat. Immunol. 5:1061-1068. [DOI] [PubMed] [Google Scholar]
- 12.Kawane, K., M. Ohtani, K. Miwa, T. Kizawa, Y. Kanbara, Y. Yoshioka, H. Yoshikawa, and S. Nagata. 2006. Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature 443:998-1002. [DOI] [PubMed] [Google Scholar]
- 13.Kimura, H. 2005. Histone dynamics in living cells revealed by photobleaching. DNA Repair (Amst.) 4:939-950. [DOI] [PubMed] [Google Scholar]
- 14.Konishi, A., S. Shimizu, J. Hirota, T. Takao, Y. Fan, Y. Matsuoka, L. Zhang, Y. Yoneda, Y. Fujii, A. I. Skoultchi, and Y. Tsujimoto. 2003. Involvement of histone H1.2 in apoptosis induced by DNA double-strand breaks. Cell 114:673-688. [DOI] [PubMed] [Google Scholar]
- 15.Kumar, H., T. Kawai, H. Kato, S. Sato, K. Takahashi, C. Coban, M. Yamamoto, S. Uematsu, K. J. Ishii, O. Takeuchi, and S. Akira. 2006. Essential role of IPS-1 in innate immune responses against RNA viruses. J. Exp. Med. 203:1795-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mosammaparast, N., K. R. Jackson, Y. Guo, C. J. Brame, J. Shabanowitz, D. F. Hunt, and L. F. Pemberton. 2001. Nuclear import of histone H2A and H2B is mediated by a network of karyopherins. J. Cell Biol. 153:251-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muhlhausser, P., E. C. Muller, A. Otto, and U. Kutay. 2001. Multiple pathways contribute to nuclear import of core histones. EMBO Rep. 2:690-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Napirei, M., H. Karsunky, B. Zevnik, H. Stephan, H. G. Mannherz, and T. Moroy. 2000. Features of systemic lupus erythematosus in Dnase 1-deficient mice. Nat. Genet. 25:177-181. [DOI] [PubMed] [Google Scholar]
- 19.Redon, C., D. Pilch, E. Rogakou, O. Sedelnikova, K. Newrock, and W. Bonner. 2002. Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 12:162-169. [DOI] [PubMed] [Google Scholar]
- 20.Roberts, T. L., A. Idris, J. A. Dunn, G. M. Kelly, C. M. Burnton, S. Hodgson, L. L. Hardy, V. Garceau, M. J. Sweet, I. L. Ross, D. A. Hum, and K. J. Stacey. 2009. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science 323:1057-1060. [DOI] [PubMed] [Google Scholar]
- 21.Stetson, D. B., and R. Medzhitov. 2006. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity 24:93-103. [DOI] [PubMed] [Google Scholar]
- 22.Sun, Q., L. Sun, H. H. Liu, X. Chen, R. B. Seth, J. Forman, and Z. J. Chen. 2006. The specific and essential role of MAVS in antiviral innate immune responses. Immunity 24:633-642. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki, K., A. Mori, K. J. Ishii, J. Saito, D. S. Singer, D. M. Klinman, P. R. Krause, and L. D. Kohn. 1999. Activation of target-tissue immune-recognition molecules by double-stranded polynucleotides. Proc. Natl. Acad. Sci. U. S. A. 96:2285-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takaoka, A., Z. Wang, M. K. Choi, H. Yanai, H. Negishi, T. Ban, Y. Lu, M. Miyagishi, T. Kodama, K. Honda, Y. Ohba, and T. Taniguchi. 2007. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448:501-505. [DOI] [PubMed] [Google Scholar]
- 25.Takeshita, F., K. J. Ishii, K. Kobiyama, Y. Kojima, C. Coban, S. Sasaki, N. Ishii, D. M. Klinman, K. Okuda, S. Akira, and K. Suzuki. 2005. TRAF4 acts as a silencer in TLR-mediated signaling through the association with TRAF6 and TRIF. Eur. J. Immunol. 35:2477-2485. [DOI] [PubMed] [Google Scholar]
- 26.Wang, Z., M. K. Choi, T. Ban, H. Yanai, H. Negishi, Y. Lu, T. Tamura, A. Takaoka, K. Nishikura, and T. Taniguchi. 2008. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc. Natl. Acad. Sci. U. S. A. 105:5477-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yasutomo, K., T. Horiuchi, S. Kagami, H. Tsukamoto, C. Hashimura, M. Urushihara, and Y. Kuroda. 2001. Mutation of DNASE1 in people with systemic lupus erythematosus. Nat. Genet. 28:313-314. [DOI] [PubMed] [Google Scholar]
- 28.Yoshida, H., Y. Okabe, K. Kawane, H. Fukuyama, and S. Nagata. 2005. Lethal anemia caused by interferon-beta produced in mouse embryos carrying undigested DNA. Nat. Immunol. 6:49-56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.