Abstract
Two novel paramyxoviruses, 81-19252 (Texas81) and 92-7783 (ISU92), isolated from the brains of pigs in the United States in the 1980s and 1990s, were characterized. The complete genome of Texas81 virus was 15,456 nucleotides (nt) in length, that of ISU92 was 15,480 nt, and both genomes consisted of six nonoverlapping genes, predicted to encode nine proteins, with conserved and complementary 3′ leader and 5′ trailer regions and conserved gene starts, gene stops, and trinucleotide intergenic sequences similar to those in paramyxoviruses. The corresponding genes from these two viruses were similar in length, except for the F genes, of which the ISU92 form had an additional 24-nt U-rich 3′ untranslated region. The P genes of swine viruses were predicted to produce V and D mRNAs by RNA editing (one to four G insertions in Texas81 and one to nine G insertions in ISU92) or C mRNA by alternative translation initiation. Sequence-specific features related to virus replication and host-specific amino acid signatures indicated that these viruses originated from bovine parainfluenzavirus 3 (bPIV3). Phylogenetic analysis of individual genes suggested that these viruses are novel members of the genus Respirovirus of the Paramyxovirinae subfamily and may be grouped into two subgenotypes of genotype A of bPIV3. Our comprehensive studies revealed that these swine PIV3 are variants of bPIV3 and were possibly transferred from cattle to pigs but failed to establish an active enzootic state. These two viruses were mildly pathogenic to conventionally reared pigs, and results from a limited enzyme-linked immunosorbent assay-based serosurvey of swine farms in Minnesota and Iowa in 2007 and 2008 were negative.
Outbreaks of infections with many novel paramyxoviruses causing catastrophic illnesses have been reported all over the world in the last few decades. A large number of diverse host species have been involved, including avian, porcine, canine, bovine, equine, ovine, human, reptilian, and aquatic species (22, 29, 40, 50, 51). Cases of cross-species transmission and pathogen jumping to humans were also reported (10, 20), demonstrating the value of characterizing new animal pathogens, even if their pathogenic potential is currently unknown. Prior to the 1990s, only La Piedad Michoacán paramyxovirus had been well studied as a neurotropic paramyxovirus isolated from pigs. Many paramyxovirus porcine pathogens have been reported since the 1950s in numerous countries, including Japan (55), Canada (18), and Israel (32), as well as the United States (25, 32). There was also a case of concurrent infection with a porcine reproductive and respiratory syndrome virus and a paramyxovirus which was subsequently named SER virus (70) in Germany in the 1990s (28). Four bat-associated paramyxoviruses were reported to cause disease in animals and humans in 1994 (72). Hendra virus and Nipah virus, which caused severe respiratory disease and death in horses and their trainer and severe febrile encephalitis and death in pigs and farmers, respectively, have been classified as members of the genus Henipavirus in the subfamily Paramyxovirinae (7, 9, 20, 30, 48). Some recently isolated viruses, such as Menangle virus (55), Tupaia paramyxovirus (69), Tioman virus (11), Mossman virus (47), J-virus (31, 33), Beilong virus (42), Mapuera virus (34), Tursiops truncatus parainfluenzavirus 1 (PIV1), isolated from bottlenose dolphins (50), and Atlantic salmon paramyxovirus (51), remain unclassified below the subfamily level. All members of the subfamily Paramyxovirinae have six genes in the following order: 3′-N-P-M-F-A-L-5′, where N, P, M, F, A, and L indicate the genes for the nucleocapsid protein, the phosphoprotein, and the matrix, fusion, attachment, and large polymerase proteins, respectively (40).
Recently, we reported the antigenic and molecular characterization of glycoprotein genes from two novel swine PIV3 (sPIV3) isolates from the brains of pigs that experienced respiratory and central nervous system disease (57). These two sPIV3 strains were antigenically and genetically very closely related to bovine PIV3 (bPIV3) in the genus Respirovirus (57). However, the pathogenicity of these sPIV3 strains in conventionally reared pigs and the complete genome sequences of these isolates are presently unknown.
In bovines, bPIV3 infection results in asymptomatic to severe respiratory disease, but no neurological disease has been reported (16). Limited sequence polymorphism among the bPIV3 strains was detected previously (12, 66). Recently, after an analysis of Australian isolates of bPIV3, two distinct genotypes of bPIV3, A and B, were proposed (29). In this study, we have performed a complete genome sequence analysis of these swine isolates and determined their pathogenicity in conventionally reared pigs. Our analysis indicated that there are two distinct genetic groupings discernible within genotype A, represented by bPIV3 shipping fever strain (bPIV3-SF)-like and bPIV3 strain 910N (bPIV3-910N)-like viruses, with one swine isolate in each of these groups. Several amino acid residues that may reflect the minor population variations in the new host due to cross-species infection were identified. But both swine viruses induced a very mild respiratory illness without any neurological signs in young piglets, suggesting that coinfection with other infectious agents or the presence of other environmental factors may be required to precipitate clinical disease.
MATERIALS AND METHODS
Viruses and cells.
Texas 81-19252 (Texas81) and ISU-92-Minnesota isolate 92-7783 (ISU92) viruses and bovine anti-bPIV3-SF polyclonal antiserum were obtained from the National Veterinary Services Laboratory, Animal and Plant Health Inspection Service, U.S. Department of Agriculture, Ames, IA. Porcine kidney (PK15) cells, tested for and found to be free of porcine circovirus, were obtained from X. J. Meng, Virginia Polytechnic Institute and State University, and used to propagate these viruses for the first three passages. Vero cells (ATCC CCL-81) were used for all tests in this study as described earlier (57).
RT-PCR and sequencing.
RNA extraction and reverse transcription (RT) reactions were performed using standard procedures. Briefly, purified virus-infected PK15 cells were scraped into the medium and subjected to three cycles of freezing and thawing. After initial clarification at 3,000 × g for 15 min, polyethylene glycol 8000 (Sigma) was added to the cell lysate to a concentration of 10% and the lysate was incubated for 4 h at 4°C. The virus was pelleted at 12,000 × g for 60 min at 4°C, and the viral genomic RNA was extracted from the virus pellet by using an RNeasy minikit (Qiagen). Oligonucleotide primers were designed based on consensus bPIV3 and human PIV3 (hPIV3) nucleotide sequences (primer sequences are available upon request) for amplification of the fragments. Based on the sequencing results, new sets of oligonucleotide primers were designed and a primer-walking strategy was employed to sequence the gap areas. The leader and trailer sequences were determined according to the method of Li et al. for the rapid amplification of cDNA ends (RACE) (43). Briefly, a single set of adaptors and identical sets of reagents were used. The methods for 5′ and 3′ RACE differ only in the order of adaptor ligation and cDNA synthesis. In 5′ RACE, adaptor ligation was carried out after cDNA synthesis, whereas in 3′ RACE, the order was reversed.
The cDNA copies obtained from the genomic RNAs of the two virus strains were synthesized using the specific oligonucleotide primers and SuperScript III reverse transcriptase (Invitrogen). Subsequently, Platinum Taq DNA polymerase (Invitrogen) was used to amplify each fragment with the following cycling parameters: 94°C for 4 min, followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min and a final 7-min extension step at 72°C. The PCR products were purified using a Qiaquick PCR purification kit (Qiagen) and cloned using a TOPO TA cloning kit per the instructions of the manufacturer (Invitrogen). To obtain the consensus sequence, every nucleotide in the genome was identified once in purified PCR products and in at least three independent TA clones sequenced in both directions with an automated sequencer at the core laboratory facility of the Virginia Bioinformatics Institute, Virginia Tech.
Studies of pathogenicity in pigs.
A total of 18 6-week-old Yorkshire crossbred pigs (each with less than 25 lbs of body weight) were obtained from the swine facility at Virginia Tech and randomly divided into three groups of 6 piglets. These piglets were determined to be free of antibodies to porcine reproductive and respiratory syndrome virus, swine influenza virus, and porcine circovirus 2. The piglets were acclimatized for 1 week before the experiment. The rectal temperatures were recorded two times a day (in the morning and evening). Clinical signs were observed three times a day (every 8 h) after inoculation until day 10. Treatment groups were inoculated intranasally with 2 ml (5 × 107 50% tissue culture infective doses) of virus stock. The control group was inoculated intranasally with 2 ml phosphate-buffered saline (PBS). From each of these groups, three pigs were euthanized on days 6 and 10. The following clinical signs were scored on a scale of 0 to 3: physical appearance (normal appearance, dullness, lameness, or recumbency), activity (alertness, anorexia, mild response, or no response), respiratory disease signs (none, sneezing, heavy nasal discharge, or dyspnea), and other signs/body weight loss (none, <10% weight loss, 10 to 15% weight loss, or >15% weight loss).
Blood was collected before inoculation and at necropsy. Trachea, lung, heart, brain, liver, spleen, and other internal organ tissues were collected at necropsy, fixed in 10% neutral buffered formalin, and used for histopathologic examination. Paraffin-embedded tissue sections were employed for immunohistochemistry analyses using bovine anti-bPIV3-SF polyclonal antiserum. Nasal and fecal swabs, blood samples, and 20% suspensions of lung and brain tissues were subjected to virus isolation in Vero cells. The serum samples were examined by a virus neutralization test in 96-well plates by using homologous viruses.
Serum neutralization test.
Preinoculation sera and sera collected at necropsy were tested against the ISU92 virus strain by a serum neutralization test as described previously (4). Briefly, 1,000 50% tissue culture infective doses of virus stock and a serially diluted serum sample inactivated by heat treatment (56°C for 1 h) were incubated at 37°C for 1 h. The virus-serum mixtures were added to confluent Vero cell monolayers, and the monolayers were incubated at 37°C for 2 h in a 5% CO2 incubator. The inoculum was then removed, and after three washes with PBS, Dulbecco's modified Eagle's medium with 2% fetal bovine serum (Atlanta Biologicals) was added and the monolayers were further incubated. Cells were fixed and stained with 1% crystal violet at 72 h. The neutralization titer was calculated as the highest dilution of serum that inhibited virus-specific cytopathic effects.
Development of indirect ELISA for serology.
The ISU92 virus stock (108 PFU/ml) was used as an antigen to coat the polystyrene enzyme-linked immunosorbent assay (ELISA) plates (Nunclon). The optimum dilution of antigen and the serum dilution were determined by checkerboard titration. Briefly, a predetermined concentration of antigen in coating buffer (Kirkegaard & Perry Laboratories, Inc. [KPL]) was used to coat each well. The plates were then covered with Parafilm and incubated overnight at 4°C. The plates were washed once with wash buffer (KPL) in an ELISA plate washer (HydroFLEX; Tecan). After the unbound sites were blocked with blocking buffer (KPL) for 1 h, 100 μl of a 1-in-50 dilution of serum was added and the plates were incubated at 37°C for 1 h. Duplicate wells were set up for each sample, and blank wells were set up with only diluent buffer. The plates were then washed three times with a brief soak and shake between each wash. The bound antigen-antibody complexes were probed with anti-species IgG horseradish peroxidase secondary antibody diluted 1:500, and the plates were incubated for a further 1 h at 37°C. After the plates were washed three times, 100 μl of a TMB (3,3′,5,5′-tetramethylbenzidene) microwell substrate (KPL) was used to develop color for 10 min and then 100 μl of BlueSTOP solution (KPL) was added to each well to stop the reaction. The absorbances at 650 nm were read immediately with a Safire2 instrument (Tecan). A cutoff value of two times the mean negative-serum absorbance +1 standard deviation was chosen based on the results of checkerboard titrations with known positive and negative serum samples. One hundred randomly selected serum samples obtained from pigs in different age groups on five farms in Minnesota and Iowa in 2007 and 2008 were screened for the presence of sPIV3 antibodies. The serum samples from pigs experimentally inoculated with ISU92 or Texas81 were also subjected to ELISA. The mean values from duplicate wells were used for analysis.
Data analysis.
Nucleotide sequence editing and alignment, prediction of amino acid sequences, and sequence analyses were conducted using the software package DNASTAR (Lasergene). Phylogenetic reconstruction of the Texas81 and ISU92 viruses was performed using PAUP 4.01 software (67) with 1,000 bootstrap replicates.
Accession numbers for sequence analysis.
The accession numbers for viral sequences used for phylogenetic analysis are as follows: Atlantic salmon paramyxovirus, EF646380; avian metapneumovirus, NC_007652; avian paramyxovirus 2, EU338414; avian paramyxovirus 6, NC_003043; Beilong virus, NC_007803; bPIV3-910N, D84095; bPIV3 strain Kansas/15626/84 (bPIV3-Ka), AF178654; bPIV3 strain Q5592, EU277658; bPIV3-SF, AF178655; bovine respiratory syncytial virus (bRSV), NC_001989; canine distemper virus, NC_001921; dolphin morbillivirus, NC_005283; Fer-de-Lance virus, NC_005084; Hendra virus, AF017149; human metapneumovirus, NC_004148; hPIV1 strain Washington/1964 (hPIV1-Wa), NC_003461; hPIV2, NC_003443; hPIV3, AB012132; hPIV3-GPv, NC_001796; hPIV3-JS, Z11575; human RSV (hRSV), NC_001781; J-virus, NC_007454; measles virus, NC_001498; Menangle virus, NC_007620; Mossman virus, NC_005339; mumps virus, NC_002200; Newcastle disease virus, NC_002617; Nipah virus, NC_002728; peste-des-petits-ruminants virus, NC_006383; pneumonia virus of mice, AY573818; porcine rubulavirus (La Piedad Michoacán paramyxovirus), NC_009640; rinderpest virus (strain Kabete O), NC_006296; Sendai virus (SeV), NC_001552; simian PIV5, NC_006430; Tioman virus, NC_004074; and Tupaia paramyxovirus, NC_002199.
The consensus nucleotide sequence of hPIV3 Wash/47885/57 (hPIV3-Wa) was assembled from individual gene sequences in GenBank (2), including those with accession numbers M11849 (N gene), M14552 (N gene), X04612 (N gene), M14890 (P gene), X04721 (P gene), D00130 (M gene), M16458 (M gene), M16569 (M gene), Y00119 (M gene), M14892 (F gene), S82195 (F gene), M21649 (F, hemagglutinin-neuraminidase [HN], and L genes and 5′ genomic terminus), Z26523 (HN gene), M17641 (HN gene), M20402 (HN and partial L genes), and X03967 (3′ genomic terminus).
Nucleotide sequence accession numbers.
The sequences of ISU92 and Texas81 were deposited in GenBank under accession numbers EU439428 and EU439429, respectively.
RESULTS
Genome features.
The complete genome of Texas81 is 15,456 nucleotides (nt) in length, and that of ISU92 is 15,480 nt. The genome lengths are divisible by six and consistent with the “rule of six” as described for most other members of the Paramyxoviridae (6, 37, 38). The coding capacity of the genome is 93.3% in Texas81 and 93.2% in ISU92. Both viruses have conserved gene starts (GS) and gene ends (GE) and a strictly conserved trinucleotide intergenic sequence, AAG (genomic sense), for all six genes compared to bPIV3. The GS and GE of ISU92 and Texas81 are identical to those of bPIV3-910N and bPIV3-SF/bPIV3-Ka, respectively, except for the GS of the L gene of Texas81, which is identical to that of bPIV3-910N. In addition, the GS and GE follow the consensus patterns in all PIV3, which are UCCUNNUUNC for GS and NUNNUNNUUUUU for GE (genomic sequence). Genomic sequence comparison between sPIV3 strains and bPIV3 strains shows that each of the swine strains has a closest bovine relative in bPIV3. Specifically, Texas81 is most closely related to bPIV3-SF, and ISU92 is most closely related to bPIV3-910N. The overall degrees of identity in nucleotide sequences were 98.2% between Texas81 and bPIV3-SF and 98.1% between ISU92 and bPIV3-910N.
The 3′ and 5′ genomic termini of members of the Paramyxoviridae show complementarity (43). These conserved terminal sequences, especially the first 12 to ∼13 nt, are believed to contain the genome and antigenome promoters essential for replication and transcription (40). The swine viruses have a 55-nt 3′ leader before the transcription start site for the N gene, and the length of this leader is conserved among almost all of members of the subfamily Paramyxovirinae. The 5′ trailer following the L gene transcription stop site is 44 nt long in swine viruses. This length is variable among Paramyxovirinae subfamily members but conserved among PIV3 strains. Overall, in the leader region, the swine viruses exhibit 96.4% identity (53 of 55 nt are identical) to the consensus bPIV3 sequence and 92.7% identity (51 of 55 nt are identical) to the hPIV3 sequence. In the trailer region, there is 97.7% identity (43 identical nucleotide pairs among 44 total nucleotide pairs) between swine viruses and the consensus bPIV3 sequence and 90.9% identity (40 identical nucleotide pairs among 44 total nucleotide pairs) between swine viruses and hPIV3. The exact complementarity of the first 14 nt of the 3′ genomic leader and the 5 ′ trailer and the overall degrees of complementarity of 65.9% (29 complementary nucleotide pairs among 44 nucleotide pairs in ISU92) and 63.6% (28 complementary nucleotide pairs among 44 nucleotide pairs in Texas81) between the 3′ leader and 5′ trailer termini suggest conserved elements in the 3′ promoter regions of the genome and antigenome.
Genome organization.
The genome organization of swine viruses can best be described as 3′-N gene-P/V/D gene-M gene-F gene-HN gene-L gene-5′, and the genome can potentially encode nine proteins. The identity of the Texas81 genome to the ISU92 genome is 94.1% at the nucleotide level. The identity between swine viruses and bPIV3 is 98.2% at the highest level (between Texas81 and bPIV3-SF), while the identity between swine viruses and hPIV3 is 80.1% at the highest level (between Texas81 and hPIV3-JS). The start sites of each gene and the P gene-editing sites in Texas81 and ISU92 are identical to those in hPIV3 and bPIV3, as are the hexamer-phasing positions of 2, 1, 1, 1, 1, 2, and 2. The hexamer phasing is genus specific within the Paramyxovirinae (27, 37).
N gene.
The nucleoprotein (N) genes in both Texas81 and ISU92 are 1,646 nt long and encode proteins of 515 amino acids (aa), with predicted molecular masses of 57.3 kDa and pIs of 5.0 (Texas81 protein) and 5.2 (ISU92 protein). A highly conserved motif located near the middle of the N proteins from all members of the Paramyxovirinae and thought to be essential in N-N self-assembly and the N-RNA interaction process, F-X4-Y-X3-Ø-S-Ø-A-M (where X is any residue and Ø is an aromatic amino acid) (40, 49), is found in the central domain of the N proteins from the swine viruses, as 323FAPGNYPALWSYAM336. In SeV and other paramyxoviruses, the first residue of this motif, F324 (in SeV), is needed for correct self-assembly and another residue, Y260 (in SeV) (49), is critical for N-viral RNA binding. These residues are conserved in swine viruses as F323 and Y259, respectively. The carboxy-terminal 24% (aa 394 to 515) of the N protein has a very low level of identity to corresponding regions from other members of the Paramyxoviridae. This low-similarity region in the C-terminal 25% of the molecule is where most of the phosphorylation and antigenic sites of the protein (35, 40) are located.
P gene and P/C/V editing.
The phosphoprotein (P) genes in the two strains of sPIV3 are 1,995 nt long, with a major open reading frame (ORF) of 1,788 nt encoding the large P protein of 596 aa, with calculated sizes of 66.3 kDa (for the ISU92 protein) and 69.2 kDa (for the Texas81 protein) and a pI of 5.7. The sequence identity between the two sPIV3 P genes is 87.6%. sPIV3 has 87.6 to 100% identity to bPIV3 and 64.3 to 65.4% identity to hPIV3 (Table 1).
TABLE 1.
Amino acid identities between proteins from swine parainfluenzaviruses and analogous proteins from other paramyxovirusesa
| Speciesc | % Amino acid identity between sPIV3b protein and other paramyxovirus protein: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N |
P |
M |
F |
HN |
L |
|||||||
| T | I | T | I | T | I | T | I | T | I | T | I | |
| sPIV3 | 97.1 | 97.1 | 87.6 | 87.6 | 96.6 | 96.6 | 95.0 | 95.0 | 96.0 | 96.0 | 99.2 | 99.2 |
| bPIV3 | 100 | 97.1 | 100 | 87.6 | 100 | 96.6 | 100 | 95.0 | 99.8 | 95.8 | 98.7 | 98.2 |
| hPIV3 | 86.2 | 86.4 | 64.3 | 65.4 | 92.6 | 92.0 | 82.6 | 81.9 | 77.1 | 77.6 | 90.6 | 90.6 |
| hPIV1 | 60.6 | 61.0 | 24.1 | 24.5 | 64.6 | 63.1 | 42.7 | 42.7 | 45.8 | 45.6 | 60.7 | 60.7 |
| SeV | 62.6 | 62.6 | 23.7 | 24.5 | 64.0 | 63.7 | 43.1 | 42.9 | 46.7 | 46.0 | 59.7 | 59.7 |
All paramyxoviruses listed belong to the genus Respirovirus.
“T” indicates Texas81, and “I” indicates ISU92; the identities in the sPIV3 row are the identities between the two swine strains.
bPIV3 indicates bPIV3-SF, and hPIV3 indicates hPIV3-JS.
The mRNA editing in the P/C/V gene is a common feature in respiroviruses, morbilliviruses, henipaviruses, and avulaviruses and is considered to be a remarkable example of the exploitation of virus coding capacity (40). The P genes in both Texas81 and ISU92 contain an mRNA editing site, 5′-794AAAAAAGGG802-3′ (mRNA sense), which is identical to those in other respiroviruses. To determine the editing patterns in the swine viruses, at least 40 clones of cDNA from the mRNA of each strain were sequenced. We found that Texas81 had one to four G insertions and that ISU92 had a broader distribution of G insertions (one to nine Gs) at the editing sites. The insertion of G residues during mRNA synthesis can shift the translational reading frame and thus potentially generate a V protein 412 aa long (with the addition of one, four, or seven Gs) and a D protein 367 aa long (with the insertion of two, five, or eight Gs). The frequencies of V protein mRNA are 34.1% in Texas81 and 35.0% in ISU92, and the frequencies of D protein mRNA are 4.9% in Texas81 and 17.5% in ISU92. The identity of V proteins from the two strains is 83.3%, and the D proteins from the two strains have 84.0% identity. The predicted V and D proteins have amino termini identical to that of the P protein (generated via the insertion of zero, three, six, or nine Gs) for the first 241 aa. The V protein plays important roles in virus replication (3, 14), functioning as a negative regulator to inhibit RNA synthesis (3, 15, 17) and inhibiting the host cell antiviral response by interacting with cellular proteins (1, 44). The C-terminal V-specific domain is highly conserved among paramyxoviruses, with invariantly spaced histidine and cysteine residues forming a domain (the Cys-rich domain) that binds two zinc molecules per V protein (23, 41, 45, 52). In sPIV3, the C terminus of V protein contains all seven conserved cysteine residues and the conserved motifs H-R-R-E and W-C-N-P. The presence of these domains suggests a function for the V proteins of swine viruses similar to those of V proteins of other paramyxoviruses. In sPIV3, the C protein is predicted to initiate 10 nt downstream of the P protein start codon at an AUG codon in an alternate frame and is predicted to be 201 aa long, with the C proteins from the two strains having 91.1% identity. The C protein ORF is commonly observed in respiroviruses and morbilliviruses (40). C proteins are small, basic polypeptides that may be involved in the viral growth cycle, control of viral RNA synthesis (40), counteraction of host cell antiviral pathways (39), and facilitation of the release of virus from infected cells (24, 36). Both the editing site and the sizes of accessory proteins of sPIV3 highly resemble those of accessory proteins of hPIV3 and bPIV3, and the proteins may have similar functional roles.
M protein gene.
The matrix (M) gene is 1,149 nt long with a single ORF of 1,053 nt. The encoded protein is 351 aa long with a predicted molecular mass of 39.3 kDa and pIs of 9.5 (for the Texas81 protein) and 9.6 (for the ISU92 protein). The M protein is the most abundant and conserved virion structural protein and is located in the inner surface of the envelope. It interacts with the cytoplasmic tails of the integral membrane proteins, the lipid bilayer, and the nucleocapsids and plays an important role in virion assembly, budding, and release, as well as the transport of viral components (35, 40, 64). The nuclear localization signal (245KMGRMYSVEYCKQKIEK261) of the M proteins in swine viruses is highly conserved in PIV3 strains (13, 53). The ISU92 virus M protein has 96.6% amino acid sequence identity to the Texas81 M protein. The levels of amino acid sequence identity to members of the Respirovirus genus were 63 to 100%.
Fusion (F) protein gene and HN protein gene.
We have recently reported the molecular characterization of the envelope glycoprotein genes of these two swine viruses (57). In the putative antigenic sites of the F protein, there were several amino acid differences from hPIV3, including E101N, V/T367I (only in ISU92), S418Q, T492A, and T513V, but the amino acids were conserved in the swine viruses and bPIV3.
L protein gene.
The large polymerase (L) genes in both sPIV3 isolates are 6,795 nt long, with a major 6,699-nt-long ORF encoding a 2,233-aa protein with a pI of 6.3 for Texas81 or 6.2 for ISU92 and a molecular mass of 256 kDa. The L proteins of parainfluenzaviruses are the major RNA polymerase component, and they are responsible for nucleotide polymerization, mRNA capping and methylation, and polyadenylation of viral mRNAs (40). There are six highly conserved domains (domain I [DI] to DVI) in the L proteins of nonsegmented negative-strand RNA viruses, and each domain may be individually responsible for each of the L proteins' multiple functions (56, 61, 65). Pairwise alignments of sequences from sPIV3 with those from the other paramyxoviruses reveal highly conserved subdomains A to D within DIII and a highly variable hinge region between DII and DIII. A highly conserved stretch (positions 543 to 562) within DII, proposed to be a template recognition site, is present in sPIV3 L proteins and matches the pattern of basic and hydrophobic amino acid pairs repeated every four residues as described previously (56). The highly conserved 772GDNQ775 motif, the active site for nucleotide polymerization (8, 46), is present in subdomain C in DIII. The L proteins of swine viruses also contain a putative ATP binding site with the motif 1786K-X21-G-E-G-A-G1810 (56) in DVI. The L protein of Texas81 has 99.2% amino acid sequence identity to that of ISU92 and 59.7 to 98.7% to those of respiroviruses.
Phylogenetic analysis.
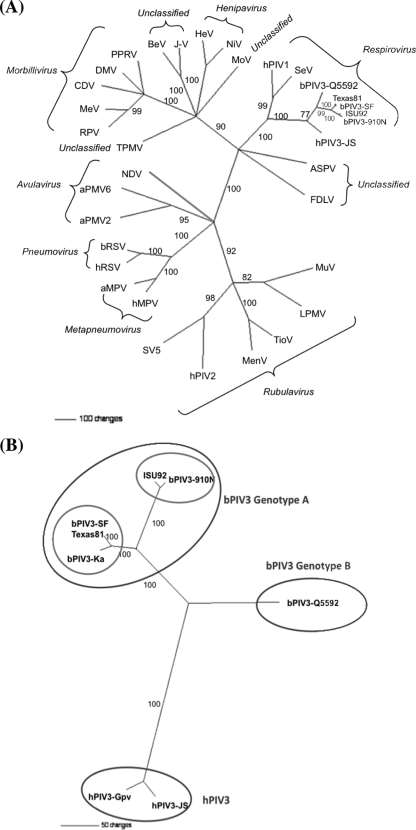
Phylogenetic trees were generated based on N, P, M, F, HN, and L protein sequences and complete genome sequences of sPIV3 and other representative members of all five genera of the family Paramyxoviridae, and the trees were found to be similar. The sPIV3 strains are phylogenetically closely related to the genus Respirovirus within the Paramyxovirinae subfamily. The phylogenetic reconstructions based on the M proteins only are shown (Fig. 1A). The phylogenetic reconstructions based on the complete M protein amino acid sequences reveal two distinct genetic groupings formed by bPIV3-910N-like and bPIV3-SF-like viruses. In addition, deduced amino acid sequences of M proteins (this study) and the F and HN proteins (57) indicate that Texas81 and ISU92 belong to two subgenotypes of genotype A and that the Australian bPIV3 strain Q5592 belongs to a separate genotype, genotype B (Fig. 1B).
FIG. 1.
Phylogenetic analysis was performed using parsimony (PAUP 4.01) with 1,000 bootstrap replicates, based on the M gene. (A) Analysis of deduced sPIV3 M protein amino acid sequences compared with sequences from other members of the Paramyxoviridae. (B) Phylogenetic analysis of PIV3 genotypes based on deduced M protein amino acid sequences. The numbers over the branches indicate the percentage of 1,000 bootstrap replicates that supports each phylogenetic branch. Strain information and GenBank accession numbers are presented in Materials and Methods. HeV, Hendra virus; BeV, Beilong virus; NiV, Nipah virus; J-V, J-virus; PPRV, peste-des-petits-ruminants virus; MoV, Mossman virus; DMV, dolphin morbillivirus; CDV, canine distemper virus; MeV, measles virus; RPV, rinderpest virus; TPMV, Tupaia paramyxovirus; NDV, Newcastle disease virus; aPMV6, avian paramyxovirus 6; aPMV2, avian paramyxovirus 2; aMPV, avian metapneumovirus; hMPV, human metapneumovirus; SV5, simian virus 5; MenV, Menangle virus; TioV, Tioman virus; LPMV, La Piedad Michoacán paramyxovirus; MuV, mumps virus; FDLV, Fer-de-Lance virus; and ASPV, Atlantic salmon paramyxovirus.
Pathogenicity.
To determine whether the swine viruses were able to induce clinical disease in conventionally reared pigs, a pathogenicity study was performed. Detailed pathogenicity studies with germfree pigs infected with ISU92 have been carried out earlier, and a report has been published (32). In the earlier studies, 3-day-old germfree pigs were inoculated intranasally with cell-culture-propagated ISU92 virus and were euthanized at 4, 8, 12, and 29 days postinoculation (dpi). Inoculated pigs developed mild clinical illness with symptoms such as mild diarrhea and rectal temperatures that were slightly elevated (by ∼1.5°F) at 2 and 3 dpi; they exhibited less vigorous, increased respiratory efforts and were stressed by handling at 2 dpi. In addition, one pig had eyelid edema at 6 dpi. Virus was isolated from lungs starting at 4 dpi. Only mild lesions like irregular foci of gray-tan lobular consolidations were observed at necropsy. Microscopically, there was mild to moderate inflammation in the lungs and brains of inoculated pigs at 4 dpi, and this inflammation was nearly resolved at 12 dpi. Focal gliosis was observed in the midbrain of only one pig at 8 dpi. Antibody conversion in the sera at a titer of 210 was detected by a neutralization test at 29 dpi.
Conventionally reared pigs infected intranasally with either of the two viruses in this study also showed mild respiratory signs only. None of the six infected pigs in either virus inoculation group developed neurological signs or elevated body temperatures throughout the experimental period. Two pigs from each of the infected groups developed mild respiratory signs at 2 dpi (mean clinical score, 1). One pig from the Texas81-infected group had diarrhea on day 2, and another pig from this group had diarrhea on day 3 (mean clinical score for the two pigs, 1). The gross lesions from virus-infected pigs upon necropsy at 6 and 10 dpi were unremarkable, including segmented, thickened small intestine tissues and greenish milky intestinal contents. Histologically, no microscopic lesions were observed. Only one nasal swab sample from a Texas81 virus-infected pig on day 6 yielded virus. None of the lung tissues examined by immunohistochemistry using polyclonal anti-bPIV3-SF serum revealed the presence of virus-specific antigen in necropsied pigs at 6 and 10 dpi. A serum neutralization test employing ISU92 virus was conducted to quantitate the neutralizing antibodies produced in pigs from virus-infected groups. Preinoculation serum samples were negative for sPIV3-specific antibodies by the neutralization test. At 6 dpi, all the inoculated pigs remained negative for neutralizing antibodies against sPIV3. By 10 dpi, the serum neutralization titers of serum samples ranged from 23 to 24.
ELISA.
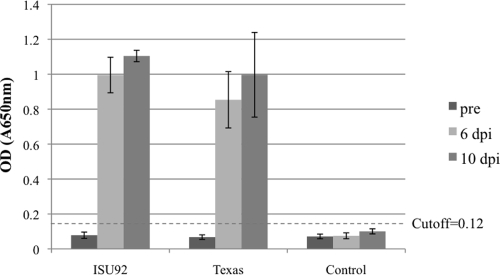
To determine the prevalence of sPIV3 in pigs, we established an indirect ELISA using ISU92 virus as the antigen. This ELISA can be used as a simple screening test for the presence of sPIV3 antibodies. None of the serum samples obtained from healthy or sick pigs aged 19 to 70 days from the five swine farms designated A, B, C, D, and E in Minnesota and Iowa in 2007 and 2008 were positive by ELISA, suggesting that sPIV3 is not prevalent in these two states. The serum samples from experimentally inoculated pigs were positive at 6 and 10 dpi, and the preinoculation samples and sera from mock-infected pigs were negative (Fig. 2). The ELISA results confirmed the neutralization test results for preinoculation and 10-dpi sera from experimentally infected pigs. However, the fact that 6-dpi samples were positive by the ELISA but not by the neutralization test suggests that the ELISA may be marginally more sensitive than the virus neutralization test.
FIG. 2.
ELISA results for experimentally inoculated pigs. The optical density (OD) values of serum samples from experimentally inoculated pigs as determined by indirect ELISA were plotted. The cutoff value was set at two times the mean of the negative-serum OD value +1 standard deviation based on checkerboard titration of known positive and negative serum samples against ISU92 virus antigen. Pre, preinoculation.
DISCUSSION
The recent insurgence of novel paramyxoviruses of zoonotic importance necessitates the close monitoring of viral pathogens with respiratory and neurological tropisms in animals. Although several paramyxoviruses have been isolated from pigs in the United States and other parts of the world, only a few have shown the potential to be extremely virulent and infect across species (21). The epidemiologic significance of many of the swine paramyxovirus isolates has never been examined beyond the initial case reports (25, 32, 55).
In this study, we have performed detailed analyses of two pigs viruses isolated in the United States between 1980 and the early 1990s. Our preliminary study of the morphology, antigenic characteristics, and sequences of the envelope glycoproteins indicated that these two viruses were very closely related to bPIV3 and belonged to the genus Respirovirus, subfamily Paramyxovirinae (57). We have further characterized these isolates with regard to their complete genomic sequences and features and their pathogenicity to conventionally reared pigs. Our comprehensive studies revealed that these sPIV3 strains are variants of bPIV3, possibly transferred from cattle to pigs but having failed to establish an active enzootic state.
The complete genome of sPIV3 Texas81 is 15,456 nt, and that of sPIV3 ISU92 is 15480 nt, which are typical genome sizes (approximately 15,500 nt) for paramyxoviruses. The genomes of sPIV3 conform to the rule of six, which is consistent in members of the subfamily Paramyxovirinae (6), and this pattern is thought to be associated with nucleocapsid organization in which each N protein monomer binds to six nucleotides of the viral genome. Recombinant PIV with a genome length that is not an even multiple of six has been shown to mutate to conform to the rule (62, 63). The genome organization of these two strains of sPIV3 is consistent with that of members of the subfamily Paramyxovirinae. The total coding percentage of the sPIV3 genome is 93%, which is similar to the percentages found in other Paramyxovirinae, with an average 92% coding capacity (40).
The amino acid identities to the corresponding proteins of other paramyxoviruses clearly place the sPIV3 strains as novel viruses in the genus Respirovirus, with high levels of identity to bPIV3. The overall conservation of homologous proteins in paramyxoviruses is generally consistent with the description in the most recent International Committee on Taxonomy of Viruses report, with the proteins ranked in the following orders on the basis of levels of conservation: V/C ≥ L > M > F > N and HN > V > P (19). Limited sequence polymorphism among strains of bPIV3 has been reported previously (2, 12, 66), and the host-specific amino acids in bPIV3 compared to hPIV3 were identified (2). The host-specific amino acid differences from bPIV3 noticed in the sPIV3 F protein (H/N100Y, T/F437A, I501V), HN protein (R/N70K, G/N88S, and G387S) (57), P protein (L/T270S, K/T271E, G/T303I, and P/S473H), and L protein (R/K79G, A/S326T, V336A, P/S721L, Y883F, I/P924M, S/T1355N, and K1565E) suggest that the corresponding amino acids are sPIV3 specific due to minor population variations attained in the new host. bPIV3-910N is a plaque-type variant of bPIV3 isolated from cattle in Japan but avirulent in mice (60). bPIV3-SF was originally obtained from a calf showing signs of shipping fever (5). The putative antigenic-site amino acid residues in F protein were conserved in both strains of sPIV3 and bPIV3 (12). The fusogenicities of Texas81 and ISU92 were comparable (57). Detailed epitope mapping studies with F and HN monoclonal antibodies are needed to elucidate host-specific antigenic-site variations.
We suggest that the host-specific amino acid differences between sPIV3 and bPIV3 are due to attempted host adaptation by bPIV3 in the novel swine host. But bPIV3 failed to establish a niche in swine, as can be seen from earlier serological surveys (4) and the limited serological survey performed in this study. Nevertheless, the sequence modifications suggest that sPIV3 was adapting to the new host after cross-species infection. Future cross-infection studies in bovines with sPIV3 may provide better insight into the host specificity of sPIV3. The genomic organization, amino acid identities of homologous proteins, phylogenetic analyses based on the genome sequences, and antigenic analyses all support the classification of these two novel strains of sPIV3 into the Respirovirus genus in the Paramyxovirinae subfamily and the Paramyxoviridae family. The International Committee on Taxonomy of Viruses suggests that 7% amino acid diversity in the glycoprotein and nucleoprotein of a related negative-strand RNA virus, hantavirus, should justify segregation of species (19). Based on our results, Texas81 appears to be a true spillover and the result of cross-species infection of pigs with bPIV3 but ISU92 may be considered a subspecies.
Our pathogenicity study with conventionally reared pigs indicated that these viruses induce mild clinical disease. We were able to show virus excretion in the nasal swab from at least one of the pigs by day 6, but all of the virus-infected pigs developed virus-neutralizing antibodies by day 10. It has been reported previously that bPIV3 can be isolated from clinically normal cattle (59). It is generally accepted that coinfection with bPIV3 or with other viruses and Mannheimia/Pasteurella species precipitates the classical shipping fever syndrome (4). A variety of factors such as environmental temperature, transportation, hygiene, stocking density, comingling, and host immune status can contribute to increased susceptibility to secondary bacterial infections and severity of clinical disease (4). The outbreaks of disease in pigs infected with these sPIV3 strains in the 1980s and 1990s, therefore, might have resulted from confection with other pathogens. Initial pathogenicity studies with germfree piglets infected with the ISU92 strain resulted only in mild respiratory disease and microscopic pathology (32), suggesting this possibility. Further, our limited serological prevalence studies with serum samples collected in 2007 and 2008 also indicated that sPIV3 strains are not prevalent in the field. An earlier study examining the seroprevalence in pigs from Iowa also indicated that there was very low seroprevalence. Among 876 serum samples collected from 36 swine farms in 1988 and 1989, only 6 samples, representing five swine herds, were found to have anti-ISU92 antibody titers by a neutralization test (4). Another study examining the prevalence of bPIV3 antibodies in 1,392 pig sera from 195 farms in Minnesota in 1988 by a hemagglutination inhibition test indicated that only 12.4% of sera tested positive at dilutions of 1:40 and above (68).
The sPIV3s may serve as good vaccine vector alternatives to bPIV3, as they do not cause any serious illness in infected pigs, are antigenically distinct from bPIV3, and have limited seroprevalence. Further, there is rapid virus clearance and limited virus replication, which are ideal qualities for a vaccine vector. A construct with bPIV3/hPIV3 chimeric virus (2, 71) as a vector backbone with the F and HN glycoproteins from hPIV3 has been evaluated as a vaccine against hPIV3 (26, 54). A construct consisting of bPIV3 as a backbone with F and HN proteins from hPIV3 and F protein from RSV was also generated. The effectiveness of this vaccine against both PIV3 and RSV challenge has been demonstrated in African green monkeys and is being evaluated in clinical trials (59). However, bPIV3 has never been tested as a vaccine vector in swine. The promising outcome of the vectored vaccines using paramyxoviruses and the biological and molecular characteristics of swine viruses described here suggest great potential for sPIV3 to be developed as a vaccine vector. The screening ELISA method developed by us with sPIV3 antigen can be used to detect seroprevalence in human and animal populations.
Acknowledgments
We thank X. J. Meng, Lijuan Yuan, Kevin Myles, and Chris Roberts for their constructive criticism and Laure Deflube, Shobana Raghunath, Vrushali Chavan, Thomas Rogers-Cotrone, and Gopakumar Moorkanat for their excellent technical assistance and help.
Footnotes
Published ahead of print on 11 November 2009.
REFERENCES
- 1.Andrejeva, J., E. Poole, D. F. Young, S. Goodbourn, and R. E. Randall. 2002. The p127 subunit (DDB1) of the UV-DNA damage repair binding protein is essential for the targeted degradation of STAT1 by the V protein of the paramyxovirus simian virus 5. J. Virol. 76:11379-11386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailly, J. E., J. M. McAuliffe, M. H. Skiadopoulos, P. L. Collins, and B. R. Murphy. 2000. Sequence determination and molecular analysis of two strains of bovine parainfluenza virus type 3 that are attenuated for primates. Virus Genes 20:173-182. [DOI] [PubMed] [Google Scholar]
- 3.Baron, M. D., and T. Barrett. 2000. Rinderpest viruses lacking the C and V proteins show specific defects in growth and transcription of viral RNAs. J. Virol. 74:2603-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battrell, M. A. 1995. Cultivation, preliminary characterization, and prevalence in swine herds of porcine paramyxovirus. Iowa State University, Ames.
- 5.Breker-Klassen, M. M., D. Yoo, and L. A. Babiuk. 1996. Comparisons of the F and HN gene sequences of different strains of bovine parainfluenza virus type 3: relationship to phenotype and pathogenicity. Can. J. Vet. Res. 60:228-236. [PMC free article] [PubMed] [Google Scholar]
- 6.Calain, P., and L. Roux. 1993. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J. Virol. 67:4822-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chadha, M. S., J. A. Comer, L. Lowe, P. A. Rota, P. E. Rollin, W. J. Bellini, T. G. Ksiazek, and A. Mishra. 2006. Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg. Infect. Dis. 12:235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chattopadhyay, A., and M. S. Shaila. 2004. Rinderpest virus RNA polymerase subunits: mapping of mutual interacting domains on the large protein L and phosphoprotein P. Virus Genes. 28:169-178. [DOI] [PubMed] [Google Scholar]
- 9.Chua, K. B., W. J. Bellini, P. A. Rota, B. H. Harcourt, A. Tamin, S. K. Lam, T. G. Ksiazek, P. E. Rollin, S. R. Zaki, W. Shieh, C. S. Goldsmith, D. J. Gubler, J. T. Roehrig, B. Eaton, A. R. Gould, J. Olson, H. Field, P. Daniels, A. E. Ling, C. J. Peters, L. J. Anderson, and B. W. Mahy. 2000. Nipah virus: a recently emergent deadly paramyxovirus. Science 288:1432-1435. [DOI] [PubMed] [Google Scholar]
- 10.Chua, K. B., K. J. Goh, K. T. Wong, A. Kamarulzaman, P. S. Tan, T. G. Ksiazek, S. R. Zaki, G. Paul, S. K. Lam, and C. T. Tan. 1999. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet 354:1257-1259. [DOI] [PubMed] [Google Scholar]
- 11.Chua, K. B., L. F. Wang, S. K. Lam, G. Crameri, M. Yu, T. Wise, D. Boyle, A. D. Hyatt, and B. T. Eaton. 2001. Tioman virus, a novel paramyxovirus isolated from fruit bats in Malaysia. Virology 283:215-229. [DOI] [PubMed] [Google Scholar]
- 12.Coelingh, K. V., and C. C. Winter. 1990. Naturally occurring human parainfluenza type 3 viruses exhibit divergence in amino acid sequence of their fusion protein neutralization epitopes and cleavage sites. J. Virol. 64:1329-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coleman, N. A., and M. E. Peeples. 1993. The matrix protein of Newcastle disease virus localizes to the nucleus via a bipartite nuclear localization signal. Virology 195:596-607. [DOI] [PubMed] [Google Scholar]
- 14.Curran, J., M. de Melo, S. Moyer, and D. Kolakofsky. 1991. Characterization of the Sendai virus V protein with an anti-peptide antiserum. Virology 184:108-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delenda, C., S. Hausmann, D. Garcin, and D. Kolakofsky. 1997. Normal cellular replication of Sendai virus without the trans-frame, nonstructural V protein. Virology 228:55-62. [DOI] [PubMed] [Google Scholar]
- 16.Dinter, Z., and B. Morein. 1990. Virus infections of ruminants. Elsevier Science & Technology, Amsterdam, The Netherlands.
- 17.Durbin, A. P., J. M. McAuliffe, P. L. Collins, and B. R. Murphy. 1999. Mutations in the C, D, and V open reading frames of human parainfluenza virus type 3 attenuate replication in rodents and primates. Virology 261:319-330. [DOI] [PubMed] [Google Scholar]
- 18.Ellis, J., L. Hassard, E. Clark, J. Harding, G. Allan, P. Willson, J. Strokappe, K. Martin, F. McNeilly, B. Meehan, D. Todd, and D. Haines. 1998. Isolation of circovirus from lesions of pigs with postweaning multisystemic wasting syndrome. Can. Vet. J. 39:44-51. [PMC free article] [PubMed] [Google Scholar]
- 19.Fauquet, C. M., M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.). 2005. Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA.
- 20.Field, H. E., A. C. Breed, J. Shield, R. M. Hedlefs, K. Pittard, B. Pott, and P. M. Summers. 2007. Epidemiological perspectives on Hendra virus infection in horses and flying foxes. Aust. Vet. J. 85:268-270. [DOI] [PubMed] [Google Scholar]
- 21.Field, H. E., J. S. Mackenzie, and P. Daszak. 2007. Henipaviruses: emerging paramyxoviruses associated with fruit bats. Curr. Top. Microbiol. Immunol. 315:133-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franke, J., S. Essbauer, W. Ahne, and S. Blahak. 2001. Identification and molecular characterization of 18 paramyxoviruses isolated from snakes. Virus Res. 80:67-74. [DOI] [PubMed] [Google Scholar]
- 23.Fukuhara, N., C. Huang, K. Kiyotani, T. Yoshida, and T. Sakaguchi. 2002. Mutational analysis of the Sendai virus V protein: importance of the conserved residues for Zn binding, virus pathogenesis, and efficient RNA editing. Virology 299:172-181. [DOI] [PubMed] [Google Scholar]
- 24.Garcin, D., M. Itoh, and D. Kolakofsky. 1997. A point mutation in the Sendai virus accessory C proteins attenuates virulence for mice, but not virus growth in cell culture. Virology 238:424-431. [DOI] [PubMed] [Google Scholar]
- 25.Goyal, S. M., R. Drolet, S. McPherson, and M. A. Khan. 1986. Parainfluenza virus type 3 in pigs. Vet. Rec. 119:363. [DOI] [PubMed] [Google Scholar]
- 26.Haller, A. A., T. Miller, M. Mitiku, and K. Coelingh. 2000. Expression of the surface glycoproteins of human parainfluenza virus type 3 by bovine parainfluenza virus type 3, a novel attenuated virus vaccine vector. J. Virol. 74:11626-11635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harcourt, B. H., A. Tamin, K. Halpin, T. G. Ksiazek, P. E. Rollin, W. J. Bellini, and P. A. Rota. 2001. Molecular characterization of the polymerase gene and genomic termini of Nipah virus. Virology 287:192-201. [DOI] [PubMed] [Google Scholar]
- 28.Heinen, E., W. Herbst, and N. Schmeer. 1998. Isolation of a cytopathogenic virus from a case of porcine reproductive and respiratory syndrome (PRRS) and its characterization as parainfluenza virus type 2. Arch. Virol. 143:2233-2239. [DOI] [PubMed] [Google Scholar]
- 29.Horwood, P. F., J. L. Gravel, and T. J. Mahony. 2008. Identification of two distinct bovine parainfluenza virus type 3 genotypes. J. Gen. Virol. 89:1643-1648. [DOI] [PubMed] [Google Scholar]
- 30.Hsu, V. P., M. J. Hossain, U. D. Parashar, M. M. Ali, T. G. Ksiazek, I. Kuzmin, M. Niezgoda, C. Rupprecht, J. Bresee, and R. F. Breiman. 2004. Nipah virus encephalitis reemergence, Bangladesh. Emerg. Infect. Dis. 10:2082-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jack, P. J., D. B. Boyle, B. T. Eaton, and L. F. Wang. 2005. The complete genome sequence of J virus reveals a unique genome structure in the family Paramyxoviridae. J. Virol. 79:10690-10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janke, B. H., P. S. Paul, J. G. Landgraf, P. G. Halbur, and C. D. Huinker. 2001. Paramyxovirus infection in pigs with interstitial pneumonia and encephalitis in the United States. J. Vet. Diagn. Invest. 13:428-433. [DOI] [PubMed] [Google Scholar]
- 33.Jun, M. H., N. Karabatsos, and R. H. Johnson. 1977. A new mouse paramyxovirus (J virus). Aust. J. Exp. Biol. Med. Sci. 55:645-647. [DOI] [PubMed] [Google Scholar]
- 34.Karabatsos, N. 1985. International catalogue of arboviruses, 3rd ed. American Society of Tropical Medicine and Hygiene, San Antonio, TX.
- 35.Karron, R. A., and P. L. Collins. 2007. Parainfluenza viruses, p. 1497-1526. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 36.Kato, A., Y. Ohnishi, M. Kohase, S. Saito, M. Tashiro, and Y. Nagai. 2001. Y2, the smallest of the Sendai virus C proteins, is fully capable of both counteracting the antiviral action of interferons and inhibiting viral RNA synthesis. J. Virol. 75:3802-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kolakofsky, D., T. Pelet, D. Garcin, S. Hausmann, J. Curran, and L. Roux. 1998. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J. Virol. 72:891-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolakofsky, D., L. Roux, D. Garcin, and R. W. Ruigrok. 2005. Paramyxovirus mRNA editing, the “rule of six” and error catastrophe: a hypothesis. J. Gen. Virol. 86:1869-1877. [DOI] [PubMed] [Google Scholar]
- 39.Komatsu, T., K. Takeuchi, J. Yokoo, and B. Gotoh. 2004. C and V proteins of Sendai virus target signaling pathways leading to IRF-3 activation for the negative regulation of interferon-beta production. Virology 325:137-148. [DOI] [PubMed] [Google Scholar]
- 40.Lamb, R. A., and G. D. Parks. 2007. Paramyxoviridae: the viruses and their replication, p. 1449-1496. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 41.Li, T., X. Chen, K. C. Garbutt, P. Zhou, and N. Zheng. 2006. Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase. Cell 124:105-117. [DOI] [PubMed] [Google Scholar]
- 42.Li, Z., M. Yu, H. Zhang, D. E. Magoffin, P. J. Jack, A. Hyatt, H. Y. Wang, and L. F. Wang. 2006. Beilong virus, a novel paramyxovirus with the largest genome of non-segmented negative-stranded RNA viruses. Virology 346:219-228. [DOI] [PubMed] [Google Scholar]
- 43.Li, Z., M. Yu, H. Zhang, H. Y. Wang, and L. F. Wang. 2005. Improved rapid amplification of cDNA ends (RACE) for mapping both the 5′ and 3′ terminal sequences of paramyxovirus genomes. J. Virol. Methods 130:154-156. [DOI] [PubMed] [Google Scholar]
- 44.Lin, G. Y., R. G. Paterson, C. D. Richardson, and R. A. Lamb. 1998. The V protein of the paramyxovirus SV5 interacts with damage-specific DNA binding protein. Virology 249:189-200. [DOI] [PubMed] [Google Scholar]
- 45.Liston, P., and D. J. Briedis. 1994. Measles virus V protein binds zinc. Virology 198:399-404. [DOI] [PubMed] [Google Scholar]
- 46.Malur, A. G., N. K. Gupta, P. De Bishnu, and A. K. Banerjee. 2002. Analysis of the mutations in the active site of the RNA-dependent RNA polymerase of human parainfluenza virus type 3 (HPIV3). Gene Expr. 10:93-100. [PMC free article] [PubMed] [Google Scholar]
- 47.Miller, P. J., D. B. Boyle, B. T. Eaton, and L. F. Wang. 2003. Full-length genome sequence of Mossman virus, a novel paramyxovirus isolated from rodents in Australia. Virology 317:330-344. [DOI] [PubMed] [Google Scholar]
- 48.Murray, K., P. Selleck, P. Hooper, A. Hyatt, A. Gould, L. Gleeson, H. Westbury, L. Hiley, L. Selvey, B. Rodwell, et al. 1995. A morbillivirus that caused fatal disease in horses and humans. Science 268:94-97. [DOI] [PubMed] [Google Scholar]
- 49.Myers, T. M., A. Pieters, and S. A. Moyer. 1997. A highly conserved region of the Sendai virus nucleocapsid protein contributes to the NP-NP binding domain. Virology 229:322-335. [DOI] [PubMed] [Google Scholar]
- 50.Nollens, H. H., J. F. Wellehan, J. T. Saliki, S. L. Caseltine, E. D. Jensen, W. Van Bonn, and S. Venn-Watson. 2008. Characterization of a parainfluenza virus isolated from a bottlenose dolphin (Tursiops truncatus). Vet. Microbiol. 128:231-242. [DOI] [PubMed] [Google Scholar]
- 51.Nylund, S., M. Karlsen, and A. Nylund. 2008. The complete genome sequence of the Atlantic salmon paramyxovirus (ASPV). Virology 373:137-148. [DOI] [PubMed] [Google Scholar]
- 52.Paterson, R. G., G. P. Leser, M. A. Shaughnessy, and R. A. Lamb. 1995. The paramyxovirus SV5 V protein binds two atoms of zinc and is a structural component of virions. Virology 208:121-131. [DOI] [PubMed] [Google Scholar]
- 53.Peeples, M. E., C. Wang, K. C. Gupta, and N. Coleman. 1992. Nuclear entry and nucleolar localization of the Newcastle disease virus (NDV) matrix protein occur early in infection and do not require other NDV proteins. J. Virol. 66:3263-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pennathur, S., A. A. Haller, M. MacPhail, T. Rizzi, S. Kaderi, F. Fernandes, L. Bicha, J. H. Schickli, R. S. Tang, W. Chen, N. Nguyen, S. Mathie, H. Mehta, and K. L. Coelingh. 2003. Evaluation of attenuation, immunogenicity and efficacy of a bovine parainfluenza virus type 3 (PIV-3) vaccine and a recombinant chimeric bovine/human PIV-3 vaccine vector in rhesus monkeys. J. Gen. Virol. 84:3253-3261. [DOI] [PubMed] [Google Scholar]
- 55.Philbey, A. W., P. D. Kirkland, A. D. Ross, R. J. Davis, A. B. Gleeson, R. J. Love, P. W. Daniels, A. R. Gould, and A. D. Hyatt. 1998. An apparently new virus (family Paramyxoviridae) infectious for pigs, humans, and fruit bats. Emerg. Infect. Dis. 4:269-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poch, O., B. M. Blumberg, L. Bougueleret, and N. Tordo. 1990. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J. Gen. Virol. 71(Pt. 5):1153-1162. [DOI] [PubMed] [Google Scholar]
- 57.Qiao, D., B. H. Janke, and S. Elankumaran. 2009. Molecular characterization of glycoprotein genes and phylogenetic analysis of two swine paramyxoviruses isolated from United States. Virus Genes 39:53-65. [DOI] [PubMed] [Google Scholar]
- 58.Reference deleted.
- 59.Sato, M., and P. F. Wright. 2008. Current status of vaccines for parainfluenza virus infections. Pediatr. Infect. Dis. J. 27:S123-S125. [DOI] [PubMed] [Google Scholar]
- 60.Shibuta, H., T. Kanda, A. Hazama, A. Adachi, and M. Matumoto. 1981. Parainfluenza 3 virus: plaque-type variants lacking neuraminidase activity. Infect. Immun. 34:262-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sidhu, M. S., J. P. Menonna, S. D. Cook, P. C. Dowling, and S. A. Udem. 1993. Canine distemper virus L gene: sequence and comparison with related viruses. Virology 193:50-65. [DOI] [PubMed] [Google Scholar]
- 62.Skiadopoulos, M. H., S. R. Surman, J. M. Riggs, C. Orvell, P. L. Collins, and B. R. Murphy. 2002. Evaluation of the replication and immunogenicity of recombinant human parainfluenza virus type 3 vectors expressing up to three foreign glycoproteins. Virology 297:136-152. [DOI] [PubMed] [Google Scholar]
- 63.Skiadopoulos, M. H., L. Vogel, J. M. Riggs, S. R. Surman, P. L. Collins, and B. R. Murphy. 2003. The genome length of human parainfluenza virus type 2 follows the rule of six, and recombinant viruses recovered from non-polyhexameric-length antigenomic cDNAs contain a biased distribution of correcting mutations. J. Virol. 77:270-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spriggs, M. K., P. R. Johnson, and P. L. Collins. 1987. Sequence analysis of the matrix protein gene of human parainfluenza virus type 3: extensive sequence homology among paramyxoviruses. J. Gen. Virol. 68(Pt. 5):1491-1497. [DOI] [PubMed] [Google Scholar]
- 65.Svenda, M., M. Berg, J. Moreno-Lopez, and T. Linne. 1997. Analysis of the large (L) protein gene of the porcine rubulavirus LPMV: identification of possible functional domains. Virus Res. 48:57-70. [DOI] [PubMed] [Google Scholar]
- 66.Swierkosz, E. M., D. D. Erdman, T. Bonnot, C. Schneiderheinze, and J. L. Waner. 1995. Isolation and characterization of a naturally occurring parainfluenza 3 virus variant. J. Clin. Microbiol. 33:1839-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Swofford, D. L. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, MA.
- 68.Tehteh, E., and S. M. Goyal. 1988. Antibodies to parainfluenza virus type 3 in Minnesota swine. Br. Vet. J. 144:613-615. [DOI] [PubMed] [Google Scholar]
- 69.Tidona, C. A., H. W. Kurz, H. R. Gelderblom, and G. Darai. 1999. Isolation and molecular characterization of a novel cytopathogenic paramyxovirus from tree shrews. Virology 258:425-434. [DOI] [PubMed] [Google Scholar]
- 70.Tong, S., M. Li, A. Vincent, R. W. Compans, E. Fritsch, R. Beier, C. Klenk, M. Ohuchi, and H. D. Klenk. 2002. Regulation of fusion activity by the cytoplasmic domain of a paramyxovirus F protein. Virology 301:322-333. [DOI] [PubMed] [Google Scholar]
- 71.van Wyke Coelingh, K. L., C. C. Winter, E. L. Tierney, W. T. London, and B. R. Murphy. 1988. Attenuation of bovine parainfluenza virus type 3 in nonhuman primates and its ability to confer immunity to human parainfluenza virus type 3 challenge. J. Infect. Dis. 157:655-662. [DOI] [PubMed] [Google Scholar]
- 72.Wang, L., B. H. Harcourt, M. Yu, A. Tamin, P. A. Rota, W. J. Bellini, and B. T. Eaton. 2001. Molecular biology of Hendra and Nipah viruses. Microbes Infect. 3:279-287. [DOI] [PubMed] [Google Scholar]