Abstract
A fundamental problem in studying the latent-to-lytic switch of Epstein-Barr virus (EBV) and the viral lytic cycle itself is the lack of a culture system fully permissive to lytic cycle induction. Strategies to target EBV-positive tumors by inducing the viral lytic cycle with chemical agents are hindered by inefficient responses to stimuli. In vitro, even in the most susceptible cell lines, more than 50% of cells latently infected with EBV are refractory to induction of the lytic cycle. The mechanisms underlying the refractory state are not understood. We separated lytic from refractory Burkitt lymphoma-derived HH514-16 cells after treatment with an HDAC inhibitor, sodium butyrate. Both refractory- and lytic-cell populations responded to the inducing stimulus by hyperacetylation of histone H3. However, analysis of host cell gene expression showed that specific cellular transcripts Stat3, Fos, and interleukin-8 (IL-8) were preferentially upregulated in the refractory-cell population, while IL-6 was upregulated in the lytic population. STAT3 protein levels were also substantially increased in refractory cells relative to untreated or lytic cells. This increase in de novo expression resulted primarily in unphosphorylated STAT3. Examination of single cells revealed that high levels of STAT3 were strongly associated with the refractory state. The refractory state is manifest in a unique subpopulation of cells that exhibits different cellular responses than do lytic cells exposed to the same stimulus. Identifying characteristics of cells refractory to lytic induction relative to cells that undergo lytic activation will be an important step in developing a better understanding of the regulation of the EBV latent to lytic switch.
Epstein-Barr virus (EBV) is a gammaherpesvirus that persists as a lifelong infection by remaining in the latent phase of its life cycle within B lymphocytes (17). EBV is associated with human cancers such as Burkitt lymphoma, nasopharyngeal carcinoma, Hodgkin's disease, and EBV-associated lymphoproliferative disease in immunocompromised individuals (11). Efforts to eliminate EBV-positive tumor cells by nucleoside analogue antiviral agents following induction of the viral lytic cycle have shown promising results (15, 16, 18, 38, 44, 52). These efforts have been preceded by extensive studies on the switch from latency to the EBV lytic cycle in lymphoid cell lines. A fundamental problem in studying the latent to lytic switch and the lytic cycle itself is the lack of a culture system fully permissive to lytic cycle induction (45). In cell culture, EBV can be induced into the lytic cycle by a variety of chemical stimuli, including agents currently being used or investigated as chemotherapeutic drugs, such as the HDAC inhibitors trichostatin A (TSA) and sodium butyrate (NaB) and the DNA methyltransferase inhibitor azacytidine (Aza) (3, 57). However, following treatment of cells latently infected with EBV, only a fraction of cells enter the lytic cycle; the remainder of the population is refractory to lytic induction. The refractory phenomenon is observed in all cell lines and for all inducing stimuli tested thus far (2, 22) and likely applies to lytic cycle induction in vivo. Understanding the refractory phenomenon will be an important step in elucidating the regulation of the EBV latent to lytic switch.
The EBV lytic genes BZLF1 and BRLF1 encode transcriptional activators responsible for initiating the cascade of viral gene expression that ultimately results in replication and virion production (9, 25). We previously demonstrated that treatment of HH514-16 cells with cycloheximide (CHX) blocks the production of the BZLF1 and BRLF1 transcripts following treatment with lytic cycle-inducing stimuli. Thus, de novo protein synthesis is required for EBV lytic cycle reactivation (56). EBV lytic cycle induction became resistant to CHX treatment between 4 and 6 h after application of the inducing stimuli. Therefore, events that determine whether a particular cell enters the lytic cycle or remains refractory to lytic induction likely occur at early times after treatment with inducing agents.
Studying the physiology underlying refractoriness of cells to a particular inducing agent is not possible in the mixed population of refractory and lytic cells that results from the stimulus. Whether refractory cells fail to respond or respond in a different manner to an inducing agent cannot be determined due to the background of lytic cells in the population. To overcome this obstacle, we used a technique to separate refractory and lytic Burkitt lymphoma-derived HH514-16 cells following induction of the lytic cycle with NaB (2). The efficient separation of refractory and lytic cells using this technique enabled an analysis of changes that occur in each population relative to each other or to untreated cells.
We show here that both the lytic and the refractory subpopulations exhibited effects consistent with drug exposure, as evidenced by increased site-specific histone acetylation. Gene expression profiling identified cellular genes that exhibited altered expression at early times (3 and 6 h) after treatment with inducing stimuli. Analysis of sorted cell populations revealed that cellular immediate-early transcripts Fos and Stat3 were preferentially upregulated in the refractory population. Using the HH514-16 cell line, we show that substantial upregulation of STAT3 protein, existing primarily in the unphosphorylated state, is characteristic of refractory cells upon treatment with the HDAC inhibitor NaB. These results show that treatment of a Burkitt lymphoma-derived EBV-positive cell line with a lytic cycle-inducing agent results in heterogeneous changes in cellular gene expression in refractory and lytic cells and identify STAT3 as one marker of the cells refractory to EBV lytic cycle induction.
MATERIALS AND METHODS
Cell line.
HH514-16 is a subclone of the P3J-HR1K Burkitt lymphoma cell line. This subclone does not exhibit spontaneous EBV lytic replication, but high levels of EBV lytic cycle gene expression can be induced following chemical treatment (26). Cells were cultured in RPMI 1640 medium containing 8% fetal bovine serum and antibiotics.
Chemical induction of the EBV lytic cycle.
HH514-16 cells were subcultured at 3 × 105 ml−1. After 48 h of subculture, 3 mM sodium butyrate (Sigma), 5 μM 5-aza-2′-deoxycytidine (azacytidine; Sigma), or 5 μM trichostatin A (TSA; Wako BioProducts) was added. For experiments in which cycloheximide was used to block protein synthesis, 33 μg/ml CHX was added.
Western blot analysis.
Total cell extracts were resolved using 8% SDS-polyacrylamide gels and transferred to nitrocellulose membranes (Bio-Rad). Rabbit polyclonal antibodies were used to detect phospho-STAT3 (Y705) (Cell Signaling) and acetylated histone 3 (K9,K14) (Upstate). Mouse monoclonal antibodies were used to detect total STAT3 (Cell Signaling), EA-D (R3.1 [43]), LMP1 (Dako), and β-actin (Sigma). Antibody-protein complexes were detected using 125I-labeled protein A or ECL Plus (GE Healthcare).
ChIP.
Chromatin immunoprecipitation (ChIP) was performed as previously described (14). Analysis was performed on HH514-16 cells untreated or treated with NaB for 24 h in the presence of phosphonoacetic acid (PAA). The relative amount of BZLF1 promoter DNA immunoprecipitated by anti-acetyl-H3 antibody was determined using real-time PCR.
Microarray analysis.
Affymetrix U133 Plus 2.0 arrays were used for all array experiments. HH514-16 cells were untreated or treated with NaB and harvested after 3 or 6 h. Total RNA was extracted using the RNeasy RNA extraction kit (Qiagen). RNA collected from three independent experiments at each time point was pooled prior to array analysis. RNA processing, hybridization, and initial data analysis were performed by the Keck Microarray Resource at Yale University by following recommended Affymetrix protocols. Data were analyzed using Affymetrix GeneChip operating software (GCOS).
Real-time RT-PCR.
The relative transcript levels of selected genes were determined by real-time reverse transcription-PCR (RT-PCR) with gene-specific primers using the iScript SYBR green RT-PCR kit (Bio-Rad). Relative expression levels were calculated using the ΔΔCT method, normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) or 18S RNA. Assays on individual samples were performed in triplicate. RT-PCR primers were designed using Primer3 (47) or have been described previously (33).
Cell sorting.
Separation of lytic and refractory cell populations was performed as described previously using reference EBV-positive and EBV-negative human sera (2).
Fluorescence-activated cell sorter (FACS) analysis.
HH514-16 cells were treated with NaB for 48 h followed by fixation and permeabilization using Cytofix/Cytoperm (BD Pharmingen). Cells were incubated with saturating amounts of rabbit anti-STAT3 antibody (C20; Santa Cruz Biotechnologies) and reference EBV-seropositive serum from healthy subjects (2). Bound STAT3 antibody and bound human anti-lytic IgG were detected using Texas Red-conjugated anti-rabbit IgG and fluorescein isothiocyanate (FITC)-conjugated anti-human IgG, respectively. Preimmune rabbit serum and reference EBV-seronegative serum from healthy subjects (2) were used to determine specific binding to STAT3 and lytic antigens. One hundred thousand events were acquired using FACSCalibur L (Becton Dickinson), and data were analyzed using WinMDI software.
RESULTS
Refractoriness of cells to lytic cycle induction is independent of stimulus and is transient.
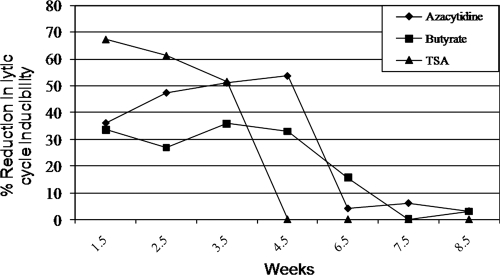
We previously described a robust technique to separate lytically induced from refractory cells following exposure to lytic cycle-inducing stimuli such as NaB (2). Although cells that remained in the latent state after treatment with NaB were found to be partially refractory to NaB for several weeks after reintroduction to culture, they regained susceptibility to lytic induction over time, indicating that permanent genetic changes, cellular or viral, were not likely responsible for the refractory state (2). The response of refractory cells to other inducing stimuli with different mechanisms of action remained unknown. We determined whether cells refractory to NaB were also refractory to lytic induction by TSA, a member of a different structural class of HDAC inhibitors (HDACi) (3), and Aza, a lytic cycle-inducing stimulus with a different mechanism of action (57). HH514-16 cells susceptible or refractory to lytic cycle induction by NaB were sorted by FACS following incubation with reference EBV-seropositive and EBV-seronegative human sera. Lytic cells did not survive when placed back into culture, but when refractory cells were reintroduced into culture, they proliferated. These cells were assessed periodically for lytic cycle inducibility by NaB, TSA, or Aza over 8.5 weeks (Fig. 1). At 1.5 weeks after reintroduction into culture, there were reductions in response to lytic cycle induction of 67.4% to TSA, 36% to Aza, and 33.6% to NaB compared to lytic cycle induction by the same agents of sorted but previously untreated cells (control cells). Relative refractoriness of cells to all three agents was maintained through 3.5 weeks (51.6% reduction in response to TSA, 51% to Aza, and 36% to NaB). At 4.5 weeks, the lytic response continued to be reduced upon exposure to Aza (53.6%) and NaB (33%) but not to TSA compared to control cells. Reacquisition of susceptibility to lytic cycle induction by Aza and NaB reached levels that were comparable to control cells after 6.5 to 7.5 weeks (Fig. 1). Full susceptibility to all three stimuli was regained after approximately 8 weeks in culture. Thus, cells refractory to an initial lytic cycle-inducing stimulus remained refractory for several cell divisions to lytic cycle induction by agents that function via different modes of action. This experiment suggested that nonpermanent changes, such as alterations in chromatin modification or gene expression, may influence the refractory state.
FIG. 1.
Refractoriness of cells to lytic cycle-inducing stimuli is transient. HH514-16 cells refractory to lytic cycle induction by sodium butyrate (NaB) were sorted by FACS and reintroduced into culture. Simultaneously, untreated cells were sorted and reintroduced into culture as a control. Lytic cycle inducibility of refractory cells was compared to that of untreated but sorted cells after exposure to azacytidine, NaB, and trichostatin A (TSA) at weekly intervals after reintroduction into culture.
HDAC inhibition results in hyperacetylation of histone H3 associated with Zp in both refractory and lytic HH514-16 cells.
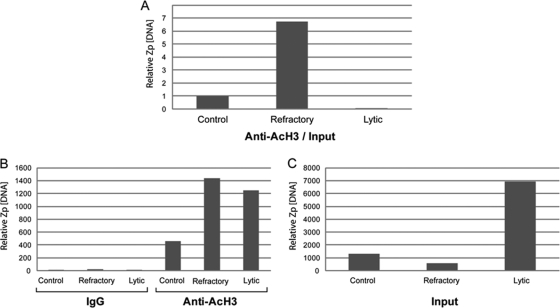
To determine whether one subpopulation of cells responded with chromatin modification and another subpopulation was unresponsive following application of a lytic cycle-inducing stimulus, we examined whether all cells exposed to NaB, an HDACi, manifested hyperacetylation of histones. Western blots of total cell extracts from untreated, sorted refractory cells and sorted lytic cells probed with an anti-acetylated histone H3 (K9,K14) antibody (Ac-H3) demonstrated an increase in global Ac-H3 in both refractory and lytic cells compared to untreated cells (data not shown). We also examined whether the lack of response of refractory cells was due to a failure to modify chromatin at the promoter (Zp) of the BZLF1 gene, whose product is the EBV lytic cycle switch protein ZEBRA. Chromatin immunoprecipitation (ChIP) with an antibody to acetylated H3 was performed using lysates from untreated, sorted refractory cells or sorted lytic HH514-16 cells (Fig. 2). Phosphonoacetic acid (PAA) was used to block lytic viral DNA replication (40) to reduce the background of newly synthesized viral DNA. After correcting for input, the amount of Zp immunoprecipitated with anti-acetylated H3 antibody from refractory cells was substantially greater than the amount recovered from untreated or lytic cells (Fig. 2A). Without correction, increased amounts of Zp were immunoprecipitated in both refractory and lytic cells following treatment with NaB compared to untreated cells (Fig. 2B). Thus, treatment with NaB induced histone hyperacetylation at Zp in refractory cells despite a failure to induce lytic activation. The relative low level of Zp immunoprecipitated from lytic cells following correction for input likely resulted from the presence of newly replicated viral genomes in the input due to a failure of PAA to completely block viral DNA synthesis (Fig. 2C). We previously demonstrated by ChIP that Zp becomes hyperacetylated in cell lines that are not lytically induced by HDAC inhibitors (10). Taken together, these results demonstrate that the failure of NaB to activate the lytic cycle in refractory cells was not due to a lack of response to the HDAC inhibitor in these cells, either globally or at Zp.
FIG. 2.
HDAC inhibition results in increased hyperacetylation of histone H3 associated with Zp in both refractory and lytic HH514-16 cells. (A, B, C) Chromatin immunoprecipitation was performed on untreated, refractory, and lytic cells using antibody to acetylated H3 after treating HH514-16 cells with NaB for 24 h in the presence of phosphonoacetic acid. The amount of Zp immunoprecipitated by anti-acetylated H3 is shown after correction (A) and before correction (B) for the total level of input DNA present in each sample (C). Immunoprecipitation of Zp by preimmune IgG is shown as a control (B).
HDAC inhibitors induce specific changes in cellular gene expression in HH514-16 cells.
As an epigenetic modifier, treatment with NaB results in considerable changes in gene expression (3, 20, 29). To identify potential target genes that may contribute to a lytic or refractory response, we examined early changes in cellular gene expression following treatment of HH514-16 cells with HDAC inhibitors. Since BZLF1 transcripts become resistant to CHX between 4 and 6 h and can be detected between 5 and 8 h after treatment with NaB or TSA in this cell line (10, 56), we analyzed cellular gene expression changes at 3 and 6 h after treatment with NaB or TSA using Affymetrix U133 Plus 2.0 human genome arrays. Substantial changes in cellular gene expression were observed at both 3 and 6 h after treatment with either NaB or TSA (Table 1). A subset of genes, reproducibly identified by array analysis, was selected for validation using quantitative RT-PCR (qRT-PCR) (Table 2). The criteria used to select these genes included either substantial changes in expression level or potential association with either the EBV life cycle or oncogenesis. Changes in gene expression were reproducible, whether assessed by array or qRT-PCR analysis. The degrees of alteration in RNA levels identified by the techniques were comparable. Genes exhibiting an increase in expression level included the IER3, Stat3, Fos, SEPP1, and C1ORF38 genes. Genes with decreased transcript levels in treated cells relative to untreated controls included the C13ORF25 (or MIRHG1) and ZFP91 genes. Comparison of array or qRT-PCR data showed that increases or decreases in transcript levels progressed in a time-dependent manner. These experiments identified a subset of genes in HH514-16 cells that exhibited altered expression in response to HDAC inhibitors at early times that preceded detectable EBV lytic activation.
TABLE 1.
Number of probe sets increased or decreased on Affymetrix U133 Plus 2.0 arrays
| HDAC inhibitor and treatment time (h) | No. of probe sets that: |
|||||
|---|---|---|---|---|---|---|
| Increased |
Decreased |
|||||
| Total | By ≥10-fold | By ≥2-fold | Total | By ≥10-fold | By ≥2-fold | |
| TSA | ||||||
| 3 | 2,091 | 32 | 877 | 3,548 | 22 | 707 |
| 3a | 3,864 | 26 | 911 | 3,078 | 10 | 437 |
| 6 | 4,097 | 115 | 1,893 | 4,363 | 39 | 1,615 |
| NaB | ||||||
| 3 | 3,733 | 25 | 690 | 2,498 | 0 | 466 |
| 6 | 5,411 | 71 | 1,747 | 2,995 | 15 | 1,024 |
Biological duplicate of 3-hour array experiment for TSA.
TABLE 2.
Fold change in mRNA levels for selected target genes
| Gene | Fold change |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 3-h arraysb |
3-h qRT-PCRc |
6-h arraysb |
6-h qRT-PCRc |
||||||
| NaB | TSA | TSA dupd | NaB | TSA | NaB | TSA | NaB | TSA | |
| IER3 gene | 4.9 | 5.7 | 2.8 | 4.6 | 4.1 | 6.1 | 7.5 | 12.5 | 9.7 |
| Stat3 gene | 2.8 | 3 | 2.8 | 2.2 | 2.6 | 5.6 | 6.1 | 5.1 | 8.1 |
| Fos gene | 18.4a | 18.4a | 9.2 | 6.7 | 11.1 | 9.2a | 8.6 | 10.4 | 12.4 |
| SEPP1 gene | 3.7 | 4.9 | 4.6 | 4.3 | 6 | 9.2 | 11.3 | 12.2 | 16.8 |
| C1ORF38 gene | 4 | 4 | 5.6 | 4.3 | 2.1 | 9.2a | 9.2a | 6.4 | 8.2 |
| C13ORF25 gene | −6.1 | −7 | −5.6 | −4.3 | −4 | −6.5 | −7 | −9.3 | −10.1 |
| ZFP91 gene | −3.5 | −3.7 | −2.3 | −1.4 | −1.1 | −4.9 | −7 | −3 | −2.9 |
Detection of transcript was an absent to present (untreated versus treated) call based on Affymetrix GCOS analysis. All other values were determined as present in both untreated and treated samples.
Pooled biological triplicates.
Mean values of technical triplicates.
TSA dup, biological duplicate of a 3-hour array experiment for TSA.
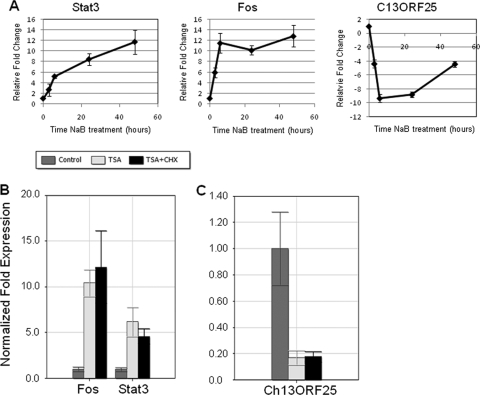
The expression profiles of Stat3, Fos, and C13ORF25 were selected for further investigation based on their potential roles in the viral life cycle. STAT3 has been shown to activate certain EBV latency promoters (5-7), while FOS has been implicated in activation of Zp in response to certain stimuli (36). C13ORF25, now termed MIRHG1, is a microRNA (miRNA) locus often upregulated in hematopoietic and other malignancies (41). Suppression of the miRNA cluster expressed from this locus by HIV-1 is required for efficient viral replication (51). The transcript levels of Stat3, Fos, and C13ORF25 were evaluated by qRT-PCR (Fig. 3A). Stat3 exhibited a continuous increase in transcript levels following treatment with NaB, with mean increases of 2.7-fold (3 h), 5.2-fold (6 h), 8.4-fold (24 h), and 11.7-fold (48 h) compared to untreated cells. The transcript levels of Fos changed rapidly, increasing to 6-fold at 3 h and 11.5-fold at 6 h, before leveling off at 10.2-fold at 24 h and 12.9-fold at 48 h postinduction. C13ORF25 transcript levels decreased 4.4-fold by 3 h and 9.4-fold by 6 h. The reduction in C13ORF25 transcript levels reversed course over time; this transcript was reduced 8.8-fold at 24 h and 4.4-fold at 48 h. Thus, treatment of cells with HDAC inhibitors results in specific temporal changes in cellular gene expression.
FIG. 3.
Kinetics of expression for selected cellular genes induced by treatment of HH514-16 cells with HDAC inhibitors NaB and trichostatin A (TSA). (A) Total RNA was extracted from cells harvested 3, 6, 24, or 48 h following treatment with NaB. qRT-PCR data for Stat3, Fos, and C13ORF25 are shown relative to untreated controls using the ΔΔCT method. Each data point represents the mean of biological triplicates ± 1 standard deviation. (B, C) Total RNA was extracted from HH514-16 cells at 3 h following treatment with either TSA or TSA and CHX. Total RNA from untreated cells was used as a control. qRT-PCR was performed as for panel A.
Since the changes in Stat3, Fos, and Ch13ORF25 transcripts were observed at the earliest time point (3 h) after treatment, we tested whether the associated genes were responding in an immediate early fashion or if they required prior protein synthesis. HH514-16 cells were untreated or treated with the HDAC inhibitor TSA or TSA and cycloheximide (CHX) and harvested 3 h after treatment (Fig. 3B, C). Both Stat3 and Fos transcripts were increased in TSA-treated cells in the presence of CHX, indicating that the increased expression of these genes was occurring independent of new protein synthesis (Fig. 3B). The decrease in Ch13ORF25 transcript was also an immediate early response (Fig. 3C). Under the same conditions, TSA and NaB induction of the viral BZLF1 and BRLF1 lytic activator genes required de novo protein synthesis (56).
Refractory and lytic cell populations exhibit different changes in cellular gene expression.
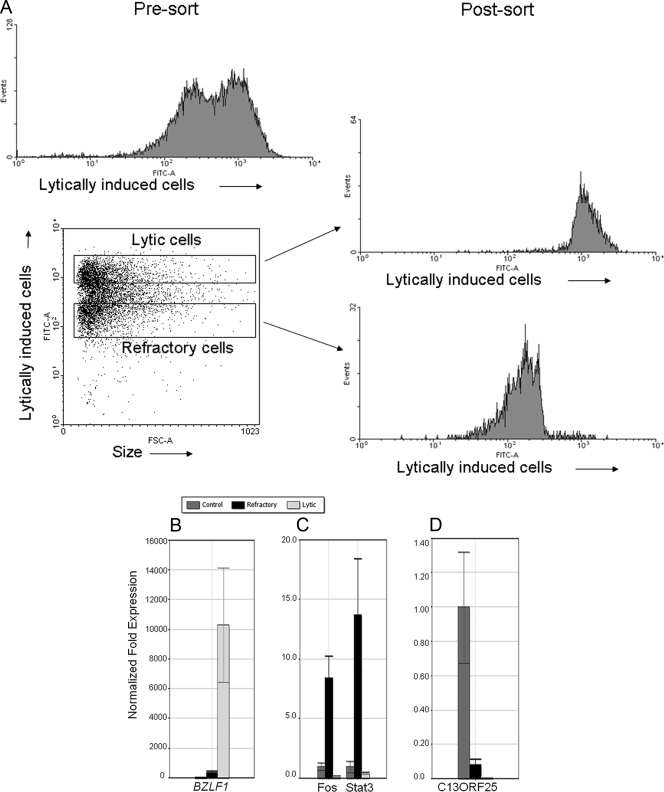
An issue crucial to the understanding of processes that occur in cells upon exposure to lytic cycle-inducing agents is whether observed changes in gene expression occur in all cells or preferentially in the refractory or lytic population. Genes required for lytic activation may fail to be activated in refractory cells. Alternatively, genes that actively repress or reduce the susceptibility to lytic cycle induction may be upregulated in refractory cells. To address this question, HH514-16 cells treated with NaB were separated into refractory and lytic subpopulations. Cells were reexamined by FACS to confirm the purity of each subpopulation (Fig. 4A). Total RNA isolated from refractory, lytic, or untreated HH514-16 cells was used for qRT-PCR analysis using gene-specific primers. Expression of RNA from BZLF1 was highly upregulated in the lytic cell population relative to refractory or untreated cells (Fig. 4B). This result confirmed that there was an effective separation of refractory and lytic cells. By contrast, Stat3 and Fos transcripts were preferentially upregulated in the refractory population. Stat3 expression was increased 12.7-fold, while Fos was increased 8.3-fold compared to untreated cells (Fig. 4C). In comparison, Stat3 was decreased 2-fold while Fos was reduced 4-fold in lytic cells. The level of transcripts from C13ORF25 was reduced approximately 11-fold in refractory cells and 100-fold in lytic cells relative to untreated cells (Fig. 4D).
FIG. 4.
HH516-16 cells refractory to lytic activation following treatment with HDAC inhibitors exhibit unique alterations in cellular gene expression. (A) HH514-16 cells treated with NaB for 48 h followed by incubation with EBV-seropositive human serum from a healthy donor and FITC-conjugated anti-human IgG were sorted into lytic and refractory populations using a FACSVantage cell sorter. Gating strategy for lytic and refractory cells as well as populations of lytic and refractory cells obtained after the sort is shown. (B, C, D) Total RNA was extracted from untreated cells and sorted refractory and sorted lytic HH514-16 cells after treatment with NaB for 48 h. Levels of RNA measured by qRT-PCR relative to untreated controls are shown for BZLF1 (B), Stat3 and Fos (C), and C13ORF25 (D).
Elevated levels of STAT3 protein are present in the refractory population.
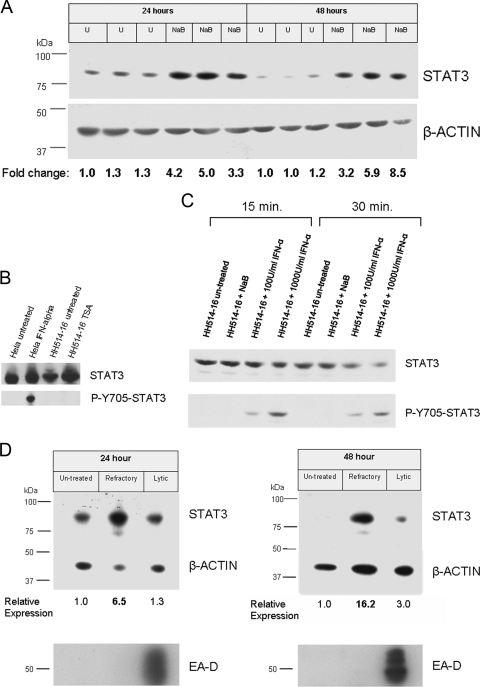
STAT3 was of particular interest as it has previously been linked to the activation of EBV latent gene promoters (5-7). STAT3 activates LMP1 (5, 7), a latent EBV protein that has been shown to block induction of the lytic cycle (45). In addition, LMP1 reciprocally activates Stat3 (5, 31, 48). STAT3 also activates Qp, one of the viral latency promoters (5, 6). We tested whether a change in the level of expression observed by the arrays and by qRT-PCR translated into a corresponding alteration in protein level. Extracts from untreated- or NaB-treated HH514-16 cells were subjected to immunoblot analysis using an anti-STAT3 monoclonal antibody (Fig. 5A). The levels of STAT3 protein in NaB-treated cells were increased relative to untreated controls at both 24 (average, 3.5-fold) and 48 h (average, 5.3-fold) posttreatment.
FIG. 5.
Elevated levels of nonphosphorylated STAT3 protein occur in refractory cells. (A) HH514-16 cells were untreated (U) or treated with NaB; total cell extracts harvested 24 or 48 h after induction were resolved on an SDS-PAGE gel. The immunoblot was probed with mouse anti-STAT3 or anti-β-actin monoclonal antibody followed by rabbit anti-mouse IgG and detection with 125I-conjugated protein A. STAT3 protein levels were normalized to β-actin, and fold changes were expressed relative to the first control lane of the respective time point (24 or 48 h). (B) STAT3 is unphosphorylated at Y705 in HH514-16 cells after treatment with HDAC inhibitors. HH514-16 cells were untreated or treated with TSA for 24 h, and total cell extracts were resolved on SDS-PAGE gels. STAT3 control cell extracts (Cell Signaling) were used as controls for STAT3-Y705 phosphorylation. Duplicate immunoblots were probed with either anti-STAT3 or anti-phospho-Y705 STAT3 antibodies. ECL Plus was used for detection. (C) STAT3 phosphorylation at Y705 after treatment with IFN-α. HH514-16 cells were untreated or treated with 3 mM NaB, 100 U/ml IFN-α, or 1,000 U/ml IFN-α. Cells were harvested 15 or 30 min after treatment. Immunoblots were probed as for panel B. (D) HH514-16 cells untreated or treated with NaB for 24 or 48 h were sorted into refractory and lytic populations. Total cell extracts were electrophoresed on 8% SDS-PAGE gels, and immunoblots were probed as for panel A. STAT3 protein levels were normalized to β-actin and expressed relative to untreated cells. Duplicate immunoblots probed with anti-EAD (a viral lytic protein) antibodies demonstrated effective cell separation.
Phosphorylation of STAT3 was not detected at either Y705 or S727 after treatment with HDAC inhibitor TSA or NaB for 24 h (Fig. 5B and data not shown); thus, the majority of STAT3 protein was present in the unphosphorylated form (UP-STAT3). HH514-16 cells were treated with alpha interferon (IFN-α) to determine if STAT3 could be phosphorylated at Y705 in this cell line or if this pathway was defective. Phosphorylation of STAT3 Y705 was detected 15 min and 30 min after treatment with IFN-α (Fig. 5C), indicating that STAT3 can be modified at this site in HH514-16 cells. Phosphorylated Y705-STAT3 was not detected in HH514-16 cells from 15 min to 6 h after treatment with NaB (Fig. 5C and data not shown). Thus, latently infected cells exhibited higher levels of expression of STAT3 protein, predominantly in the unphosphorylated state, following exposure to NaB, a prototype HDAC inhibitor lytic cycle-inducing agent.
Since the increase of Stat3 transcript occurred primarily in refractory cells (Fig. 4C), we examined whether increased levels in STAT3 protein were correspondingly present in this population. Western blot analysis of cell extracts from untreated, refractory, and lytic HH514-16 cells demonstrated substantial increases in STAT3 protein in the refractory population relative to the untreated cells at 24 (6.5-fold) and 48 h (16.2-fold) after treatment with NaB (Fig. 5D). STAT3 protein was not significantly upregulated in the lytic subpopulation. The EBV lytic protein EA-D was explicitly upregulated in lytic cells (Fig. 5D).
Elevated levels of LMP1 are present in the lytic cell population.
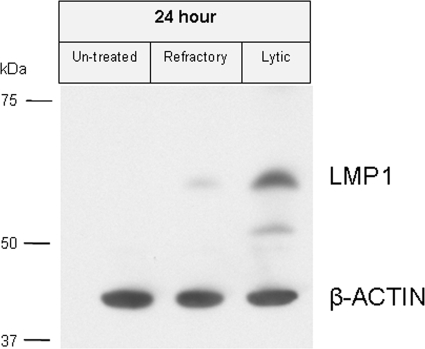
It has been demonstrated that STAT3 can activate EBV LMP1. LMP1 is a viral protein which promotes cell survival during latency (5, 13). LMP1 has been shown to inhibit EBV lytic cycle induction (45). While LMP1 is not normally expressed in P3HR-1 derived cell lines (including HH514-16 cells) during latency due to a deletion of the viral EBNA2 gene, activation of LMP1 expression is known to occur in P3HR-1 cells treated with HDAC inhibitors (42). Since refractory cells showed increased expression of STAT3, we considered the possibility that LMP1 might be upregulated in the refractory population, thereby helping to maintain a latent phenotype. However, as shown in Fig. 6, LMP1 was markedly upregulated only in the lytic population. This result suggests that the refractory state was not attributable to LMP1 expression.
FIG. 6.
LMP1 is induced in the lytic cell population. HH514-16 cells untreated or treated with NaB for 24 h were sorted into refractory and lytic populations. Total cell extracts were electrophoresed on 8% SDS-PAGE gels. Immunoblots of the sorted HH514-16 cell extracts were probed with anti-LMP1 and anti-β-actin antibodies. ECL Plus was used for detection.
High levels of expression of STAT3 characterize cells refractory to lytic cycle induction.
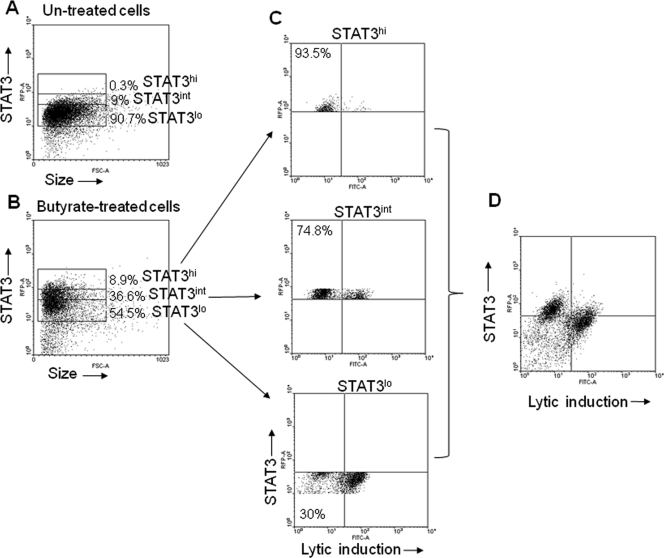
While our previous results indicated that STAT3 level was preferentially increased in the refractory population at the transcriptional and translational levels, it was unclear what fraction of the individual refractory cells expressed higher levels of STAT3. We therefore examined the levels of STAT3 in cells treated with NaB using FACS (Fig. 7A to D). We gated on three groups of cells based on the relative levels of expression of STAT3, high (STAT3hi), intermediate (STAT3int), and low (STAT3lo). In HH514-16 cells treated with NaB for 48 h (Fig. 7B), 8.9% of cells expressed STAT3hi, 36.6% STAT3int, and 54.5% STAT3lo, compared to 0.3% (STAT3hi), 9% (STAT3int), and 90.7% (STAT3lo) in untreated cells (Fig. 7A). When NaB-treated HH514-16 cells were examined simultaneously for STAT3 expression and EBV lytic cycle induction, higher levels of STAT3 were characteristic of the refractory cell population (Fig. 7D). Approximately 30% of cells expressing STAT3lo were refractory to lytic cycle induction compared to 74.8% of STAT3int cells, and 93.5% of cells expressing STAT3hi (Fig. 7C). Thus, with increasing levels of expression of STAT3, there was a progressive increase in refractoriness to lytic cycle induction. These results indicate that high levels of STAT3 are strongly correlated with cells refractory to lytic cycle induction by NaB.
FIG. 7.
Elevated levels of STAT3 protein are characteristic of the refractory state. (A, B) HH514-16 cells untreated (A) or treated with NaB (B) for 48 h were stained with Texas Red-conjugated anti-STAT3 antibody, EBV-seropositive human serum, and FITC-conjugated anti-human IgG. Cells expressing low (STAT3lo), intermediate (STAT3int), or high (STAT3hi) levels of STAT3 were gated on, and percentages of each are shown for untreated (A) and treated (B) cells. (C) Responsiveness of STAT3lo, STAT3int, and STAT3hi cells to lytic cycle induction. (D) FACS plot demonstrating overall lytic induction relative to STAT3 expression.
Differential expression of cellular cytokines in refractory and lytic populations.
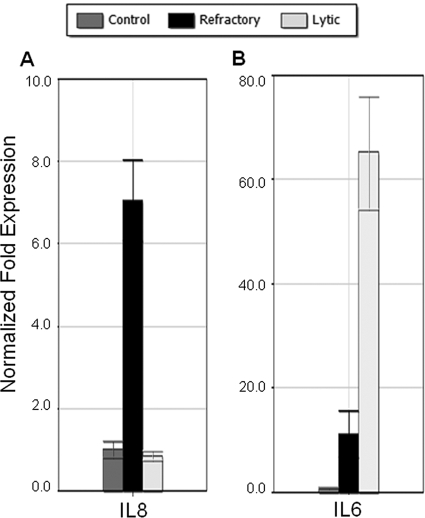
In the classical STAT3 pathway, phosphorylated STAT3 (P-STAT3) dimerizes and activates downstream genes. Recently it has been shown that unphosphorylated STAT3 can also play a role in transcriptional activation, influencing the expression of a distinct set of genes (54). Using preliminary data from sorted cell arrays as a guide, we sought to identify potential targets of UP-STAT3 that exhibit increased levels of expression in the refractory population. By qRT-PCR, we found that expression of the IL-8 gene, a known UP-STAT3 target gene, was specifically upregulated in refractory cells (Fig. 8A).
FIG. 8.
Differential cytokine gene expression in lytic and refractory cells. (A, B) HH514-16 cells untreated or treated with NaB for 48 h were sorted into refractory and lytic populations. Levels of RNA measured by qRT-PCR relative to untreated controls are shown for IL-8 (A) and IL-6 (B).
One concern regarding these observed expression patterns was the ability of EBV lytic gene activation to induce host cell shutoff through increased mRNA turnover (46). To investigate whether all cellular transcripts in lytic cells were suppressed, we sought to identify cellular genes that were increased in expression in lytic cells. In experiments to be presented separately, we have found that expression of specific cellular genes is upregulated in the subpopulation undergoing EBV lytic reactivation (data not shown). One example of the genes identified is the IL-6 gene (Fig. 8B). This is in agreement with work on Kaposi's sarcoma virus (KSHV), where it has been shown that the IL-6 transcript is resistant to the cell shutoff mechanism induced by the KSHV lytic gene product SOX (21). Thus, EBV-induced host cell shutoff appears to be selective in terms of the cellular transcripts affected. These results indicate that differential changes in cellular gene expression occur in refractory and lytic cells following treatment with a lytic cycle-inducing agent.
DISCUSSION
The refractory phenomenon is of general biological interest. Do permanent genetic changes mark cells that are refractory to lytic cycle induction? Are there features that distinguish cells that fail to respond from those that respond to lytic cycle-inducing stimuli? Are such distinguishing features characterized by changes in cellular gene expression? The refractory phenomenon requires acknowledging that all cells within a population do not necessarily respond uniformly to inducing stimuli to which all cells are exposed. Rather, subpopulations exist that respond to stimuli in distinct ways. Observations of changes in the total population (for example, in gene expression) can be misleading, as the source of those changes is not always clear. Here, using a strategy to separate lytic from refractory cells (2), we show that the refractory and lytic subpopulations exhibit different changes in cellular gene expression in response to the lytic cycle-inducing agent NaB. We provide novel insights into cellular gene expression unique to the refractory population (Fig. 4, 5, and 7) by demonstrating that UP-STAT3 is a marker for cells refractory to EBV lytic induction using the HH514-16 cell line as a model system.
Relationship of UP-STAT3 expression to the refractory state.
Identified as a critical oncoprotein in a wide variety of cancers (1, 12), STAT3 acts as a prosurvival, antiapoptotic transcription factor via its phosphorylated “active” form (34). Recent evidence suggests that unphosphorylated STAT3 may also exert oncogenic effects (53-55). While increased levels of STAT3 transcript and protein have been observed in tumor tissues (23, 32), it is not yet known whether STAT3 expression characterizes tumor or stromal cells (53).
Dimerized P-STAT3 activates transcription of UP-STAT3 and thereby creates a positive autoregulatory loop (53). However, since phosphorylated STAT3 is not detected following treatment with HDAC inhibitors (Fig. 5B, C; data not shown), increased de novo expression of UP-STAT3 in refractory BL cells most likely occurs through a distinct mechanism. STAT3 has widespread impact on the control of cellular gene expression (1, 34). While the “classical” STAT3 pathway requires phosphorylation of STAT3 (34), recent studies have emphasized the biological role of UP-STAT3. UP-STAT3 can dimerize (4), shuttle between nuclear and cytoplasmic compartments (37), and play a key role in transcriptional activation (53-55) and in non-transcription-related functions (19, 39). By binding to unphosphorylated NF-κB and recruiting it to the nucleus, UP-STAT3 activates a distinct set of genes via κB elements within their promoters (24, 54, 58).
In support of a functional role of UP-STAT3 as a transcriptional activator, our data demonstrate that the transcript of IL-8 is specifically increased in the refractory population (Fig. 8A), Our preliminary analysis of sorted cell arrays indicates that other genes thought to be upregulated by UP-STAT3, such as the RANTES (CCL5) and ICAM1(54) genes, are indeed elevated in refractory cells (data not shown). Further experiments will be needed to determine if the observed expression changes in these genes are indeed a direct result of upregulation of UP-STAT3.
Based on our findings, we cannot yet conclude that UP-STAT3 plays a direct role in the refractory phenomenon in HH514-16 cells. The upregulation of UP-STAT3 may lead to expression of downstream genes that repress components required for lytic activation, either cellular or viral. Alternatively, UP-STAT3 may directly repress the expression of necessary genes. A role of STAT3 in transcriptional repression has been demonstrated previously (28). Another possibility is that the expression of UP-STAT3 is an indicator of the unique responses occurring in the refractory population, potentially downstream of key events that control the refractory state. Additional efforts are ongoing to elucidate the nature of the relationship between UP-STAT3 expression and the refractory phenomenon.
Relationship of findings to EBV-induced host cell shutoff.
While STAT3 is actively upregulated in refractory cells, no increase in STAT3 expression is observed in lytic cells. While EBV-induced host cell shutoff is suggested to act by increasing cellular mRNA turnover in general, we found that several transcripts, including IL-6, are specifically increased in lytic cells (Fig. 8B and data not shown). This suggests that the shutoff mechanism is to some degree selective. Unlike EBV early lytic genes, induction of Stat3 by HDAC inhibitors does not require prior protein synthesis (Fig. 3B). In addition, induction of Stat3 is detectable prior to induction of EBV lytic transcripts. Therefore, a viral host cell shutoff mechanism would have to be extensive enough to affect already accrued Stat3 mRNA in addition to blocking ongoing accumulation of Stat3 transcript. While we cannot rule out that Stat3 transcripts are affected by EBV host cell shutoff, the idea that Stat3 is specifically upregulated in the refractory population independent of any host cell shutoff mechanism is a more tenable conclusion.
The refractory phenomenon is relevant to oncolytic therapy for EBV.
Since EBV is present in tumor cells in cancers associated with the virus, virus-targeted therapies are an intriguing option (11, 27). Although induction of the lytic cycle in conjunction with ganciclovir has shown promising results, a major drawback to this strategy is the resistance of cells to lytic activation (15, 16, 52). Bystander killing via phosphorylated nucleotide analogue exchange between cells exerts limited effects (18, 52). While use of radiolabeled nucleoside analogs may enhance localized destruction of tumor cells (18), any strategy that depends on prodrug activation by viral kinases would benefit from an enhancement in the number of cells induced into the lytic cycle. Understanding why most cells do not respond lytically could be paramount in making these strategies more effective. Also, the ability to correlate responsiveness to oncolytic therapy with expression of specific markers such as UP-STAT3 may be of therapeutic consequence.
Refractoriness to EBV lytic induction may provide clues to resistance to chemotherapeutic agents.
The phenomenon of a state refractory to viral lytic activation is a specific example of a more general enigma that is just beginning to be addressed in cancer therapy research. Seemingly identical cancer cells respond differently to the same drug. New techniques to observe variation in protein dynamics on a broad scale have led to identification of genes that may enhance a cell's ability to survive chemotherapeutic treatment (8). It has been suggested that STAT3 is an important marker for drug resistance (1). Increased expression of STAT3 has been found in several cisplatin-resistant cell lines (30). Downstream targets of UP-STAT3, such as IL-8, can also play antiapoptotic, proproliferative roles (35). One possible scenario is that cells refractory to lytic induction, via unique patterns of cellular gene expression, are those protected from cell cycle arrest induced by NaB through activation of p21 (49, 50). The refractory state is manifest in a unique population of cells that exhibit different cellular responses than do lytic cells exposed to the same stimulus. Identifying and comparing the molecular changes that occur in cells refractory or susceptible to EBV lytic induction will not only be an important step in developing a better understanding of the regulation of the EBV latent to lytic switch but may also contribute to understanding differential responses to chemotherapeutic agents.
Acknowledgments
This study was supported by NIH grants CA16038 and CA12055 (G.M.); K08 AI062732, K12 HD001401, and 1UL1RR024139-02 (S.B.-M.); 5 T32 GM07223 and the Anna Fuller Fellowship for Cancer Research (D.D.); the Yale Center of Excellence in Molecular Hematology and NIH 1 U24 NS051869 Neuroscience Microarray Consortium.
We thank the Keck lab at Yale University and Shrikant Mane and Sheila Westman for their generous assistance in microarray analysis.
Footnotes
Published ahead of print on 4 November 2009.
REFERENCES
- 1.Barre, B., A. Vigneron, N. Perkins, I. B. Roninson, E. Gamelin, and O. Coqueret. 2007. The STAT3 oncogene as a predictive marker of drug resistance. Trends Mol. Med. 13:4-11. [DOI] [PubMed] [Google Scholar]
- 2.Bhaduri-McIntosh, S., and G. Miller. 2006. Cells lytically infected with Epstein-Barr virus are detected and separable by immunoglobulins from EBV-seropositive individuals. J. Virol. Methods 137:103-114. [DOI] [PubMed] [Google Scholar]
- 3.Bolden, J. E., M. J. Peart, and R. W. Johnstone. 2006. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 5:769-784. [DOI] [PubMed] [Google Scholar]
- 4.Braunstein, J., S. Brutsaert, R. Olson, and C. Schindler. 2003. STATs dimerize in the absence of phosphorylation. J. Biol. Chem. 278:34133-34140. [DOI] [PubMed] [Google Scholar]
- 5.Chen, H., L. Hutt-Fletcher, L. Cao, and S. D. Hayward. 2003. A positive autoregulatory loop of LMP1 expression and STAT activation in epithelial cells latently infected with Epstein-Barr virus. J. Virol. 77:4139-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, H., J. M. Lee, Y. Wang, D. P. Huang, R. F. Ambinder, and S. D. Hayward. 1999. The Epstein-Barr virus latency BamHI-Q. promoter is positively regulated by STATs and Zta interference with JAK/STAT activation leads to loss of BamHI-Q. promoter activity. Proc. Natl. Acad. Sci. USA 96:9339-9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, H., J. M. Lee, Y. Zong, M. Borowitz, M. H. Ng, R. F. Ambinder, and S. D. Hayward. 2001. Linkage between STAT regulation and Epstein-Barr virus gene expression in tumors. J. Virol. 75:2929-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen, A. A., N. Geva-Zatorsky, E. Eden, M. Frenkel-Morgenstern, I. Issaeva, A. Sigal, R. Milo, C. Cohen-Saidon, Y. Liron, Z. Kam, L. Cohen, T. Danon, N. Perzov, and U. Alon. 2008. Dynamic proteomics of individual cancer cells in response to a drug. Science 322:1511-1516. [DOI] [PubMed] [Google Scholar]
- 9.Countryman, J., H. Jenson, R. Seibl, H. Wolf, and G. Miller. 1987. Polymorphic proteins encoded within BZLF1 of defective and standard Epstein-Barr viruses disrupt latency. J. Virol. 61:3672-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Countryman, J. K., L. Gradoville, and G. Miller. 2008. Histone hyperacetylation occurs on promoters of lytic cycle regulatory genes in Epstein-Barr virus-infected cell lines which are refractory to disruption of latency by histone deacetylase inhibitors. J. Virol. 82:4706-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crawford, D. H. 2001. Biology and disease associations of Epstein-Barr virus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:461-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darnell, J. E. 2005. Validating Stat3 in cancer therapy. Nat. Med. 11:595-596. [DOI] [PubMed] [Google Scholar]
- 13.D'Souza, B., M. Rowe, and D. Walls. 2000. The bfl-1 gene is transcriptionally upregulated by the Epstein-Barr virus LMP1, and its expression promotes the survival of a Burkitt's lymphoma cell line. J. Virol. 74:6652-6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Guindy, A., L. Heston, H. J. Delecluse, and G. Miller. 2007. Phosphoacceptor site S173 in the regulatory domain of Epstein-Barr Virus ZEBRA protein is required for lytic DNA replication but not for activation of viral early genes. J. Virol. 81:3303-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng, W. H., G. Hong, H. J. Delecluse, and S. C. Kenney. 2004. Lytic induction therapy for Epstein-Barr virus-positive B-cell lymphomas. J. Virol. 78:1893-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng, W. H., B. Israel, N. Raab-Traub, P. Busson, and S. C. Kenney. 2002. Chemotherapy induces lytic EBV replication and confers ganciclovir susceptibility to EBV-positive epithelial cell tumors. Cancer Res. 62:1920-1926. [PubMed] [Google Scholar]
- 17.Fields, B. N., D. M. Knipe, and P. M. Howley. 1996. Fields virology, 3rd ed. Lippincott-Raven Press, Philadelphia, PA.
- 18.Fu, D. X., Y. Tanhehco, J. Chen, C. A. Foss, J. J. Fox, J. M. Chong, R. F. Hobbs, M. Fukayama, G. Sgouros, J. Kowalski, M. G. Pomper, and R. F. Ambinder. 2008. Bortezomib-induced enzyme-targeted radiation therapy in herpesvirus-associated tumors. Nat. Med. 14:1118-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao, S. P., and J. F. Bromberg. 2006. Touched and moved by STAT3. Sci. STKE 2006:pe30. [DOI] [PubMed] [Google Scholar]
- 20.Glaser, K. B., M. J. Staver, J. F. Waring, J. Stender, R. G. Ulrich, and S. K. Davidsen. 2003. Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol. Cancer Ther. 2:151-163. [PubMed] [Google Scholar]
- 21.Glaunsinger, B., and D. Ganem. 2004. Highly selective escape from KSHV-mediated host mRNA shutoff and its implications for viral pathogenesis. J. Exp. Med. 200:391-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gradoville, L., D. Kwa, A. El-Guindy, and G. Miller. 2002. Protein kinase C-independent activation of the Epstein-Barr virus lytic cycle. J. Virol. 76:5612-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grandis, J. R., S. D. Drenning, Q. Zeng, S. C. Watkins, M. F. Melhem, S. Endo, D. E. Johnson, L. Huang, Y. He, and J. D. Kim. 2000. Constitutive activation of Stat3 signaling abrogates apoptosis in squamous cell carcinogenesis in vivo. Proc. Natl. Acad. Sci. USA 97:4227-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagihara, K., T. Nishikawa, Y. Sugamata, J. Song, T. Isobe, T. Taga, and K. Yoshizaki. 2005. Essential role of STAT3 in cytokine-driven NF-kappaB-mediated serum amyloid A gene expression. Genes Cells 10:1051-1063. [DOI] [PubMed] [Google Scholar]
- 25.Hardwick, J. M., P. M. Lieberman, and S. D. Hayward. 1988. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J. Virol. 62:2274-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heston, L., M. Rabson, N. Brown, and G. Miller. 1982. New Epstein-Barr virus variants from cellular subclones of P3J-HR-1 Burkitt lymphoma. Nature 295:160-163. [DOI] [PubMed] [Google Scholar]
- 27.Israel, B. F., and S. C. Kenney. 2003. Virally targeted therapies for EBV-associated malignancies. Oncogene 22:5122-5130. [DOI] [PubMed] [Google Scholar]
- 28.Ivanov, V. N., A. Bhoumik, M. Krasilnikov, R. Raz, L. B. Owen-Schaub, D. Levy, C. M. Horvath, and Z. Ronai. 2001. Cooperation between STAT3 and c-jun suppresses Fas transcription. Mol. Cell 7:517-528. [DOI] [PubMed] [Google Scholar]
- 29.Joseph, J., G. Mudduluru, S. Antony, S. Vashistha, P. Ajitkumar, and K. Somasundaram. 2004. Expression profiling of sodium butyrate (NaB)-treated cells: identification of regulation of genes related to cytokine signaling and cancer metastasis by NaB. Oncogene 23:6304-6315. [DOI] [PubMed] [Google Scholar]
- 30.Kato, K., M. Nomoto, H. Izumi, T. Ise, S. Nakano, Y. Niho, and K. Kohno. 2000. Structure and functional analysis of the human STAT3 gene promoter: alteration of chromatin structure as a possible mechanism for the upregulation in cisplatin-resistant cells. Biochim. Biophys. Acta 1493:91-100. [DOI] [PubMed] [Google Scholar]
- 31.Kung, C. P., and N. Raab-Traub. 2008. Epstein-Barr virus latent membrane protein 1 induces expression of the epidermal growth factor receptor through effects on Bcl-3 and STAT3. J. Virol. 82:5486-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lassmann, S., I. Schuster, A. Walch, H. Gobel, U. Jutting, F. Makowiec, U. Hopt, and M. Werner. 2007. STAT3 mRNA and protein expression in colorectal cancer: effects on STAT3-inducible targets linked to cell survival and proliferation. J. Clin. Pathol. 60:173-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lefever, S., J. Vandesompele, F. Speleman, and F. Pattyn. 2009. RTPrimerDB: the portal for real-time PCR primers and probes. Nucleic Acids Res. 37:D942-D945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levy, D. E., and C. K. Lee. 2002. What does Stat3 do? J. Clin. Invest. 109:1143-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, A., S. Dubey, M. L. Varney, B. J. Dave, and R. K. Singh. 2003. IL-8 directly enhanced endothelial cell survival, proliferation, and matrix metalloproteinases production and regulated angiogenesis. J. Immunol. 170:3369-3376. [DOI] [PubMed] [Google Scholar]
- 36.Liang, C. L., J. L. Chen, Y. P. Hsu, J. T. Ou, and Y. S. Chang. 2002. Epstein-Barr virus BZLF1 gene is activated by transforming growth factor-beta through cooperativity of Smads and c-Jun/c-Fos proteins. J. Biol. Chem. 277:23345-23357. [DOI] [PubMed] [Google Scholar]
- 37.Liu, L., K. M. McBride, and N. C. Reich. 2005. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-alpha3. Proc. Natl. Acad. Sci. USA 102:8150-8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore, S. M., J. S. Cannon, Y. C. Tanhehco, F. M. Hamzeh, and R. F. Ambinder. 2001. Induction of Epstein-Barr virus kinases to sensitize tumor cells to nucleoside analogues. Antimicrob. Agents Chemother. 45:2082-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng, D. C., B. H. Lin, C. P. Lim, G. Huang, T. Zhang, V. Poli, and X. Cao. 2006. Stat3 regulates microtubules by antagonizing the depolymerization activity of stathmin. J. Cell Biol. 172:245-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nyormoi, O., D. A. Thorley-Lawson, J. Elkington, and J. L. Strominger. 1976. Differential effect of phosphonoacetic acid on the expression of Epstein-Barr viral antigens and virus production. Proc. Natl. Acad. Sci. USA 73:1745-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ota, A., H. Tagawa, S. Karnan, S. Tsuzuki, A. Karpas, S. Kira, Y. Yoshida, and M. Seto. 2004. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 64:3087-3095. [DOI] [PubMed] [Google Scholar]
- 42.Park, J. H., and D. V. Faller. 2002. Epstein-Barr virus latent membrane protein-1 induction by histone deacetylase inhibitors mediates induction of intercellular adhesion molecule-1 expression and homotypic aggregation. Virology 303:345-363. [DOI] [PubMed] [Google Scholar]
- 43.Pearson, G. R., B. Vroman, B. Chase, T. Sculley, M. Hummel, and E. Kieff. 1983. Identification of polypeptide components of the Epstein-Barr virus early antigen complex with monoclonal antibodies. J. Virol. 47:193-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perrine, S. P., O. Hermine, T. Small, F. Suarez, R. O'Reilly, F. Boulad, J. Fingeroth, M. Askin, A. Levy, S. J. Mentzer, M. Di Nicola, A. M. Gianni, C. Klein, S. Horwitz, and D. V. Faller. 2007. A phase 1/2 trial of arginine butyrate and ganciclovir in patients with Epstein-Barr virus-associated lymphoid malignancies. Blood 109:2571-2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prince, S., S. Keating, C. Fielding, P. Brennan, E. Floettmann, and M. Rowe. 2003. Latent membrane protein 1 inhibits Epstein-Barr virus lytic cycle induction and progress via different mechanisms. J. Virol. 77:5000-5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rowe, M., B. Glaunsinger, D. van Leeuwen, J. Zuo, D. Sweetman, D. Ganem, J. Middeldorp, E. J. Wiertz, and M. E. Ressing. 2007. Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc. Natl. Acad. Sci. USA 104:3366-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 48.Shair, K. H., K. M. Bendt, R. H. Edwards, E. C. Bedford, J. N. Nielsen, and N. Raab-Traub. 2007. EBV latent membrane protein 1 activates Akt, NFkappaB, and Stat3 in B cell lymphomas. PLoS Pathog. 3:e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siavoshian, S., J. P. Segain, M. Kornprobst, C. Bonnet, C. Cherbut, J. P. Galmiche, and H. M. Blottiere. 2000. Butyrate and trichostatin A effects on the proliferation/differentiation of human intestinal epithelial cells: induction of cyclin D3 and p21 expression. Gut 46:507-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sowa, Y., T. Orita, S. Minamikawa-Hiranabe, T. Mizuno, H. Nomura, and T. Sakai. 1999. Sp3, but not Sp1, mediates the transcriptional activation of the p21/WAF1/Cip1 gene promoter by histone deacetylase inhibitor. Cancer Res. 59:4266-4270. [PubMed] [Google Scholar]
- 51.Triboulet, R., B. Mari, Y. L. Lin, C. Chable-Bessia, Y. Bennasser, K. Lebrigand, B. Cardinaud, T. Maurin, P. Barbry, V. Baillat, J. Reynes, P. Corbeau, K. T. Jeang, and M. Benkirane. 2007. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 315:1579-1582. [DOI] [PubMed] [Google Scholar]
- 52.Westphal, E. M., W. Blackstock, W. Feng, B. Israel, and S. C. Kenney. 2000. Activation of lytic Epstein-Barr virus (EBV) infection by radiation and sodium butyrate in vitro and in vivo: a potential method for treating EBV-positive malignancies. Cancer Res. 60:5781-5788. [PubMed] [Google Scholar]
- 53.Yang, J., M. Chatterjee-Kishore, S. M. Staugaitis, H. Nguyen, K. Schlessinger, D. E. Levy, and G. R. Stark. 2005. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res. 65:939-947. [PubMed] [Google Scholar]
- 54.Yang, J., X. Liao, M. K. Agarwal, L. Barnes, P. E. Auron, and G. R. Stark. 2007. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB. Genes Dev. 21:1396-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang, J., and G. R. Stark. 2008. Roles of unphosphorylated STATs in signaling. Cell Res. 18:443-451. [DOI] [PubMed] [Google Scholar]
- 56.Ye, J., L. Gradoville, D. Daigle, and G. Miller. 2007. De novo protein synthesis is required for lytic cycle reactivation of Epstein-Barr virus, but not Kaposi's sarcoma-associated herpesvirus, in response to histone deacetylase inhibitors and protein kinase C agonists. J. Virol. 81:9279-9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoo, C. B., and P. A. Jones. 2006. Epigenetic therapy of cancer: past, present and future. Nat. Rev. Drug Discov. 5:37-50. [DOI] [PubMed] [Google Scholar]
- 58.Yoshida, Y., A. Kumar, Y. Koyama, H. Peng, A. Arman, J. A. Boch, and P. E. Auron. 2004. Interleukin 1 activates STAT3/nuclear factor-kappaB cross-talk via a unique TRAF6- and p65-dependent mechanism. J. Biol. Chem. 279:1768-1776. [DOI] [PubMed] [Google Scholar]