Abstract
The virophage Sputnik is a satellite virus of the giant mimivirus and is the only satellite virus reported to date whose propagation adversely affects its host virus' production. Genome sequence analysis showed that Sputnik has genes related to viruses infecting all three domains of life. Here, we report structural studies of Sputnik, which show that it is about 740 Å in diameter, has a T=27 icosahedral capsid, and has a lipid membrane inside the protein shell. Structural analyses suggest that the major capsid protein of Sputnik is likely to have a double jelly-roll fold, although sequence alignments do not show any detectable similarity with other viral double jelly-roll capsid proteins. Hence, the origin of Sputnik's capsid might have been derived from other viruses prior to its association with mimivirus.
Mimivirus is the largest virus known to date and has a 1.2-Mbp genome. It was discovered in a British water tower while searching for the cause of a hospital-acquired pneumonia outbreak (14). Although mimivirus' natural host is amoeba, it could be a potential human pathogen (2, 7, 8, 15). Recently, a smaller virus named Sputnik was isolated from amoeba infected with mamavirus, a new strain of mimivirus (10). Sputnik utilizes the virus factory formed by mamavirus for replication and cannot reproduce on its own in amoeba. Furthermore, the coinfection of Sputnik with mamavirus reduces the yield of mamavirus by about 70% and causes the formation of many types of defective mamavirus virions.
Sputnik has an 18-kbp, double-stranded, circular, highly AT-rich genome, which is predicted to encode 21 proteins ranging from 88 to 779 amino acids in size. Of these 21 proteins, 13 do not have detectable homologues in current sequence databases. The other eight genes have homologues in viruses whose hosts are from all three domains of life, the Eukarya, Archaea, and Bacteria. The chimeric characteristics of the Sputnik genome implies that it is involved in lateral gene transfer between viruses. It was proposed that Sputnik represents a new family of viruses termed virophage (10).
Because of the mosaic nature of viral genomes and the lack of 16S rRNA for traditional phylogenetic tree analysis, the classification of viruses has been difficult. Recent structural studies of viral capsid proteins have led to the idea of structure-based viral lineages, which classify viruses based on the organization and structure of the viral capsids (3, 9). One of these lineages is the PRD1-adenovirus lineage, which is comprised of icosahedral double-stranded DNA (dsDNA) viruses including adenovirus, bacteriophage PRD1, Sulfolobus turreted icosahedral virus, the marine bacteriophage PM2, and the nucleocytoplasmic large DNA viruses (NCLDVs) such as mimivirus and Paramecium bursaria Chlorella virus 1 (PBCV-1). All these viruses have major capsid proteins (MCPs) whose polypeptides have a similar fold and in some cases, such as the NCLDVs, have significant sequence similarity. The MCP structures of the above-mentioned viruses, excluding mimivirus, have been determined to atomic resolution and were shown to have two consecutive “jelly-roll” domains (double jelly-roll fold) (1, 4, 13, 16, 17). A jelly-roll domain is an antiparallel β barrel consisting of eight β strands named B, C, …, I. The MCPs are organized into “capsomers” that are arranged into hexagonal arrays. Each viral capsomer contains three monomers that have a double jelly-roll fold, resulting in a pseudohexameric shape at the base, appropriate for packing into the hexagonal arrays. However, there are often large insertions in the loops between β strands D and E as well as between strands F and G of each jelly-roll fold (loops “DE” and “FG”). This gives the capsomers a triangular appearance on the surface. The thickness of capsomers is about 75 Å, and the diameter varies between 74 Å and 85 Å.
Here, we report the cryo-electron microscopy (cryoEM) three-dimensional (3D) reconstruction of Sputnik to 10.7-Å resolution. We show that the MCP is organized into a hexagonal surface lattice characterized by a T=27 triangulation number. We also show that the capsomer structure in Sputnik is trimeric and that the MCP structure of PBCV-1 can be fitted into the cryoEM map of Sputnik. Thus, the MCP of Sputnik is probably a double jelly-roll fold as in viruses belonging to the PRD1-adenovirus lineage. However, there is no significant sequence similarity between the MCP of Sputnik and other members of the PRD1-adenovirus lineage, suggesting that Sputnik is a member of a separate branch from the NCLDVs (mimivirus and PBCV-1, etc.).
MATERIALS AND METHODS
Sample preparation.
Acanthamoeba polyphaga cells were coinfected with mamavirus and Sputnik. After most of the amoeba cells were lysed, the culture was passed through a 0.8-μm filter to remove amoeba debris and then through a 0.22-μm filter to remove mamavirus. Sputnik was pelleted by polyethylene glycol (PEG) precipitation and further purified on a CsCl gradient. The virus band was collected, and buffer was exchanged to phosphate-buffered saline (PBS) buffer and concentrated to about 1 mg/ml.
CryoEM.
Sputnik particles were flash-frozen on holey grids in liquid ethane. Images were recorded at a 39K magnification with a CM200 FEG microscope with electron dose levels of approximately 20 e−/Å2. All micrographs were digitized at 6.35 μm pixel−1 by using a Nikon scanner. Individual particle images were boxed, floated, and preprocessed to normalize mean intensities and variances and to remove linear background gradients by using the program ROBEM (http://cryoem.ucsd.edu/programs.shtm). Contrast transfer function (CTF) parameters were determined and phases were flipped by using the program CTFIT from the EMAN package (12). An initial model was constructed with EMAN using assigned 2-, 3-, and 5-fold views and was refined with EMAN assuming icosahedral symmetry. The number of particles incorporated into the final reconstruction was 6,780, giving a final resolution of 10.7 Å.
Mass spectroscopy.
A 50-μl aqueous sample of Sputnik was mixed with 300 μl methanol and 100 μl chloroform. Subsequently, 200 μl water was added and mixed, followed by 5 min of centrifugation at 10,000 × g to separate phases. The lower phase was analyzed by size-exclusion chromatography (SW2000 XL; Tosoh Biosciences) with a buffer composed of chloroform, methanol, and 1% formic acid in water (4/4/1, vol/vol/vol) at 40°C. The major peak was then analyzed by electron spray ionization mass spectroscopy (19). The magnitudes of individual fractions were compared to a standard lipid sample (dipalmitoyl phospatidyl choline [DPPC]) in order to determine the amount of lipid in the sample. A chloroform blank was used as a negative control.
The percentage of weight that the putative lipid membrane would represent as a fraction of the weight of the whole virus was calculated as follows. The mean diameter of the virus was taken as the weighted average of the 5-, 3-, and 2-fold diameters and found to be 741 Å. Thus, the volume of the whole virus (V1) is 4π(370)3/3. The thickness of the outer protein coat is 75 Å, and the thickness of the lipid membrane is 40 Å. Thus, the mean radius of the lipid is 370 − 75 − 20 = 275 Å. Hence, the volume of the lipid membrane (V2) would be about 4π(275)2 × 40. The lipid density is approximately 0.9 g/ml, whereas protein and DNA densities are approximately 1.2 g/ml. The ratio of the mass of lipid/mass of the whole virus would then be 0.9V2/[1.2(V1 − V2) + 0.9V2], which gives about 14%.
According to the mass spectroscopy results, the amount of the ionized lipid was between 5 and 10 μg in a starting Sputnik sample of about 40 μg, corresponding to between 12.5% and 25% lipid.
Fitting of PBCV-1 Vp54 into the Sputnik cryoEM map.
A trimer of PBCV-1 Vp54 was reconstituted from the deposited Protein Data Bank (PDB) coordinates (accession number 1M3Y) by applying crystallographic symmetry. When using the Colores program, the five independent capsomers (four in general positions and one associated with a 3-fold axis in the icosahedral asymmetric unit) were fitted separately as trimers. In EMfit, the starting position of the center of mass for each capsomer was taken from the best fit obtained by the Colores program. The best fits from the four generally positioned capsomers and one-third of the icosahedral 3-fold capsomer were then combined into one PDB file representing the fitted icosahedral asymmetric unit. Icosahedral 2-, 3-, and 5-fold matrices were applied to the fitted asymmetric unit to generate the model for the whole virus. The fitting results are summarized in Table 1.
TABLE 1.
Fit of the crystal structure of PBCV-1 Vp54 to the Sputnik cryoEM densitya
| Capsomer | Best fit |
Second-best fit |
||||
|---|---|---|---|---|---|---|
| Colores best peak (σ) | EMfit |
Colores highest-noise peak (σ) | EMfit |
|||
| Sumf (%) | Negative density | Sumf (%) | Negative density | |||
| 1 | 11.05 | 34.2 | 1.3 | 7.31 | 32.1 | 1.5 |
| 2 | 8.96 | 32.8 | 1.4 | 6.4 | 30.8 | 2.9 |
| 3 | 10.19 | 32.2 | 1.9 | 6.87 | 31.6 | 2.9 |
| 4 | 9.08 | 32.6 | 1.9 | 6.77 | 30.5 | 2.3 |
| 5 | 12.18 | 32.7 | 0.7 | 8.45 | 31.5 | 1.9 |
The program EMfit measures the fit (Sumf) in terms of the average density at all fitted Cα atoms normalized by the maximum density fixed at 100 (18).
Electron microscopy data accession number.
The cryoEM map has been deposited with the Electron Microscopy Data Bank (accession number EMD-1662).
RESULTS AND DISCUSSION
Icosahedral reconstruction of Sputnik.
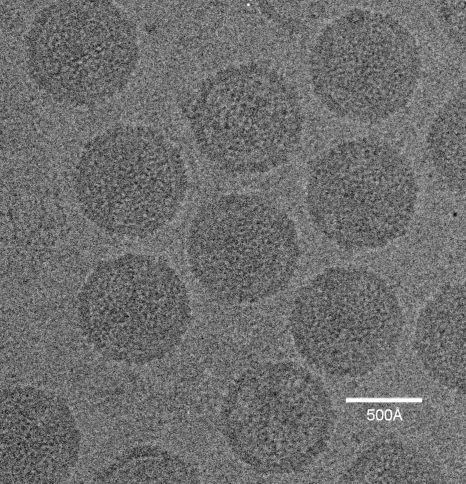
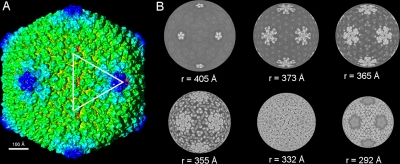
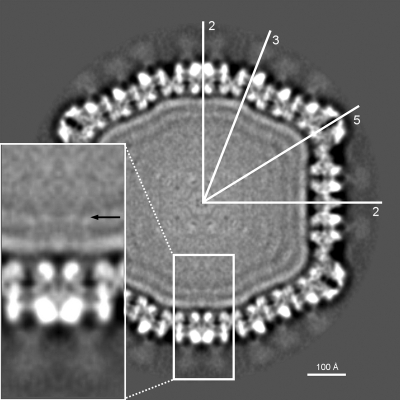
Sputnik particles (∼1 mg/ml) were purified as described previously (10) and frozen in vitrified ice, and data were recorded on film and processed (see Materials and Methods) (Fig. 1). The resolution of the icosahedrally averaged 3D map was estimated to be ∼10.7 Å, based on a 0.5 Fourier shell correlation threshold. The dimensions of Sputnik measured 840 Å, 730 Å, and 710 Å along the 5-, 3-, and 2-fold axes, respectively (Fig. 2A). The MCP is organized into a hexagonal lattice with a triangulation number of T=27 (h=3, k=3). Each icosahedral asymmetric unit contains four generally positioned, pseudohexameric capsomers, one-third of one pseudohexameric capsomer on the icosahedral 3-fold axis, and one-fifth of one pentameric capsomer at the 5-fold axis. The distance of the capsomers from the center of the virus decreases successively from the pentameric capsomers, the capsomers surrounding the pentameric capsomers, and the capsomers at the icosahedral 3-fold axis to the rest of the capsomers in general positions (Fig. 2B). The pseudohexameric capsomers are about 75 Å thick, and the distance between the center of adjacent capsomers is about 75 Å, both consistent with capsomer dimensions in viruses of the PRD1-adenovirus lineage. There is a 55-Å-long “mushroom”-like fiber with a triangular head protruding from the center of each pseudohexameric capsomer but not from the pentameric capsomers (Fig. 3). The density of the fibers is lower than that of the capsid, suggesting either that the fibers are not fully occupied in each capsomer or that the fibers are flexible. There are “cavities” in the middle of the pentameric capsomers (Fig. 3), one of which could serve as a gate for DNA entry or exit. Indeed, the pentameric vertices of many viruses, especially in bacteriophages, act as genome entry or exit gates (5, 11, 21, 22).
FIG. 1.
Micrograph of Sputnik particles embedded in vitreous ice, recorded using a Philips CM200 FEG electron microscope.
FIG. 2.
CryoEM reconstruction of Sputnik at 10.7-Å resolution. (A) Shaded-surface representation of the Sputnik 3D density map viewed along an icosahedral 2-fold axis. The white triangle marks the boundary of an icosahedral asymmetric unit. The numbers show the positions of icosahedral 2-, 3-, and 5-fold axes limiting an asymmetric unit. (B) Radial sections of the cryoEM map at different radii, with gray scale used to denote density levels, with white being the highest density.
FIG. 3.
Cross-section through the Sputnik cryoEM map. The orientation of the icosahedral (2-, 3-, and 5-fold) axes is shown with white lines. Note the probable lipid bilayer under the protein capsid and the “mushroom”-like fibers on the trimeric capsomers. In the zoomed view, a black arrow is pointing toward possible transmembrane protein densities.
Inside the capsid shell, there are two layers of densities that together are about 40 Å thick, consistent with the dimensions of a lipid bilayer (Fig. 3). There are weak interactions between the protein capsid shell and the likely lipid bilayer, represented by “foggy” densities (Fig. 3). The putative membrane surrounds a series of layers separated by about 25 Å that become progressively more fused toward the center of the virus. These are presumably layers of packaged DNA that are distinct from the membrane in being less resolved from each other than are the inside and outside lipid leaflets of the membrane. The volume enclosed by the lipid layer is about 3.6 × 107 Å3, and as there are 18,343 bp in the genome, the density of the packaged DNA is about 1,966 Å3/bp. This is comparable to other viruses such as T4, phiKZ, PRD1, and adenovirus, which have densities of 2,907, 2,100, 2,148, and 2,057 Å3/bp, respectively. Underneath the icosahedral 2-fold axes, there is some weak density that spans across the putative membrane, representing possibly a transmembrane protein (Fig. 3).
Lipid content of Sputnik.
Lipid was extracted with chloroform from a Sputnik sample of known mass. Mass spectroscopy showed that the sample contained between 12% and 24% lipid, by weight, and that there were a number of types of lipids in the extract with the dominant species (764.7 mass units) having a molecular mass corresponding to phosphatidylserine. This can be compared with the anticipated volume of the membrane relative to the volume of the virus assuming appropriate densities for lipid, protein, and DNA showing that the putative lipid membrane would represent about 14% of the mass of the virus. Judging from the cryoEM images of the sample, the contamination from host amoeba debris was not significant. Thus, the measured amount of lipid corresponds well with the structural information, confirming the presence of a lipid membrane within Sputnik.
Surface fibers.
Quite a few viruses have fiber-like structures on their surface. Among viruses with double jelly-roll capsid structures, PBCV-1 has fibered capsomers in special positions (5); adenovirus has long fibers emanating from the pentameric vertices, which function to recognize the host (6); Chilo iridescent virus (CIV) has short fibers emanating from every capsomer whose function is unknown; and mimivirus has a forest of fibers whose function may be to act as a decoy for amoeba phagocytosis (21). Other viruses, especially bacteriophages such as T4 and phi29, have hexameric capsomers with protruding fibers whose function is uncertain. Equally, it is unclear what the function is of the fibers protruding from the Sputnik surface. Some possibilities are that the fibers of Sputnik, as perhaps also the fibers of other viruses, help to stabilize each individual capsomer or that the fibers are associated with host cell recognition or entry.
Fitting of the PBCV-1 Vp54 structure into the Sputnik cryoEM map.
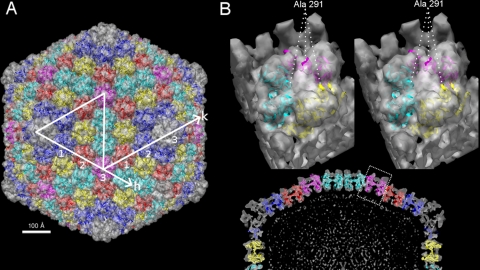
The thickness of the Sputnik capsomer and the distance between adjacent capsomers suggest that the MCP of Sputnik is similar to the MCPs from the PRD1-adenovirus lineage. Therefore, the crystal structure of PBCV-1 MCP Vp54 was fitted into the density map of Sputnik by using both the Colores (20) and EMfit (18) programs (Fig. 4A). The results obtained from both programs were essentially the same and produced reasonably good fits for each of the five independent capsomers (Table 1).
FIG. 4.
Structure of PBCV-1 Vp54 fitted into the Sputnik cryoEM density. (A) Shaded-surface representation of the Sputnik 3D density map viewed along an icosahedral 2-fold axis. The fitted Vp54 structures are shown as ribbons. Symmetry-related capsomers are shown in the same color. The capsomers counted along the h and k axes demonstrate T=27 quasisymmetry. (B, bottom) Central sliced view of half of the Sputnik 3D map fitted with Vp54. (Top) A zoomed view of one of the capsomers is shown as a stereo pair. The position of Ala 291 shows that the uninterpreted density corresponds to an insertion in the DE loop of PBCV-1 Vp54.
The protein encoded by open reading frame 20 (ORF20), gene product 20 (gp20), is the most abundant protein in Sputnik and, therefore, is probably the MCP. Its length (595 amino acids) is comparable to that of the mimivirus MCP (591 amino acids) and is 158 amino acids longer than PBCV-1 Vp54 (437 amino acids). Therefore, there should be unoccupied capsomer densities in the Vp54-fitted Sputnik map. Indeed, this is what was observed, and the unoccupied densities are mostly on the external side of the capsomers. Upon further examination, the unoccupied densities are close to Ala 291 in the Vp54 structure, which is in the DE loop of the second jelly-roll fold (Fig. 4B). It was shown by sequence alignment that mimivirus is likely to have a large insertion around this position. This would account for most of the length difference in MCPs between PBCV-1 on the one hand and mimivirus or Sputnik on the other (21).
Sputnik gp20 does not have any detectable sequence identity to the mimivirus MCP, nor does gp20 have any homologues in current sequence databases. Hence, Sputnik is probably not a result of divergent evolution from the Mimiviridae but, rather, diverged from the NCLDVs at an earlier time. Thus, possibly, this virophage had been associated with another viral host before being associated with mimivirus. Genome sequence analysis showed Sputnik to represent an unknown family of viruses (10). The structural work reported here supports this conclusion and shows that Sputnik would be a new branch in the PRD1-adenovirus lineage.
Acknowledgments
We thank Ye Xiang for helpful advice on cryoEM reconstruction. We thank L. Barassi and I. Pagnier for their help. We thank Sheryl Kelly for help in the preparation of the manuscript.
This work was supported by a National Institutes of Health grant to M.G.R. (grant AI11219).
We declare that we have no conflict of interest.
Footnotes
Published ahead of print on 4 November 2009.
REFERENCES
- 1.Abrescia, N. G. A., J. M. Grimes, H. M. Kivela, R. Assenberg, G. C. Sutton, S. J. Butcher, J. K. H. Bamford, D. H. Bamford, and D. I. Stuart. 2008. Insights into virus evolution and membrane biogenesis from the structure of the marine lipid-containing bacteriophage PM2. Mol. Cell 31:749-761. [DOI] [PubMed] [Google Scholar]
- 2.Agnès, V., B. La Scola, M. D. Bernard, J.-M. Forel, V. Pauly, D. Raoult, and L. Papazian. 2009. Clinical significance of a positive serology for mimivirus in patients presenting a suspicion of ventilator-associated pneumonia. Crit. Care Med. 37:111-118. [DOI] [PubMed] [Google Scholar]
- 3.Bamford, D. H., J. M. Grimes, and D. I. Stuart. 2005. What does structure tell us about virus evolution? Curr. Opin. Struct. Biol. 15:655-663. [DOI] [PubMed] [Google Scholar]
- 4.Benson, S. D., J. K. H. Bamford, D. H. Bamford, and R. M. Burnett. 1999. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell 98:825-833. [DOI] [PubMed] [Google Scholar]
- 5.Cherrier, M. V., V. A. Kostyuchenko, C. Xiao, V. D. Bowman, A. J. Battisti, X. Yan, P. R. Chipman, T. S. Baker, J. L. Van Etten, and M. G. Rossmann. 2009. An icosahedral algal virus has a complex unique vertex decorated by a spike. Proc. Natl. Acad. Sci. U. S. A. 106:11085-11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chroboczek, J., E. Gout, A. L. Favier, and R. Galinier. 2003. Novel partner proteins of adenovirus penton. Curr. Top. Microbiol. Immunol. 272:37-55. [DOI] [PubMed] [Google Scholar]
- 7.Ghigo, E., J. Kartenbeck, P. Lien, L. Pelkmans, C. Capo, J.-L. Mege, and D. Raoult. 2008. Amoebal pathogen mimivirus infects macrophages through phagocytosis. PLoS Pathog. 4:e1000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khan, M., B. La Scola, H. Lepidi, and D. Raoult. 2007. Pneumonia in mice inoculated experimentally with Acanthamoeba polyphaga mimivirus. Microb. Pathog. 42:56-61. [DOI] [PubMed] [Google Scholar]
- 9.Krupovic, M., and D. H. Bamford. 2008. Virus evolution: how far does the double β-barrel viral lineage extend? Nat. Rev. Microbiol. 6:941-948. [DOI] [PubMed] [Google Scholar]
- 10.La Scola, B., C. Desnues, I. Pagnier, C. Robert, L. Barrassi, G. Fournous, M. Merchat, M. Suzan-Monti, P. Forterre, E. Koonin, and D. Raoult. 2008. The virophage, a unique parasite of the giant mimivirus. Nature 455:100-104. [DOI] [PubMed] [Google Scholar]
- 11.Leiman, P. G., P. R. Chipman, V. A. Kostyuchenko, V. V. Mesyanzhinov, and M. G. Rossmann. 2004. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell 118:419-429. [DOI] [PubMed] [Google Scholar]
- 12.Ludtke, S. J., P. R. Baldwin, and W. Chiu. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82-97. [DOI] [PubMed] [Google Scholar]
- 13.Nandhagopal, N., A. A. Simpson, J. R. Gurnon, X. Yan, T. S. Baker, M. V. Graves, J. L. Van Etten, and M. G. Rossmann. 2002. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc. Natl. Acad. Sci. U. S. A. 99:14758-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raoult, D., S. Audic, C. Robert, C. Abergel, P. Renesto, H. Ogata, B. La Scola, M. Suzan, and J. M. Claverie. 2004. The 1.2-megabase genome sequence of Mimivirus. Science 306:1344-1350. [DOI] [PubMed] [Google Scholar]
- 15.Raoult, D., P. Renesto, and P. Brouqui. 2006. Laboratory infection of a technician by mimivirus. Ann. Intern. Med. 144:702-703. [DOI] [PubMed] [Google Scholar]
- 16.Rice, G., L. Tang, K. Stedman, F. Roberto, J. Spuhler, E. Gillitzer, J. E. Johnson, T. Douglas, and M. Young. 2004. The structure of a thermophilic archaeal virus shows a double-stranded DNA viral capsid type that spans all domains of life. Proc. Natl. Acad. Sci. U. S. A. 101:7716-7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts, M. M., J. L. White, M. G. Grutter, and R. M. Burnett. 1986. Three-dimensional structure of the adenovirus major coat protein hexon. Science 232:1148-1151. [DOI] [PubMed] [Google Scholar]
- 18.Rossmann, M. G., R. Bernal, and S. V. Pletnev. 2001. Combining electron microscopic with X-ray crystallographic structures. J. Struct. Biol. 136:190-200. [DOI] [PubMed] [Google Scholar]
- 19.Whitelegge, J. P., J. le Coutre, J. C. Lee, C. K. Engel, G. G. Privé, and K. F. Faull. 1999. Toward the bilayer proteome, electrospray ionization-mass spectrometry of large, intact transmembrane proteins. Proc. Natl. Acad. Sci. U. S. A. 96:10695-10698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wriggers, W., R. A. Milligan, and J. A. McCammon. 1999. Situs: a package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 125:185-189. [DOI] [PubMed] [Google Scholar]
- 21.Xiao, C., Y. G. Kuznetsov, S. Sun, S. L. Hafenstein, V. A. Kostyuchenko, P. R. Chipman, M. Suzan-Monti, D. Raoult, A. McPherson, and M. G. Rossmann. 2009. Structural studies of the giant Mimivirus. PLoS Biol. 7:e1000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zauberman, N., Y. Mutsafi, D. B. Halevy, E. Shimoni, E. Klein, C. Xiao, S. Sun, and A. Minsky. 2008. Distinct DNA exit and packaging portals in the virus Acanthamoeba polyphaga mimivirus. PLoS Biol. 6:e114. doi: 10.1371/journal.pbio.0060114. [DOI] [PMC free article] [PubMed] [Google Scholar]