Abstract
Although yellow fever has historically been one of the most important viral infections of humans, relatively little is known about the evolutionary processes that shape its genetic diversity. Similarly, there is limited information on the molecular epidemiology of yellow fever virus (YFV) in Africa even though it most likely first emerged on this continent. Through an analysis of complete E gene sequences, including a newly acquired viral collection from Central and West Africa (Senegal, Cameroon, Central African Republic, Côte d'Ivoire, Mali, and Mauritania), we show that YFV exhibits markedly lower rates of evolutionary change than dengue virus, despite numerous biological similarities between these two viruses. From this observation, along with a lack of clock-like evolutionary behavior in YFV, we suggest that vertical transmission, itself characterized by lower replication rates, may play an important role in the evolution of YFV in its enzootic setting. Despite a reduced rate of nucleotide substitution, phylogenetic patterns and estimates of times to common ancestry in YFV still accord well with the dual histories of colonialism and the slave trade, with areas of sylvatic transmission (such as Kedougou, Senegal) acting as enzootic/epidemic foci.
Historically, yellow fever has been one of the most important viral infections of humans, causing considerable morbidity and mortality when it encounters a sufficient number and density of susceptible hosts and where the environment facilitates transmission by the principal Aedes (Stegomyia) aegypti mosquito vector (2, 26). It was these conditions that enabled the virus to cause regular and debilitating epidemics throughout the Americas during the 18th and 19th centuries. For example, the Philadelphia yellow fever epidemic of 1793 resulted in 17,000 cases and 5,000 deaths, approximately 10% of the total population. So notorious were the yellow fever epidemics of the Americas that they even drew the attention of Charles Darwin, who speculated that the virus was most likely of African origin (9). Although fewer than 5,000 cases of human yellow fever were reported in Africa and South America during the period 2000 to 2005, these numbers are likely to be large underestimates (2), such that yellow fever may still represent a significant public health threat.
Yellow fever is caused by a single-strand positive sense RNA virus (yellow fever virus [YFV]) that belongs to the family Flaviviridae (genus Flavivirus). Although a great deal is known about the basic biology of YFV, including its likely origin (4) and extent of global genetic diversity (11, 24, 29, 30, 41), other aspects of its evolution remain uncertain. These unresolved questions become particularly apparent relative to information about dengue virus (DENV), where gene sequence data are far more abundant. These two viruses are both classified within the genus Flavivirus and possess a transmission cycle that involves primates (and perhaps other mammals) (10) and various species of Aedes mosquitoes and an epidemiological history that appears to be closely linked to the slave trade (4, 45). However, despite these similarities, YFV and DENV display a number of important differences that shed light on their emergence and evolution (27). Most notably, YFV can still be considered a sylvatic disease of nonhuman primates that causes sporadic outbreaks in humans, which often act as little more than spill-over hosts. In addition, YFV has only a single serotype, unlike DENV with four antigenically distinct species (denoted DENV-1 to DENV-4), and is notoriously absent from Asia (and countries of the Pacific) even though both the hosts and vectors are present in this region. Finally, YFV is far more virulent than DENV, with reported case fatality rates of ∼25% in Africa (2). There may be a direct relationship between the inability of YFV to evolve into a human pathogen with areas of endemicity and its virulence. Specifically, an elevated virulence means that YFV will require a larger critical community size (CCS) to sustain its transmission in humans (i.e., to achieve a value of the basic reproductive number, R0, that is greater than unity). Historically, YFV spread rapidly through human populations, killing hosts before the susceptible pool could be replenished. Consequently, YFV would not establish sustained transmission networks unless it encountered relatively large and dense human populations, such as those found in the Americas during past centuries. It is therefore no surprise that the rise of yellow fever is closely tied to urbanization and the slave trade, both of which would have greatly increased the number of susceptible hosts. However, relatively little is known about the evolutionary dynamics of YFV in its enzootic situation in Africa and what this may tell us about how the virus interacts with its various host and vector species. In particular, although most yellow fever outbreaks have occurred in West Africa, few studies of YFV genetic diversity at the country level have been conducted in this region. As a consequence, little is known about the spatial and temporal dynamics of YFV within countries where vaccination campaigns in reaction to localized outbreaks have been the main control strategy.
Arthropod-borne viruses like YFV and DENV are characterized by increased purifying selection pressure on nonsynonymous nucleotide sites compared to RNA viruses transmitted by other routes, which are likely a function of a life cycle that involves phylogenetically divergent hosts (44). However, whether YFV and DENV differ in evolutionary dynamics and how this relates to their contrasting epidemiological profiles in human populations are unknown. While previous work has suggested a lower rate of evolutionary change in YFV than in DENV (4), this analysis was based on an analysis of partial prM/E sequences, whereas most studies of DENV evolution have considered the E gene.
To explore the evolution of YFV in more detail, and particularly in an African setting, we obtained the complete E gene sequences of 37 YFV isolates sampled from a variety of host (principally mosquito) species and epidemiological contexts in three sampling localities in Senegal, as well as a small number of sequences from Cameroon, Central African Republic (CAR), Cote d'Ivoire, Mali, and Mauritania. With these and published sequence data in hand, we conducted a descriptive analysis of YFV circulation within Senegal and West Africa, as well as a comparative analysis of the evolutionary dynamics of DENV and YFV using Bayesian Markov chain Monte Carlo (MCMC) methods. Although our analysis revealed multiple circulating YFV lineages within Senegal and West Africa, we also observed a marked and consistent difference between the evolutionary rates of YFV and DENV.
MATERIALS AND METHODS
New YFV isolates.
A total of 37 YFV E gene sequences were newly acquired as part of this study (Table 1). Twenty-eight sequences came from three different sampling locations in Senegal: 8 sequences from Koungheul; 2 sequences from Kaffrine, central Senegal, where two outbreaks occurred in 1995 and 1996 (37, 38); and 18 sequences from Kedougou, an area of sylvatic YFV circulation in southeast Senegal (see Fig. S1 in the supplemental material for a map of Senegal depicting sampling locations). The Senegalese viruses also came from a variety of different host species: human (n = 2), Erythrocebus patas monkeys (n = 1), A. aegypti mosquitoes (n = 5), Aedes furcifer mosquitoes (n = 8), Aedes luteocephalus mosquitoes (n = 5), Aedes metallicus mosquitoes (n = 1), Aedes taylori mosquitoes (n = 3), Aedes vittatus mosquitoes (n = 1), and uncertain mosquito species (n = 2).
TABLE 1.
Isolates of YFV newly sequenced as part of this study
| Isolate | Year of sampling | Place of sampling | Hosta |
|---|---|---|---|
| Ka_ArD122522 | 1996 | Senegal (Kafffrine) | A. aegypti |
| Ka_HD122030 | 1996 | Senegal (Kafffrine) | Human |
| Ke_ArD156583 | 2001 | Senegal (Kedougou) | A. taylori |
| Ke_AnD26923 | 1978 | Senegal (Kedougou) | Erythrocebus patas |
| Ke_ArD121040 | 1996 | Senegal (Kedougou) | A. furcifer |
| Ke_ArD149179 | 2000 | Senegal (Kedougou) | A. luteocephalus |
| Ke_ArD149194 | 2000 | Senegal (Kedougou) | A. taylori |
| Ke_ArD149213 | 2000 | Senegal (Kedougou) | A. luteocephalus |
| Ke_ArD149214 | 2000 | Senegal (Kedougou) | A. furcifer |
| Ke_ArD149215 | 2000 | Senegal (Kedougou) | A. vittatus |
| Ke_ArD149791 | 2000 | Senegal (Kedougou) | A. furcifer* |
| Ke_ArD149815 | 2000 | Senegal (Kedougou) | A. furcifer |
| Ke_ArD149887 | 2000 | Senegal (Kedougou) | A. taylori |
| Ke_ArD156029 | 2001 | Senegal (Kedougou) | A. furcifer |
| Ke_ArD156468 | 2001 | Senegal (Kedougou) | A. furcifer |
| Ke_ArD157928 | 2001 | Senegal (Kedougou) | A. luteocephalus |
| Ke_ArD24553 | 1976 | Senegal (Kedougou) | A. furcifer/taylori |
| Ke_ArD25112 | 1977 | Senegal (Kedougou) | A. luteocephalus |
| Ke_ArD99740 | 1993 | Senegal (Kedougou) | A. furcifer |
| Ko_ARDX | 2000 | Senegal (Kedougou) | Mosquitoes |
| Ko_ArD114891 | 1995 | Senegal (Koungheul) | A. aegypti |
| Ko_ArD114896 | 1995 | Senegal (Koungheul) | A. aegypti* |
| Ko_ArD114970 | 1995 | Senegal (Koungheul) | A. aegypti* |
| Ko_ArD114972 | 1995 | Senegal (Koungheul) | A. aegypti* |
| Ko_ArD114987 | 1995 | Senegal (Koungheul) | A. luteocephalus |
| Ko_ArD114988 | 1995 | Senegal (Koungheul) | A. furcifer |
| Ko_ArD114989 | 1995 | Senegal (Koungheul) | A. metallicus |
| Ko_HD117294 | 1995 | Senegal (Koungheul) | Human |
| HD78359 | 1990 | Cameroon | Human |
| Bo_ArB5656 | 1974 | Central African Republic | A. africanus |
| To_DakArAmt7 | 1973 | Cote d'Ivoire | A. africanus |
| SS_ARM154 | 1995 | Cote d'Ivoire | A. aegypti |
| De_ArA523 | 1996 | Cote d'Ivoire | A. aegypti |
| MK_ArD27797 | 1979 | Gambia | A. aegypti |
| Ba_HA872 | 1987 | Mali | Human |
| Sa_ArA20267 | 1987 | Mali | A. furcifer |
| Ro_HD47471b | 1987 | Mauritania | Human |
Asterisk, collected from male mosquitoes or female neonates.
Adverse reaction following vaccination.
Sequence data.
For the YFV isolates newly sequenced here, RNA was extracted from virus-infected AP61 cells or freeze-dried mouse brains using a QiaAmp Viral RNA kit (Qiagen, Inc., Chatsworth, California), as instructed by the manufacturer, and amplified and sequenced as previously described (7).
All complete E gene sequences of YFV were downloaded from GenBank and combined with those sequenced here. This resulted in a data set of 72 E gene sequences (1,521 nucleotides [nt] in length) sampled between 1927 and 2004 (see Table S1 in the supplemental material). All sequences were manually aligned using SE-AL (http://tree.bio.ed.ac.uk/software/seal/). In a subsequent analysis, sequences collected from vaccine strains (Ghana_Asibi_1927, Senegal_FrenchViscerotropic_1927, and 17D), as well as those sampled from patients experiencing an adverse effect following vaccination (Mauritania_Ro_HD47471_1987 and Spain_YF_AVD2791_93F_2004) and that cluster closely with the vaccine strains (see Results) (see Fig. S2 in the supplemental material) were excluded so as not to bias estimates of evolutionary dynamics. Importantly, their inclusion would have resulted in even lower mean substitution rates (with greater variances) than those estimated here. This pruning process resulted in a data set of 67 year-dated sequences sampled between 1940 and 2003.
To place the evolutionary dynamics of YFV in a comparative context, we undertook equivalent analyses of DENV. However, because of the very large potential size of DENV data sets, our analysis in each case (i.e., DENV-1 to DENV-4) was undertaken using a randomly sampled subset of 67 E gene sequences that contained the full range of phylogeographic diversity (i.e., viruses were randomly sampled from every human genotype to generate data sets that matched in size those of YFV). In the case of DENV-2, 15 E gene sequences were available from sylvatic isolates of this virus sampled in West Africa and Malaysia (5), and these were analyzed separately. A full list of the DENV sequences analyzed is provided in Table S1 in the supplemental material.
Bayesian MCMC analysis.
We estimated both the rate of nucleotide substitution per site and the time to the most recent common ancestor (TMRCA) for each data set using a Bayesian MCMC approach available in the BEAST package (http://beast.bio.ed.ac.uk/) (15). In each case we used both strict and relaxed (uncorrelated lognormal) molecular clocks and a substitution model employing the general time reversible (GTR) substitution matrix with a different rate assigned to each codon position (although different substitution models produced similar results [see Results]). We also employed the Bayesian skyline population coalescent prior in all cases as this is clearly the best descriptor of the complex population dynamics of both YFV and DENV, and inferring demographic processes was not the aim of this study. In each case, MCMC chains were run for sufficient time to achieve convergence (assessed using the TRACER program [http://tree.bio.ed.ac.uk/software/tracer/]), with uncertainty in parameter estimates reflected in values of the 95% highest probability density (HPD). Finally, for the relaxed clock analysis of the YFV data set, we also used BEAST to compute the maximum clade credibility (MCC) tree from all the plausible trees using the TreeAnnotator program, with the first 10% trees removed as burn-in.
Additional phylogenetic analysis.
As well as inferring time-structured phylogenetic trees, we performed an initial phylogenetic analysis on the complete 72-sequence data set using the Bayesian method available in the MrBayes package (version 3) (34). This analysis utilized the most general GTR+I+Γ4 (GTR with a proportion of invariant sites and gamma-distributed rate variation across sites) model of nucleotide substitution and a chain length of 10 million generations to ensure that stationary solution had been achieved (again as assessed using TRACER). Statistical support for nodes of interest is given as Bayesian posterior probability (BPP) values.
Phylogeographic analysis.
To determine the extent of geographic structure of YFV both globally (by country) and in Senegal (by sampling locality), we computed the association index (AI) (42) and parsimony score (PS) (36) statistics of clustering strength using the BaTS (Bayesian tip-association significance testing) method (31) that examines all the plausible (relaxed clock) trees produced by BEAST and therefore accounts for phylogenetic uncertainty (10% of trees removed as burn-in; 1,000 replicates).
Analysis of selection pressures.
To determine the overall nature of natural selection acting on the E gene of both YFV and DENV, we computed the mean ratio of nonsynonymous to synonymous nucleotide substitutions (dN/dS) per site using the single-likelihood ancestor counting (SLAC) method available in the Datamonkey web interface of the HY-PHY package (23). In each case this analysis utilized the GTR model of nucleotide substitution and an input neighbor-joining tree.
Nucleotide sequence accession numbers.
All sequences of the E gene generated here have been deposited in the GenBank database under accession numbers GU073130 to GU073166.
RESULTS
Phylogenetic relationships of YFV.
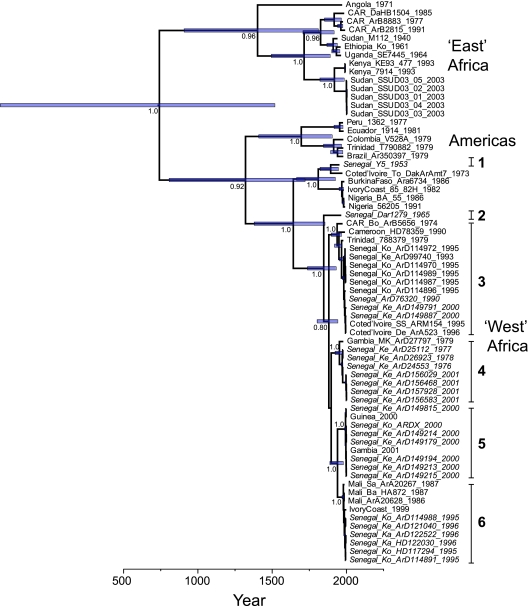
Our phylogenetic analysis of the E gene of YFV with a particular focus on isolates of African origin accords with those trees presented previously (4, 7, 30). The MCC tree for 67 year-dated E gene sequences is presented in Fig. 1, while the MrBayes tree of the same data, with the addition of a tight cluster of five vaccine and vaccine-related strains, is available as Fig. S2 in the supplemental material. The major phylogenetic division observed was between the sequences sampled in countries of East Africa (notably Sudan) and those from West Africa, including Senegal. The only geographical overlap among these two clades was a small number of sequences from the Central African Republic (CAR). Notably, all the sequences of American origin fall into a separate group that shares a close relationship with viruses from West Africa, thereby supporting previous phylogenetic studies which suggested that YFV was imported into the Americas from West Africa (4). A single sequence from Trinidad (Trinidad_788379_1979) clusters closely with sequences sampled from Senegal and therefore represents either a recent importation into the Caribbean from Africa (7) or a laboratory contamination (41).
FIG. 1.
Maximum clade credibility tree of 67 complete E gene sequences of YFV. The major geographical groupings of viruses are indicated, as are the six lineages (numbered) that have circulated in Senegal since 1953. Senegalese viruses are shown in italics. All tip times on the tree correspond to the year of sampling (as reflected in the timescale on the x axis). The shaded bars at each node represent the 95% HPD values for node height (age), while posterior probability values, a measure of clustering support, are shown for key nodes. The tree was rooted on the assumption of a relaxed molecular clock. Abbreviations in isolate designations are as follows: Ko, Koungheul; Ka, Kaffrine; Ke, Kedougou.
The sequences sampled from Senegal fall into six discrete lineages within the larger West African group (Fig. 1; see also Fig. S2 in the supplemental material). These lineages cocirculate within Senegal during human outbreaks (i.e., lineages 3 and 6 in Koungheul in 1995) or during sylvatic amplifications (lineages 3 to 5 in Kedougou in 2000). In addition, it is striking that all lineages except 1 and 2 (which have only one member each) have been sampled in Kedougou during the last 35 years of surveillance, while lineage 4 was twice sampled from a sylvatic context more than 2 decades apart (1976 to 1978 and 2001). Finally, it is noteworthy that all YFV isolates for which there was clear evidence for vertical transmission (i.e., the viruses were isolated from male mosquitoes or female neonates) belong to lineage 3.
Rates of evolutionary change.
Our Bayesian MCMC analysis revealed strikingly lower rates of nucleotide substitution in YFV than in DENV (Fig. 2; Table 2). Specifically, the mean evolutionary rate for the 67 E gene sequences of YFV assuming a relaxed molecular clock was 2.1 × 10−4 substitutions/site/year (95% HPD, 1.0 × 10−4 to 3.3 × 10−4 substitutions /site/year), whereas the equivalent mean rates for the four human DENV serotypes were 7.0 × 10−4 to 8.7 × 10−4 substitutions/site/year (range of 95% HPD, 5.8 × 10−4 to 9.8 × 10−4 substitutions/site/year). Rate estimates of DENV under a strict clock were almost identical to those estimated with a relaxed clock (range of 95% HPD values, 5.6 × 10−4 to 9.6 × 10−4 substitutions/site/year), while rather narrower 95% HPD values were observed with YFV under the strict clock (1.0 × 10−4 to 2.0 × 10−4 substitutions/site/year). Hence, there is no overlap in the distribution of substitution rates among YFV and the human DENV serotypes. Although a much smaller sample size led to far wider HPD values for the 15 sylvatic DENV-2 sequences, the mean substitution rates in this case (5.5 × 10−4 and 5.8 × 10−4 substitutions/site/year under the strict and relaxed molecular clocks, respectively) were again higher than those observed in YFV yet overlapped with the values seen in human DENV. Importantly, the mean substitution rate of YFV fell outside of the 95% HPD values for sylvatic DENV-2, suggesting that these rates have been drawn from different distributions. These results seem robust to both the particular substitution and demographic model employed. For example, the mean substitution rate for YFV estimated using the HKY plus Γ4 substitution model, a strict molecular clock, and constant population size coalescent prior was 1.2 × 10−4 substitutions/site/year (95% HPD, 0.6 × 10−4 to 1.9 × 10−4 substitutions/site/year).
FIG. 2.
Comparison of rates of nucleotide substitution among YFV and DENVs. The 95% HPD values of the substitution rates are shown in each case. Light bars represent estimates based on the relaxed (uncorrelated lognormal) molecular clock while dark bars are those values estimated assuming a strict molecular clock.
TABLE 2.
Estimates of evolutionary parameters in YFV and DENV
| Data set (n)a | Substitution rate (10−4 substitutions/site/year [95% HPD]) | TMRCA (no. of years [95% HPD]) | CoV (95% HPD) |
|---|---|---|---|
| All YFV (67) | |||
| Strictclock | 1.5 (1.0-2.0) | 1,574 (1,010-2,203) | NAb |
| Relaxed clock | 2.1 (1.0-3.3) | 1,262 (485-2,332) | 1.0 (0.6-1.3) |
| YFV post-1970 (62) | |||
| Strict clock | 1.4 (0.9-2.0) | 1,677 (1,045-2,390) | NA |
| Relaxed clock | 2.1 (0.9-3.5) | 1,332 (372-2,476) | 1.0 (0.6-1.5) |
| YFV Africa only (61) | |||
| Strict clock | 1.7 (1.1-2.4) | 1,601 (993-2,330) | NA |
| Relaxed clock | 2.6 (1.0-4.3) | 1,145 (328-2,193) | 1.0 (0.6-1.5) |
| DENV-1 (67) | |||
| Strict clock | 6.9 (6.0-7.8) | 101 (90-112) | NA |
| Relaxed clock | 7.0 (6.0-8.0) | 100 (88-112) | 0.1 (0-0.2) |
| DENV-2 (67) | |||
| Strict clock | 7.1 (6.0-8.1) | 116 (98-134) | NA |
| Relaxed clock | 7.1 (6.0-8.2) | 116 (94-138) | 0.1 (0-0.3) |
| DENV-3 (67) | |||
| Strict clock | 8.6 (7.6-9.6) | 65 (58-71) | NA |
| Relaxed clock | 8.7 (7.6-9.8) | 65 (56-74) | 0.2 (0-0.4) |
| DENV-4 (67) | |||
| Strict clock | 6.6 (5.6-7.5) | 111 (93-131) | NA |
| Relaxed clock | 7.2 (5.8-8.8) | 100 (69-141) | 0.5 (0.3-0.7) |
| DENV-2 sylvatic (15) | |||
| Strict clock | 5.5 (3.0-8.1) | 220 (136-323) | NA |
| Relaxed clock | 5.8 (2.7-9.1) | 210 (112-330) | 0.1 (0-0.4) |
n, number of sequences.
NA, not applicable.
To assess whether these low rates were robust to laboratory history, we repeated the analysis and arbitrarily removed the sequences of all isolates sampled before 1970 (Table 2). This also revealed relatively low substitution rates, with a range of 95% HPD values across both the strict and relaxed molecular clocks of 0.9 × 10−4 to 3.5 × 10−4 substitutions/site/year and, therefore, again markedly lower than the substitution rates seen in human DENV. Finally, similar rates (95% HPD, 1.0 × 10−4 to 4.3 × 10−4 substitutions/site/year) were also obtained excluding the six isolates sampled from the Americas (Table 2).
It is also striking how clock-like the estimates of substitution rate are in DENV compared to YFV, as reflected in estimates of the coefficient of variation (CoV) obtained under the relaxed molecular clock (Table 2). In the case of DENV-1, DENV-2 (both human and sylvatic), and DENV-3, not only does the low mean value of CoV suggest little non-clock-like behavior (i.e., rate variation among lineages), but also the lower 95% HPD value encompasses zero, indicating that a strict molecular clock cannot be rejected. In contrast, very high CoV values (mean values of 1.0 in both cases) are observed in all YFV data sets and are hence indicative of large-scale rate variation among viral lineages.
Times of common ancestry.
Our mean estimate under a relaxed molecular clock for the TMRCA of the YFV data set analyzed here, corresponding to the divergence time of the “East” and “West” African lineages, was almost twice that estimated previously using prM/E sequences (4), with mean dates of 1262 and 742 years, respectively (Table 2). However, the 95% HPD values of our YFV date estimate are very broad and encompass both the mean and much of the 95% HPD value of the prM/E estimate (and the current study used slightly different substitution and demographic models from those of Bryant et al.[4]). In addition, our time estimates for other nodes on the YFV tree are compatible with those estimated previously using the prM/E gene, again displaying broad HPD values. For example, the mean age of American viruses estimated under the prM/E gene was 306 years (95% HPD value, 120 to 590 years) while our E gene-based estimate for the TMRCA of this node was 307 years (95% HPD, 99 to 593 years). The mean TMRCA estimate for the joint West African and American group based on the prM/E gene data was 470 years (95% HPD, 186 to 869 years) while our mean estimate was 682 years (95% HPD, 282 to 1,196 years). Also of note was that our mean estimate of the TMRCA for groups 2 to 6 from Senegal, a distinct cluster representing the majority of genetic diversity in this country, was 156 years ago (range of 95% HPD values, 72 to 265 years), which is in good accord with a history of French colonialism (see Discussion).
The phylogeography of YFV.
Across the YFV tree as a whole there is more phylogenetic clustering of YFV sequences by country than expected by chance alone (AI, P = 0; PS, P = 0). Hence, although YFV has clearly spread to other regions both locally (for example, between Senegal and Gambia) (Fig. 1; see also Fig. S2 in the supplemental material) and globally, the strongest signal in the E gene data is still that of clustering by country. A similar pattern of strong spatial clustering was observed within Senegal (AI, P = 0; PS, P = 0.027), such that viruses from the three sampling localities—Kaffrine, Kedougou, and Koungheul—are more phylogenetically distinct than expected by chance alone although such clustering is less marked than that observed on the global scale. Together, these results indicate that most YFV evolution in Africa occurs within a localized geographical area, as might be expected from a largely sylvatic pathogen with limited host and vector mobility.
Selection pressures.
Mean dN/dS values were low in both YFV (0.043) and DENV (0.070, 0.062, 0.068, 0.075, and 0.067 for DENV-1, DENV-2, sylvatic DENV-2, DENV-3, and DENV-4, respectively). It is therefore clear that E gene evolution in both of these flaviviruses is predominantly controlled by purifying selection acting on deleterious mutations, as noted previously (44). Indeed, it is striking how few amino acid changes are fixed across the YFV phylogeny. For example, of the four groups of Senegalese YFV sequences that contain more than a single sequence, only two are defined by amino acid changes: group 3 by T13A, N255D, K337R, S456N, and M463I and group 4 by S208N. Similarly, no amino acid changes are found on the branch leading to the West African cluster of viruses.
DISCUSSION
Epidemiological history and phylogeography of yellow fever.
The timescale of YFV evolution we depict is compatible with spread during the colonial history of West Africa and its connections to the rest of the world through the slave trade. It is particularly notable that no East African lineages of YFV are found in the Americas, again supporting the idea that it was the movement of slaves from West Africa that was largely responsible for the emergence of yellow fever in the Americas (4). In this context, it is noteworthy that the two major phylogenetic lineages (West and East Africa) both contain isolates from the Central African Republic. Indeed, this is the first time that an isolate from Central Africa (CAR_Bo_ArB5656_1974) has been found to be closely related to isolates from West Africa. Such a finding suggests that Central Africa may contain more YFV genetic diversity than is usually recognized and that this region may even represent its place of origin although this will need to be confirmed with larger data sets.
In general, the European colonization of Africa had two main impacts for the epidemiology of human arboviruses. First, there was a process of progressive urbanization, which increased contact networks between the countryside, city, and coastal areas, in turn facilitating the spread of YFV. For example, the first well-documented outbreak of yellow fever in West Africa occurred in 1775 among British soldiers stationed in St. Louis, Senegal (6). Similarly, the major epidemic of 1927 (that was central to the development of the yellow fever vaccine), started in Senegal (St. Louis and Dakar) and moved among coastal cities down to Angola. The second major impact of colonization was a change in the composition of the susceptible population in West Africa. In particular, European colonists were highly susceptible to yellow fever and experienced severe levels of mortality; for example, case fatality rates during the early outbreaks in Goree and St. Louis were as high as 64% (13). Although the vaccination campaigns during the 1940s to late 1950s reduced the burden of yellow fever in West Africa (28), these campaigns were halted when Senegal became independent in 1960. Within 5 years, populations of unvaccinated children were large enough to again sustain major outbreaks.
From an African perspective, it is noteworthy that six phylogenetically distinct lineages of YFV circulate in Senegal, and this number is likely to increase with more intensive sampling. It is also interesting that the DAR1279/1965 sequence occupies a more basal lineage than the viruses sampled more recently in Senegal; 1965 was the year of the first major urban outbreak in West Africa since Senegalese independence in 1960, during which vaccination campaigns were halted or reduced in a number of African countries.
It is also striking that four of the six YFV Senegalese lineages circulate in Kedougou although this in part reflects the more intensive sampling in the region. This observation is compatible with the notion that Kedougou, with a forest habitat conducive to the transmission of YFV, acts as a key reservoir population for the virus in Senegal and perhaps West Africa more generally. Indeed, since 1972, uninterrupted entomological surveillance has revealed a major amplification of YFV in the area of Kedougou, which has been associated with outbreaks in both Senegal and neighboring countries (39). Such regional spatial diffusion is clearly depicted in the phylogenetic analysis presented here, in which the same viral lineages circulated in Senegal and Gambia in 1978 to 1979 and 2000 to 2001 and in Senegal and Guinea in 2000 (Fig. 1; see also Fig. S2 in the supplemental material). Similarly, it is important to note (i) that a large amplification of sylvatic YFV occurred in 1993 to 1994 in Kedougou, prior to major outbreaks in 1995 in Koungheul 400 km to the northeast that were halted by a local vaccination campaign (37); (ii) that lineages 3 and 6 were present during both the sylvatic amplification in Kedougou and the Koungheul outbreak; and (iii) that a lineage 6-associated outbreak occurred in Kaffrine (100 km west of Koungheul) in 1996. Indeed, one can speculate that YFV moves along the main road from Kedougou to Dakar and that the limited vaccination campaign was insufficient to prevent its diffusion from Koungheul to Kaffrine. Taken together, these observations suggest that entomological surveillance in Kedougou, as well as the thorough follow-up of circulating viral lineages, can be used as tools to monitor and perhaps predict YFV activity in West Africa. At the very least, this study further highlights the need for additional sampling and sequencing of wild isolates of YFV in West Africa.
Evolutionary dynamics of YFV.
The most notable result of our study was that YFV evolves significantly more slowly than DENV and with far more variance in substitution rate among lineages so that it does not conform to a strict molecular clock; on average, the substitution rate of the E gene of DENV is approximately five times that seen in YFV. Although slightly higher substitution rates have been observed in other regions of the YFV genome (4), the lower HPDs on these estimates overlapped the rates obtained here, and these rates are still lower than those observed in DENV (41).
What factors might be responsible for both the low rate and non-clock-like behavior of evolutionary change in YFV compared to DENV? One possibility is that YFV has an intrinsically lower rate of mutation than DENV. Indeed, a polymerase error rate as low as 0.0021 to 0.0025 mutations/genome/replication has been reported for YFV (33). Although such a low rate would at face value provide a simple explanation for the low rate of nucleotide substitution in YFV, this estimate of mutation rate is some 400 times lower than that usually associated with RNA polymerase (14). Additionally, the YFV substitution rate estimated here clearly falls within the “normal” range associated with RNA viruses (19, 21) and, hence, is incompatible with such infrequent mutation. It is therefore likely that this estimate of the YFV polymerase error rate is inaccurate, particularly as the experimental assay used ignored the critical consequences of natural selection on the mutational spectrum, including that on lethal mutations, and was based on the 17D vaccine virus. In sum, it is uncertain whether the mutation rate of YFV is lower than that of DENV.
Another possibility is that the E genes of YFV and DENV differ greatly in selection pressure, such that the substitution rates in DENV have been elevated by positive selection or that nonsynonymous sites in YFV are subject to stronger purifying selection, thereby reducing substitution rates in this case. However, our analysis of the relative numbers of nonsynonymous and synonymous changes argues against this hypothesis. Although the mean dN/dS ratio is always lower in YFV than in DENV, indicative of stronger purifying selection in the former virus, mean dN/dS ratios are very low in both viruses (dN/dS ≪ 1.0), signifying that the vast majority of mutations fixed in these viruses are synonymous. In addition, the difference in substitution rates between YFV and DENV is greater than the estimated difference in dN/dS.
We therefore suggest that a viable explanation for the difference in substitution rates between YFV and DENV is that the generation time (i.e., replication rate) of YFV is reduced relative to DENV such that there are fewer opportunities to mutate per unit time. In theory, such lower replication rates could occur in either the mammalian host or the mosquito vector. As both YFV and DENV appear to use largely the same mammalian hosts, particularly nonhuman primates, this explanation seems unlikely. Indeed, it is notable that the substitution rate of YFV is still lower than that of sylvatic DENV, in which humans probably play a negligible role in transmission. Therefore, differences in the time spent in mosquitoes are likely to constitute the main reason for differences in evolutionary dynamics. In particular, it is possible that a mechanism of vertical transmission, such as transovarial transmission where the virus may remain quiescent in mosquito eggs for many months, plays a more important role in YFV than in DENV; in the latter virus the rapid rates of transmission associated with specific DENV outbreaks are likely to swamp any reduction in rate due to vertical transmission. Indeed, there is compelling, and growing, evidence for transovarial transmission in YFV, including in Senegal (1, 3, 8, 12, 17, 18, 25). Importantly, not only would vertical transmission result in a lower mean substitution rate caused by a lack of active replication, but it would also increase the variance in rate estimates, exactly as observed here. Such a hypothesis is directly supported by observations that the vertical transmission rate (31.5%) of YFV in A. aegypti in nature, the principle epidemic vector (18), is up to 12,000 times higher than that in DENV-2 and DENV-4 (0.0053 to 0.0026%) (20, 22, 35). It is clear that such a mechanism should be further investigated for YFV in relation to wild vectors (i.e., A. furcifer, A. taylori, or A. luteocephalus) and perhaps for specific YFV lineages (such as lineage 3 described here). Similarly, it will be important to determine whether the RNA interference response in enzootic mosquito vectors is better able to modulate YFV than DENV replication as this may also act to reduce the overall substitution rate.
Finally, it is intriguing that the overall age of YFV (emergence within the last 2,500 years) is broadly similar to the time of origin of the four DEN viruses (16, 40, 43). Hence, YFV and DENV seem to have radiated at approximately the same time. However, since this time, DENV has differentiated into four antigenically distinct viruses while YFV is still classified as a single serotype. It is therefore tempting to speculate that the lower rate of evolutionary change in the E gene of YFV, the key viral antigen, may in part explain the lower level of antigenic diversity in this virus and, in turn, the easier development of a vaccine against YFV than DENV.
Supplementary Material
Acknowledgments
This work was in part funded by NIH grant GM080533.
We thank three reviewers for insightful comments.
Footnotes
Published ahead of print on 4 November 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Aitken, T. H. G., R. B. Tesh, B. J. Beaty, and L. Rosen. 1979. Transovarial transmission of yellow fever virus by mosquitoes (Aedes aegypti). Am. J. Trop. Med. Hyg. 28:119-121. [DOI] [PubMed] [Google Scholar]
- 2.Barrett, A. D. T., and S. Higgs. 2007. Yellow fever: a disease that has yet to be conquered. Annu. Rev. Entomol. 52:209-229. [DOI] [PubMed] [Google Scholar]
- 3.Beaty, B. J., R. B. Tesh, and T. H. G. Aitken. 1980. Transovarial transmission of yellow fever virus in Stegomyia mosquitoes. Am. J. Trop. Med. Hyg. 29:125-132. [DOI] [PubMed] [Google Scholar]
- 4.Bryant, J. E., E. C. Holmes, and A. D. T. Barrett. 2007. Out of Africa: a molecular perspective on the introduction of yellow fever virus into the Americas. PLoS Pathog. 3:e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cardosa, J., M. H. Ooi, P. H. Tio, D. Perera, E. C. Holmes, K. Bibi, and Z. A. Manap. 2009. Dengue virus serotype 2 from a sylvatic linage isolated a patient with dengue haemorrhagic fever. PLoS Neglect. Trop. Dis. 3:e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter, H. R. 1931. Yellow fever: an epidemiological and historical study of its place of origin. Williams and Wilkins Co., Baltimore, MD.
- 7.Chang, G. J., B. C. Cropp, R. M. Kinney, D. W. Trent, and D. J. Gubler. 1995. Nucleotide sequence variation of the envelope protein gene identifies two distinct genotypes of yellow fever virus. J. Virol. 69:5773-5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornet, M., Y. Robin, G. Heme, C. Adam, J. Renaudet, M. Valade, and M. Eyraud. 1979. Une poussée épizootique de fièvre jaune selvatique au Sénégal Oriental. Isolement du virus de lots de moustiques adultes mâles et femelles. Med. Mal. Infect. 9:63-66. [Google Scholar]
- 9.Darwin, C. 1871. The descent of man, and selection in relation to sex. John Murray, London, United Kingdom.
- 10.de Thoisy, B., P. Dussart, and M. Kazanji. 2004. Wild terrestrial rainforest mammals as potential reservoirs for flaviviruses (yellow fever, dengue 2 and St. Louis encephalitis viruses) in French Guiana. Trans. R. Soc. Trop. Med. Hyg. 98:409-412. [DOI] [PubMed] [Google Scholar]
- 11.Deubel, V., J.-P. Digoutte, T. P. Monath, and M. Girard. 1986. Genetic heterogeneity of yellow fever strains from Africa and the Americas. J. Gen. Virol. 67:209-213. [DOI] [PubMed] [Google Scholar]
- 12.Diallo, M., J. Thonnon, and D. Fontenille. 2000. Vertical transmission of the yellow fever virus by Aedes aegypti (Diptera, Culicidae): dynamics of infection in F1 adult progeny of orally infected females. Am. J. Trop. Med. Hyg. 62:151-156. [DOI] [PubMed] [Google Scholar]
- 13.Digoutte, J.-P. 1985. Dengue et fièvre jaune en Afrique de l'ouest; introduction et historique. Etudes Med. 3:111-114. [Google Scholar]
- 14.Drake, J. W., B. Charlesworth, D. Charlesworth, and J. F. Crow. 1998. Rates of spontaneous mutation. Genetics 148:1667-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drummond, A. J., and A. Rambaut. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunham, E. J., and E. C. Holmes. 2007. Inferring the time-scale of dengue virus evolution under realistic models of DNA substitution. J. Mol. Evol. 64:656-661. [DOI] [PubMed] [Google Scholar]
- 17.Dutary, B. E., and J. W. Leduc. 1981. Transovarial transmission of yellow fever virus by a sylvatic vector Haemagogus equinus. Trans. R. Soc. Trop. Med. Hyg. 75:128. [DOI] [PubMed] [Google Scholar]
- 18.Fontenille, D., M. Diallo, M. Mondo, M. Ndlaye, and J. Thonnon. 1997. First evidence of natural vertical transmission of yellow fever virus in Aedes aegypti, its epidemic vector. Trans. R. Soc. Trop. Med. Hyg. 91:533-535. [DOI] [PubMed] [Google Scholar]
- 19.Hanada, K., Y. Suzuki, and T. Gojobori. 2004. A large variation in the rates of synonymous substitution for RNA viruses and its relationship to a diversity of viral infection and transmission modes. Mol. Biol. Evol. 21:1074-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hull, B., E. Tikasingh, M. de Souza, and R. Martinez. 1984. Natural transovarial transmission of dengue 4 virus in Aedes aegypti in Trinidad. Am. J. Trop. Med. Hyg. 33:1248-1250. [DOI] [PubMed] [Google Scholar]
- 21.Jenkins, G. M., A. Rambaut, O. G. Pybus, and E. C. Holmes. 2002. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J. Mol. Evol. 54:152-161. [DOI] [PubMed] [Google Scholar]
- 22.Khin, M. M., and K. A. Than. 1983. Transovarial transmission of dengue 2 virus by Aedes aegypti in nature. Am. J. Trop. Med. Hyg. 32:590-594. [DOI] [PubMed] [Google Scholar]
- 23.Kosakovsky Pond, S. L., and S. D. W. Frost. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531-2533. [DOI] [PubMed] [Google Scholar]
- 24.Lepiniec, L., L. Dalgarno, V. T. Q. Huong, T. P. Monath, J.-P. Digoutte, and V. Deubel. 1994. Geographic distribution and evolution of yellow fever viruses based on direct sequencing of genomic cDNA fragments. J. Gen. Virol. 75:417-423. [DOI] [PubMed] [Google Scholar]
- 25.Marchoux, E., and P. L. Simmond. 1905. La transmission héréditaire du virus de la fièvre jaune chez le Stegomyia fasciata. C. R. Soc. Biol. (Paris) 59:259-260. [Google Scholar]
- 26.Monath, T. P. 1991. Yellow fever: Victor, Victoria? Conqueror, conquest? Epidemics and research in the last forty years and prospects for the future. Am. J. Trop. Med. Hyg. 45:1-43. [DOI] [PubMed] [Google Scholar]
- 27.Monath, T. P. 1994. Yellow fever and dengue—the interactions of virus, vector and host in the re-emergence of epidemic disease. Semin. Virol. 5:133-145. [Google Scholar]
- 28.Mutebi, J. P., and A. D. T. Barrett. 2002. The epidemiology of yellow fever in Africa. Microbes Infect. 4:1459-1468. [DOI] [PubMed] [Google Scholar]
- 29.Mutebi, J.-P., R. C. Rijnbrand, H. Wang, K. D. Ryman, E. Wang, L. D. Fulop, R. Titball, and A. D. T. Barrett. 2004. Genetic relationships and evolution of genotypes of yellow fever virus and other members of the yellow fever virus group within the Flavivirus genus based on the 3′ noncoding region. J. Virol. 78:9652-9665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mutebi, J.-P., H. M. Wang, L. Li, J. E. Bryant, and A. D. T. Barrett. 2001. Phylogenetic and evolutionary relationships among yellow fever virus isolated in Africa. J. Virol. 75:6999-7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker, J., A. Rambaut, and O. G. Pybus. 2008. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect. Genet. Evol. 8:239-246. [DOI] [PubMed] [Google Scholar]
- 32.Reference deleted.
- 33.Pugachev, K. V., F. Guirakhoo, S. W. Ocran, F. Mitchell, M. Parsons, C. Penal, S. Girakhoo, S. O. Pougatcheva, J. Arroyo, D. W. Trent, and T. P. Monath. 2004. High fidelity of yellow fever virus RNA polymerase. J. Virol. 78:1032-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 35.Sabin, A. B. 1952. Research on dengue during World War II. Am. J. Trop. Med. Hyg. 1:30-50. [DOI] [PubMed] [Google Scholar]
- 36.Slatkin, M., and W. P. Maddison. 1989. A cladistic measure of gene flow measured from phylogenies of alleles. Genetics 123:603-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thonnon, J., D. Fontenille, A. Tall, M. Diallo, Y. Renaudineau, B. Baudez, and G. Raphenon. 1998. Re-emergence of yellow fever in Senegal in 1995. Am. J. Trop. Med. Hyg. 59:108-114. [DOI] [PubMed] [Google Scholar]
- 38.Thonnon, J., A. Spiegel, M. Diallo, R. Sylla, A. Fall, M. Mondo, and D. Fontenille. 1998. Yellow fever outbreak in Kaffrine, Senegal 1996: epidemiological and entomological findings. Trop. Med. Int. Health 3:872-877. [DOI] [PubMed] [Google Scholar]
- 39.Traoré-Lamizana, M., D. Fontenille, H. G. Zeller, M. Mondo, M. Diallo, F. Adam, M. Eyraud, A. Maiga, and J.-P. Digoutte. 1996. Surveillance for yellow fever virus in eastern Senegal during 1993. J. Med. Entomol. 33:760-765. [DOI] [PubMed] [Google Scholar]
- 40.Twiddy, S. S., E. C. Holmes, and A. Rambaut. 2003. Inferring the rate and time-scale of dengue virus evolution. Mol. Biol. Evol. 20:122-129. [DOI] [PubMed] [Google Scholar]
- 41.Wang, E., S. C. Weaver, R. E. Shope, R. B. Tesh, D. M. Watts, and A. D. T. Barrett. 1996. Genetic variation in yellow fever virus: duplication in the 3′ noncoding region of strains from Africa. Virology 225:274-281. [DOI] [PubMed] [Google Scholar]
- 42.Wang, T. H., Y. K. Donaldson, R. P. Brettle, J. E. Bell, and P. Simmonds. 2001. Identification of shared populations of human immunodeficiency virus type 1 infecting microglia and tissue macrophages outside the central nervous system. J. Virol. 75:11686-11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weaver, S. C., and A. D. T. Barrett. 2004. Transmission cycles, host range, evolution and emergence of arboviral disease. Nat. Rev. Microbiol. 2:789-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woelk, C. H., and E. C. Holmes. 2002. Reduced positive selection in vector-borne RNA viruses. Mol. Biol. Evol. 19:2333-2336. [DOI] [PubMed] [Google Scholar]
- 45.Zanotto, P. M., E. A. Gould, G. F. Gao, P. H. Harvey, and E. C. Holmes. 1996. Population dynamics of flaviviruses revealed by molecular phylogenies. Proc. Natl. Acad. Sci. U. S. A. 93:548-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.