Abstract
Rift Valley fever virus (RVFV) is an emerging, highly pathogenic virus; RVFV infection can lead to encephalitis, retinitis, or fatal hepatitis associated with hemorrhagic fever in humans, as well as death, abortions, and fetal deformities in animals. RVFV nonstructural NSs protein, a major factor of the virulence, forms filamentous structures in the nuclei of infected cells. In order to further understand RVFV pathology, we investigated, by chromatin immunoprecipitation, immunofluorescence, fluorescence in situ hybridization, and confocal microscopy, the capacity of NSs to interact with the host genome. Our results demonstrate that even though cellular DNA is predominantly excluded from NSs filaments, NSs interacts with some specific DNA regions of the host genome such as clusters of pericentromeric γ-satellite sequence. Targeting of these sequences by NSs was correlated with the induction of chromosome cohesion and segregation defects in RVFV-infected murine, as well as sheep cells. Using recombinant nonpathogenic virus rZHΔNSs210-230, expressing a NSs protein deleted of its region of interaction with cellular factor SAP30, we showed that the NSs-SAP30 interaction was essential for NSs to target pericentromeric sequences, as well as for induction of chromosome segregation defects. The effect of RVFV upon the inheritance of genetic information is discussed with respect to the pathology associated with fetal deformities and abortions, highlighting the main role played by cellular cofactor SAP30 on the establishment of NSs interactions with host DNA sequences and RVFV pathogenesis.
Rift Valley fever virus (RVFV) is a highly pathogenic arthropod-borne virus transmitted by mosquitoes that infects a wide range of vertebrate hosts. In humans RVFV infection can lead to encephalitis, retinitis, or fatal hepatitis associated with hemorrhagic fevers, and in ruminants it can lead to high mortality rates, abortion, and fetal deformities. RVFV is an emerging zoonotic disease endemic in many countries of sub-Saharan Africa and in Egypt. For the first time in 2000, RVFV manifested itself outside of Africa, causing two simultaneous outbreaks in Yemen and Saudi Arabia (10, 9). The number of devastating outbreaks has increased progressively since then, the latest ones occurring in Kenya, Somalia, and Tanzania in 2007 and in Sudan and Madagascar in 2008.
RVFV is a Phlebovirus of the Bunyaviridae family that has a tripartite single stranded RNA genome consisting of large (L), medium (M), and small (S) segments (7, 29). The L and M segments are of negative polarity and express, respectively, the RNA-dependent RNA polymerase L and the precursor to the glycoproteins GN and GC, the cleavage of which generates also a nonstructural protein (NSm) that has been recently identified as a suppressor of virus-induced apoptosis (37, 4). The S segment utilizes an ambisense strategy and encodes the nonstructural protein NSs in genome orientation and the nucleoprotein N in antigenome orientation.
RVFV nonstructural protein NSs was identified as a main factor of virulence (34). Consequently, natural RVFV clone 13, which possesses a truncated defective NSs protein which is rapidly degraded by the proteasome in the cytoplasm of infected cells, is avirulent (35). NSs is not necessary for the viral cycle since recombinant RVFVΔNSs produced by reverse genetics, in which the NSs gene is completely deleted, is viable (16, 5, 3, 12). Whereas all of the steps of replication occur in the cytoplasm, NSs accumulates in the nuclei of infected cells, where it polymerizes and forms filamentous structures (32, 38) interacting with several cellular nuclear proteins that are trapped within these structures. Among the cellular nuclear proteins colocalizing with the NSs filaments, some, such as the p44 and XPB subunits of the RNA polymerase II TFIIH factor (20), are associated with the transcription machinery, whereas others, such as SAP30, Sin3A, and HDAC3 (21), are associated with chromatin remodeling events. Sequestration of p44 and XPB along the NSs filament was correlated with the general inhibition of RNA synthesis that occurs at late times after infection beyond 8 h postinfection (p.i.), whereas colocalization with SAP30, Sin3A, and HDAC3 was linked to the inhibition of the expression of the host beta interferon (IFN-β) gene, blocking the cellular antiviral response, that occurred early after infection (starting 3 to 4 h p.i.).
In spite of the fact that several of the nuclear proteins colocalizing with NSs filaments directly or indirectly bind to DNA regulatory sequences, the capacity of NSs filamentous structures to establish an interaction with the genome of the host cell has until now not been analyzed. To further decipher RVFV pathology and host-pathogen interactions, we have investigated here for the first time the capacity of NSs filaments to interact with the genome of the infected cell. This was carried out by using chromatin immunoprecipitation (ChIP), immunofluorescence, and immunofluorescence in situ hybridization (immuno-FISH) techniques, as well as Amira 3D image reconstruction. Our results show that even though DNA was mostly excluded from NSs filaments, constitutive heterochromatin clusters of pericentromeric DNA sequences appeared intimately associated with the NSs filamentous structures. Establishment of interaction of NSs with pericentromeric γ-satellite sequences, detected early after infection before filament formation, was progressively reinforced as the size of the filament was enhanced and resulted in the induction of a high incidence of RVFV-induced nuclear anomalies translating chromosome cohesion and segregation defects. Such an effect of RVFV infection upon inheritance of genetic information, which was observed on murine fibroblasts as well as on sheep kidney fetal cells, could account for the high rate of fetal deformities and abortions induced after RVFV infection.
Experiments carried out with the recombinant virus expressing protein NSsΔ210-230, deleted of its region of interaction with SAP30, demonstrated that interaction of NSs with SAP30 was essential for NSs to interact with these host DNA sequences and induce genetic instability. This finding reinforces our previous observations concerning the main role of NSs-SAP30 interaction (21) on RVFV pathogenicity.
MATERIALS AND METHODS
Antibodies.
Mouse anti-NSs polyclonal antibodies raised against the entire NSs protein (38) were used for ChIP or immunofluorescence analyses. Other primary antibodies used for immunofluorescence included anti-SAP30 C-18 (sc-8471) from Santa Cruz; anti-phosphoSer10H3 (06-570), anti-trimethyl-HistoneH3Lys9 (07-523), and anti-acetyl-HistoneH3Lys14 (06-911) from Upstate; and anti-biotin (catalog no. 43861) from Enzo Life Sciences. The secondary antibodies used for immunofluorescence and immuno-FISH were the Alexa 488-conjugated chicken anti-goat (A21467), Alexa 488-conjugated chicken anti-mouse (A21200), Alexa 555-conjugated donkey anti-mouse (A31570), and Alexa 488-conjugated chicken anti-rabbit (A 21441) antibodies from Molecular Probes.
Viruses and cells.
Stocks of RVFV ZH548, clone 13, and rZHNSsΔ210-230 (19, 25, 21) were produced under biosafety level 3 conditions by infecting Vero cells at a multiplicity of infection of 10−3 and by harvesting the medium at 72 h p.i. Murine fibroblastic L929 cells were described previously (21). Murine BF cells were as described elsewhere (2). Sheep kidney fetal cells (OA4.K/S1) were from the American Type Culture Collection.
Immunofluorescence.
For immunofluorescence, cells grown in 12-well plates on coverslips were fixed with 4% formaldehyde in phosphate-buffered saline (PBS) for 15 min and permeabilized with 1% Triton X-100 in PBS for 30 min. The cells were then incubated for 1 h at room temperature with corresponding antibodies diluted in PBS-5% bovine serum albumin. Cells were next washed with PBS and incubated for 45 min at room temperature with the corresponding secondary antibodies. TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) assays were carried out according to the manufacturer's instructions with an apoptotic detection kit (Chemicon International).
Immuno-FISH.
For immuno-FISH, cells were treated for immunofluorescence before in situ DNA hybridization (FISH). Plasmid pBluescript-γ-satellite (a generous gift from Niall Dillon), which contains eight mouse γ-satellite repeats cloned into the EcoRI site of a low-copy version of pBluescript (22), was used as a pericentromeric γ-satellite probe for Fig. 2, 3, and 6 and labeled for nick translation (Amersham) with either rhodamine-5-dUTP (Enzo Life Sciences) or biotine-11-dUTP (Enzo Life Sciences) as indicated in the figure legends. Plasmid pCR4 Min5-1 (11) was used as a centromeric α-satellite probe for Fig. 3 and labeled by nick translation (Amersham) with rhodamine-5-dUTP (Enzo Life Sciences).
FIG. 2.
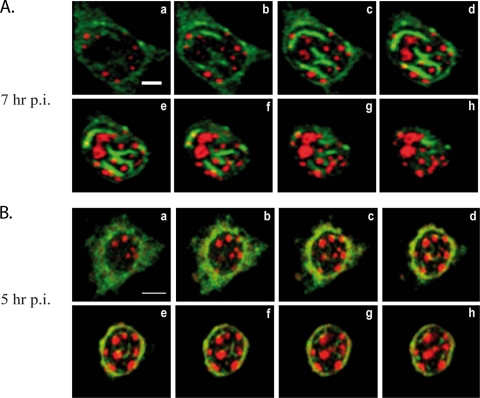
NSs filaments colocalize with clusters of pericentromeric γ-satellite sequences. (A and B) Merge images of single confocal sections taken from the bottom (a) to the top (h) of the nucleus at 0.4-μm intervals through the z axis of murine L929 cells infected by RVFV ZH548 strain collected at 7 (A) or 5 (B) h p.i. displaying NSs immunostaining (green) combined with rhodamine-labeled pericentromeric γ-satellite FISH (red). Bars: A, 5 μm; B, 10 μm.
FIG. 3.
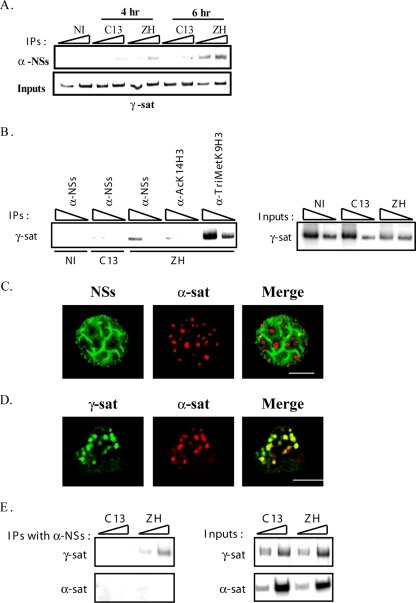
NSs filaments interact with pericentromeric γ-satellite sequences rather than with centromeric α-satellite sequences. (A) DNA immunoprecipitated (IP) (top) with anti-NSs antibody (α-NSs) and inputs corresponding to nonimmunoprecipitated genomic DNA (bottom), collected from murine L929 cells either not infected (NI) or 4 and 6 h after infection by pathogenic ZH548 (ZH) or nonpathogenic clone 13 (C13) strain was amplified with primers specific for pericentromeric γ-satellite DNA sequences (γ-sat). (B) DNA immunoprecipitated (IP) (left) with antibodies directed against NSs (α-NSs), the acetylated form of lysine 14 of histone H3 (α-AcK14H3), and the trimethylated form of lysine 9 of histone H3 (α-TriMetK9H3) and inputs corresponding to nonimmunoprecipitated genomic DNA (right), collected from murine L929 cells either not infected (NI) or 5 h after infection by pathogenic ZH548 (ZH) or nonpathogenic clone 13 (C13) strain, was amplified with primers specific for pericentromeric γ-satellite DNA sequences (γ-sat). (C) Single confocal section taken through the z axis of murine L929 cells infected by RVFV ZH548 strain displaying NSs immunostaining (left panel, green) combined with rhodamine-labeled centromeric minor α-satellite FISH (middle panel, red) and the corresponding merge image (right panel). (D) Single confocal section taken through the z axis of uninfected murine L929 cells displaying biotin-labeled pericentromeric γ-satellite FISH (left panel, green), rhodamine-labeled centromeric minor α-satellite FISH (middle panel, red), and the corresponding merge image (right panel). (E) DNA immunoprecipitated (IP) with anti-NSs antibody (α-NSs) (top) and inputs corresponding to nonimmunoprecipitated genomic DNA (bottom), collected from murine L929 cells 6 h p.i. by pathogenic ZH548 (ZH) or nonpathogenic clone 13 (C13) strain was amplified with primers specific for either pericentromeric γ-satellite (γ-sat) or centromeric minor α-satellite (α-sat) DNA sequences. In panels A, B, and E the triangles indicate increasing amounts of IPs or inputs amplified in pair of samples. The experiments whose results are shown in panel A were performed more than five times. The experiments whose results are shown in panels B and E were performed two times. Bars in panels C and D, 10 μm.
FIG. 6.
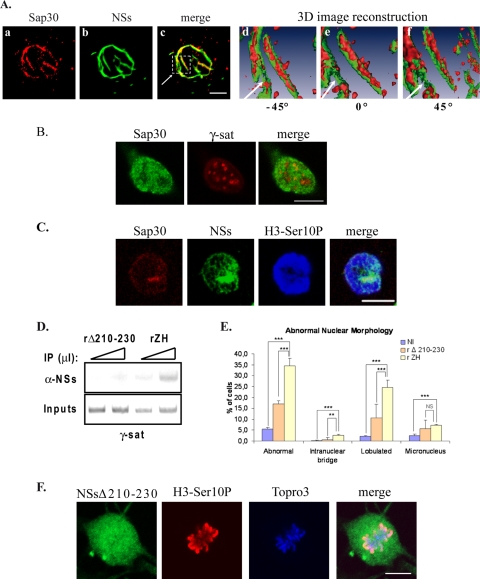
Interaction of NSs with SAP30 is required for NSs to target γ-satellite DNA sequences. (A) The distribution of SAP30 within NSs filaments was analyzed by using confocal microscopy (a to c) and three-dimensional (3D) image reconstruction (d to f) in murine L929 cells infected by RVFV ZH548 strain. In panels a to c, each row represents a single optical section through the z axis of an individual RVFV ZH548-infected murine fibroblastic L929 nucleus with SAP30 in red (a), NSs filament in green (b), and the corresponding merge image (c). The three-dimensional image reconstruction from confocal image stacks, one of which is shown in panels a to c, was performed by using Amira software. In panels d to f is shown the enlarged three-dimensional image of the region selected in panel c with the surface of SAP30 displayed in red and the surface of NSs displayed in green. (B) Single confocal section of uninfected murine L929 cells displaying SAP30 immunostaining (left panel, green) combined with rhodamine-labeled pericentromeric γ-satellite FISH (middle panel, red) and the corresponding merge image (right panel). (C) Single optical section of a ZH-infected mitotic L929 cell displaying immunostaining with anti-SAP30 in red, anti-NSs in green, anti-H3-Ser10P in blue, and the corresponding merge image (far right panel). (D) DNA immunoprecipitated (IP) (top) with anti-NSs antibody (α-NSs) and inputs corresponding to nonimmunoprecipitated genomic DNA (bottom), collected from murine L929 cells 6 h after infection by recombinant RVFV strain expressing either NSsΔ210-230 (rΔ210-230) or wild-type NSs (rZH) was amplified with primers specific for pericentromeric γ-satellite DNA sequences (γ-sat). Triangles indicate increasing amounts of IPs or inputs amplified in pair of samples. (E) The number of nuclei displaying abnormalities was quantified in uninfected cells (NI), as well as in cells infected by recombinant RVFV strains expressing either deleted NSsΔ210-230 (rΔ210-230) or wild-type NSs (rZH). The incidence of abnormal nuclei in rZH-infected cells was compared to NI or rΔ210-230-infected cells by using a chi-square test. **, P < 0.01; ***, P < 0.001; NS, not significant. (F) Single confocal sections taken through the z axis of a ZH-infected mitotic kidney sheep cell displaying immunostaining with anti-NSs in green, immunostaining with anti-H3-Ser10P in red, DNA counterstained with ToPro3 in blue, and the corresponding merge images. Bars, 10 μm.
For immunofluorescence, cells grown on coverslips were fixed 10 min at room temperature with paraformaldehyde 4% in PBS and permeabilized with 1% Triton X-100 in PBS. After three 5-min washes in PBS, the cells were incubated for 1 h at room temperature with mouse anti-NSs polyclonal antibodies as the primary antibody. The cells were then washed three times in PBS and incubated for 1 h at room temperature with Alexa 488-conjugated chicken anti-mouse antibodies as the secondary antibody. After three washes in PBS, the coverslips were kept overnight in PBS at 4°C. For FISH treatment after immunofluorescence, the cells were postfixed for 10 min at room temperature with paraformaldehyde 4% in PBS, washed for 5 min in PBS, and permeabilized with 1% Triton X-100 in PBS for 10 min at room temperature. After an initial wash for 2 min in PBS-0.1 M Tris-HCl (pH 7) and then two washes in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), the cells were dehydrated in 70, 80, 90, and 100% ethanol at 4°C for 2 min for each step and air dried. The cells were then treated with 100 μg of RNase A/ml for 45 min at 37°C, washed in PBS, dehydrated, dried, and subjected to in situ hybridization with the corresponding rhodamine-labeled probes. Cells were hybridized with 2 ng of plasmids in 50% formamide, 10% dextran sulfate, 2× SSC, and 50 μg/ml of single-stranded salmon sperm DNA (Boehringer). Before hybridization, the probe was denatured at 95°C for 5 min. Hybridization was performed on slides for 5 min at 80°C (to denature DNA in cells) and then overnight at 37°C. After hybridization, the coverslips were washed in 2× SSC for 30 min at 37°C, 1× SSC for 30 min at room temperature, and 0.5× SSC for 30 min at room temperature. Finally, the coverslips were mounted in Mowiol (500 μl with 12.5 mg of DABCO for antibleaching).
ChIP.
ChIP experiments were carried out as previously described (21). PCR analysis of inputs or immunoprecipitated DNAs was performed with the sense oligonucleotide 5′-TAT GGC GAG AAC CTG AAA-3′ as the 5′ primer and the antisense oligonucleotide 5′-TTC ACG TCC TAA AGT GTG TAT-3′ as the 3′ primer to reveal the pericentromeric γ-satellite sequence and the sense oligonucleotide 5′-GGT TTT TAT CAT TTT CCA TG-3′ as the 5′ primer and the antisense oligonucleotide 5′-CCA CAC TGT AGA ACA TAT TAG ATG-3′ as the 3′ primer to reveal the centromeric minor α-satellite sequence. For both sets of primers, PCR conditions were as follows: 1 cycle of 94°C for 5 min; 11 cycles of 94°C for 30 s, 54°C for 30 s, and 72°C for 30 s; and 1 cycle of 72°C for 10 min. A first “cold” PCR was carried out in the presence of 25 pmol of each primer; 1.5 μl of the product of the first PCR was subjected to a second “hot” PCR carried out in the presence of 0.1 μl of [α-32P]dATP (6,000 Ci/mmol) and 25 pmol of each primer.
Image acquisition and manipulation.
Samples were analyzed at room temperature by confocal laser scanning microscopy using a Axiovert 200M (Zeiss LSM510 confocal system). This system is equipped with a 63× lens, 1.4-numerical-aperture oil immersion lens (Plan Neofluor). For oil immersion microscopy, we used oil with refractive index of 1.518 (Zeiss). Images were collected in the direction of the z-axis corresponding to the optical axis of the microscope at 0.37 μm intervals with the z-axis going through the image planes. LSM 510 imaging software was used for image capture (512x512 pixels, 8 bit data). The images were analyzed by the LSM5 Image browser or Image J sofware. Double-labeled pixels were displayed in yellow on the merge images. Three-dimensional image reconstruction of confocal image stacks was performed by using Amira software from Mercury Computer Systems.
Statistical analyses.
Statistical analyses of the incidence of abnormal nuclei in Fig. 4C, 5C, and 6E were performed by applying the χ2 test using R software (26). The data were considered as statistically significant at P < 0.05, very significant at P < 0.01, and highly statistically significant at P < 0.001.
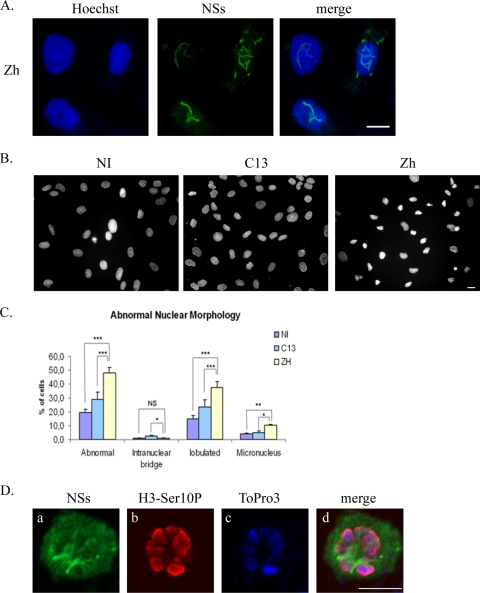
FIG. 4.
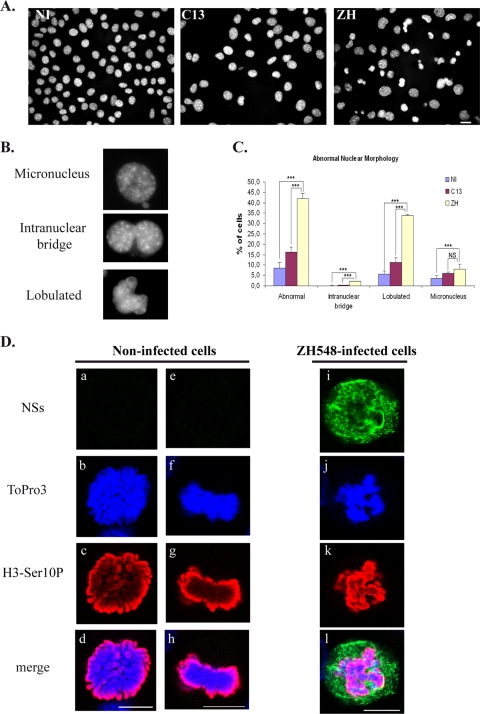
Infection with pathogenic RVFV strain ZH548 induces abnormal nuclear morphologies in murine fibroblasts. (A) Nonconfocal conventional fluorescence microscopy of L929 cells that were either not infected (NI) or infected by RVFV ZH548 strain (ZH) or by nonpathogenic clone 13 (C13) strain displaying DNA stained with Hoechst. (B) Common defects include micronuclei, intranuclear bridges, and lobulated nuclei. (C) The number of nuclei displaying abnormalities was quantified in cells that were not infected (NI) or in clone 13 (C13)- and ZH548 (ZH)-infected cells from two different experiments with n > 1,000 for each condition. The incidence of abnormal nuclei in ZH-infected was compared to uninfected cells or C13-infected cells by using the chi-square test. ***, P < 0.001; NS, not significant. (D) Single confocal sections taken through the z axis of uninfected or ZH-infected mitotic L929 cells displaying immunostaining with anti-NSs in green (a, e, and i), DNA counterstained with ToPro3 in blue (b, f, and j), immunostaining with anti-H3-Ser10P in red (c, g, and k), and the corresponding merge images (d, h, and l). Bars, 10 μm.
FIG. 5.
Infection with pathogenic RVFV strain ZH548 induces abnormal nuclear morphologies in kidney sheep cells. (A) Nonconfocal conventional fluorescence microscopy of kidney sheep cells of fetal origin infected by the RVFV ZH548 strain displaying total DNA distribution (Hoechst), NSs detected with anti-NSs antibody (green), and the corresponding merge image. (B) Nonconfocal conventional fluorescence microscopy of kidney sheep cells that were either not infected (NI) or infected by the RVFV ZH548 strain (ZH) or the nonpathogenic clone 13 (C13) strain displaying DNA stained with Hoechst. (C) The number of nuclei displaying abnormalities was quantified in uninfected (NI) and clone 13 (C13)- and ZH548 (ZH)-infected cells from two different experiments, with n > 800 at each condition. The incidence of abnormal nuclei in ZH-infected was compared to uninfected or C13-infected cells by using a chi-square test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant. (D) Single confocal sections taken through the z axis of uninfected or ZH-infected mitotic kidney sheep cells displaying immunostaining with anti-NSs in green (a), immunostaining with anti-H3-Ser10P in red (b), DNA counterstained with ToPro3 in blue (c), and the corresponding merge images (d). Bars, 10 μm.
RESULTS
Euchromatic host DNA is predominantly excluded from viral NSs filamentous structures.
RVFV nonstructural NSs protein, which has been identified as a major factor responsible of RVFV pathogenesis (34), has the characteristic to form filamentous structures in the nuclei of many different infected cell types (32, 38, 21). In a recently published work, we identified the IFN-β promoter as a region of the host genome interacting with the NSs protein (21), demonstrating that viral NSs protein could establish an interaction with a particular DNA sequence of the host.
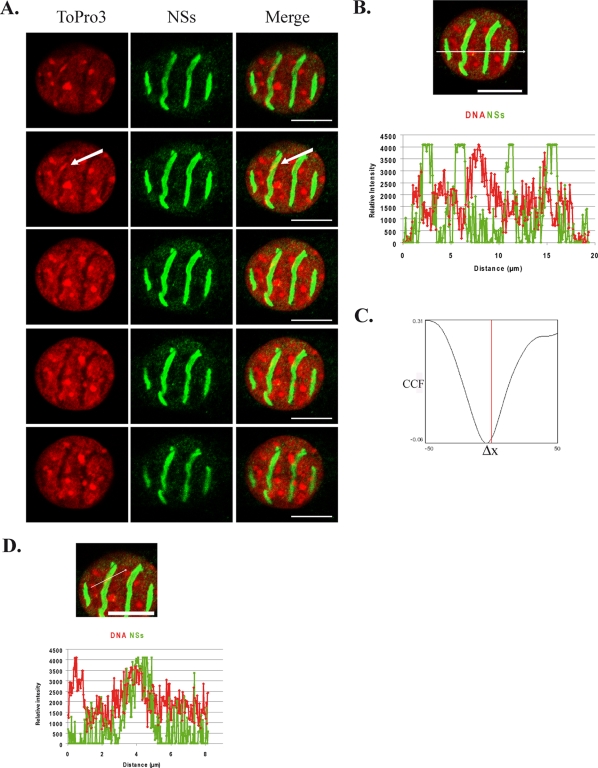
In order to further investigate the interaction of NSs with host DNA, we analyzed here the general distribution of cellular DNA with respect to viral NSs filaments, carrying out confocal microscopy analysis of the nuclei of murine fibroblastic L929 cells infected by virulent RVFV ZH548 strain. In Fig. 1A is shown a series of confocal sections along the z axis, corresponding to the optical axis of the microscope, from the bottom to the top of an individual nucleus representative of the population of murine L929 cells infected with RVFV ZH548. Cellular DNA labeled with intercalating ToPro3 dye is shown in red (left column), and NSs filament is shown in green (middle column), with the corresponding merge images displayed in the right column. Analysis of the general distribution of DNA in this series of confocal sections showed that cellular DNA is mainly excluded from NSs filaments in all of the sections analyzed. The line scan displaying the profile of the relative fluorescence intensities of DNA and NSs along a line traced across the nucleus (Fig. 1B) confirms this observation. It shows that an increase of the intensity of NSs fluorescence (green) corresponds to a decrease of the intensity of ToPro3-labeled DNA fluorescence (red) and vice versa. Also, the profile of van Steensel's cross-correlation function (CCF) shown in Fig. 1C, displayed a dip immediately aside of Δx = 0, which is indicative of an almost total exclusion (33, 6), further confirming the predominant exclusion of cellular DNA from NSs filaments.
FIG. 1.
Cellular DNA is predominantly excluded from NSs filaments. Colocalization of cellular DNA with viral NSs filaments was analyzed by confocal microscopy in murine L929 cells infected by RVFV ZH548 strain at 16 h p.i. (A) Single confocal sections taken at 0.4-μm intervals through the z axis from the bottom (top row) to the top (bottom row) of the nucleus. Each row represents a single optical section of the same nucleus. Left panels correspond to cellular DNA counterstained with ToPro3 (red). Middle panels show NSs distribution detected with anti-NSs mouse polyclonal antibody (green). Merge images of ToPro3 and NSs are shown in right panels with colocalization displayed in yellow. An arrow indicates a dense DNA region colocalizing with NSs filaments. (B and C) Profile of the line scan quantifying the relative intensity of the two fluorochromes along the indicated line (B), as well as the corresponding CCF analysis (C). (D) Profile of the line scan through the region indicated by an arrow in panel A with ToPro3 displayed in red and NSs displayed in green. Bars, 10 μm.
Margination of DNA resulting from the formation of a chromatin-free zone was accompanied by a significant increase of the nuclear volume whose diameter reached almost 20 μm compared to an average value of 10 μm in uninfected cells or in cells infected with nonvirulent RVFV C13 strain that does not induce NSs filament formation (data not shown). Such an enlargement of the nuclear volume after virus infection has been described in the case of other viruses, such as herpes simplex virus type 1, that also induce the formation of new nuclear compartments of viral origin, excluding host chromatin (24). Nevertheless, despite the general exclusion of cellular DNA from NSs filamentous structures, some DNA regions densely labeled by ToPro3 (such as the one indicated by an arrow in Fig. 1A) appeared colocalizing with the NSs filament. The profile of the line scan of the relative fluorescence intensities of DNA and NSs along a line traced across this region showed strong overlapping of the respective intensities (Fig. 1D). The overlap distance of the corresponding fluorescence intensities at mid-height was of over 1 μm, which is largely superior to the resolution of the objective used for image acquisition (200 nm) and therefore corresponds to a true colocalization (6).
NSs filaments interact with clusters of cellular pericentromeric DNA.
Centromeres are constituted of highly repeated sequences corresponding to pericentromeric major γ-satellite sequences and centromeric minor α-satellite sequences (8). During interphase, centromeres from different chromosomes associate together to form chromocenters that are easily detectable cytologically as highly dense DNA structures (36), like the one colocalizing with NSs in Fig. 1D. In order to determine whether the colocalization of NSs with Topro3-dense DNA regions corresponded to an interaction of NSs filaments with clusters of satellite DNA sequences, we carried out immuno-FISH and ChIP experiments.
FISH of DNA of RVFV ZH548-infected murine L929 cells carried out using rhodamine-labeled major pericentromeric γ-satellite DNA probe (red) was coupled to NSs fluorescence (green). Figure 2A shows the corresponding merge images of a series of confocal sections at 0.4 μm intervals through the z axis of a nucleus representative of the population of RVFV ZH-infected L929 nuclei at 7 h p.i. The confocal sections are shown from the bottom (Fig. 2Aa) to the top (Fig. Ah) of the nucleus. A strong colocalization (displayed in yellow) of NSs filaments with clusters of pericentromeric γ-satellite DNA was clearly observed, especially in the middle sections (Fig. 2Ad to f). More than 100 ZH548-infected cells, from four different experiments equivalent to the one described in Fig. 2A, were analyzed for NSs γ-satellite colocalization events. On average, each nucleus had 2.6 filaments, on which 1.6 colocalization events per filament were counted corresponding on average to 4.16 colocalization events per cell. Equivalent experiments were carried out with a nonpathogenic RVFV C13 strain that codes for a truncated form of NSs that is immediately degraded in the cytoplasm of infected cells (34). As previously published (21), no NSs was visible in the nuclei of C13-infected cells (data not shown).
In order to analyze to what extent the organization of NSs in fully formed filamentous structures was required for NSs to colocalize with γ-satellite sequences, the same experiment was performed at earlier times after infection before NSs filaments were fully formed. In Fig. 2B are shown the corresponding merge images of another series of confocal sections, from the bottom (Fig. 2Ba) to the top (Fig. 2Bh) of the nucleus of a RVFV ZH548-infected cell collected at 5 h p.i. Even though at this time, most of NSs (green) was still diffusely distributed outside the nucleus, some NSs could be observed inside the nucleus, with a fraction of NSs clearly colocalizing (displayed in yellow) with clusters of γ-satellite pericentromeric DNA. A total of 42 ZH548-infected cells, from two different experiments equivalent to the one described in Fig. 2B, were analyzed for NSs γ-satellite colocalization events. The number of nascent thin filaments observed at early times postinfection was superior (i.e., 7.5) to that observed at later times after infection and, in contrast to what was observed in Fig. 2A, not all of the nascent filaments established colocalization events with γ-satellite clusters. For filaments that displayed colocalization, mostly 1 colocalization event was detected per filament with, on average, 2.5 colocalization events per cell. These data indicate that colocalization of NSs filaments with chromocenters is an event that can be observed starting early times after infection, that does not appear to require full filament formation, but that is reinforced as the presence of NSs inside the nucleus is enhanced.
To further analyze the capacity of NSs to interact with pericentromeric γ-satellite, we carried out ChIP experiments. For this, genomic DNA from uninfected L929 cells or from L929 cells infected with either pathogenic RVFV ZH548 strain (ZH) or nonpathogenic RVFV C13 strain (C13) lacking nuclear NSs protein (25, 34) was immunoprecipitated with anti-NSs antibodies (α-NSs). Precipitated DNA was amplified by PCR using primers specific for γ-satellite pericentromeric DNA sequences. The results shown in Fig. 3A indicate that NSs had the capacity to interact with γ-satellite sequences in ZH548-infected cells. Amplification of γ-satellite sequences detected in ZH-infected cells as early as 4 h p.i. was enhanced from 4 to 6 h p.i., whereas the slight amplification observed in C13-infected cells corresponding to a nonspecific event remained constant. To assess the specificity of the ChIP assay shown in Fig. 3A, genomic DNA from ZH-infected cells was immunoprecipitated in a second independent experiment with antibodies directed either against the acetylated form of residue lysine 14 of H3 (AcK14H3, absent from pericentromeric γ-satellite sequences used here as a negative control) or directed against the trimethylated form of residue lysine 9 of histone H3 (TriMetK9H3, specifically present on pericentromeric γ-satellite sequences used here as a positive control). As shown in Fig. 3B, no amplification of γ-satellite DNA sequences was observed after immunoprecipitation of genomic DNA from ZH548-infected cells (5 h p.i.) with anti-AcK14H3 antibodies, whereas a strong amplification was observed after immunoprecipitation with anti-TriMetK9H3 confirming the specificity of the ChIP assay. In this experiment, amplification of γ-satellite DNA sequences was again clearly observed after immunoprecipitation with anti-NSs antibodies of genomic DNA from ZH548-infected cells (5 h p.i.) but not from uninfected or C13-infected (5 h p.i.) cells. The amount of DNA immunoprecipitated with anti-NSs antibodies was weaker than the amount immunoprecipitated with anti-TriMetK9H3, indicating that not all pericentromeric γ-satellite sequences were contacted by NSs in agreement with results shown in Fig. 2A and B.
Highly repeated centromeric α-satellite sequences are, as pericentromeric γ-satellite DNA, embedded within chromocenters (36, 8). In order to analyze the capacity of NSs to colocalize with centromeric α-satellite sequences, immuno-FISH experiments equivalent to those shown in Fig. 2A were carried out using a probe specific for centromeric sequences. As seen in Fig. 3C, some discrete colocalization events were observed between NSs and centromeric α-satellite sequences. Nevertheless, the results obtained during double FISH experiments carried out with a biotin-labeled pericentromeric probe (green) and a rhodamine-labeled centromeric probe (red) indicated that, as expected, all centromeric sequences either totally or partially overlap with clusters of pericentromeric sequences (Fig. 3D). Because of this overlap, these immuno-FISH experiments did not allow to distinguish colocalization of NSs with centromeric from pericentromeric sequences. Therefore, in order to determine the degree of specificity of the interaction of NSs between pericentromeric and centromeric sequences, DNA immunoprecipitated with anti-NSs antibodies was amplified with primers specific for either γ-satellite sequences or centromeric α-satellite sequences. As shown in Fig. 3E, whereas pericentromeric γ-satellite sequences were amplified from α-NSs immunoprecipitates, no amplification of centromeric α-satellite sequences was detected, indicating that NSs specifically interacts with pericentromeric γ-satellite sequences compared to centromeric α-satellite sequences.
RVFV ZH548-infected cells display nuclear anomalies translating chromosome cohesion and segregation defects.
Pericentromeric sequences correspond to the sequences where the ring-shaped cohesin multiprotein is retained until anaphase and therefore are essential for the cohesion and segregation of chromosomes during mitosis (11). Cells with chromosome cohesion and segregation defects display nuclear anomalies such as the presence of lobulated nuclei, as well as micronuclei and intranuclear bridges (27). In order to investigate whether the interaction of NSs with pericentromeric γ-satellite sequences could induce chromosome cohesion and segregation defects, the morphology of the nuclei of uninfected, as well as RVFV C13- and ZH548-infected, cells was analyzed by using classical fluorescence microscopy. As shown in Fig. 4A, nuclei with abnormal morphologies were frequently observed among cells infected with pathogenic RVFV ZH548 strain compared to cells either not infected or infected with nonpathogenic RVFV strain lacking nuclear NSs. Most common defects, shown in Fig. 4B, corresponded to lobulated nuclei, intranuclear DNA bridges, and micronuclei. The number of abnormal nuclei was scored among uninfected cells (1,750 total cells counted), C13-infected cells (total of 1,232 cells counted), and ZH-infected cells (1,264 total cells counted) from two different experiments. As shown in Fig. 4C, an increased incidence of nuclear abnormalities, mainly lobulated nuclei, was scored among ZH-infected cells.
Micronuclei, bridges and lobulated nuclei can also be observed during programmed cell death, which is triggered after infection by most viruses (28). In order to determine the eventual contribution of apoptosis to the amount of nuclear anomalies observed after RVFV infection, we measured the percentage of apoptotic cells, using TUNEL assays, among L929 cells either not infected or infected at 19 h p.i. by the RVFV ZH548 strain. For each condition a minimum of 1,000 cells was counted from two independent experiments. No induction of apoptosis was detected in ZH548-infected cells (1.19% ± 0.7%) compared to uninfected cells (1% ± 0.7%), ruling out the possibility that a fraction of nuclear anomalies observed after ZH infection could result from virus-induced programmed cell death. The absence of apoptosis in ZH-infected cells is in agreement with recent observations showing, on the one hand, that nonstructural NSm protein, encoded by RVFV M segment, suppresses virus induced apoptosis (37) and, on the other hand, that NSs protein induces the specific degradation of double-stranded RNA-dependent protein kinase (PKR) (13, 17), which otherwise is known to induce apoptosis when activated after infection by RNA viruses (31). C13-infected cells displayed a slightly higher incidence of abnormal nuclei compared to uninfected cells. This probably results from the induction of the host antiviral response, leading to IFN-β synthesis and PKR activation, which have been described to induce apoptosis (28), a phenomenon that occurs after infection by RVFV C13 but not after infection by the ZH548 strain (21). In agreement with this hypothesis, a slightly higher incidence of apoptotic cells (4.9% ± 1.41%) was observed among cells infected by nonpathogenic C13 strain lacking stable nuclear NSs protein than in uninfected cells (1% ± 0.7%).
A higher incidence of nuclear anomalies could also partly result from a higher rate of cells going through mitosis. In order to investigate this possibility, the percentage of cells engaged into mitosis was measured in uninfected cells and in RVFV ZH548- or C13-infected cells using as a mitotic marker the antibody directed against the phosphorylated form of Ser10 of histone H3 (H3-Ser10P) (14). A total of over 4,000 cells for each condition, from at least two different experiments, were counted. The percentage of cells engaged into mitosis was strongly reduced in RVFV ZH548-infected cells (1.12%) compared to uninfected (11.65%) or C13-infected cells (4.75%). This result not only indicated that infection by pathogenic ZH548 strain strongly affected the host cell cycle, but it also indicated that the higher incidence of nuclear anomalies scored in ZH548-infected cells was not the result of a higher incidence of mitotic events occurring in this particular cell population.
Overall, these results indicated that there was no contribution of virus-induced programmed cell death or of a higher incidence of mitotic events upon the establishment of nuclear anomalies observed after pathogenic RVFV ZH548 infection, which can therefore be considered as translating chromosome cohesion and segregation defects. Normal mitotic stages such as late prophase (Fig. 4Da to d) or metaphase (Fig. 4De to h) could be observed among uninfected or C13-infected cells ongoing mitosis. However, these stages were never observed among ZH-infected cells. Instead, all ZH-infected cells ongoing mitosis displayed cohesion and segregation defects as the one shown in Fig. 4Di-l. In such cells, NSs formed a “cagelike” structure around mitotic DNA and, even though NSs appeared less filamentous (a possible consequence of the disappearance of the nuclear membrane during mitosis), it still partially colocalized with cellular DNA.
To analyze whether the capacity of RVFV ZH548 infection to induce nuclear anomalies could be enlarged to other mammalian species, the same experiment as the one described in Fig. 4A with murine cells was carried out on kidney sheep cells of fetal origin. Infection with the pathogenic ZH-548 RVFV strain induced the formation of NSs filaments in nuclei of sheep cells (Fig. 5A) as in murine cells. Also as in murine cells, a higher rate of nuclei with abnormal morphologies was observed among sheep cells infected by ZH compared to uninfected or C13-infected cells (Fig. 5B and C). As shown in Fig. 5D, the distribution of NSs with respect to DNA in a sheep cell undergoing mitosis was very similar to that observed in murine cells, with NSs forming a cagelike structure around DNA labeled with H3-Ser10P. Overall, these results indicated that induction of abnormal nuclear morphologies by RVFV ZH infection is not a phenomenon restricted to murine cells but can be enlarged to other species, particularly to ovines.
Interaction of NSs with cellular cofactor SAP30 was required for NSs filaments to target pericentromeric DNA and induce nuclear anomalies.
Using the two-hybrid system as well as coimmunoprecipitation experiments, NSs was demonstrated to directly interact with cellular corepressor SAP30 (21). In order to visualize the distribution of SAP30 respect to NSs filaments, we performed three-dimensional image reconstruction of confocal immunofluorescence image stacks using Amira software, which allows the visualization of the volume occupied by one object with respect to another. Only the surfaces of each object are represented. In Fig. 6Ad to f the surfaces of SAP30 and NSs are displayed, respectively, in red and green. The corresponding colors are not superposed; therefore, in contrast to classical confocal microscopy, colocalization is not displayed. In Fig. 6Ad to f, only the interior of the filament is shown, which became visible after performing a section of the three-dimensional image through the z axis. Protein SAP30 appeared strongly present within the interior of the filament in a continuous way, and this was evident throughout the filament except for a short, less-structured region (indicated by an arrow). Overall, the results shown in Fig. 6A indicated that SAP30 not only colocalizes with NSs but also is strongly present concentrated within, at the interior of the NSs filaments. SAP30 can either directly (35) or indirectly (18) interact with DNA. In particular, SAP30 has been shown to directly interact with transcription factor YY1 (15), which itself directly interacts with several regions of the genome containing YY1 DNA-binding sites, among which are pericentromeric γ-satellite sequences rich in YY1 DNA-binding sites (30). In a previous study we demonstrated that interaction of NSs with SAP30 was required for NSs to establish an interaction with the IFN-β promoter (21). We hypothesized that this phenomenon could be enlarged to the interaction of NSs with γ-satellite sequences. As shown in Fig. 6B, SAP30 displayed the capacity to partially colocalize with some clusters of γ-satellite DNA, as was also the case with NSs filaments and, during mitosis, cellular SAP30 protein still colocalized with NSs protein (Fig. 6C), being part of the “cagelike” structure surrounding mitotic DNA previously observed in Fig. 4D with the NSs protein.
In order to check whether the interaction of NSs with SAP30 was required for NSs to interact with clusters of pericentromeric γ-satellite sequences and induce nuclear anomalies, we analyzed the capacity of NSsΔ210-230 protein, which is deleted of its region of interaction with cellular corepressor SAP30 (21), to interact with pericentromeric γ-satellite DNA. In order to do this, genomic DNA isolated from L929 cells infected with either the previously described nonpathogenic RVFV recombinant virus rZHΔNSs210-230 (21) expressing NSsΔ210-230 (rΔ210-230) or with the corresponding recombinant strain rZH expressing wild-type NSs was immunoprecipitated with anti-NSs antibody and analyzed by ChIP. The results (Fig. 6D) indicated that NSsΔ210-230 had lost its capacity to interact with γ-satellite DNA sequences, whereas recombinant rZH NSs interacted with these sequences similarly to wild-type NSs from the ZH548 strain. This result indicated that, indeed, NSs interaction with SAP30 was required for NSs to interact with γ-satellite sequences.
To analyze the eventual correlation between the establishment of an interaction between NSs and pericentromeric γ-satellite sequences and the incidence of nuclear defects, we analyzed the morphology of nuclei after infection with nonpathogenic recombinant RVFV NSsΔ210-230 strain. As shown in Fig. 6E, the presence of nuclear anomalies was strongly reduced after infection with the nonpathogenic rZHNSsΔ210-230 strain that behaved like C13 strain compared to cells infected with the pathogenic rZH strain that induced a high percentage of abnormal nuclei behaving as the wild-type ZH548 strain. In contrast to ZH-infected cells, normal mitotic stages such as metaphase could be observed after infection with rZHNSsΔ210-230 (Fig. 6F), demonstrating that the presence of NSsΔ210-230 does not affect chromosome cohesion. Overall, these observations strongly suggested that NSs interaction with γ-satellite sequences, through SAP30, was necessary for nuclear anomalies to be induced after RVFV infection.
DISCUSSION
NSs targeting of host DNA and RVFV pathology.
The results presented here focus on cellular DNA targeting by RVFV NSs filaments. In agreement with early work describing the margination of chromatin as one of the characteristic histological signs of RVFV infection (32), we found that most cellular DNA was excluded from the filament except for some specific DNA regions of the host genome that closely interacted with NSs filamentous structures. These data raise the question of a possible link between the capacity of NSs to target specific cellular DNA sequences and RVF pathogenesis.
Using immuno-FISH and ChIP assays, we have shown here that NSs interacts with pericentromeric γ-satellite sequences of the host, correlating with the induction of a high rate of nuclear anomalies translating chromosome cohesion and segregation defects of fetal origin. In animals, RVFV has been shown to be present in the visceral organs and brains of aborted and malformed fetuses (9), and a case of human vertical RVF transmission was recently described (1). Even though we have no direct evidence that the induction of nuclear anomalies, observed here in murine fibroblasts as well as in sheep kidney cells of fetal origin, leads to abortion and teratogenesis, it is reasonable to postulate that an abnormally high accumulation of defects in chromosome cohesion and segregation during embryonic development could at least partly account for the high rate of abortions and teratogenic disorders predominantly observed after RVFV infection among ovines.
Altogether, previous (21) and present work on NSs showed that NSs can target some DNA regions of the host's genome, such as the IFN-β promoter and pericentromeric DNA. For the moment, we cannot exclude that other regions of the cellular genome could also interact with NSs. Nevertheless, we observed that DNA carrying triMetK4H3 as an epigenetic mark was predominantly excluded from NSs filaments (data not shown), and no interaction between NSs and centromeric α-satellite sequences was detected during ChIP experiments, demonstrating that no cellular DNA sequence could interact with NSs.
The NSs-SAP30 interaction plays a main role during the establishment of NSs-host DNA interactions.
Until now, NSs has never been described as a DNA-binding protein. Nevertheless, for the time being we have no data allowing us to clearly determine whether NSs can directly interact either with DNA in general or particularly with γ-satellite DNA sequences. Meanwhile, the results obtained during the present study clearly demonstrated that interaction of NSs with SAP30 was necessary for NSs to establish interaction with host DNA, be it pericentromeric γ-satellite sequences or the IFN-β promoter (21). In the case of the IFN-β promoter, transcription factor YY1 served as an intermediary between the NSs/SAP30 protein complex and the IFN-β promoter that contains two YY1 DNA-binding sites (23). In the case of γ-satellite DNA, YY1 could also serve as an intermediary factor since γ-satellite sequences are also rich in YY1 DNA-binding sites (30). Interestingly, whereas γ-satellite sequences contain nine YY1-binding sites (30), only two sequences potentially matching with YY1 consensus binding site could be identified within murine centromeric α-satellite sequences. This difference could explain the preferential interaction of NSs with γ- rather than α-satellite sequences.
RVFV NSs protein has been identified as a main factor of virulence. Recombinant RVFV virus rZHNSsΔ210-230 that expresses a NSs protein deleted of its region of interaction with NSs protein was shown to have completely lost its pathogenicity (21). Interestingly, this nonpathogenic RVFV expresses a stable NSs protein located in the nucleus. A situation that contrasts with the nonpathogenic RVF viruses known to date, that either do not express NSs (rZHΔNSs) or express an unstable cytoplasmic NSs (C13). This suggests that NSs-SAP30 interaction probably plays a crucial role during the development of RVFV pathogenicity, making SAP30 and NSs-SAP30 interaction interesting potential targets for possible antiviral compounds.
Acknowledgments
We are grateful to Guylaine Hoffner for critical reading of the manuscript and helpful comments.
T.J. and P.L. were financed by the Agence Nationale de la Recherche (ANR) and by the Fondation pour la Recherché Médicale (FRM), respectively. This study was supported by grants ANR-MIIM-033-01 and ANR-08-MIE-022 to E.B. and M.B.
Footnotes
Published ahead of print on 4 November 2009.
REFERENCES
- 1.Adam, I., and M. S. Karsany. 2008. Case report: Rift Valley fever with vertical transmission in a pregnant Sudanese woman. J. Virol. 80:929. [DOI] [PubMed] [Google Scholar]
- 2.Billecocq, A., M. Spiegel, P. Vialat, A. Kohl, F. Weber, M. Bouloy, and O. Haller. 2004. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J. Virol. 78:9798-9806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billecocq, A., N. Gauliard, N. Le May, R. M. Elliott, R. Flick, and M. Bouloy. 2008. RNA polymerase I-mediated expression of viral RNA for the rescue of infectious virulent and avirulent Rift Valley fever viruses. Virology 378:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bird, B. H., C. G. Albariño, and S. T. Nichol. 2007. Rift valley fever virus lacking NSm proteins retains high virulence in vivo and may provide a model of human delayed onset neurologic disease. Virology 362:10-15. [DOI] [PubMed] [Google Scholar]
- 5.Bird, B. H., C. G. Albariño, A. L. Hartman, B. R. Erickson, T. G. Ksiazek, and S. T. Nichol. 2008. Rift valley fever virus lacking the NSs and NSm genes is highly attenuated, confers protective immunity from virulent virus challenge and allows for differential identification of infected and vaccinated animals. J. Virol. 82:2681-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolte, S., and F. P. Cordelières. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224:213-232. [DOI] [PubMed] [Google Scholar]
- 7.Elliott, R. M., M. Bouloy, C. H. Calisher, R. Goldbach, J. T. Moyer, S. T. Nichol, R. F. Petterson, A. Plyusnin, and C. S. Schmaljohn. 2000. Family Bunyaviridae, p. 614-616. In M. H. V. Van Regenmortel, C. Fauquet, D. H. L. Bishop, E. B. Cartens, M. K. Estes, et al. (ed.), Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, Inc., San Diego, CA.
- 8.Eymery, A., M. Callanan, and C. Vourc'h. 2009. The secret message of heterochromatin: new insights into the mechanisms and function of centromeric and pericentromeric repeat sequence transcription. Int. J. Dev. Biol. 53:259-268. [DOI] [PubMed] [Google Scholar]
- 9.Flick, R., and M. Bouloy. 2005. Rift Valley fever virus. Curr. Mol. Med. 5:827-834. [DOI] [PubMed] [Google Scholar]
- 10.Gerdes, G. H. 2002. Rift valley fever. Vet. Clin. N. Am. Food Anim. Pract. 18:549-555. [DOI] [PubMed] [Google Scholar]
- 11.Guenatri, M., D. Bailly, C. Maison, and G. Almouzni. 2004. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 166:493-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Habjan, M., N. Penski, M. Spiegel, and F. Weber. 2008. T7 RNA polymerase-dependent and independent systems for cDNA-based rescue of Rift Valley fever virus. J. Gen. Virol. 89:2157-2166. [DOI] [PubMed] [Google Scholar]
- 13.Habjan, M., A. Pichlmair, R. M. Elliott, A. K. Overby, T. Glatter, M. Gstaiger, G. Superti-Furga, H. Unger, and F. Weber. 2009. NSs protein of Rift Valley Fever Virus induces the specific degradation of the double-stranded RNA-dependent protein kinase (PKR). J. Virol. 83:4365-4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendzel, M. J., Y. Wei, M. A. Mancini, A. Van Hooser, T. Ranalli, B. R. Brinkley, D. P. Bazett-Jones, and C. D. Allis. 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106:348-360. [DOI] [PubMed] [Google Scholar]
- 15.Huang, N. E., C. H. Lin, Y. S. Lin, and W. C. Yu. 2003. Modulation of YY1 activity by SAP30. Biochem. Biophys. Res. Commun. 306:267-275. [DOI] [PubMed] [Google Scholar]
- 16.Ikegami, T., S. Won, C. J. Peters, and S. Makino. 2006. Rescue of infectious Rift Valley fever virus entirely from cDNA, analysis of virus lacking the NSs gene and expression of a foreign gene. J. Virol. 80:2933-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikegami, T., K. Narayanan, S. Won, W. Kamitani, C. J. Peters, and S. Makino. 2009. Rift Valley fever virus NSs protein promotes posttranscriptional downregulation of protein kinase PKR and inhibits eIF2a phosphorylation. PLoS Pathog. 5:e1000287. doi: 10.1371/journal.ppat.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laherty, C. D., A. N. Billin, R. M. Lavinsky, G. S. Yochum, A. C. Bush, J.-M. Sun, T.-M. Mullen, J. R. Davie, D. W. Rose, C. K. Glass, M. G. Rosenfeld, D. E. Ayer, and R. N. Eisenman. 1998. SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol. Cell 2:33-42. [DOI] [PubMed] [Google Scholar]
- 19.Laughlin, L. W., J. M. Meegan, L. J. Strausbaugh, D. M. Morens, and R. H. Watten. 1979. Epidemic Rift Valley fever in Egypt: observations of the spectrum of human illness. Trans. R. Soc. Trop. Med. Hyg. 73:630-633. [DOI] [PubMed] [Google Scholar]
- 20.Le May, N., S. Dubaele, L. P. De Santis, A. M. Billecocq, and M. Bouloy. 2004. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell 116:541-550. [DOI] [PubMed] [Google Scholar]
- 21.Le May, N., Z. Mansuroglu, P. Léger, T. Josse, G. Blot, A. Billecocq, R. Flick, Y. Jacob, E. Bonnefoy, and M. Bouloy. 2008. A SAP30 complex inhibits IFN-beta expression in Rift Valley fever virus-infected cells. PLoS Pathog. 4:134-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lundgren, M., C.-M. Chow, P. Sabbattini, A. Georgiou, S. Minaee, and N. Dillon. 2000. Transcription factor dosage affects changes in higher order chromatin structure associated with activation of a heterochromatic gene. Cell 103:733-743. [DOI] [PubMed] [Google Scholar]
- 23.Mokrani, H., O. Sharaf el Dein, Z. Mansuroglu, and E. Bonnefoy. 2006. Binding of YY1 to the proximal region of the murine beta interferon promoter is essential to allow CBP recruitment and K8H4/K14H3 acetylation on the promoter region after virus infection. Mol. Cell. Biol. 26:8551-8561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monier, K., J. C. Gonzalez Armas, S. Etteldorf, P. Ghazal, and K. F. Sullivan. 2000. Annexation of the interchromosomal space during viral infection. Nat. Cell Biol. 2:661-665. [DOI] [PubMed] [Google Scholar]
- 25.Muller, R., J. F. Saluzzo, N. Lopez, T. Dreier, M. Turell, J. Smith, and M. Bouloy. 1995. Characterization of clone 13, a naturally attenuated avirulent isolate of Rift Valley fever virus, which is altered in the small segment. Am. J. Trop. Med. Hyg. 53:405-411. [DOI] [PubMed] [Google Scholar]
- 26.R Development Core Team. 2009. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- 27.Ritchie, K., C. Seah, J. Moulin, C. Issac, F. Dick, and N. G. Bérubé. 2008. Loss of ATRX leads to chromosome cohesion and congression defects. J. Cell Biol. 180:315-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roulston, A., R. C. Marcellus, and P. E. Branton. 1999. Viruses and apoptosis. Annu. Rev. Microbiol. 53:577-628. [DOI] [PubMed] [Google Scholar]
- 29.Schmaljohn, C., and J. W. Hooper. 2001. Bunyaviridae: the viruses and their replication, p.1581-1602. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 30.Shestakova, E., Z. Mansuroglu, H. Mokrani, N. Ghinea, and E. Bonnefoy. 2004. Transcription factor YY1 associates with pericentromeric γ-satellite DNA in cycling but not in quiescent (G0) cells. Nucleic Acids Res. 32:4390-4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srivastava, S. P., K. U. Kumar, and R. J. Kaufman. 1998. Phosphorylation of eukaryotic translation initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J. Biol. Chem. 273:2416-2423. [DOI] [PubMed] [Google Scholar]
- 32.Swanepoel, R., and N. K. Blackburn. 1977. Demonstration of nuclear immunofluorescence in Rift Valley fever infected cells. J. Gen. Virol. 34:557-561. [DOI] [PubMed] [Google Scholar]
- 33.van Steensel, B., E. P. van Binnendijk, C. Diane Hornsby, H. T. M. van der Voort, Z. S. Krozowski, E. R. de Kloet, and R. van Driel. 1996. Partial colocalization of glucocorticoid and mineralocorticoid receptors in discrete compartments in nuclei of rat hippocampus neurons. J. Cell Sci. 109:787-792. [DOI] [PubMed] [Google Scholar]
- 34.Vialat, P., A. Billecocq, A. Kohl, and M. Bouloy. 2000. The S segment of rift valley fever phlebovirus (Bunyaviridae) carries determinants for attenuation and virulence in mice. J. Virol. 74:1538-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Viiri, K. M., J. Jänis, T. Siggers, T. Y. K. Heinonen, J. Valjakka, M. L. Bulyk, M. Mäki, and O. Lohi. 2009. DNA-binding and -bending activities of SAP30L and SAP30 are mediated by a zinc-dependent module and monophosphoinositides. Mol. Cell. Biol. 29:342-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vos, L. J., J. K. Famulski, and G. K. T. Chan. 2006. How to build a centromere: from centromeric and pericentromeric chromatin to kinetochore assembly. Biochem. Cell Biol. 84:619-639. [DOI] [PubMed] [Google Scholar]
- 37.Won, S., T. Ikegami, C. J. Peters, and S. Makino. 2007. NSm protein of Rift valley fever virus suppresses virus-induced apoptosis. J. Virol. 81:13335-13345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yadani, F. Z., A. Kohl, C. Prehaud, A. Billecocq, and M. Bouloy. 1999. The carboxy-terminal acidic domain of Rift Valley Fever virus NSs protein is essential for the formation of filamentous structures but not for the nuclear localization of the protein. J. Virol. 73:5018-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]