Abstract
To accommodate its RNA synthesis in the infected cell, severe acute respiratory syndrome coronavirus (SARS-CoV) induces a cytoplasmic reticulovesicular network (RVN) that is derived from endoplasmic reticulum (ER) membranes. We set out to investigate how the early secretory pathway interacts with the RVN and the viral replication/transcription complex (RTC) that is anchored to it. When the secretory pathway was disrupted by brefeldin A (BFA) treatment at the start of infection, RVN formation and viral RTC activity were not blocked and continued up to 11 h postinfection, although RNA synthesis was reduced by ca. 80%. In vitro RTC assays, using membrane fractions from infected cells, demonstrated that BFA does not directly interfere with the activity of the viral RNA-synthesizing enzymes. Confocal microscopy studies showed that early secretory pathway components are not associated with SARS-CoV-induced replication sites, although our studies revealed that infection induces a remarkable redistribution of the translocon subunit Sec61α. Ultrastructural studies, including electron tomography, revealed that the formation of the RVN and all its previously documented features can occur in the presence of BFA, despite differences in the volume and morphology of the network. We therefore conclude that early secretory pathway proteins do not play a direct role in RVN morphogenesis or the functionality of the SARS-CoV RTC. The BFA-induced disruption of ER integrity and functionality probably affects the overall quality of the membrane scaffold that is needed to support the viral RTC and/or the availability of specific host factors, which in turn compromises viral RNA synthesis.
In eukaryotic cells, the RNA replication of plus-stranded RNA (+RNA) viruses occurs exclusively in the cytoplasm and is inextricably associated with modified host membranes. Depending on the virus group, membrane modifications can range from small invaginations in the (outer) membrane of the target organelle to multiple, physically connected membrane compartments, including vesicular and reticular structures (for reviews, see references 31, 33, 37, and 45). The microenvironment created in this manner presumably benefits the activities of the viral replication complex. The membrane structures probably promote efficient RNA synthesis by concentrating the molecular players and likely also shield the viral machinery from host defense mechanisms recognizing viral proteins and/or intermediates of viral RNA synthesis. Little is known about the morphogenesis, activities, and maintenance of these viral RNA factories. Their structural and functional dissection will enhance our understanding of +RNA virus replication strategies and may reveal new opportunities for antiviral strategies.
Among +RNA viruses, coronaviruses are unique for their exceptionally large polycistronic genome of ∼30 kb (for a recent review, see reference 42). The large open reading frames (ORFs) 1a and 1ab are translated from the genomic mRNA, yielding the replicase precursor polyproteins pp1a and pp1ab, whereas downstream ORFs, encoding structural and “accessory” proteins, are expressed from a nested set of subgenomic mRNAs. Two or three proteinases encoded in ORF1a process pp1a and pp1ab into 15 or 16 nonstructural protein subunits (nsp's), most of which are known or presumed to be functionally associated with the viral replication/transcription complex (RTC) that drives the production of new genomes and subgenomic mRNAs (18). Three nsp's (3, 4, and 6) contain multiple membrane-spanning domains and are likely involved in the modification of intracellular membranes into the unusual membrane structures to which the coronavirus RTC is anchored (20, 39, 49, 50).
Following the 2003 severe acute respiratory syndrome (SARS) outbreak (for a review, see reference 41), SARS coronavirus (SARS-CoV) has rapidly become one of the best-studied members of the coronavirus family. Like mouse hepatitis coronavirus (MHV) (20), SARS-CoV replication induces cytoplasmic membrane alterations, with the most conspicuous structures being large numbers of double-membrane vesicles (DMVs) with diameters of 250 to 300 nm (17, 49). The modified membranes are concentrated in the perinuclear area of the infected cell and label for a variety of coronavirus RTC subunits (21, 43, 49). Using electron tomography (ET), we recently established that SARS-CoV DMVs are not free-floating vesicles but instead are interconnected through their outer membranes via narrow necklike connections and can thus be described as single-membrane vesicles surrounded by a common outer membrane (25). In fact, these interconnected DMVs are part of a membranous reticulovesicular network (RVN) that also includes convoluted membranes (CM) and is physically connected to the endoplasmic reticulum (ER). Ribosomes can be found on both CM and DMV outer membranes. Late in infection, the interconnected DMVs transform into so-called vesicle packets (VPs), in which multiple inner vesicles are surrounded by a more dilated outer membrane. Frequently, virus particles can be seen budding from VP outer membranes into the lumen. The interior of DMV inner vesicles labels extensively for dsRNA, presumably representing intermediates of viral replication and transcription. However, the bulk of various replicase proteins (nsp3, nsp5, and nsp8) localizes to the CM, and not to DMVs (25). In addition, even with the resolution of ET, visible connections between the double-stranded RNA (dsRNA)-containing interior of DMVs and the cytoplasm were not detected, raising a number of questions regarding RVN morphogenesis, function, and the exact position and orientation of the RTC in relation to the modified membranes (25).
Besides viral transmembrane nsp's, cellular pathways or factors likely play a role in the morphogenesis and/or function of membrane structures associated with +RNA virus replication, which was in particular investigated for various picornaviruses. Picornavirus “replication vesicles” usually are of nonuniform shape and size (ranging from 70 to 400 nm), appear to have a double membrane, and lack obvious connections to the cytosol (3, 12, 16, 31, 35, 46, 51). Again, the ER appears to be the primary source for vesicle formation, although cellular ER proteins are excluded from these structures, except for several secretory pathway markers (44, 51). Late in infection, also markers of other organelles, such as lysosomes and the Golgi complex, can be found in these picornavirus-induced structures (31, 46). Although not yet addressed with the resolution of ET, the available light and electron microscopy data suggest that these picornavirus-induced membrane structures may resemble the coronavirus-induced RVN (16, 35, 46, 51).
The connection between picornavirus replication vesicles, RNA synthesis, and the early secretory pathway has been investigated in considerable detail. It was proposed that vesicle formation is triggered by the assembly of COPII coats, which direct vesicular transport from the ER to the ER-Golgi intermediate compartment (ERGIC) (44). The involvement of the COPI coat complex was investigated by using the drug brefeldin A (BFA), which forms stable trimeric complexes with one of three guanine nucleotide exchange factors (GEFs)—GBF1, BIG1, or BIG2—and an ADP-ribosylation factor (Arf) belonging to a family of small GTPases. As a result, Arf cannot be activated to recruit subunits of the COPI complex to the membrane, which in turn blocks COPI-mediated vesicular transport between ERGIC and the Golgi complex and retrograde transport between the Golgi complex and the ER (11, 30, 36, 40). Picornaviruses display highly variable sensitivity to BFA treatment. For example, poliovirus and echovirus 11 replication is completely blocked by BFA (5, 16, 22, 32), whereas other family members are partially sensitive or not sensitive at all (16, 35, 38). The nonsensitive picornaviruses do not seem to require or recruit COPI, whereas the replication complexes of sensitive viruses clearly associate with COPI-coated membranes and depend on COPI function, which is reflected in a strong sensitivity to BFA (16). Poliovirus protein 3A appears to hijack GBF1, which also inhibits secretory pathway function (2, 10, 62, 63). In support of these findings, the sensitivity of poliovirus replication to BFA can in part be overcome by GBF1 overexpression (2). Protein 3CD recruits BIG1 and BIG2, possibly with similar effects (2). As also suggested by in vitro experiments in which BFA was added, the COPI machinery is likely involved in the replication process itself but is not a key player in vesicle biogenesis (1, 5).
Two recent studies have addressed the involvement of the early secretory pathway in coronavirus infection. Oostra et al. disrupted COPII-driven protein export from the ER and found that this impaired MHV replication (39). Verheije et al. (59) then analyzed the effect of BFA treatment and GBF1/Arf knockdown and again noticed a significant reduction in MHV replication. However, the relation between MHV replication and the processes targeted by BFA treatment seemed to be of a very different nature than the intimate involvement of GEFs and Arfs in the replication of BFA-sensitive picornaviruses. Whereas the latter are completely blocked by the presence of the drug, MHV replication was merely inhibited, and virus-induced membrane alterations could still be observed. Furthermore, no significant colocalization was observed between the MHV replicase, represented by nsp8, and any of the BFA targets, which is in strong contrast to the results obtained for the above-mentioned picornaviruses.
The present study addresses the relation between SARS-CoV RNA synthesis and the host cell's early secretory pathway. The addition of BFA to SARS-CoV-infected cells early in infection inhibited RVN formation and viral RNA synthesis, but the drug did not completely block viral RNA synthesis, which continued for up to 10 h. In vitro RTC assays were used to exclude the possibility that the drug might directly affect the activity of SARS-CoV key enzymes. By confocal microscopy, we compared the subcellular localization of the SARS-CoV RVN to that of early secretory pathway factors and other ER markers. Although none of the secretory pathway markers colocalized with the RVN, infection induced a remarkable redistribution of the translocon subunit Sec61α. Ultrastructural studies, including ET, revealed that a reduced level of RVN formation can be maintained in the presence of BFA, suggesting that it is unlikely that the early secretory pathway is intimately involved in coronavirus replication.
MATERIALS AND METHODS
Virus, cell culture, antisera, and cellular markers.
SARS-CoV strain Frankfurt-1 was used to infect Vero-E6 cells (multiplicity of infection [MOI] of 10) as described previously (23). All work with live SARS-CoV was performed inside biosafety cabinets in the biosafety level 3 facility at Leiden University Medical Center. Polyclonal rabbit antisera recognizing SARS-CoV nsp3, nsp5, and nsp8 and M protein have been described previously (49). A rabbit antiserum against the N protein was raised as described previously (49) using a recombinant full-length N protein as antigen. Mouse monoclonal antibody (MAb) J2 (47) recognizing dsRNA was purchased from Scicons (Hungary). To visualize host cell marker proteins, a variety of antibodies and yellow fluorescent protein (YFP)-tagged proteins were used. Mouse MAbs recognizing protein disulfide isomerase (PDI) (58), Giantin (G1/133; Alexis Biochemicals), GBF1 (BD Transduction Labs), and Sec13 (52) were used. Mouse MAb against human β-actin (clone AC74) was obtained from Sigma-Aldrich, and rabbit anti-human sec61α was purchased from Upstate. Rabbit anti-syntaxin 5 was kindly provided by Matsuo Tagaya (Tokyo, Japan), and expression constructs encoding wtArf1, Arf1T31N, and Arf1Q71L (57) were kindly made available by Guillermo Romero (Pittsburg, PA). Transfections in combination with SARS-CoV infections were done as described before (23). TUNEL assay kits were from Roche (In Situ Cell Death Detection kit TMR Red), and a kit for MTS assays was obtained from Promega.
Metabolic labeling of viral RNA synthesis and RNA hybridization analysis.
At different time points after infection, in the presence or absence of BFA, infected cells were given 10 μg of actinomycin D (ActD; Sigma-Aldrich)/ml for 1 h to block cellular transcription. Subsequently, viral RNA synthesis was labeled for 1 h using medium containing 73 μCi of [3H]uridine and 10 μg of ActD/ml. For RNA isolation, cells were washed with phosphate-buffered saline (PBS) and lysed for 5 min in a buffer containing 20 mM Tris-HCl (pH 7.6), 150 mM NaCl, 0.5% deoxycholine, 1.0% Nonidet P-40, 0.1% sodium dodecyl sulfate, and 1.25 U of DNase I (Invitrogen)/ml. Lysates were mixed with an equal volume of 100 mM Tris-HCl (pH 8), 500 mM LiCl, 10 mM EDTA (pH 8), 5 mM dithiothreitol, and 5% lithium dodecyl sulfate containing 100 μg of proteinase K/ml, followed by incubation for 10 min at 37°C. Aliquots (5 μl), equaling approximately 1,500 cells, were spotted in duplicate on Whatman filter paper. After precipitation by washing with TCA-P (5% TCA, 100 mM Na4P2O7, 95 mM NaH2PO4) and drying, the incorporation of [3H]uridine was measured in a Beckman liquid scintillation counter (LS6500 IC). Alternatively, intracellular RNA was isolated by phenol extraction and ethanol precipitation, separated in a formaldehyde-agarose gel, and hybridized to a 32P-labeled oligonucleotide probe complementary to the 3′ end of the genome, as described previously (48).
SDS-PAGE and Western blotting.
Cells were lysed in Laemmli sample buffer (28), boiled, separated in SDS-10% PAGE gels, and electroblotted onto polyvinylidene difluoride membrane according to standard protocols. Blots were blocked with 5% skimmed milk and 0.02% Tween 20 in PBS prior to incubation with antisera dilutions. Between antibody incubations, blots were washed with 0.5% skimmed milk and 0.02% Tween 20 in PBS and before ECL-Plus chemiluminescence detection (GE Healthcare) washing was done with PBS containing 0.02% Tween 20.
IF microscopy.
Infected cells on glass coverslips were fixed with 3% paraformaldehyde at the indicated time points postinfection (p.i.) and processed for immunofluorescence (IF) microscopy essentially as described by van der Meer et al. (55). As before, an Alexa 488-coupled SARS-CoV nsp3 antiserum was used to visualize the viral replicase. After permeabilization, single- or dual-labeling IF assays were carried out with rabbit antisera and/or mouse monoclonal antibodies, which were detected by using indocarbocyanine (Cy3)-conjugated donkey anti-rabbit immunoglobulin and Alexa Fluor 488-conjugated goat anti-mouse immunoglobulin secondary antibodies, respectively (Molecular Probes/Invitrogen). Samples were examined with a Leica SP5 confocal laser scanning microscope, using a pinhole size of 1 airy unit. Images were optimized and analyzed for colocalization with the WCIF version of ImageJ (http://www.uhnresearch.ca/facilities/wcif/imagej/). The product of the differences from the mean (PDM) images and the Manders' overlap coefficients were calculated as described by Li et al. (29).
In vitro RNA synthesis assay.
In vitro assays to measure the RTC activity of membrane fractions isolated from infected cells (10 h p.i.) were performed as described previously (56). Activity was measured on the basis of [α-32P]CTP incorporation into RNA products. Assays were done in the presence or absence of 6 μg of BFA/ml and were terminated after 40 or 100 min. Subsequently, RNA was isolated by acid-phenol extraction and isopropanol precipitation. Reaction products were separated on 1% denaturing formaldehyde agarose gels and visualized by using a Typhoon variable mode imager (GE Healthcare, Chalfont St. Giles, United Kingdom). Phosphorimager data were analyzed and quantified by using ImageQuant TL software (GE Healthcare).
Electron microscopy.
For ultrastructural morphological investigations, SARS-CoV-infected Vero-E6 cells, with or without BFA treatment, were prepared as described before (25). After prefixation, cryofixation by plunging into liquid ethane, and freeze substitution, the samples were embedded in epoxy LX-12 resin. Thin sections (100 nm) were viewed with a FEI Tecnai 12 Biotwin transmission electron microscope operating at 120 kV.
DMVs were quantified in the cell slice showing the largest nuclear diameter as described before (25). Briefly, electron micrographs covering the entire cross-section of the cell were recorded and digitally merged to produce an image composition representing a 100-nm-thick plane through the center of the cell. Merged images were analyzed using ImageJ software (http://rsb.info.nih.gov/ij/). Examples of representative images used for DMV counting have been deposited in the Cell Centered Database (http://ccdb.ucsd.edu) under the accession numbers 6935 to 6938.
For immuno-electron microscopy, infected cell monolayers were prefixed, cryofixed by high-pressure freezing using a Leica EM PACT2, and freeze substituted using anhydrous acetone containing 0.2% uranyl acetate. After being washed with ethanol, the samples were infiltrated with Lowicryl HM20 and polymerized under UV light at −50°C. Thin sections were labeled with anti-dsRNA mouse MAb J2 (47), a bridging rabbit- anti-mouse IgG antibody (Dako Cytomation) and protein A-gold particles (10 or 15 nm). Grids were contrasted with uranyl acetate and lead hydroxide.
Electron tomography.
Freeze-substituted cell samples, processed for morphological investigation as described above, were used to cut 200-nm-thick sections. Then, 10-nm gold particles were layered on top of the sections as fiducial markers. For dual-axis tomography, two single-axis tilt series were recorded with a FEI T12 transmission EM operating at 120 kV. Per single-axis tilt series, 141 images were recorded with 1° tilt increments by using automated tomography acquisition software (Xplore 3D; FEI Company). Images were acquired with a cooled slow-scan charge-coupled device camera (4k Eagle; FEI Company). To obtain electron tomograms, the dual-axis tilt series were aligned by means of the fiducial markers and processed by using the IMOD software package (27). The size of the voxels in the tomogram corresponds to 1.2 nm. The full data set has been deposited in the Cell Centered Database (http://ccdb.ucsd.edu) under accession number 6939. Three-dimensional surface-rendered reconstructions of the RVN and adjacent cellular features were produced using the AMIRA visualization package (TSG Europe) by masking structures of interest and subsequent thresholding. For this process, volumes were denoised by using nonlinear anisotropic diffusion filtering (13). Videos were made with QuickTime Pro (http://www.apple.com/quicktime).
RESULTS
SARS-CoV replication is inhibited, but not abrogated, by BFA treatment.
Before treating SARS-CoV-infected cells with BFA, the drug was titrated to establish the dose to use (range tested, 1 to 12 μg/ml). By IF microscopy 6 μg of BFA/ml was found to suffice to completely disperse the Golgi complex in all cells. TUNEL assays did not show apoptosis at this BFA concentration and also MTS cell viability assays did not reveal adverse effects of this drug concentration during treatment for up to 10 h, the maximum incubation time used in our experiments.
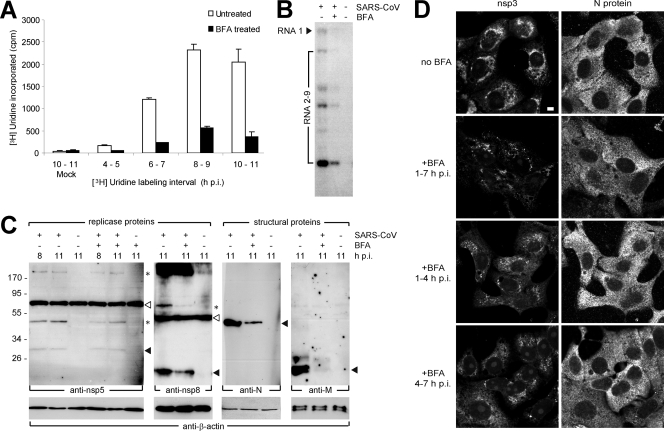
Vero-E6 cells were infected with SARS-CoV (MOI of 10) and, upon removal of the inoculum at 1 h p.i., medium containing BFA was given to the cells. Subsequently, viral RNA synthesis was monitored at different time points after infection by performing 1-h metabolic labeling with [3H]uridine. ActD was added to inhibit cellular transcription. Tritium incorporation into RNA was measured by scintillation counting and, as illustrated in Fig. 1A, SARS-CoV RNA synthesis was considerably affected, but certainly not blocked by BFA treatment. Compared to the untreated control, ca. 20% of residual activity was measured throughout infection. Essentially similar data were obtained using a second cell line, mouse L cells expressing the human ACE2 receptor used by SARS-CoV (data not shown). In line with these metabolic labeling data, a hybridization analysis of viral RNA produced in the presence of BFA confirmed that at 9 h p.i. both genome and subgenomic mRNAs had accumulated to ca. 20% of the amount observed in the untreated control (Fig. 1B), indicating that replication and transcription were equally affected by the presence of the drug.
FIG. 1.
SARS-CoV replication is inhibited, but not abrogated, by BFA treatment. SARS-CoV-infected Vero-E6 cells were BFA treated or mock treated from 1 h p.i. until the time of harvesting or fixation. (A) Viral RNA synthesis at different time points p.i. in the presence or absence of BFA. At 1 h prior to metabolic labeling of viral RNA synthesis, cells were given 10 μg of ActD/ml to inhibit host cell transcription. After a 1-h [3H]uridine labeling in the presence of ActD, cells were harvested and incorporation of radiolabel was measured by scintillation counting. (B) Hybridization analysis to compare the viral RNA content (genome and subgenomic mRNAs) from infected control cells and BFA-treated infected cells lysed at 9 h p.i. RNA isolated from uninfected cells was included as a negative control. (C) Western blot showing the accumulation of SARS-CoV nsp5, nsp8, nucleocapsid (N), and membrane (M) protein (indicated by the respective black arrowheads) in the presence or absence of BFA. β-Actin was included as a loading control (lower panels). The asterisks indicate possible SARS-CoV replicase processing intermediates, and the white arrowheads indicate nonspecific bands that were also detected in mock-infected cell lysates. (D) IF assays showing nsp3 and N protein accumulation in SARS-CoV-infected cells that were fixed at 7 h p.i and BFA treated for the indicated time intervals. Bar, 5 μm.
To assess the effect of BFA treatment on viral protein synthesis, cell extracts were analyzed by Western blotting (Fig. 1C). A clear reduction in the amount of the structural proteins M and N, expressed from subgenomic mRNAs 8 and 9, was observed, but nsp levels seemed to be only moderately affected. Although we cannot formally exclude major stability differences between nsp's and structural proteins, these observations suggest that—under normal conditions—there is an excess of genome molecules and that part of these may not be engaged in translation.
IF microscopy revealed that the effect of BFA, when present from 1 to 7 h p.i., can differ substantially between individual infected cells. Most of the cells showed reduced signal for nsp3 and N protein and a more punctate and dispersed nsp3 labeling pattern compared to the dense perinuclear staining in untreated control cells (Fig. 1D, compare the top four panels). However, in some cells the difference with untreated cells was barely detectable (not shown). When analyzed at 7 h p.i., the overall effect of BFA treatment from 1 to 4 h p.i. was similar, i.e., the labeling was more punctate and dispersed, although compared to samples treated 1 to 7 h p.i. the reduction of the signal was less obvious (Fig. 1D). The sustained effects observed at 7 h p.i. after 1 to 4 h p.i. treatment may be explained by the observation that after removal of the drug, the secretory pathway of Vero-E6 cells needed several hours to recover from BFA treatment (data not shown). When BFA was added later in infection (at 4 h p.i.), the staining was generally more indistinguishable from that in untreated control cells at 7 h p.i. (Fig. 1D). This implies that in particular the early phase of SARS-CoV replication is likely to be sensitive to the effects of BFA treatment.
BFA does not inhibit the RNA-synthesizing enzymes of the SARS-CoV RTC.
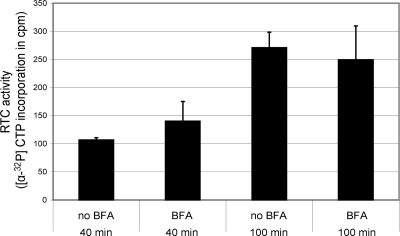
To investigate the possibility that BFA might inhibit one of the many enzyme functions of the SARS-CoV RTC, we made use of an in vitro RTC assay recently developed in our laboratory. The assay is based on isolation of membrane fractions and the associated RTCs from infected cells and in vitro RTC activity is measured on the basis of incorporation of [α-32P]CTP into viral RNA products (56). BFA was added to this type of assay at the same concentration (6 μg/ml) as that used for the in vivo experiments, and the RTC activity was measured 40 and 100 min into the reaction. At these time points, we could not establish a negative influence of BFA on RTC activity (Fig. 2), indicating that the observed reduction of viral RNA synthesis in vivo was probably not due to the direct inhibition of viral RNA-synthesizing enzymes by the presence of the drug.
FIG. 2.
BFA does not inhibit the RNA-synthesizing enzymes of the SARS-CoV RTC. Using membrane fractions isolated from SARS-CoV-infected cells, an in vitro RNA synthesis assay (56) was performed to assess the effect of BFA on SARS-CoV RTC activity. The incorporation of [α-32P]CTP into viral RNA products was allowed to continue for 40 or 100 min, either with or without 6 μg of BFA/ml being present in the reaction. No significant inhibition of RTC activity by BFA was observed.
Early secretory pathway factors, including those targeted by BFA, do not colocalize with the SARS-CoV replicase.
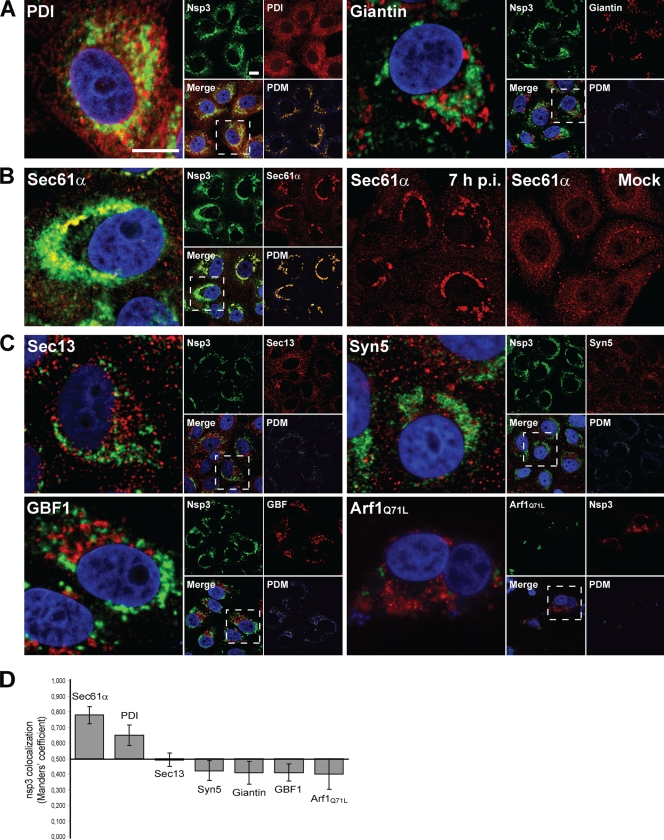
In contrast to observations made for certain picornaviruses, BFA did not completely block SARS-CoV virus replication and had no direct effect on the in vitro activity of isolated RTCs, as described above. We subsequently sought to determine whether the proteins targeted by BFA or other early secretory pathway factors would colocalize with the SARS-CoV replicase, possibly revealing their involvement in RVN formation or RTC function. This was assessed by using confocal laser scanning microscopy (Fig. 3A to C), which was supplemented with quantifications of colocalization levels of SARS-CoV replicase subunit nsp3 and a variety of cellular marker proteins (Fig. 3D; see also Materials and Methods and the figure legend). nsp3 was previously shown to colocalize with the majority of replicase subunits, including several of the key enzymes of the viral RTC (25, 49).
FIG. 3.
Early secretory pathway factors are not directly associated with the SARS-CoV-induced RVN. Colocalization analysis of the SARS-CoV replicase and a variety of cellular marker proteins by indirect confocal microscopy. SARS-CoV-infected Vero-E6 cells were fixed at 7 h p.i. and processed for IF assays. A cytomegalovirus (CMV) promoter-containing expression vector for YFP-tagged Arf1Q71L was transfected to visualize this Arf mutant. The subpanels marked PDM (lower right corner of each set in panels A to C) show the product of the differences from the mean (PDM) (29), a method for colocalization analysis in which positive values (displayed in yellow) represent signal in both channels, whereas negative values (blue) represent signal in only one of the channels. Bars, 10 μm. (A) As positive and negative controls for colocalization, the ER resident protein PDI and Golgi marker Giantin were included, respectively. As reported previously, PDI labeling partially overlapped with the replicase, whereas Giantin showed no colocalization. (B) Redistribution of Sec61α, a major component of the translocon, upon SARS-CoV infection. The double labeling on the left shows the extensive overlap of the signal for nsp3 and Sec61α in the perinuclear area of SARS-CoV-infected cells. The two panels on the right illustrate the virus-induced redistribution of Sec61α, which normally shows a typical reticular labeling pattern throughout the cytoplasm. (C) Colocalization analysis for a number of important early secretory pathway markers, including factors targeted by BFA treatment. Colocalization with SARS-CoV nsp3 was not observed for Sec13, Syntaxin 5 (Syn5), GBF1, and Arf1. (D) Graph showing the average Manders' overlap coefficients of the host proteins used in this analysis and SARS-CoV nsp3 (n = 18 cells per pair). By definition, Manders' overlap coefficients range from 0 to 1, representing full separation and complete colocalization of signals, respectively. We interpreted values greater than 0.5 as indicative of (a certain level of) colocalization and values less than 0.5 as indicative of a lack of colocalization.
We first analyzed the colocalization of nsp3 and resident ER proteins not involved in vesicular trafficking. Earlier studies have reported that the luminal ER PDI partially colocalizes with the SARS-CoV replicase (23), which was confirmed in the present study (Fig. 3A and D). As a negative control for colocalization, we used the Golgi marker Giantin, which indeed did not colocalize (3A and D), in line with previous reports that SARS-CoV nsp's and Golgi markers are separated in the cell (23).
Various other ER markers showed a generally very limited colocalization with nsp3 (data not shown), but the labeling for one marker protein strikingly overlapped with the SARS-CoV replicase. This protein is Sec61α, a key subunit of the translocon that is involved in the docking of ribosomes on the ER and translocation of the nascent polypeptide chain into the lumen. In uninfected cells, Sec61α labeling was dispersed throughout the cytoplasm (Fig. 3B), but upon SARS-CoV infection the protein became concentrated around the nucleus and colocalized extensively with the characteristic perinuclear labeling pattern of the SARS-CoV replicase (Fig. 3B and D). To exclude that the Sec61α antiserum cross-reacted with any of the viral nsp's, which could also have explained our observation, we checked its specificity by Western blot analysis. In both uninfected and SARS-CoV-infected Vero-E6 cell lysates, the serum recognized only a single protein species of ∼40 kDa, the reported molecular mass of Sec61α (19; data not shown). This makes cross-reaction with viral proteins unlikely, and we therefore believe that in infected cells Sec61α may indeed be recruited to the SARS-CoV-induced RVN.
No similar recruitment to the SARS-CoV RVN was observed for any of the ER-associated secretory pathway markers that were tested. In fact, neither the COPII coat subunit Sec13 nor the COPI-associated proteins GBF1 and Arf1, which are targeted by BFA treatment, showed any colocalization with SARS-CoV nsp3 (Fig. 3C and D). Colocalization with Arf1 was tested by the transfection of infected cells with a plasmid expressing a YFP-tagged Q71L mutant of Arf1 (57, 65). This mutant has increased affinity for GTP, which prolongs its active and membrane-associated state, in contrast to wild-type Arf1 that continuously shuttles between cytosol and membranes (65). Expression of this mutant protein facilitated the colocalization analysis of Arf1 and nsp3, but no overlap was observed (Fig. 3C and D). Also wild-type Arf1 and the inactive T31N Arf1 mutant, which is predominantly present in the cytosol (6), did not colocalize with nsp3 (data not shown). Finally, we included syntaxin 5, an essential t-SNARE protein of the ER and cis-Golgi involved in vesicular transport between these compartments (7), but also this early secretory pathway marker did not colocalize with SARS-CoV nsp3 (Fig. 3C and D). We therefore conclude that early secretory pathway factors, including those targeted by BFA, are not abundantly associated with the SARS-CoV-induced RVN.
BFA treatment appears to accelerate RVN maturation.
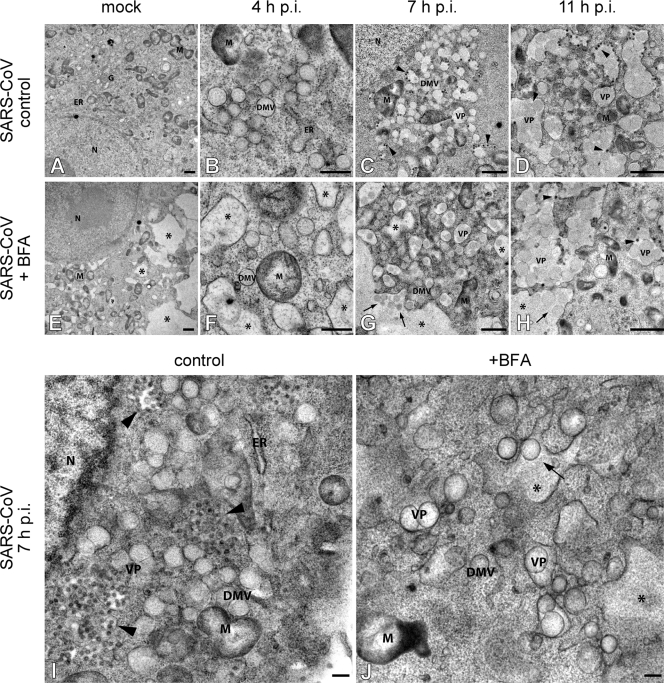
Since we concluded that it is unlikely that cellular targets of BFA are directly involved in SARS-CoV replication, we sought to find another explanation for the effect of the drug. BFA disrupts the early secretory pathway and causes massive rearrangements of intracellular membrane compartments, including the ER that is the membrane donor of the SARS-CoV-induced RVN. We therefore analyzed the development and morphology of the RVN during BFA treatment. SARS-CoV-infected Vero-E6 cells were again given BFA at 1 h p.i. and were fixed and processed for EM analysis at 4, 7, or 11 h p.i. The presence of dilated ER (14, 34) and tubulovesicular clusters (15), documented effects of BFA treatment in sensitive cells, was apparent in both infected samples and mock-infected controls (Fig. 4E to H and J). When we compared the morphology of the RVN in the presence or absence of BFA, the early stage of infection (4 h p.i.) did not show striking differences, since regular DMVs were also observed in BFA-treated cells, often adjacent to typically swollen ER stacks (compare Fig. 4B and F). However, by 7 h p.i. the effect of BFA treatment had become very pronounced. Whereas we predominantly observed DMVs in untreated cells (Fig. 4C and I), BFA-treated cells mainly contained VPs, multiple inner vesicles sharing a dilated outer membrane, which usually becomes the dominant structure only later in infection (25) (Fig. 4G and J [compare with Fig. 4D]). Moreover, the luminal space between the inner vesicle and outer membranes of the VPs and remaining DMVs was often markedly increased compared to the structures in untreated cells, an observation reminiscent of what can be seen at later stages of infection in the absence of BFA treatment (e.g., see Fig. 4D, top left [see also below]). Therefore, we conclude that the drug apparently expedites the maturation of delimited though interconnected DMVs into VPs and thus accelerates the morphological maturation of the RVN, as observed during regular SARS-CoV infection.
FIG. 4.
BFA treatment appears to accelerate RVN maturation. Transmission EM analysis of the influence of BFA treatment on the morphogenesis of the SARS-CoV-induced RVN. Vero-E6 cells were mock infected (A and E) or SARS-CoV infected (B to D, F to H, I, and J) and were either not treated (A to D, and I) or treated with BFA (E to H and J) starting at 1 h p.i. Samples were processed for electron microscopy by cryofixation and freeze substitution as described in Materials and Methods. In all BFA-treated samples (E to H and J), the collapse of the Golgi complex and its merger with the ER resulted in the absence of Golgi stacks and the formation of large vacuoles (E to H and J; indicated by asterisks). Untreated SARS-CoV-infected cells showed the normal clusters of DMVs at 4 h p.i. (B), which gradually transformed into VPs from 7 h p.i. on (C, D, and I). When BFA was present, small DMV clusters were observed in close proximity of swollen ER at 4 h p.i. (F). By 7 h p.i., VPs had become the most prominent component of the RVN and seemed to merge with the dilated ER-Golgi vacuoles, which is a striking difference from the control cells (G and J, indicated by arrows). At 11 h p.i., many large VPs were seen, both in untreated and in BFA-treated cells (Fig. 3D and H). Virus particles were observed budding from VP outer membranes (indicated by arrowheads) in both untreated and BFA-treated cells. Panels I and J show higher-magnification examples of Vero-E6 cells fixed at 7 h p.i., with or without BFA treatment. Abbreviations: ER, endoplasmic reticulum; G, Golgi apparatus; M, mitochondrion; DMV, double membrane vesicle; VP, vesicle packet. Bars represent 500 nm (A, C to E, and G to H) or 250 nm (B, F, and I to J).
Clearly, the swollen ER induced by BFA treatment and RVN were integrated at 7 h p.i. and later time points of infection (arrows in Fig. 4G and H); this was probably due to the fusion of the two structures. By 11 h p.i., many large VPs were seen, both in untreated and in BFA-treated infected cells (Fig. 4D and H). Also, in both samples virus budding into RVN membranes or nearby compartments was apparent at 7 and 11 h p.i. (Fig. 4C and D and G to I). Examination of photo mosaics representing complete cross-sections of BFA-treated, infected cells showed that they contain many intracellular virions. However, in contrast to untreated infected cells, these were not observed near the plasma-membrane or outside the cells (data not shown), which was not unexpected since a functional exocytic pathway is known to be required for coronavirus maturation and release. Plaque assays using medium harvested from BFA-treated SARS-CoV-infected cells confirmed the very limited release of infectious progeny.
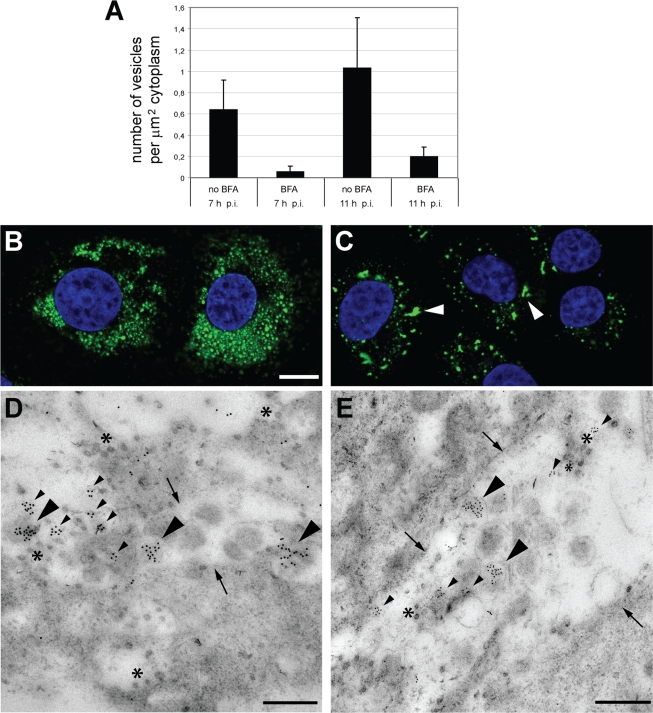
The same photo mosaics allowed us to accurately assess the effect of BFA treatment on the number of inner vesicles (either in individual DMVs or in VPs) present at 7 and 11 h p.i. We counted a total of 15 untreated and 15 BFA-treated infected cells for both time points and calculated the average number of inner vesicles per μm2 of cytoplasm. As illustrated by Fig. 5A, BFA treatment caused a major reduction in the number of vesicles: ca. 9 and 20% of the normal number were counted at 7 and 11 h p.i., respectively. We next sought to determine whether, despite their altered morphology and reduced number, the vesicles in BFA-treated cells still labeled for dsRNA, as described for regular infection samples (25). Indeed, both IF microscopy and immuno-electron microscopy confirmed the presence of dsRNA in the vesicles in BFA-treated cells (Fig. 5). IF analysis revealed a higher degree of clustering of the labeling after BFA treatment (compare Fig. 5B and C), which matched the larger number of VPs in these samples described above (Fig. 4). Immuno-electron microscopy showed that in BFA-treated cells dsRNA was still concentrated within the inner vesicles of VPs (Fig. 5D), as well as in vesicles inside dilated ER membranes (Fig. 5E). Although the exact significance of the dsRNA labeling inside SARS-CoV vesicles remains to be elucidated, our joint observations suggest that the primary difference induced by BFA is the accelerated maturation of the RVN, which may well contribute to the drop in RNA synthesis observed upon drug treatment.
FIG. 5.
BFA treatment affects RVN morphogenesis and volume, but vesicles still contain dsRNA. (A) Thin sections of SARS-CoV-infected cells were cut in the direction parallel to the substrate and, for each condition (with or without BFA; 7 or 11 h p.i.), the RVN inner vesicles in DMVs or VPs were counted. For this purpose, images covering a complete slice through the center of the cell were stitched into large mosaics to facilitate the analysis. Per sample, 15 cells were analyzed. The graph represents the average number of SARS-CoV-induced vesicles per μm2 of cytoplasm and illustrates how BFA treatment reduces the number of vesicles to ca. 9 and 20% at 7 and 11 h p.i., respectively. (B and C) SARS-CoV-infected cells were BFA treated (C) or left untreated (B), fixed at 11 h p.i., and processed for IF assays with a MAb specific for dsRNA. Imaging was done by using a confocal laser scanning microscope. Although both treated and untreated cells contained dsRNA products, the labeling seemed less intense in BFA-treated cells (C) compared to untreated cells (B). Also, the distribution of dsRNA was clearly different in BFA-treated samples, which contained fewer and larger clusters of label (C; arrowheads). Bars, 5 μm. (D and E) SARS-CoV-infected Vero-E6 cells were treated with BFA from 1 h p.i. until fixation at 11 h p.i. Immunogold labeling was performed with an antibody recognizing dsRNA. Two representative micrographs are shown. Large arrowheads indicate dsRNA labeling inside DMV inner vesicles; smaller arrowheads point to clusters of labeling not obviously associated with vesicles. Arrows indicate outer membranes of the large VPs that are characteristic of BFA-treated SARS-CoV-infected samples (see also Fig. 4G and H). Asterisks indicate examples of virus particles. Bars, 500 nm.
Electron tomography confirms the general integrity of the SARS-CoV-induced RVN upon BFA treatment.
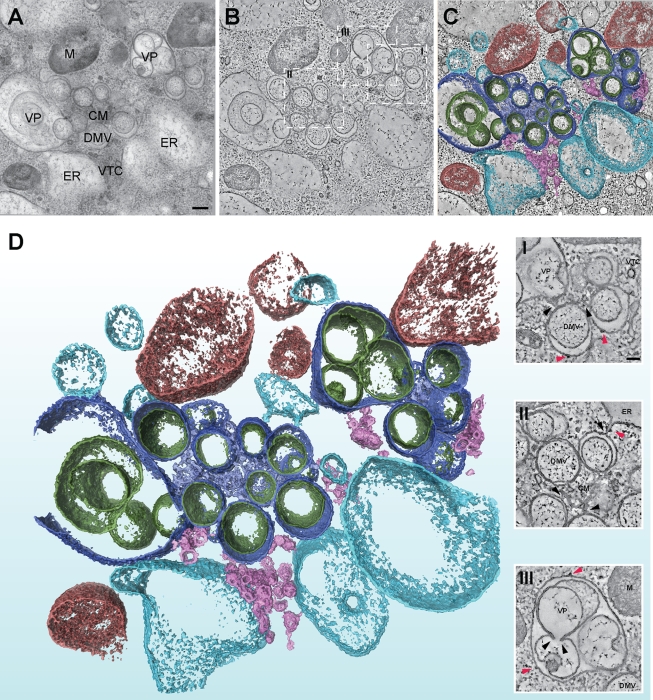
The EM analysis presented in Fig. 4 revealed interesting differences in RVN morphogenesis in the presence of BFA, particularly at 7 h p.i. (Fig. 4C and G). However, as described previously (25), various aspects of the three-dimensional organization of the RVN can only be properly discerned by ET. These include the structure of the CM, the narrow connections between DMVs and between DMVs and the ER, and the integrity of inner vesicle membranes. Therefore, ET was applied to BFA-treated samples fixed at 7 h p.i. (Fig. 6 and 7; see also Videos S1 and S2 in the supplemental material). Untreated control samples were included in all ET experiments and were processed for conventional EM to confirm the overall quality of infection experiments. The images in Fig. 4I and J were derived from the same experiment as used for the ET analysis shown in Fig. 6 and 7. An extensive ET analysis of untreated samples was described in a prior publication from our laboratory (25). Analysis of BFA-treated samples by ET revealed that the presence of the drug did not affect the necklike connections between DMVs and between DMVs and ER (Fig. 6D [closeups I and II], Video S2 in the supplemental material, and Fig. 7). The appearance of the CM compartment of the RVN, the intertwined membranes that are most abundantly labeled for viral nsp's (25), was also unchanged after BFA treatment (Fig. 6D, closeups I and II). As in untreated controls (25), ribosomes were associated with the outer membranes of the RVN upon BFA treatment (Fig. 6D, closeups I to III). The accelerated development of VPs, two or more inner vesicles sharing an outer membrane, was confirmed when BFA-treated cells and untreated controls were compared at 7 h p.i. (Fig. 6A to D, closeup III). The integrity of the inner vesicles quite often seemed disrupted and peculiar vesicle fusions were observed, resulting in complicated vesicular structures that seemed to have partially single and partially double membranes (Fig. 6A to D, closeup III; Video S2 in the supplemental material; and Fig. 7). Similar observations, although possibly with a lower frequency, were previously made late in the infection of non-BFA-treated cells (25). However, the meaning of these membrane disruptions is unclear, particularly in view of the documented extreme fragility of the inner vesicles, which might even be enhanced upon BFA treatment. The outer membranes, on the other hand, seemed intact and unchanged.
FIG. 6.
ET confirms the general integrity of the SARS-CoV-induced RVN upon BFA treatment. ET was applied for the three-dimensional analysis of the RVN in BFA-treated cells cryofixed at 7 h p.i. The top three panels illustrate how a three-dimensional surface-rendered model was derived by applying ET to a semithick section of a SARS-CoV-infected Vero-E6 cell, which had been treated with BFA from 1 to 7 h p.i. (A) A 0°-tilt transmission EM image of a 200-nm-thick resin-embedded section showing part of a SARS-CoV RVN after BFA treatment. Scale bar, 250 nm. (B) Using the IMOD software package, tomograms were computed from dual-axis tilt series of the 200-nm-thick section shown in panel A. The tomographic slice shown was taken from the central plane of the section and represents a thickness of 1.2 nm. The dashed squares mark the insets shown below in panel D. (C) The improved image from panel B after anisotropic diffusion filtering. The optimized signal-to-noise ratio facilitates masking, thresholding, and surface rendering. (D) Final three-dimensional surface-rendered model showing the RVN against a background of mitochondria (M, red), dilated ER and tubulo-vesicular clusters (both resulting from BFA treatment; depicted in light blue and pink, respectively). Furthermore, all substructures normally encountered in the SARS-CoV-induced RVN were observed after BFA treatment, including DMVs and VPs (outer membranes, dark blue; inner membranes, dark green) and CM (depicted in lavender). The insets (I, II, and III) show tilted tomographic slices taken from panel B and highlight RVN membrane connections between DMVs, between ER and DMVs, and between CM and DMVs (black arrowheads). Ribosomes attached to the cytosolic face of RVN membranes or ER are indicated with red arrowheads. Scale bar, 100 nm. For abbreviation definitions, see Fig. 4. TVC, tubulovesicular cluster (15).
FIG. 7.
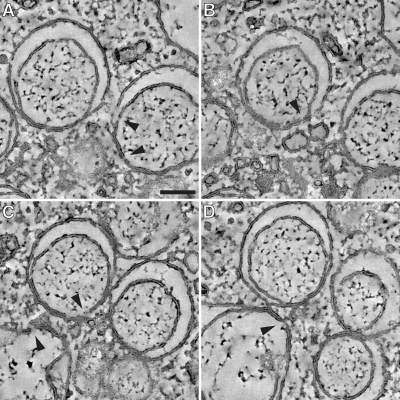
In-depth analysis of RVN membrane discontinuities after BFA treatment. The tomograms that were used for our ultrastructural analysis of RVN membranes in BFA-treated cells were scrutinized for openings that connect the DMV interior with the cytosol. (A to D) Examples of membrane discontinuities observed in our specimens (arrowheads), including a number that were present on the side of the vesicle facing the CM (e.g., in panels A and C), where double membranes remained tightly apposed. We doubt whether these discontinuities could be associated with viral RNA trafficking between vesicle interior and cytosol and consider it more likely that they may be artifacts resulting from the previously documented fragility of the RVN, which may actually be promoted by BFA treatment. Panels A to D also illustrate the separation of DMV inner and outer membranes during BFA treatment. However, the increased luminal space was generally lacking at the side of the DMV that faces the CM, where the two membranes remained tightly apposed. Panel D also shows an example of a necklike connection between the outer membranes of two DMVs. Bar, 100 nm.
In contrast to the tightly apposed DMV and type 1 VP membranes found in untreated SARS-CoV-infected cells (25), increased luminal space between the inner and outer membranes of VPs and particularly also DMVs was observed after BFA treatment (Fig. 6A to D, closeups I and II; Video S2 in the supplemental material; and Fig. 7). This implies that the treatment apparently disrupts the (unidentified) mechanism that is responsible for the close apposition of inner and outer membranes. An additional striking observation (Fig. 6A and B and Fig. 7) was the fact that the increased luminal space of many DMVs was not obvious at the side of the vesicle that faced the center of a DMV cluster, which is often adjacent to CM (Fig. 6, closeups I and II). This illustrates how the detailed analysis of the RVN under alternative conditions, like BFA treatment, may reveal additional structural features (see also the Discussion).
DISCUSSION
Our recent detailed analysis of the ultrastructure of the SARS-CoV-induced RVN revealed that the ER is the organelle from which this presumed scaffold for the coronavirus RTC arises (25). A logical next step was to investigate the relation between the morphogenesis and functionality of the RVN and the host cell's secretory pathway, for which the ER is the general starting point and membrane source. We utilized the commonly used drug BFA to disrupt the COPI pathway that directs vesicular transport between ER and Golgi complex and observed a more or less consistent 80% decrease of SARS-CoV RNA synthesis throughout infection (Fig. 1). Although this is a sizeable reduction, the effect of BFA treatment on the replication of certain picornaviruses, which appear intimately associated with and dependent on early secretory pathway factors, was found to be much more severe, rendering RNA synthesis essentially undetectable (16, 32). Our data on the effect of BFA treatment on SARS-CoV protein synthesis (Fig. 1C and D) and in vitro RTC activity (Fig. 2), the lack of colocalization between replicase and important secretory pathway factors (Fig. 3), and RVN morphogenesis in the presence of BFA (Fig. 4 and 5) are also consistent with the conclusion that the drug cannot abrogate either RTC function or RVN formation to a similar extent as reported for BFA-sensitive picornaviruses. Apparently, all critical steps in the early stages of viral replication can proceed in the presence of BFA, suggesting that the early secretory pathway does not directly participate in or contribute to these steps. Although the integrity of the ER and the secretory pathway clearly promote RVN formation and SARS-CoV RTC function, the data presented here argue against a direct connection or functional dependence.
Coronaviruses versus picornaviruses: a different relationship with the secretory pathway.
Picornaviruses may subvert the cell's secretory system without direct consequences, since they do not need this pathway for the production or release of viral progeny. In fact, they have been postulated to use this strategy to avoid certain host defense mechanisms (8, 9). In contrast, coronaviruses, as well as many other enveloped viruses, rely on the secretory pathway for virus assembly, maturation, and release. The interaction of viral proteins with GEFs such as GBF1 or BIG1 and BIG2, affecting Arf function and thereby disrupting secretory pathway functionality, may therefore be a property that is rare or lacking in viruses such as coronaviruses that depend on the secretory pathway for the release of their progeny.
For the most part, our results corroborate and extend the data of an MHV study recently published by Verheije et al. (59). Instead of directly analyzing viral RNA synthesis, these authors measured the impact of BFA treatment by using a recombinant MHV that expresses luciferase from a subgenomic mRNA. They also utilized small interfering RNA (siRNA) knockdown of GBF1 and Arf1 in MHV-infected cells, which profoundly affected MHV replication. In contrast, knockdown of BIG1 or BIG2 did not inhibit MHV. In our opinion, this confirms that the normal functionality of ER and the early secretory pathway promotes efficient coronavirus replication but that the integrity of the downstream secretory pathway (beyond cis-Golgi), which is regulated by BIG1 and BIG2, is much less important. Nonetheless, the MHV nsp's tested also did not convincingly colocalize with GBF1 or Arf1 (59). Although, in our opinion, the study by Verheije et al. insufficiently discriminates between the viral RTC itself and the modified membranes that presumably carry the viral enzyme complex, it was concluded that BFA treatment did not prevent MHV RTC formation, since both reduced numbers of DMVs and inhibition of reporter gene expression were observed. They then postulated that BFA treatment, or the depletion of Arf1 or GBF1, should in some other way interfere with replication, resulting in reduced de novo formation of viral replication complexes. Since these authors did not observe colocalization of GBF1 or Arf1 with MHV nsp's, they proposed that downstream effectors of Arf1 may be involved in MHV RNA replication. However, association with αγCOPI, a subunit of the COPI coat complex, was not observed, nor was colocalization detected with another obvious downstream effector of Arf1, phospholipase D. Finally, it was hypothesized that the role of GBF1-mediated Arf1 activation in MHV replication may be the delivery of membranes to the ER through retrograde vesicular transport, which is inhibited after BFA treatment or knockdown of GBF1 or Arf1 (59). Whereas normally there is a regulated stream of membrane delivery from ER to the Golgi complex and back through anterograde and retrograde transport, respectively, BFA treatment induces the collapse of the Golgi complex and its merger with the ER. In this way, Golgi membranes could in fact become available for inclusion in the RVN. Indeed, we observed that the colocalization of the Golgi marker Giantin and SARS-CoV nsp3 slightly increased upon BFA treatment, going from a negative correlation in untreated cells (0.386 ± 0.02, equaling no significant colocalization; Fig. 3A) to a slightly positive correlation in BFA-treated cells (0.514 ± 0.04, indicating minimal colocalization, data not shown). This suggests that Golgi membranes may indeed end up in the RVN in the presence of BFA, although it remains unclear whether their inclusion actually promotes RVN formation or RTC function.
At present, we can only speculate on the mechanism(s) behind the indirect effect of BFA treatment on SARS-CoV replication. Clearly, viral replication may be hampered by structural changes of RVN elements (e.g., the reduced number of vesicles; Fig. 5A) and accelerated maturation of the network (VP formation). The first effect may be explained by the general influence of BFA treatment on ER integrity. Much of the ER becomes dilated soon after the start of BFA treatment (14, 34), as also observed in the present study (Fig. 3F). Our unpublished data obtained with equine arteritis virus, a distant relative of SARS-CoV in the order Nidovirales, support the hypothesis that DMV morphogenesis commences with the formation of paired membranes, which are likely held together by protein-protein interactions. We do not yet have comparable information for SARS-CoV, but it is clear that the formation of paired membranes may be affected when the ER is dilated due to BFA treatment.
Separation of inner and outer membranes after BFA treatment.
The changes in RVN morphology upon BFA treatment are intriguing, since the drug seems to speed up the normally observed maturation of the structure and thereby possibly also its deterioration near the end stage of infection. The expedited maturation is best illustrated by the accelerated appearance of VPs, although their significance for coronavirus replication has remained elusive thus far (Fig. 4). BFA treatment apparently disrupts or prevents membrane pairing, leading to separation of membranes that are normally tightly apposed. This may in fact reflect the mechanism leading to VP formation during normal infection. DMV outer membranes are continuous with each other and, when the interaction between inner and outer membranes would weaken, the necklike connection between DMVs may open up, resulting in VP formation. Possibly, SARS-CoV infection and BFA treatment affect ER integrity in a comparable manner. Indeed, in the absence of BFA, large VPs with extensive “empty volume” were observed late in infection (25), resembling the dilated ER membranes now found in BFA-treated samples (Fig. 4D, large VPs on left side). Moreover, Verheije et al. reported decreased secretion of a reporter protein from MHV-infected cells (59). Thus, although at least a certain level of secretory pathway functionality is required for the transport of progeny virions, RVN formation and maturation may somehow mimic the effects of BFA treatment on the ER and early secretory pathway. This effect may be intrinsic to the transmembrane nsp's that induce RVN formation.
The increased luminal space between the RVN membranes that we observed in BFA-treated cells also relates to the unresolved issue of possible RNA transport between DMV interior and cytoplasm. Whereas dsRNA labeling in +RNA virus-infected cells is commonly taken to indicate the site of viral RNA synthesis, RNA products must reach the cytoplasm to be translated and/or packaged. Although, using ET methods, we previously scrutinized DMVs for the presence of a connection to the cytoplasm (25), these were not detected, leaving the question of the localization of the exact site of coronavirus RNA synthesis wide open.
The fact that BFA treatment can induce separation of inner and outer membranes (Fig. 6A to C, closeups I and II) strengthens the notion that the inner membrane indeed is a distinct vesicular structure. The interaction between inner and outer membrane appeared to be stronger at the DMV side that faces the CM (Fig. 6), resulting in striking images of DMVs in which the luminal space was asymmetrically distributed. Using our electron tomograms, the part of the DMV surface where membranes remained apposed was again scrutinized for connections to the cytosol. Figure 7 shows closeups of a number of membrane discontinuities, but we do not believe that these should be considered equivalent to the openings previously described for replication structures of other +RNA viruses (26, 61). We doubt whether these discontinuities could be associated with viral RNA trafficking and consider it more likely that they may be artifacts resulting from the previously documented fragility of the RVN, which may actually be promoted by BFA treatment. Consequently, the question of whether RNA synthesis indeed occurs inside DMVs, as suggested by the abundant presence of dsRNA, and the possibility of RNA transport across double membranes (25), remain intriguing and highly relevant topics for further research. In this context, it was interesting that a quantitative correlation seemed to exist between the reduction of viral RNA synthesis and the decrease of vesicle numbers upon BFA treatment (Fig. 1A and 5A). Moreover, in untreated cells both viral RNA synthesis and DMV numbers approximately doubled between 7 and 11 h p.i. (Fig. 1A and 5A), a correlation that even appears to hold up in the presence of BFA. Although these observations could be taken to support the hypothesis that RNA synthesis indeed occurs inside DMVs, they may on the other hand also be accommodated in models proposing that DMV formation is a cellular response to viral replication or a viral strategy to avoid antiviral host responses (25). According to the latter hypothesis, DMVs would essentially function as “garbage bags” to hide excess viral dsRNA from the cytosolic sensors of the innate immune system, while viral RNA synthesis would take place in association with another RVN component, for example, the CM that appear to be the primary site of nsp accumulation (25). In any case, our analysis, for the first time, revealed a correlation between RTC activity and DMV number, strengthening the connection between viral RNA synthesis and the coronavirus-induced RVN.
Which host factors are involved in SARS-CoV replication?
Various markers of the early secretory pathway such as Sec13, syntaxin 5, GBF1, and Arf1 do not seem to localize to the SARS-CoV-induced RVN (Fig. 3) and therefore are unlikely to play a prominent role in its morphogenesis or function. On the other hand, ER proteins that are associated with the RVN are PDI and Sec61α, which normally have prominent functions in the translocation and modification of proteins synthesized in association with the ER. Considering the presence of ribosomes on the RVN outer membrane and in CM, we speculate that viral protein synthesis may take place at these sites, as was also proposed for the CM induced by flavivirus infection (31). Our data suggest that cellular factors, such as PDI and Sec61α, may therefore be recruited to the network, which could also hamper cellular protein synthesis.
For both picornaviruses and the coronavirus MHV, previous studies addressed the possibility of a link between virally induced DMVs and the autophagic pathway, the only cellular mechanism that is also associated with double membrane structures. Thus far, however, contradictory data sets have been described that either confirm or reject the hypothesis for the same or similar picorna- or coronaviruses (4, 24, 43, 49, 51, 54, 64, 66). Now that we conclude that also the early secretory pathway is unlikely to be directly involved in SARS-CoV RVN formation or RTC function, it remains to be established which cellular factors do play a prominent role during the earliest stages of coronavirus replication. In addition to our observations here regarding Sec61α redistribution upon infection, recent in vitro studies have shown that an (as-yet-unidentified) cytosolic host factor is essential for in vitro activity of the SARS-CoV RTC (56). Besides the secretory and autophagic pathways, the reticulon family of membrane curvature-inducing ER proteins (60) was implicated in picornavirus replication (53), and their role in the life cycle of other +RNA viruses, including coronaviruses, should be investigated. Also, systematic large-scale approaches, such as the use of siRNA libraries, will be important in the quest for host factors and pathways involved in +RNA virus replication.
Supplementary Material
Acknowledgments
We are very grateful to Mitsuo Tagaya (Tokyo, Japan) and Wanjin Hong (Singapore) for providing the antibodies against syntaxin 5 and Sec13, respectively. Arf1 expression constructs were kindly provided by Guillermo Romero (Pittsburgh). We thank Frank van Kuppeveld (Nijmegen, The Netherlands) for the antiserum against GBF1 and for numerous helpful discussions. We thank Helene Verheije, Xander de Haan, and Peter Rottier (Utrecht, The Netherlands) for sharing unpublished data and stimulating discussions. Finally, we acknowledge the input of Martijn van Hemert and the technical assistance of our coworkers Yvonne van der Meer (IF microscopy) and Raimond Ravelli and Joke van der Meer (whole-cell mosaics).
This study was supported in part by The Netherlands Organization for Scientific Research (NWO) through ECHO grant 700.55.002 from the Council for Chemical Sciences (NWO-CW) and Toptalent grant 021.001.037.
The funders had no role in study design, data collection and analysis, the decision to publish, or the preparation of the manuscript.
Footnotes
Published ahead of print on 4 November 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Belov, G. A., Q. Feng, K. Nikovics, C. L. Jackson, and E. Ehrenfeld. 2008. A critical role of a cellular membrane traffic protein in poliovirus RNA replication. PLoS Pathog. 4:e1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belov, G. A., N. tan-Bonnet, G. Kovtunovych, C. L. Jackson, J. Lippincott-Schwartz, and E. Ehrenfeld. 2007. Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. J. Virol. 81:558-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bienz, K., D. Egger, and L. Pasamontes. 1987. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 160:220-226. [DOI] [PubMed] [Google Scholar]
- 4.Brabec-Zaruba, M., U. Berka, D. Blaas, and R. Fuchs. 2007. Induction of autophagy does not affect human rhinovirus type 2 production. J. Virol. 81:10815-10817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuconati, A., A. Molla, and E. Wimmer. 1998. Brefeldin A inhibits cell-free, de novo synthesis of poliovirus. J. Virol. 72:6456-6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dascher, C., and W. E. Balch. 1994. Dominant inhibitory mutants of ARF1 block endoplasmic reticulum to Golgi transport and trigger disassembly of the Golgi apparatus. J. Biol. Chem. 269:1437-1448. [PubMed] [Google Scholar]
- 7.Dascher, C., J. Matteson, and W. E. Balch. 1994. Syntaxin 5 regulates endoplasmic reticulum to Golgi transport. J. Biol. Chem. 269:29363-29366. [PubMed] [Google Scholar]
- 8.Deitz, S. B., D. A. Dodd, S. Cooper, P. Parham, and K. Kirkegaard. 2000. MHC I-dependent antigen presentation is inhibited by poliovirus protein 3A. Proc. Natl. Acad. Sci. USA 97:13790-13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dodd, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2001. Poliovirus 3A protein limits interleukin-6 (IL-6), IL-8, and beta interferon secretion during viral infection. J. Virol. 75:8158-8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doedens, J. R., and K. Kirkegaard. 1995. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 14:894-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donaldson, J. G., D. Finazzi, and R. D. Klausner. 1992. Brefeldin A inhibits Golgi membrane-catalyzed exchange of guanine nucleotide onto ARF protein. Nature 360:350-352. [DOI] [PubMed] [Google Scholar]
- 12.Egger, D., N. Teterina, E. Ehrenfeld, and K. Bienz. 2000. Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis. J. Virol. 74:6570-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frangakis, A. S., and R. Hegerl. 2001. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J. Struct. Biol. 135:239-250. [DOI] [PubMed] [Google Scholar]
- 14.Fujiwara, T., K. Oda, S. Yokota, A. Takatsuki, and Y. Ikehara. 1988. Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J. Biol. Chem. 263:18545-18552. [PubMed] [Google Scholar]
- 15.Fullekrug, J., B. Sönnichsen, U. Schäfer, P. Nguyen Van, H. D. Söling, and G. Mieskes. 1997. Characterization of brefeldin A induced vesicular structures containing cycling proteins of the intermediate compartment/cis-Golgi network. FEBS Lett. 3:75-81. [DOI] [PubMed] [Google Scholar]
- 16.Gazina, E. V., J. M. Mackenzie, R. J. Gorrell, and D. A. Anderson. 2002. Differential requirements for COPI coats in formation of replication complexes among three genera of Picornaviridae. J. Virol. 76:11113-11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldsmith, C. S., K. M. Tatti, T. G. Ksiazek, P. E. Rollin, J. A. Comer, W. W. Lee, P. A. Rota, B. Bankamp, W. J. Bellini, and S. R. Zaki. 2004. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 10:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorbalenya, A. E., L. Enjuanes, J. Ziebuhr, and E. J. Snijder. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res. 117:17-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorlich, D., S. Prehn, E. Hartmann, K. U. Kalies, and T. A. Rapoport. 1992. A mammalian homolog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation. Cell 71:489-503. [DOI] [PubMed] [Google Scholar]
- 20.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harcourt, B. H., D. Jukneliene, A. Kanjanahaluethai, J. Bechill, K. M. Severson, C. M. Smith, P. A. Rota, and S. C. Baker. 2004. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 78:13600-13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irurzun, A., L. Perez, and L. Carrasco. 1992. Involvement of membrane traffic in the replication of poliovirus genomes: effects of brefeldin A. Virology 191:166-175. [DOI] [PubMed] [Google Scholar]
- 23.Ivanov, K. A., V. Thiel, J. C. Dobbe, Y. van der Meer, E. J. Snijder, and J. Ziebuhr. 2004. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 78:5619-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson, W. T., T. H. Giddings, Jr., M. P. Taylor, S. Mulinyawe, M. Rabinovitch, R. R. Kopito, and K. Kirkegaard. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3:e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knoops, K., M. Kikkert, S. H. van den Worm, J. C. Zevenhoven-Dobbe, Y. van der Meer, A. J. Koster, A. M. Mommaas, and E. J. Snijder. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6:e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kopek, B. G., G. Perkins, D. J. Miller, M. H. Ellisman, and P. Ahlquist. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 5:e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kremer, J. R., D. N. Mastronarde, and J. R. McIntosh. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71-76. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 29.Li, Q., A. Lau, T. J. Morris, L. Guo, C. B. Fordyce, and E. F. Stanley. 2004. A syntaxin 1, Gα (o), and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization. J. Neurosci. 24:4070-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lippincott-Schwartz, J., L. C. Yuan, J. S. Bonifacino, and R. D. Klausner. 1989. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell 56:801-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mackenzie, J. 2005. Wrapping things up about virus RNA replication. Traffic 6:967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maynell, L. A., K. Kirkegaard, and M. W. Klymkowsky. 1992. Inhibition of poliovirus RNA synthesis by brefeldin A. J. Virol. 66:1985-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, S., and J. Krijnse Locker. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6:363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Misumi, Y., Y. Misumi, K. Miki, A. Takatsuki, G. Tamura, and Y. Ikehara. 1986. Novel blockade by brefeldin A of intracellular transport of secretory proteins in cultured rat hepatocytes. J. Biol. Chem. 261:11398-11403. [PubMed] [Google Scholar]
- 35.Monaghan, P., H. Cook, T. Jackson, M. Ryan, and T. Wileman. 2004. The ultrastructure of the developing replication site in foot-and-mouth disease virus-infected BHK-38 cells. J. Gen. Virol. 85:933-946. [DOI] [PubMed] [Google Scholar]
- 36.Mossessova, E., R. A. Corpina, and J. Goldberg. 2003. Crystal structure of ARF1*Sec7 complexed with brefeldin A and its implications for the guanine nucleotide exchange mechanism. Mol. Cell 12:1403-1411. [DOI] [PubMed] [Google Scholar]
- 37.Netherton, C., K. Moffat, E. Brooks, and T. Wileman. 2007. A guide to viral inclusions, membrane rearrangements, factories, and viroplasm produced during virus replication. Adv. Virus Res. 70:101-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Donnell, V. K., J. M. Pacheco, T. M. Henry, and P. W. Mason. 2001. Subcellular distribution of the foot-and-mouth disease virus 3A protein in cells infected with viruses encoding wild-type and bovine-attenuated forms of 3A. Virology 287:151-162. [DOI] [PubMed] [Google Scholar]
- 39.Oostra, M., E. G. te Lintelo, M. Deijs, M. H. Verheije, P. J. Rottier, and C. A. de Haan. 2007. Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. J. Virol. 81:12323-12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orci, L., M. Stamnes, M. Ravazzola, M. Amherdt, A. Perrelet, T. H. Sollner, and J. E. Rothman. 1997. Bidirectional transport by distinct populations of COPI-coated vesicles. Cell 90:335-349. [DOI] [PubMed] [Google Scholar]
- 41.Peiris, J. S., Y. Guan, and K. Y. Yuen. 2004. Severe acute respiratory syndrome. Nat. Med. 10:S88-S97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perlman, S., and J. Netland. 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 7:439-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prentice, E., J. McAuliffe, X. Lu, K. Subbarao, and M. R. Denison. 2004. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 78:9977-9986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rust, R. C., L. Landmann, R. Gosert, B. L. Tang, W. Hong, H. P. Hauri, D. Egger, and K. Bienz. 2001. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J. Virol. 75:9808-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salonen, A., T. Ahola, and L. Kaariainen. 2005. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285:139-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schlegel, A., T. H. Giddings, Jr., M. S. Ladinsky, and K. Kirkegaard. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 70:6576-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schonborn, J., J. Oberstrass, E. Breyel, J. Tittgen, J. Schumacher, and N. Lukacs. 1991. Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res. 19:2993-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snijder, E. J., Y. van der Meer, J. C. Zevenhoven-Dobbe, J. J. Onderwater, J. van der Meulen, H. K. Koerten, and A. M. Mommaas. 2006. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 80:5927-5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stertz, S., M. Reichelt, M. Spiegel, T. Kuri, L. Martinez-Sobrido, A. Garcia-Sastre, F. Weber, and G. Kochs. 2007. The intracellular sites of early replication and budding of SARS-coronavirus. Virology 361:304-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang, B. L., F. Peter, J. Krijnse Locker, S. H. Low, G. Griffiths, and W. Hong. 1997. The mammalian homolog of yeast Sec13p is enriched in the intermediate compartment and is essential for protein transport from the endoplasmic reticulum to the Golgi apparatus. Mol. Cell. Biol. 17:256-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang, W. F., S. Y. Yang, B. W. Wu, J. R. Jheng, Y. L. Chen, C. H. Shih, K. H. Lin, H. C. Lai, P. Tang, and J. T. Horng. 2007. Reticulon 3 binds the 2C protein of enterovirus 71 and is required for viral replication. J. Biol. Chem. 282:5888-5898. [DOI] [PubMed] [Google Scholar]
- 54.Taylor, M. P., and K. Kirkegaard. 2008. Potential subversion of autophagosomal pathway by picornaviruses. Autophagy 4:286-289. [DOI] [PubMed] [Google Scholar]
- 55.van der Meer, Y., H. van Tol, J. Krijnse Locker, and E. J. Snijder. 1998. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J. Virol. 72:6689-6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Hemert, M. J., S. H. van den Worm, K. Knoops, A. M. Mommaas, A. E. Gorbalenya, and E. J. Snijder. 2008. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 4:e1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vasudevan, C., W. Han, Y. Tan, Y. Nie, D. Li, K. Shome, S. C. Watkins, E. S. Levitan, and G. Romero. 1998. The distribution and translocation of the G protein ADP-ribosylation factor 1 in live cells is determined by its GTPase activity. J. Cell Sci. 111(Pt. 9):1277-1285. [DOI] [PubMed] [Google Scholar]
- 58.Vaux, D., J. Tooze, and S. Fuller. 1990. Identification by anti-idiotype antibodies of an intracellular membrane protein that recognizes a mammalian endoplasmic reticulum retention signal. Nature 345:495-502. [DOI] [PubMed] [Google Scholar]
- 59.Verheije, M. H., M. Raaben, M. Mari, E. G. te Lintelo, F. Reggiori, F. J. van Kuppeveld, P. J. Rottier, and C. A. de Haan. 2008. Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLoS Pathog. 4:e1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Voeltz, G. K., W. A. Prinz, Y. Shibata, J. M. Rist, and T. A. Rapoport. 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124:573-586. [DOI] [PubMed] [Google Scholar]
- 61.Welsch, S., S. Miller, I. Romero-Brey, A. Merz, C. K. Bleck, P. Walther, S. D. Fuller, C. Antony, J. Krijnse-Locker, and R. Bartenschlager. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wessels, E., D. Duijsings, K. H. Lanke, W. J. Melchers, C. L. Jackson, and F. J. van Kuppeveld. 2007. Molecular determinants of the interaction between coxsackievirus protein 3A and guanine nucleotide exchange factor GBF1. J. Virol. 81:5238-5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wessels, E., D. Duijsings, T. K. Niu, S. Neumann, V. M. Oorschot, F. de Lange, K. H. Lanke, J. Klumperman, A. Henke, C. L. Jackson, W. J. Melchers, and F. J. van Kuppeveld. 2006. A viral protein that blocks Arf1-mediated COP-I assembly by inhibiting the guanine nucleotide exchange factor GBF1. Dev. Cell 11:191-201. [DOI] [PubMed] [Google Scholar]
- 64.Wong, J., J. Zhang, X. Si, G. Gao, I. Mao, B. M. McManus, and H. Luo. 2008. Autophagosome supports coxsackievirus B3 replication in host cells. J. Virol. 82:9143-9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, C. J., A. G. Rosenwald, M. C. Willingham, S. Skuntz, J. Clark, and R. A. Kahn. 1994. Expression of a dominant allele of human ARF1 inhibits membrane traffic in vivo. J. Cell Biol. 124:289-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao, Z., L. B. Thackray, B. C. Miller, T. M. Lynn, M. M. Becker, E. Ward, N. N. Mizushima, M. R. Denison, and H. W. Virgin. 2007. Coronavirus replication does not require the autophagy gene ATG5. Autophagy 3:581-585. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.