Abstract
The entry of human immunodeficiency virus type 1 (HIV-1) into a target cell entails a series of conformational changes in the gp41 transmembrane glycoprotein that mediates the fusion of the viral and target cell membranes. A trimer-of-hairpins structure formed by the association of two heptad repeat (HR) regions of the gp41 ectodomain has been implicated in a late step of the fusion pathway. Earlier native and intermediate states of the protein are postulated to mediate the antiviral activity of the fusion inhibitor enfuvirtide and of broadly neutralizing monoclonal antibodies (NAbs), but the details of these structures remain unknown. Here, we report the identification and crystal structure of a dimerization domain in the C-terminal ectodomain of gp41 (residues 630 to 683, or C54). Two C54 monomers associate to form an asymmetric, antiparallel coiled coil with two distinct C-terminal α-helical overhangs. This dimer structure is conferred largely by interactions within a central core that corresponds to the sequence of enfuvirtide. The mutagenic alteration of the dimer interface severely impairs the infectivity of Env-pseudotyped viruses. Moreover, the C54 structure binds tightly to both the 2F5 and 4E10 NAbs and likely represents a potential intermediate conformation of gp41. These results should enhance our understanding of the molecular basis of the gp41 fusogenic structural transitions and thereby guide rational, structure-based efforts to design new fusion inhibitors and vaccine candidates intended to induce broadly neutralizing antibodies.
The entry of human immunodeficiency virus type 1 (HIV-1) into its target cell to establish an infection requires the fusion of viral and cellular membranes, a process that is mediated by the viral envelope glycoprotein (Env) through interactions with receptors on the target cell membrane (CD4 and a coreceptor, such as CCR-5 or CXCR-4) (14). HIV-1 Env is synthesized as the glycoprotein precursor gp160, which oligomerizes in the endoplasmic reticulum and subsequently is cleaved by the cellular furin endoprotease to create a metastable state that is primed for the induction of membrane fusion activity (19). The resulting Env complex is a trimeric structure comprising three gp120 surface glycoproteins, each associated noncovalently with one of three subunits of the gp41 transmembrane glycoprotein (24, 27, 47, 48). This native (prefusion) Env spike protrudes from the virus surface and is the target for neutralizing antibodies (NAbs) (reviewed in reference 3). It is generally accepted that HIV-1 membrane fusion is promoted by a series of receptor binding-triggered conformational changes in the Env complex, culminating in the formation of an energetically stable trimer of α-helical hairpins in gp41 (10, 14).
The core structure of the trimer-of-hairpins is an antiparallel six-helix bundle: a central, three-stranded coiled coil formed by the first heptad repeat (HRN) region of gp41 is sheathed by three α-helices derived from the second HR (HRC) (5, 27, 42, 44). HRN is immediately C terminal to the fusion peptide, while HRC is adjacent to the transmembrane helix anchored in the viral membrane. The interaction of gp120 with CD4 and a chemokine receptor is thought to alter intersubunit interactions in the native Env complex, leading to gp41 reorganization into a postulated prehairpin intermediate (reviewed in references 10 and 14). At this point, the N-terminal HRN coiled-coil trimer is formed, relocating the fusion peptides to allow them to insert into the cellular membrane. The HRC region then is thought to jackknife so as to pack against the inner coiled-coil core and form the postfusion trimer-of-hairpin structure that brings the attached target cell and viral membranes together. Evidence for the existence of these different gp41 conformational states in the fusion pathway is indirect, being inferred from the antiviral activity of peptides derived from the two HR regions of gp41 (20, 45). These peptide inhibitors likely act in a dominant-negative manner by binding to the prehairpin intermediate, preventing the formation of the trimer-of-hairpins (6, 13, 27, 31). This intermediate is relatively stable, with a half-life of many minutes, as detected by the capacity of such peptides to inhibit fusion once prefusion gp41 has undergone a conformational transition (21, 31). Although mounting evidence indicates that the prefusogenic and intermediate states are important targets for drug- and vaccine-elicited NAbs (reviewed in references 3 and 10), little is known about their structures and how they modulate gp41 fusogenicity or serve as targets for inhibition.
The C-terminal part of the gp41 ectodomain consists of HRC (or C34) and the membrane-proximal external region (MPER) (Fig. 1). The C34 peptide is intrinsically disordered in isolation and forms an outer-layer α-helix only in the six-helix bundle (27, 29). Structural studies of the trimeric coiled-coil state of the MPER and of its bent helix conformation after binding to lipid membranes have begun to provide clues regarding the function of this unusual and important NAb-associated segment (25, 41). The MPER is the established target for two very rare but broadly reactive NAbs, 2F5 and 4E10/z13, which are elicited during natural human infection (50). These neutralizing epitopes seem to be poorly exposed on the surface of both HIV-1-infected cells and virions (reviewed in reference 3). Their exposure is enhanced or triggered by receptor binding but diminishes on the formation of the trimer-of-hairpins, suggesting that both of the NAbs target a more extended intermediate conformation rather than the native gp41 structure (8, 12). Despite extensive efforts, how structural aspects of the MPER explain its antigenicity and immunogenicity remains unclear. Here, we report the identification of the C-terminal dimerization domain of gp41 and present the 1.65-Å crystal structure of this domain. We characterize the role of this antiparallel two-stranded coiled-coil structure in NAb reactivity and viral function. Our study provides a potential structure for the fusion-intermediate state of gp41 and for the future design of new HIV-1 immunogens that may elicit broad and potent NAbs.
FIG. 1.
Structural and functional domains of HIV-1 gp41. (Upper) Schematic view of gp41 showing the location of the fusion peptide (FP), the two HR regions, the MPER, the transmembrane segment (TM), and the cytoplasmic region (CP). HRC and MPER are depicted in blue and green, respectively. (Lower) Sequences of the C56, C54, C54N656L, and C39 peptides employed in the study. The Asn-656→Leu mutation in C54N656L is shown in red. The sequences of T-20 and core epitopes recognized by the human 2F5 and 4E10 MAbs are indicated.
MATERIALS AND METHODS
Protein production.
The C56, C54, and C39 constructs (Fig. 1) were prepared by subcloning the fragments containing residues 628 to 683, 630 to 683, and 636 to 674, respectively, of HIV-1 HXB2 gp160 into the pTMHa expression vector encoding an N-terminal His9-TrpLE chimeric protein (39). All mutants were created by site-directed DNA mutagenesis; the generated sequences were confirmed by DNA sequencing. The chimeric proteins were expressed in Escherichia coli strain BL21(DE3)/pLysS (Novagen) and purified from insoluble bacterial inclusion bodies by nickel-nitrilotriacetate (Ni-NTA) metal affinity chromatography by following published procedures (39). The TrpLE leader sequence subsequently was cleaved off by using a cyanogen bromide (CNBr) or hydroxylamine reaction. The N36(L6)C34 construct and its variants were expressed in E. coli BL21(DE3)/pLysS by using a modified pET3a vector (Novagen) and purified as previously described (28). All peptides were purified to homogeneity by reverse-phase high-performance liquid chromatography (Waters Corporation) on a Vydac C18 preparative column (Hesperia, CA) with a linear water-acetonitrile gradient containing 0.1% trifluoroacetic acid and then lyophilized. The identity of the peptides was confirmed by electrospray mass spectrometry (PerSeptive Biosystems Inc., Framingham, MA). Protein concentrations were determined by absorbance at 280 nm in 6 M guanidine hydrochloride (11). A selenomethionine (SeMet)-substituted C54L645M mutant (Leu-645 mutated to Met) was produced for multiwavelength anomalous diffraction (MAD) analysis in amino acid-supplemented minimal medium and purified as described above. The C56 and C54L645M peptides each contain an additional glycine residue at the N terminus after hydroxylamine cleavage at the Asn-Gly bond.
Biophysical analysis.
Circular dichroism (CD) spectra were measured on an Aviv 62A/DS CD spectrometer in TBS (5 mM Tris-HCl, pH 8.0, 5 mM NaCl). Thermal stability was assessed by monitoring [θ]222 as a function of temperature under the same conditions. A [θ]222 value of −33,000 deg cm2 dmol−1 was taken to correspond to 100% helix. Analytical ultracentrifugation (AUC) measurements were carried out on a Beckman XL-A analytical ultracentrifuge equipped with an An-60 Ti rotor. Peptide samples were dialyzed overnight against TBS and analyzed at rotor speeds of 24,000 and 27,000 rpm at 20°C for C56, C54, and C54N656L and 28,000 and 31,000 rpm at 4°C for C39. Data sets were fitted to a single-species model.
Crystallization and structure determination.
The best-diffracting crystals of C54 grew from 1 μl of 15 mg ml−1 peptide sample added to 1 μl of reservoir buffer (0.1 M morpholineethanesulfonic acid [MES], pH 6.0, 28% Jeffamine M600) and allowed to equilibrate against 500 μl of reservoir buffer at room temperature. Crystals belong to space group P3121 (a = b = 33.96 Å; c = 174.86 Å) and contain two monomers in the asymmetric unit. SeMet-C54L645M was crystallized in 0.1 M sodium acetate (pH 3.6), 0.3 M NH4H2PO4, 35% Jeffamine M600, and crystals were isomorphous to the wild-type peptide. Prior to data collection, the native and SeMet crystals were harvested in the reservoir buffer and frozen in liquid nitrogen. Native and MAD data sets were collected on beamline X4A at the National Synchrotron Light Source (Brookhaven, NY). For MAD data, the peak and inflection wavelengths of the selenium K absorption edge as well as the remote wavelength were selected based on the fluorescence spectrum of the SeMet-C54L645M crystal (Table 1). Reflections were integrated and scaled with DENZO and SCALEPACK (36). MAD phases were generated from two selenium atoms located within the asymmetric unit using SOLVE (43), and ∼70% of the polypeptide chain was traced automatically into the electron density maps (2.7-Å resolution). Density modification and phase extension to 1.65 Å with the native data using Arp/Warp (23) improved the quality of the electron density maps and allowed ∼95% of the final structure to be interpreted. Iterative rounds of model building with O, refinement with Refmac (32), and the addition of ordered solvent molecules clarified the trace, except for that of the N-terminal region of monomer B, which is not visible in the electron density maps and therefore must be disordered. The final model (Rcryst = 23.2% and Rfree = 25.9% for the resolution range 58.3 to 1.65 Å) consists of residues 2 to 54 (monomer A) and 12 to 52 (monomer B) in the asymmetric unit, 1 sodium ion, 2 Jeffamine M600 molecules, and 47 water molecules. All protein residues occupy the most favored regions of the Ramachandran plot.
TABLE 1.
X-ray data collection and phasing and refinement statistics
| Statistics | Value for: |
|||||
|---|---|---|---|---|---|---|
| C54 | SeMet λ1 | SeMet λ2 | SeMet λ3 | C54N656L | C39 | |
| Diffraction data | ||||||
| Wavelength (Å) | 0.9795 | 0.9793 | 0.9796 | 0.9681 | 0.9795 | 1.1000 |
| Resolution (Å) | 58.3-1.65 | 50-2.50 | 50-2.50 | 50-2.50 | 63.8-1.70 | 58.7-2.20 |
| Rsym (%) | 6.2 (48.5)a | 7.9 (31.6) | 8.3 (45.3) | 6.7 (26.0) | 6.1 (14.2) | 4.4 (26.9) |
| Completeness (%) | 99.3 (100.0) | 99.5 (99.8) | 99.7 (99.5) | 99.6 (100.0) | 98.7 (98.8) | 93.8 (76.2) |
| I/σ(I) | 15.6 (5.2) | 13.7 (7.3) | 12.7 (5.4) | 13.6 (7.2) | 16.2 (10.5) | 18.7 (3.1) |
| Phasing | ||||||
| Resolution (Å) | 20-2.70 | |||||
| No. of sites | 2 | |||||
| Phasing power (ano/iso) | 1.2/0.9 | 1.0/0.9 | 1.0 | |||
| Overall figure of merit | 0.61 | |||||
| Refinement | ||||||
| Resolution (Å) | 58.3-1.65 | 63.8-1.70 | 58.7-2.20 | |||
| No. of reflections | 14,912 | 14,906 | 16,151 | |||
| Total no. of atoms | 937 | 1,041 | 807 | |||
| Rcryst /Rfree (%) | 23.2/25.9 | 20.5/22.8 | 21.2/25.5 | |||
| Avg B factor (Å2) | 36.3 | 24.2 | 52.6 | |||
| Root square mean deviation | ||||||
| Bond length (Å) | 0.018 | 0.018 | 0.029 | |||
| Bond angles (°) | 1.7 | 1.3 | 2.4 | |||
| Torsion angles (°) | 3.5 | 3.4 | 8.3 | |||
The highest-resolution shell is shown in parentheses.
Crystals of C39 were grown from a 4 mg ml−1 peptide solution containing 0.1 M Tris-HCl (pH 7.8), 0.1 M cesium chloride, 3% polyethylene glycol 8000 at room temperature. Crystals belong to space group C2 (a = 120.76 Å, b = 47.50 Å, c = 74.82 Å, β = 128.3°) and contain four monomers in the asymmetric unit. The crystals were harvested in the precipitant buffer supplemented with 15% (vol/vol) glycerol and frozen in liquid nitrogen. Diffraction data were collected on beamline X25, and reflection intensities were integrated and scaled with DENZO and SCALEPACK (36) (Table 1). Initial phases were determined by molecular replacement using the aforementioned C54 structure as a search model in Phaser (40). The structure of C39 was refined using Refmac (32). Density interpretation and manual model building were done with O (22). The final model (Rcryst = 21.2% and Rfree = 25.5% for the resolution range 58.7 to 2.20 Å) consists of residues 1 to 39 (monomer A), 1 to 38 (monomer B), 1 to 39 (monomer C), and 1 to 36 (monomer D) in the asymmetric unit and 93 water molecules. All main-chain dihedral angles fall within the helical regions of the Ramanchandran space.
Crystals of C54N656L were obtained from a 10 mg ml−1 peptide solution containing 0.1 M sodium citrate (pH 5.0), 2.8 M 1,6-hexanediol at room temperature and were frozen in liquid nitrogen. Crystals belong to space group P3121 (a = b = 33.97 Å, c = 191.33 Å) and contain two monomers in the asymmetric unit. Data were collected on beamline X4A and were reduced and scaled with DENZO and SCALEPACK (36) (Table 1). The structure of C54N656L was solved by molecular replacement by using the C54 structure as a search model in Phaser (40). Electron density map interpretations and model building were carried out with O (22), and the structure was refined at 1.70-Å resolution by using Refmac (32). The final model (Rcryst = 20.5% and Rfree = 22.8% for the resolution range 63.8 to 1.70 Å) consists of residues 1 to 54 (monomer A) and 9 to 54 (monomer B) in the asymmetric unit, 8 1,6-hexanediols, and 78 water molecules. All main-chain torsional angles fall within the most preferred regions of the Ramachandran plot.
Structure analysis.
The root mean square deviations were calculated with LSQKAB in the CCP4i program suite (38). Buried surface areas were calculated from the difference in the accessible side chain surface areas of the dimer structure and of the individual helical monomers by using CNS 1.0 (2).
Surface plasmon resonance (SPR) measurements.
Biacore experiments were conducted on a Biacore 3000 with the CM5 sensor chip, which is composed of carboxymethylated dextran covalently attached to a gold surface at 20 or 25°C. The running buffers were 10 mM HEPES (pH 7.4), 150 mM NaCl, 3 mM EDTA, 0.005% (vol/vol) surfactant P20 (HBS-EP) for C54 and HBS-EP supplemented with 7 mM sodium dodecyl sulfate (SDS) for the C22 peptide (25). Approximately 1,000 response units of 2F5 or 4E10 antibody at a flow rate of 5 μl min−1 were immobilized to the sensor chip via amino coupling in two experimental flow cells. A mock-treated flow cell was used as a control. All flow cells then were blocked with 1 M ethanolamine-HCl (pH 8.0) and washed using 10 mM glycine-HCl (pH 2.5). For kinetic measurements, peptide antigens (2 μM C54, 2.5 μM C22) were injected at a flow rate of 5 μl min−1. The sensor surfaces were regenerated between each experiment with two 10-μl injections of 10 mM glycine-HCl (pH 3.0). Identical injections over blank surfaces were subtracted from the response data for the analysis of binding interactions. Antibody binding to peptide antigens was evaluated by using BiaEvaluation.
HIV-1 infectivity assay.
The Asn-656→Leu mutant of JR-FL gp160 was prepared by performing QuikChange site-directed mutagenesis on the pCI expression plasmid encoding the wild-type protein. Luciferase-expressing Env-pseudotyped viruses were produced by cotransfecting HEK 293 cells with pCI and the pNL4-3 Env(−)Luc(+) reporter plasmid (18). Two days posttransfection, virion-containing supernatants were clarified by low-speed centrifugation and filtered through a 0.45-μM membrane. The clarified, filtered supernatants were layered over a 20% sucrose cushion in phosphate-buffered saline (PBS) and centrifuged for 2 h at 100,000 × g at 4°C. The virus pellet then was resuspended in growth medium. To measure infectivity, the Env-pseudotyped viruses (50 μl) containing normalized amounts of p24 antigen were added to 3 × 103 U87-CD4+-CCR5+ cells/well (U87-CD4+-CXCR4+ cells were used as a negative control). Four days postinfection, the cells were harvested and luciferase activity was measured in relative light units.
Protein structure accession numbers.
The atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank (3GWO for C54, 3H00 for C39, and 3H01 for C54N656L).
RESULTS
The C-terminal dimerization domain of gp41.
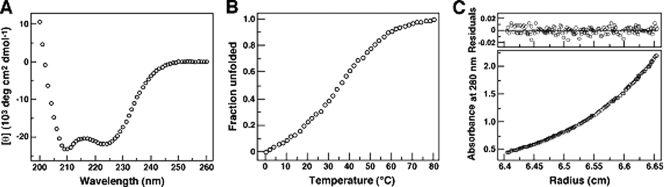
To evaluate the structural properties of the C-terminal ectodomain of gp41, we produced and characterized the protein segment comprising the entire HRC and MPER modules (residues 628 to 683, or C56) (Fig. 1). The CD spectrum of C56 is typical of an α-helix, showing the characteristic minima at 208 and 222 nm and with ∼70% of the helical content at 50 μM concentration in TBS at 0°C (Fig. 2A). Under these conditions, C56 lacks a folded baseline at low temperatures and undergoes a broad thermal unfolding transition with a melting temperature (Tm) of ∼40°C (Fig. 2B). AUC experiments indicate that C56 sediments as a discrete dimer: the apparent molecular mass is 15.1 kDa in the concentration range of 5 to 50 μM (Fig. 2C). Thus, the C56 peptide forms a labile α-helical dimerization domain.
FIG. 2.
Solution properties of C56. (A) CD spectrum at 50 μM peptide in TBS at 0°C. (B) Thermal dependence of the CD signal at 222 nm. The decrease in the fraction of a folded molecule is shown as a function of temperature. (C) AUC data of a 15 μM sample at 24,000 rpm in TBS. The data fit closely to that for a dimeric complex. (Upper) The deviation in the data from the linear fit for a dimeric model is plotted.
Crystal structure of the C54 dimer.
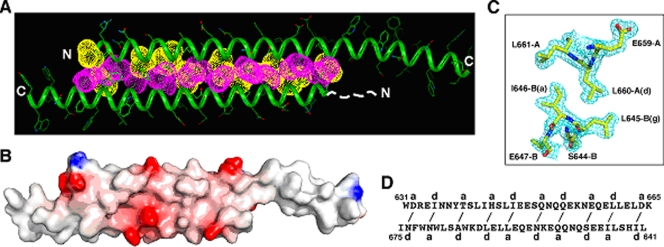
Diffraction-quality crystals were obtained only when we deleted the first two residues in C56 (denoted C54) (Fig. 1). C54 demonstrates the same helicity and Tm as C56, suggesting that the disordered Trp-628 and Met-629 side chains prevent crystallization. We solved the X-ray crystal structure by the method of MAD (17) using an SeMet-substituted C54 mutant and refined the structure to 1.65-Å resolution (Table 1). Two C54 monomers dimerize by forming an asymmetric, antiparallel coiled coil between the N-terminal region of helix A (residues 631 to 665) and the central region of helix B (residues 641 to 675), thus giving rise to two C-terminal single-helix overhangs (Fig. 3A and B). At the coiled-coil interface, residues at distinct a and d positions of the two monomers make side-by-side contacts in a typical knobs-into-holes mode (7), leading to the formation of interlocking hydrophobic seams between the supercoiled α-helical ribbons (Fig. 3C and D). The A and B helices are offset by ∼0.25 heptad with respect to each other; this interhelix offset allows for knobs-into-holes core packing by compensating for the altered Cα-Cβ vectors due to the opposing directions of these interfacial side chains in an antiparallel helical array.
FIG. 3.
Crystal structure of the C54 dimer. (A) Lateral view of the dimer. Yellow van der Waals surfaces identify residues at the a positions, and the pink surfaces identify residues at the d positions. The first 11 residues of helix B, indicated by the dashed line, are disordered in the crystal structure. (B) The MAD-phased electron density contoured at 1.0σ showing the cross-section of the duplex coiled coil. (C) Surface electrostatic potential of the dimer. Basic regions are blue, and acidic regions are red. (D) Schematic of coiled-coil packing. Residues at distinct a and d positions of the A and B helices interact (indicated by solid lines) to form the hydrophobic core of the dimer.
Serendipitously, the duplex coiled coil lacks continuous 3-4 heptad periodicity on both the A and B helices but exhibits the pairwise complementary knobs-into-holes packing interactions, as discussed above (Fig. 3A and B). These HR anomalies appear to facilitate a global restructuring of the coiled-coil interface, leading to more favorable hydrophobic and van der Waals interactions in the dimer. All of these core amino acids, except two (Gln-650 and Gln-653 of helix A), assume the preferred rotamer conformations in α-helices (26). The side chain of Gln-650 is oriented to make bifurcated hydrogen bonds with Gln-653 and Asn-656 of helix B (Nɛ2-O = 2.73 Å and Oɛ1-Nδ2 = 2.71 Å), and Gln-653 forms interstrand hydrogen bonds with Ser-649 (O-Oγ = 3.22 Å and Oɛ1-Oγ = 3.11 Å). Approximately 1,730 Å2 of solvent-accessible surface is buried in the hydrophobic interface of the coiled coil. Eight bulky apolar side chains (Ile-635, His-643, Ile-646, and Leu-660 from helix A and Ile-642, Ile-646, Leu-660, and Leu-663 from helix B) dominate this interface. The aforementioned hydrogen-bonding networks and additional four hydrogen bonds also are involved in the close packing and stabilization of interhelical contacts. In summary, the requirement to satisfy the different, interleaved core-packing interactions between the two C54 helices specifies and stabilizes an asymmetric, antiparallel homodimer structure that has not, to our knowledge, been seen before.
Dimeric association in the structure of C39.
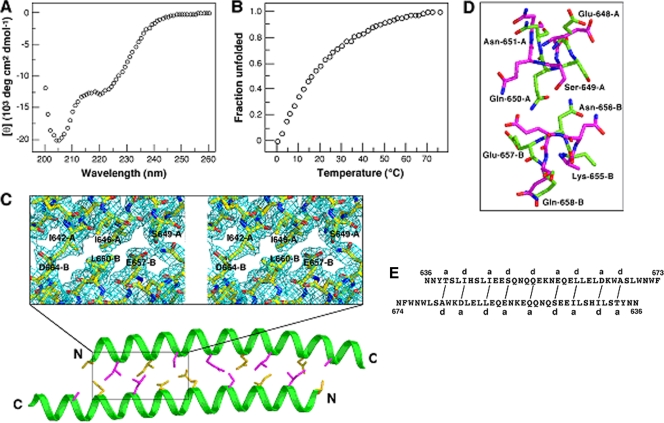
The fusion-inhibitory peptide T-20 corresponds to the central segment of the C-terminal ectodomain of gp41 (Fig. 1). The C54 structure suggests that T-20 retains key determinants that specify the overall fold of the two-helix bundle studied above (Fig. 3D). To probe the structural specificity of this dimeric coiled-coil formation, we characterized the conformational properties of a C39 peptide construct (residues 636 to 674) (Fig. 1) that includes the entire T-20 sequence together with two N-terminal and one C-terminal Asn residue; the latter are expected to increase solubility. CD measurements at 50 μM concentration show that C39 contains only approximately one-third of the α-helical structure in TBS at 0°C (Fig. 4A). Along with incomplete helix formation, C39 exhibits an ill-defined thermal-unfolding transition (Fig. 4B). AUC of C39 shows it to be predominantly monomeric at 20 μM; at higher concentrations (>100 μM), however, nonideal behavior precludes satisfactory interpretation. Taken together, the present results demonstrate that C39 cannot impart the duplex coiled-coil association specificity conferred by C54 in solution. It is likely that the loss of knobs-into-holes packing interactions around Asp-632 and Ile-635 results in the destabilization of the C54 dimerization domain.
FIG. 4.
Solution experiments and structure of C39. (A) CD spectrum of C39 at 50 μM in TBS at 0°C. (B) Thermal dependence of the CD signal at 222 nm for C39 at 50 μM. The decrease in the fraction of a folded molecule is shown as a function of temperature. (C) Packing interactions in the C39 dimer interface. The Cα backbones of the A (upper) and B (lower) helices are shown. The side chains of a (yellow) and d (pink) residues in the coiled coil are displayed. Residues 642 to 650 of helix A and 656 to 664 of helix B are boxed. The inset shows an expanded stereo view of the 2Fo-Fc electron density contoured at 1.0σ in the area of the three core positions (see the text). (D) Comparison of knobs-into-holes contacts in the C39 and C54 structures. Glu-648 to Asn-651 of helix A and Lys-655 to Gln-658 of helix B are shown. The hole of the Ser-649(g) and Gln-650(a) residues of C54 helix A (green), into which the Asn-656(d) side chain on helix B nestles, is indicated. In contrast, the Ser-649(d) side chain of C39 helix A (pink) packs inside the hole of Asn-656(g) and Glu-657(a) of helix B. (E) Schematic of core packing in the C39 dimer. Packing interactions between residues at the a and d positions of the A (upper) and B (lower) helices are indicated by solid lines.
We determined the 2.20-Å crystal structure of C39 by molecular replacement (Table 1, Fig. 4C). C39 adopts an antiparallel two-helix conformation virtually identical to that in the C54 crystal structure (root mean square deviation for equivalent Cα atoms of 1.6 Å), demonstrating the preference of this segment to form an autonomously folded dimeric subdomain. The most obvious difference between the two structures can be described as a partial rotation of the central α-helical backbone of C39 spanning residues 642 to 650 of helix A and 656 to 664 of helix B (Fig. 4D). As a result, the C39 and C54 dimers show a different set of local interactions among side chains at each of the three core positions (Fig. 3D and 4E). Evidently, the deletion of Glu-630 to Ile-635 and Ile-675 to Lys-683 as in the C39 construct enables significant side chain rearrangements in the two-stranded coiled coil that perhaps are associated with a loss in stability.
Structure-function analysis.
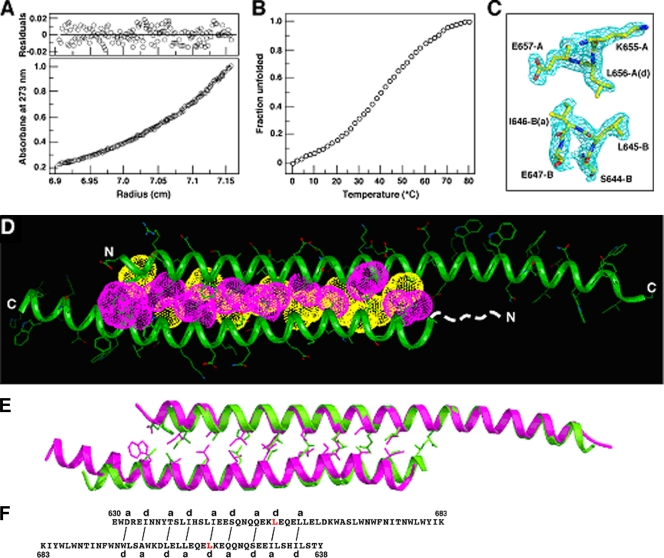
To test the hypothesis that C54 dimerization domain plays a role in HIV-1 membrane fusion, we sought to alter the stability and specificity of this newly discovered coiled-coil interaction. The dimer interface of C54 is formed by interlocking residues at the central a and d positions and by lining with the peripheral g side chains (Fig. 3D). Detailed packing interactions between the a and d amino acids can dictate both helix orientation preference and oligomerization state and therefore are a critical determinant of the overall structure (16). Our mutational strategy was to replace each of four polar or charged amino acids at the g positions of C54 (Ser-649, Gln-652, Asn-656, and Glu-659) with leucine. Leucine was selected because it is a helix-promoting residue and is favorable in stabilizing the core of coiled coils (34). Based on AUC data, we identified and expressed the Asn-656→Leu mutant (named C54N656L) that remains dimeric in aqueous solution for further study (Fig. 5A); all other leucine mutants do not form a monodisperse species or have poor solubility in physiologic buffer.
FIG. 5.
Biophysical characterization of the C54N656L mutant. (A) AUC data of C54N656L (10 μM) at 24,000 rpm in TBS. The data fit closely to that for a dimeric complex. (Upper) The deviation in the data from the linear fit for a dimeric model is plotted. (B) Thermal melt of C54N656L at 50 μM. (C) The 2Fo-Fc electron density map contoured at 1.0σ showing the cross-section of the C54N656L dimer. (D) Crystal structure of the C54N656L dimer. Yellow van der Waals surfaces identify residues at the a positions, and pink surfaces identify residues at the d positions. The first eight residues of helix B, indicated by the dashed line, are disordered in the crystal structure. (E) Superposition of the Cα backbones of the C54 (green) and C54N656L (pink) dimers. The a (yellow) and d (pink) side chains in the coiled coil are shown. (F) Schematic of core packing in the C54N656L dimer. Packing interactions between residues at the a and d positions of the A (upper) and B (lower) helices are indicated by solid lines. The Asn-656→Leu mutation in C54N656L is shown in red.
On the basis of CD measurements at 50 μM concentration, C54N656L is ∼75% helical in TBS at 0°C and has a Tm of 44°C (Fig. 5B); wild-type C54 has a Tm of 40°C at the same concentration (Fig. 2B). We determined the 1.70-Å crystal structure of C54N656L by molecular replacement (Table 1). C54N656L forms an asymmetric, antiparallel two-stranded coiled coil with long and short helical overhangs (Fig. 5C and D). The distinct HRs of the two strands are maintained in register through their entire 31-residue regions. In summary, C54N656L has a global conformation similar to that of C54 (Fig. 5E), although there are substantial differences in the interior packing and interhelix register shifts of these antiparallel dimmers (Fig. 5F). It seems that these structural differences result from specific van der Waals interactions of the substituted Leu-656 side chains in C54N656L.
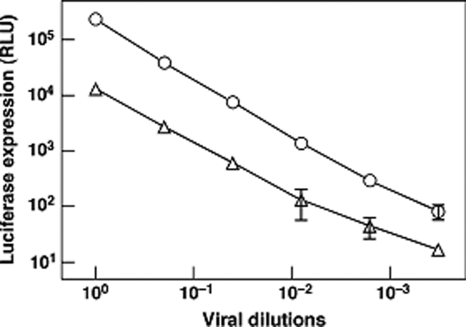
To evaluate the impact of the Asn-656→Leu mutation on the fusion function of gp41, we assessed the specific infectivity of the wild-type and mutant JR-FL Env-pseudotyped viruses in a single-cycle assay using U87-CD4+-CCR5+ target cells and a luciferase reporter gene readout 4 days postinfection. All of the buried a, d, and g residues of the HXB2 C54 dimer described above, except three (Asp-632→Glu, His-643→Tyr, and Asn-674→Asp), are absolutely conserved in the JR-FL primary isolate. The mutant virus was >10-fold less infectious than the wild-type virus (Fig. 6). Thus, the dimer-stabilizing Leu substitution for Asn-656 has a large effect on viral infectivity.
FIG. 6.
Effect of the Asn-656→Leu substitution of gp41 on Env-pseudotyped virion infectivity. Pseudovirions, containing normalized amounts of p24 antigen and bearing the wild-type (circles) or mutant (triangles) form of JR-FL Env, were serially diluted and used to infect U87-CD4-CCR5 cells. Infectivity was quantified by measuring luciferase activity 4 days postinfection.
As Asn-656 is in an a position of HRC and packs against the central trimeric coiled coil in the final trimer-of-hairpins state of gp41 (5, 27, 42, 44), we characterized the mutagenic effect of a Leu substitution at this position on six-helix bundle formation by using the N36(L6)C34 construct (30). CD and AUC studies indicate that the N36(L6)C34 mutant folds into a trimeric six-helix bundle with a Tm of 88°C, whereas the Tm of the wild-type molecule was 76°C at 10 μM. Based on the clear positive correlation between the stability properties and fusion activities of the six-helix bundle variants (28), it is probable that the reduced pseudovirion infectivity associated with the Asn-656→Leu mutation results from its impact on the C54 coiled-coil interface, suggesting that the dimerization domain is an on-pathway intermediate structure rather than a dead end.
Interaction of 2F5 and 4E10 MAbs with the C54 dimer.
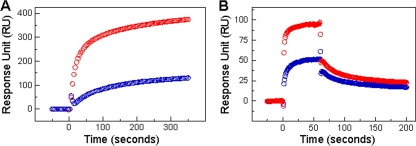
The two most broadly NAbs, 2F5 and 4E10, recognize adjacent but distinct epitopes on the MPER of gp41 (50). The identification and structure determination of the C54 dimerization domain prompted us to examine the binding of 2F5 and 4E10 to the duplex helical structure by using SPR measurements. Biacore analyses reveal that C54 interacts specifically with both immobilized 2F5 and 4E10, with 2F5 forming a more stable NAb-epitope complex than 4E10 (Fig. 7). Significantly, the binding profiles of both MAbs to C54 are dramatically different from those to linear peptide epitopes, which follow a simple Langmuir model (1, 41). The interaction of the C54 dimer with 2F5 and 4E10 is biphasic and might best be described by a two-step (encounter-docking) conformational change model. Similar behavior has been seen in the binding of 2F5 and 4E10 to membrane-embedded MPER peptides (1, 15, 33, 41). Interestingly, several nonneutralizing antibodies have been shown to bind MPER epitopes or liposome-peptide conjugates with simple Langmuir isotherms (1, 9). Taken together, these results suggest that the antiparallel dimer conformation of C54 is a potential intermediate conformation capable of interacting with 2F5 and 4E10.
FIG. 7.
Binding of MAbs 2F5 and 4E10 to C54 as assessed by SPR. (A) Sensograms for binding dimeric C54 at 2 μM to immobilized 2F5 (red) and 4E10 (blue) were acquired with a 360-s association phase at 20°C. Specific interactions of 2F5 and 4E10 with C54 were observed. Data represent two independent binding experiments, each of which was carried out in duplicate. (B) Representative sensograms for binding the monomeric MPER peptide (C22) at 2.5 μM to immobilized 2F5 (red) and 4E10 (blue) also are shown, with a 60-s association phase and a >120-s dissociation phase at 25°C (25).
DISCUSSION
Potential biological implications.
A wealth of evidence suggests that the HRN and HRC regions of gp41, initially sequestered in the prefusion form of the Env complex, undergo a programmed series of conformational rearrangements in the fusion process (reviewed in references 10 and 14). HRN and HRC are released upon receptor binding and then refold into the postulated prehairpin intermediate, which in turn collapses into the folded-back trimer-of-hairpins structure that drives membrane fusion. In this prehairpin intermediate, the inner HRN coiled-coil core in postfusion gp41 is formed (reviewed in references 10 and 14), but the structure of the C-terminal ectodomain remains unknown. The crystal structure of the C54 dimerization domain reported here suggests a plausible mechanism for the conversion of the prehairpin intermediate to the trimer-of-hairpins state. The prehairpin intermediate bridges the cellular and viral membranes, with its fusion peptides embedded in the former and its transmembrane helices anchored in the latter. In this case, the newly formed HRN trimeric coiled coil would be oriented perpendicularly to the two membranes, and the threefold axis of symmetry in the trimer-of-hairpins would lie parallel to the membranes (44). These topological constraints raise an intriguing question: how does the homotrimeric prehairpin structure access the trimer-of-hairpins conformation? The structure and thermodynamic properties of the C54 dimerization domain studied here suggest a possible route for gp41 refolding. We propose that the trimeric prehairpin structure of gp41 undergoes an asymmetric refolding event to form the C54 dimer, possibly releasing a free monomer. In this scenario, each gp41 polypeptide chain could refold independently of the other two so as to attain the postfusion trimer-of-hairpins conformation. This hypothesis is testable through further studies of mutations that modulate the C54 dimer interaction as well as its impact on the fusion function of gp41, thereby providing molecular details of a regulated sequence of structural transitions in the fusion process.
Implications for vaccine candidates.
Intense interest in the design of immunogens capable of eliciting broadly neutralizing antibodies has stimulated efforts to establish the fundamental biophysical properties of the functional modules encoded in the HIV-1 Env complex. This endeavor could provide important structural details that help account for the antigenicity and immunogenicity of the neutralizing epitopes (reviewed in reference 3). All attempts to elicit anti-gp41 NAbs by using unstructured MPER peptide immunogens have failed thus far. This suggests that a specific substructure(s) within MPER is essential for the induction of 2F5- and 4E10-like HIV-1 NAb responses (49). Thus, it is possible that the conformational state in the MPER affects the development of immune responses to the unusual 2F5/4E10/z13 epitope cluster and perhaps other important gp41 epitopes. We show that both the 2F5 and 4E10 NAbs manifest greater affinity for the C54 dimer structure than for linear MPER peptide epitopes (25). The slow dissociation reaction seen in the SPR data suggest that a conformational change accompanies the interaction of 2F5 and 4E10 with the C54 dimerization domain. This rearrangement could position the MAbs for high-affinity binding to the MPER epitopes in the C54 dimer. A large body of evidence indicates that 2F5 and 4E10 act by blocking the fusion-promoting conformational changes of gp41 necessary to achieve the trimer-of-hairpins state (8, 12). Our results provide structural details for this model, namely that the C54 dimerization domain as an intermediate conformation of gp41 is a potential target of 2F5 and 4E10 antibodies. The atomic structure of the C54 dimer may inform the design of tailored immunogens that stably present a conformation similar to that of critical MPER epitopes in a fusion-intermediate conformation of gp41. During natural infection, the neutralizing epitopes on the MPER of gp41 are not particularly immunogenic (reviewed in reference 3). The two-helix bundle could expose the hydrophobic surface of the α-helical NAb epitopes for broad immune recognition.
The instability of the native Env complex is essential to prime membrane-fusion activity. It is possible that instead of a single pathway involving discrete, sequential intermediates, a funnel-like array of nonfunctional forms arises. While the folding and refolding events are unlikely to proceed via an ensemble of nearly equally stable states, this situation could present a formidable challenge both for the elucidation of the fusion pathway and for the design of vaccines based on immunogens that mimic the native Env structure. Indeed, various nonfunctional forms of Env are present on virus particles and HIV-1-infected cells, some of which probably arise as the native complex decays (reviewed in references 4, 37, and 46). Intensive efforts have been directed toward the elimination of these nonproductive forms of Env in preparing immunogens. If the C54 dimerization domain was shown to represent a specific association motif in an off-pathway Env intermediate, our structural information could make it possible to design constructs that deplete dimers and promote authentic trimer stability. Detailed information of this kind should prove fundamental in future efforts to destabilize the C54 dimerization domain and enhance native gp41 intersubunit interactions in vaccine candidates. This would help create novel antigens that enhance the immunogenicity of neutralization epitopes.
Acknowledgments
We thank Neville Kallenbach for comments on the manuscript and John Schwanof for assistance with beamline X4A at the National Synchrotron Light Source.
This work was supported by National Institutes of Health grants R01 AI42382 (to M.L.) and R01 AI45463 and R37 AI36082 (to J.P.M.).
Footnotes
Published ahead of print on 21 October 2009.
REFERENCES
- 1.Alam, S. M., M. McAdams, D. Boren, M. Rak, R. M. Scearce, F. Gao, Z. T. Camacho, D. Gewirth, G. Kelsoe, P. Chen, and B. F. Haynes. 2007. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J. Immunol. 178:4424-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 3.Burton, D. R., R. C. Desrosiers, R. W. Doms, W. C. Koff, P. D. Kwong, J. P. Moore, G. J. Nabel, J. Sodroski, I. A. Wilson, and R. T. Wyatt. 2004. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5:233-236. [DOI] [PubMed] [Google Scholar]
- 4.Burton, D. R., and D. C. Montefiori. 1997. The antibody response in HIV-1 infection. AIDS 11(Suppl A):S87-S98. [PubMed] [Google Scholar]
- 5.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 6.Chen, C. H., T. J. Matthews, C. B. McDanal, D. P. Bolognesi, and M. L. Greenberg. 1995. A molecular clasp in the human immunodeficiency virus (HIV) type 1 TM protein determines the anti-HIV activity of gp41 derivatives: implication for viral fusion. J. Virol. 69:3771-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crick, F. H. C. 1953. The packing of α-helices: simple coiled-coils. Acta Crystallogr. 6:689-697. [Google Scholar]
- 8.Dimitrov, A. S., A. Jacobs, C. M. Finnegan, G. Stiegler, H. Katinger, and R. Blumenthal. 2007. Exposure of the membrane-proximal external region of HIV-1 gp41 in the course of HIV-1 envelope glycoprotein-mediated fusion. Biochemistry 46:1398-1401. [DOI] [PubMed] [Google Scholar]
- 9.D'Souza, M. P., D. Livnat, J. A. Bradac, and S. H. Bridges. 1997. Evaluation of monoclonal antibodies to human immunodeficiency virus type 1 primary isolates by neutralization assays: performance criteria for selecting candidate antibodies for clinical trials. AIDS Clinical Trials Group Antibody Selection Working Group. J. Infect. Dis. 175:1056-1062. [DOI] [PubMed] [Google Scholar]
- 10.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 11.Edelhoch, H. 1967. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry 6:1948-1954. [DOI] [PubMed] [Google Scholar]
- 12.Follis, K. E., S. J. Larson, M. Lu, and J. H. Nunberg. 2002. Genetic evidence that interhelical packing interactions in the gp41 core are critical for transition of the human immunodeficiency virus type 1 envelope glycoprotein to the fusion-active state. J. Virol. 76:7356-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furuta, R. A., C. T. Wild, Y. Weng, and C. D. Weiss. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5:276-279. [DOI] [PubMed] [Google Scholar]
- 14.Gallo, S. A., C. M. Finnegan, M. Viard, Y. Raviv, A. Dimitrov, S. S. Rawat, A. Puri, S. Durell, and R. Blumenthal. 2003. The HIV Env-mediated fusion reaction. Biochim. Biophys. Acta 1614:36-50. [DOI] [PubMed] [Google Scholar]
- 15.Grundner, C., T. Mirzabekov, J. Sodroski, and R. Wyatt. 2002. Solid-phase proteoliposomes containing human immunodeficiency virus envelope glycoproteins. J. Virol. 76:3511-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harbury, P. B., T. Zhang, P. S. Kim, and T. Alber. 1993. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262:1401-1407. [DOI] [PubMed] [Google Scholar]
- 17.Hendrickson, W. A. 1991. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science 254:51-58. [DOI] [PubMed] [Google Scholar]
- 18.Herrera, C., P. J. Klasse, E. Michael, S. Kake, K. Barnes, C. W. Kibler, L. Campbell-Gardener, Z. Si, J. Sodroski, J. P. Moore, and S. Beddows. 2005. The impact of envelope glycoprotein cleavage on the antigenicity, infectivity, and neutralization sensitivity of Env-pseudotyped human immunodeficiency virus type 1 particles. Virology 338:154-172. [DOI] [PubMed] [Google Scholar]
- 19.Hunter, E., and R. Swanstrom. 1990. Retrovirus envelope glycoproteins. Curr. Top. Microbiol. Immunol. 157:187-253. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, S., K. Lin, N. Strick, and A. R. Neurath. 1993. HIV-1 inhibition by a peptide. Nature 365:113. [DOI] [PubMed] [Google Scholar]
- 21.Jones, P. L., T. Korte, and R. Blumenthal. 1998. Conformational changes in cell surface HIV-1 envelope glycoproteins are triggered by cooperation between cell surface CD4 and co-receptors. J. Biol. Chem. 273:404-409. [DOI] [PubMed] [Google Scholar]
- 22.Jones, T. A., J. Y. Zou, S. W. Cowan, and Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 23.Lamzin, V. S., and K. S. Wilson. 1993. Automated refinement of protein models. Acta Crystallogr. D. 49:129-149. [DOI] [PubMed] [Google Scholar]
- 24.Liu, J., A. Bartesaghi, M. J. Borgnia, G. Sapiro, and S. Subramaniam. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, J., Y. Deng, A. Dey, J. Moore, and M. Lu. 2009. Structure of the HIV-1 gp41 membrane-proximal ectodomain region in a putative prefusion conformation. Biochemistry 48:2915-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lovell, S. C., J. M. Word, J. S. Richardson, and D. C. Richardson. 2000. The penultimate rotamer library. Proteins 40:389-408. [PubMed] [Google Scholar]
- 27.Lu, M., S. C. Blacklow, and P. S. Kim. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2:1075-1082. [DOI] [PubMed] [Google Scholar]
- 28.Lu, M., H. Ji, and S. Shen. 1999. Subdomain folding and biological activity of the core structure from human immunodeficiency virus type 1 gp41: implications for viral membrane fusion. J. Virol. 73:4433-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu, M., and P. S. Kim. 1997. A trimeric structural subdomain of the HIV-1 transmembrane glycoprotein. J. Biomol. Struct. Dyn. 15:465-471. [DOI] [PubMed] [Google Scholar]
- 30.Markosyan, R. M., X. Ma, M. Lu, F. S. Cohen, and G. B. Melikyan. 2002. The mechanism of inhibition of HIV-1 env-mediated cell-cell fusion by recombinant cores of gp41 ectodomain. Virology 302:174-184. [DOI] [PubMed] [Google Scholar]
- 31.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 33.Ofek, G., M. Tang, A. Sambor, H. Katinger, J. R. Mascola, R. Wyatt, and P. D. Kwong. 2004. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J. Virol. 78:10724-10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Neil, K. T., and W. F. DeGrado. 1990. A thermodynamic scale for the helix-forming tendencies of the commonly occurring amino acids. Science 250:646-651. [DOI] [PubMed] [Google Scholar]
- 35.Otwinowski, Z., D. Borek, W. Majewski, and W. Minor. 2003. Multiparametric scaling of diffraction intensities. Acta Crystallogr. A. 59:228-234. [DOI] [PubMed] [Google Scholar]
- 36.Otwinowski, Z., and W. Minor. 1997. Processing X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 37.Parren, P. W., D. R. Burton, and Q. J. Sattentau. 1997. HIV-1 antibody—debris or virion? Nat. Med. 3:366-367. [DOI] [PubMed] [Google Scholar]
- 38.Potterton, E., P. Briggs, M. Turkenburg, and E. Dodson. 2003. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 59:1131-1137. [DOI] [PubMed] [Google Scholar]
- 39.Shu, W., H. Ji, and M. Lu. 1999. Trimerization specificity in HIV-1 gp41: analysis with a GCN4 leucine zipper model. Biochemistry 38:5378-5385. [DOI] [PubMed] [Google Scholar]
- 40.Storoni, L. C., A. J. McCoy, and R. J. Read. 2004. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 60:432-438. [DOI] [PubMed] [Google Scholar]
- 41.Sun, Z. Y., K. J. Oh, M. Kim, J. Yu, V. Brusic, L. Song, Z. Qiao, J. H. Wang, G. Wagner, and E. L. Reinherz. 2008. HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity 28:52-63. [DOI] [PubMed] [Google Scholar]
- 42.Tan, K., J. Liu, J. Wang, S. Shen, and M. Lu. 1997. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. USA 94:12303-12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Terwilliger, T. C., and J. Berendzen. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55:849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 45.Wild, C., T. Oas, C. McDanal, D. Bolognesi, and T. Matthews. 1992. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc. Natl. Acad. Sci. USA 89:10537-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 47.Zanetti, G., J. A. Briggs, K. Grunewald, Q. J. Sattentau, and S. D. Fuller. 2006. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog. 2:e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu, P., J. Liu, J. Bess, Jr., E. Chertova, J. D. Lifson, H. Grise, G. A. Ofek, K. A. Taylor, and K. H. Roux. 2006. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441:847-852. [DOI] [PubMed] [Google Scholar]
- 49.Zwick, M. B., R. Jensen, S. Church, M. Wang, G. Stiegler, R. Kunert, H. Katinger, and D. R. Burton. 2005. Anti-human immunodeficiency virus type 1 (HIV-1) antibodies 2F5 and 4E10 require surprisingly few crucial residues in the membrane-proximal external region of glycoprotein gp41 to neutralize HIV-1. J. Virol. 79:1252-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]