Abstract
The full repertoire of hepatitis B virus (HBV) peptides that bind to the common HLA class I molecules found in areas with a high prevalence of chronic HBV infection has not been determined. This information may be useful for designing immunotherapies for chronic hepatitis B. We identified amino acid residues under positive selection pressure in the HBV core gene by phylogenetic analysis of cloned DNA sequences obtained from HBV DNA extracted from the sera of Tongan subjects with inactive, HBeAg-negative chronic HBV infections. The repertoires of positively selected sites in groups of subjects who were homozygous for either HLA-B*4001 (n = 10) or HLA-B*5602 (n = 7) were compared. We identified 13 amino acid sites under positive selection pressure. A significant association between an HLA class I allele and the presence of nonsynonymous mutations was found at five of these sites. HLA-B*4001 was associated with mutations at E77 (P = 0.05) and E113 (P = 0.002), and HLA-B*5602 was associated with mutations at S21 (P = 0.02). In addition, amino acid mutations at V13 (P = 0.03) and E14 (P = 0.01) were more common in the seven subjects with an HLA-A*02 allele. In summary, we have developed an assay that can identify associations between HLA class I alleles and HBV core gene amino acids that mutate in response to selection pressure. This is consistent with published evidence that CD8+ T cells have a role in suppressing viral replication in inactive, HBeAg-negative chronic HBV infection. This assay may be useful for identifying the clinically significant HBV peptides that bind to common HLA class I molecules.
The most potent nucleoside/nucleotide analogue drugs used to treat chronic hepatitis B reduce serum hepatitis B virus (HBV) DNA to undetectable levels in over 90% of subjects (5, 10). It was originally hoped that such a substantial reduction in viral titers would reverse T-cell tolerance for HBV antigens (17, 30) and lead to an immune response that permanently suppressed the virus, thus removing the need for expensive, lifelong drug therapy. However, HBeAg seroconversion rates of under 30% suggest that suppression of HBV replication is not sufficient to reverse the defects (4, 15) in the HBV peptide-specific CD8+ T-cell compartment that occur in these patients. A therapeutic vaccine that stimulated a diverse repertoire of functional CD8+ T cells could make a valuable contribution to management of chronic hepatitis B.
The first step in designing a therapeutic vaccine that will suppress viral replication without exacerbating chronic hepatitis B (15) is to identify the HBV peptides that stimulate functional CD8+ T cells by binding to the most common HLA class I alleles. These peptides may contribute to the antigen component of a vaccine and to the design of assays for use as correlates of immunity in trials of antiviral therapies. Although some of the HBV peptides that bind to four HLA-A alleles have been published (3, 19, 25, 28), a much wider repertoire of peptide-HLA interactions needs to be identified. There is no established method for finding them (32). Adding pools of peptides to peripheral blood mononuclear cells in enzyme-linked immunospot assays is the most commonly used technique (4), but it has disadvantages. Pools of peptides contain epitopes that are not produced by in vivo antigen-processing mechanisms (32), and the influence of these epitopes on complex mixtures of T cells with degenerate antigen receptors is unknown. False-positive and false-negative results are possible. In addition, it cannot be assumed that the ability of a T cell to secrete gamma interferon in an enzyme-linked immunospot assay correlates with its ability to place clinically significant selection pressure on the virus in vivo.
We are proposing an alternative approach, which should lead to the identification of the most clinically significant wild-type peptide antigens. This is to assess the influence of HLA class I alleles on the repertoire of escape mutations (3, 18) encoded in the HBV DNA extracted from the sera of HBeAg-negative subjects with an inactive chronic HBV infection. A functional CD8+ T-cell repertoire (15, 22) develops in these subjects at the same time the virus in their sera accumulates amino acid mutations (2). Phylogenetic analysis can distinguish those amino acid mutations that have arisen as a result of positive selection pressure from those that have arisen as a result of random processes (31). CD8+ T cells are likely to have placed selection pressure on any of the nonrandom amino acid mutations that preferentially occur in patients with a specific HLA class I allele. It should be possible to obtain the precise amino acid sequences of the peptides that contain these amino acids using immunological assays.
This study was carried out with Tongan subjects who are homozygous for one of two common HLA-B alleles. Since there is significant linkage disequilibrium within the HLA class I locus in Tongan people (1), this has allowed two groups of subjects with distinct HLA class I haplotypes to be studied. In addition, we restricted the study to subjects infected with a genotype C3 HBV.
MATERIALS AND METHODS
Patients and blood samples.
The New Zealand Hepatitis B Screening Programme monitors 1,439 Tongan adults with chronic HBV infections at 6- to 12-month intervals and checks HBeAg status, serum alanine aminotransferase (ALT) levels, and α-fetoprotein. We serially recruited 345 of these subjects into this study at the time of a routine blood test. An extra 3 ml of serum was taken for HBV DNA analyses, and genomic DNA was extracted from peripheral blood mononuclear cells for HLA class I genotyping. All 345 subjects were genotyped at the HLA-B locus using a previously published method (1). Twenty subjects who were homozygous for the HLA-B*4001 allele and 19 subjects who were homozygous for the HLA-B*5602 allele were selected for further study. Thirteen subjects in whom we could not detect HBV DNA by PCR and 5 subjects with an HBV of genotype D were excluded, leaving 21 subjects infected with an HBV of genotype C3 (20). These subjects were then genotyped at the HLA-A and HLA-C loci. The age, gender, HBeAg status, HLA-A and HLA-C allele frequencies, and ALT levels of these subjects at the time of sampling are summarized in Table 1. The HLA-B*4001 allele was strongly associated with HLA-A*02 alleles and with the HLA-C*0304 and *0403 alleles. The HLA-B*5602 allele was strongly associated with the HLA-C*0102 allele. One HLA-B*5602-homozygous subject met the criteria for specialist referral for assessment of chronic liver disease (two serum ALT levels of >60 U/liter at least 6 months apart) and was diagnosed as having nonalcoholic fatty liver disease.
TABLE 1.
Subjects with an inactive chronic HBV infection who were homozygous for either HLA-B*4001 or HLA-B*5602
| Parameter | Value |
|
|---|---|---|
| HLA-B*4001 | HLAB*5602 | |
| No. of homozygous subjects | 20 | 19 |
| No. with HBV DNA undetectable by PCR | 7 | 6 |
| No. of subjects with genotype D | 1 | 4 |
| No. of subjects with genotype C3 | 12 | 9 |
| No. HBeAg (positive/negative) | 2/10 | 2/7 |
| No. with HLA-A allele (homozygotes) | ||
| *02a | 10 (2) | 0 (0) |
| *1101 | 2 (0) | 3 (0) |
| *2402 | 10 (3) | 13 (4) |
| *2601 | 1 (0) | 0 (0) |
| *3401 | 1 (0) | 2 (0) |
| No. with HLA-C allele (homozygotes) | ||
| *0102 | 0 (0) | 17 (8) |
| *0304 | 17 (6) | 0 (0) |
| *0403 | 7 (1) | 0 (0) |
| *1203 | 0 (0) | 1 (0) |
| Mean age ± SEM (yr) | 47.3 ± 4.0 | 46.8 ± 4.1 |
| Gender (male/female) | 8/4 | 5/4 |
| Mean ALT ± SEM (normal < 45 U/liter) | 34.3 ± 6.5 | 40.9 ± 15.3b |
HLA-A*0201 (n = 5) and *0206 (n = 5).
ALT = 153 U/liter in a subject with nonalcoholic fatty liver disease.
All subjects gave written consent, and the study was approved by the Northern X Regional Ethics Committee of the New Zealand Ministry of Health.
Cloning and sequencing of HBV.
HBV DNA was extracted from 300 μl of serum using the High Pure Viral Nucleic Acid kit (Roche Diagnostics, Indianapolis, IN). There are both between-subject and within-subject differences in the sequences of the HBV in the Tongan population. Consequently, it was necessary to custom design some of the PCR primers for amplifying and sequencing the HBV from each subject, especially those who were HBeAg negative. Our initial strategy was to obtain a consensus sequence of base pairs 1700 to 2411. The primers for obtaining this sequence were initially identified by finding conserved sequences in an alignment (6) of 54 genotype C and genotype D sequences obtained from the NCBI nucleotide database (http://www.ncbi.nlm.nih.gov/sites/entrez). We obtained three forward primers from this alignment (5′-TTCACCTCTGCACGTCG [1590 to 1606], 5′-ATGTCAACGACCGACCTTGA [1680 to 1699], and 5′-GAGGCTGTAGGCATAAATTGGTCT [1777 to 1801]), which all matched two reverse primers (5′-AGGAGTGCGAATCCACACT [2287 to 2269] and 5′-CCGAGATTGAGATCTTCTGCGACGCG [2437 to 2412]). The base numbers in brackets are from a genotype C3 sequence (X75656).
We cloned the HBV genome in two fragments. The first clone was approximately 2.6 kb in length and included the S open reading frame (ORF), the terminal 2,455 bp of the P ORF, and the initial 403 bp of the X ORF. The primers for amplification of the 2.6-kb clone were chosen after analysis of the bp 1700 to 2411 consensus sequence mentioned above and were designed to amplify the minus strand of HBV. The primers for the first 20-μl PCR were custom designed for each subject, with the upper primer comprised of either bases 1819 to 1845 or bases 1847 to 1869 and the lower primer being the reverse complement of either bases 1804 to 1825 or bases 1777 to 1801. The PCR products from this reaction were purified and concentrated using polyethylene glycol (MW = 8,000; P5413; Sigma Aldrich, St. Louis, MO) and resuspended in 5 μl of water, and an aliquot of between 0.1 and 5.0 μl was used as the template for a second PCR. The upper primer for the second reaction was comprised of either bases 2359 to 2380 or bases 2370 to 2391, and the lower primer was the reverse complement of bases 1777 to 1801. The three internal sequencing primers used were bases 817 to 833 and the reverse complements of bases 903 to 924 and 94 to 117.
The second amplimer that was cloned was approximately 1.2 kb in length and included the full C ORF, the terminal 203 bp of the X ORF, and the initial 451 bp of the P ORF. It was amplified from the plus strand of the HBV. The primers for this amplimer were chosen after inspection of the sequences of the 2.6-kb clones that were obtained from that patient, with the upper primer comprised of bases 1590 to 1606 and the lower primer being the reverse complement of bases 2778 to 2802. Clones were sequenced using the external primers. Our initial strategy for cloning the C ORF was to amplify a 2.2-kb amplimer from the plus strand using bases 1590 to 1606 as the upper primer and the reverse complement of bases 547 to 573 as the lower primer. We were unable to produce amplimers from most HBeAg-negative patients using these primers, possibly because the plus strand of the virus was of insufficient length. However, a small number of clones produced from this amplimer are included in the results.
The PCR cycling conditions for each reaction were determined using Oligo Primer Analysis Software, version 6 (Molecular Biology Insights [http://www.oligo.net]). We used a three-stage touchdown protocol. The denaturation temperature was 96°C for 10 s for the first stage and 2°C above the melting temperature (Tm) of the product for 10 s for the last two stages. The first- and second-stage annealing temperatures were set at the Tm of the primers (with a maximum of 72°C) for 15 s, decreasing by 0.2°C and 0.5°C per cycle for eight cycles and four cycles for the first and second stages, respectively. The third-stage annealing temperature was set at the optimal value calculated by the Oligo Primer Analysis Software for 30 cycles. The extension temperature was 72°C, and the extension time was 50 to 60 s per kilobase of product. All amplifications of HBV DNA were performed using Accuprime Taq DNA Polymerase High Fidelity (Invitrogen Life Technologies, Carlsbad, CA). Polyethylene glycol-cleaned amplimers were A tailed (30 to 300 ng of amplimer; 0.88 μl 10× ammonium sulfate buffer [pH = 8.4], 0.8 μl 2 mM dATP, 0.33 μl 25 mM MgCl2, 3 U Taq polymerase, and water to 8.8 μl) for 30 min at 72°C, ligated into pGEM-T (Promega, Madison, WI), and used to transform Escherichia coli (DH5α). Only one full-length clone was sequenced from each PCR. The sequencing reaction mixtures contained between 20 and 40 ng of cloned DNA and 1 μl of ABI Prism BigDye Terminator v3.1 cycle-sequencing mix (Applied Biosystems, Foster City, CA). The sequencing reaction products were purified with magnetic beads (CleanSeq; Agencourt Biosciences Corp., Beverly, MA) and analyzed on an ABI Prism 3130XL genetic analyzer fitted with a 50-cm capillary array using POP7 polymer. The sequencing reactions were performed using annealing temperatures of either 56°C or 50.5°C, depending on the Tm of the sequencing primer. Sequence analysis and contig assembly were performed using Chromas 1.61 (Technelysium Pty Ltd., Queensland, Australia).
To minimize the risk of contamination between samples, all PCR cocktails were prepared in a PCR workstation (Bigneat Ltd., Hampshire, United Kingdom) and transferred to a separate room for the addition of templates. Thermal cycling and agarose gel analysis of PCR products were conducted in a third room. Transformation and cloning of PCR products were performed in an externally vented fume cabinet with an immediately adjacent, dedicated 4 to 8°C refrigerator, 37°C incubator, and water bath. Whenever possible, the PCR and cloning procedures were conducted by separate technicians. We found that there was a unique repertoire of mutations in the HBVs from most subjects, and cross-contamination between subjects could be identified from the sequences. When there was a high level of similarity between HBV sequences from two different subjects (this occurred in some HBeAg-negative subjects with very diverse viral sequences), a fresh aliquot of HBV-DNA was extracted from a fresh tube of serum at a separate time using a new extraction kit, and further clones were obtained to confirm that the sequence existed in each subject.
Genotyping the HBV.
The genotype of any HBV sequence was obtained by aligning (6) the sequence of any PCR amplimer, contig, or HBV genome produced by any one of the primer pairs against a panel of sequences for genotype A (AF297623, AY128092, and AB126580), genotype B (AY167100, AY206391, and AY206390), genotype C (AY167090, AY167096, AY167092, and X75656), genotype D (AB126581, AB104711, and AB104712), genotype E (AB091256, AB091255, and AB10654), genotype F (AB036920, AY179734, and AY179735), genotype G (AB064315 and AF405706), and genotype H (AY090460 and AY090457). The first amplimers we sequenced (containing bases 1700 to 2411; see above) contained a haplotype of 52 bases that distinguish genotype C from genotype D in the Tongan population, and we used this sequence to exclude genotype D patients. The possibility that any patient had a virus with a mixed genotype was excluded by aligning the sequences of the 2.6-kb clones with the panel of sequences described above. We have not identified any Tongan patients with a virus comprised of a mixture of genotype C and genotype D sequences.
Phylogenetic analyses of HBV clones.
The phylogenies of the viruses obtained from the different patients were constructed by maximum likelihood using PhyML (8). PAML (model 2A) (31) was then used to estimate the overall proportion of codon sites in the whole alignment that were likely to be under positive, neutral, or negative selection pressures, and an estimate of the average value of the ratio of nonsynonymous to synonymous nucleotide substitution rates (ω) was made for each of the three categories. The statistical significance of positive selection pressure at each amino acid site was then calculated using the Bayes Empirical Bayes criterion.
Prediction of peptide binding to HLA class I.
We used the artificial neural network method in the online Immune Epitope and Database Analysis Resource (14) to predict the most likely sequences of the peptides containing positively selected amino acids that might bind to the HLA-B*4001 and HLA-A*02 family alleles.
General statistics.
Numerical data are summarized as the mean ± the standard error of the mean. Ordinal data are summarized as the median with the range. Comparisons of ordinal data between groups were conducted with a Kruskal-Wallis test. Comparisons of frequency data between groups were conducted with Fisher's exact test. Statistical calculations were performed using SAS (SAS Institute, Inc., Cary, NC).
RESULTS
The phylogenetic tree.
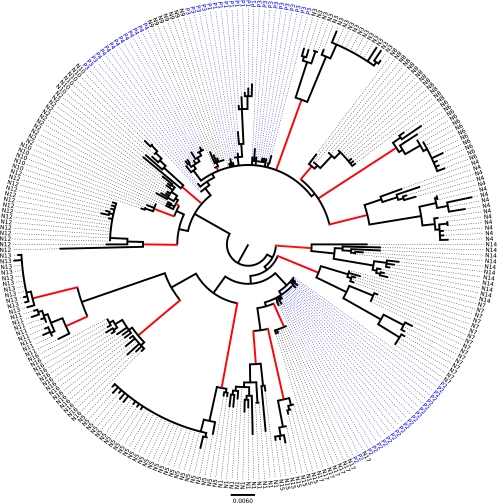
Figure 1 shows the phylogenetic tree assembled from 274 cloned sequences of HBV DNA extracted from the sera of 21 subjects with inactive, genotype C3, chronic HBV infections. Each clone contained the 1,125-bp sequence running from bases 1633 to 2757 (the numbering is from GenBank sequence X75656). Cloned sequences containing nonsense mutations within the core gene and/or large deletions of genomic material from the core gene were excluded.
FIG. 1.
Phylogeny of 274 clones containing 1,125 bp of genetic material from the HBVs extracted from the sera of 21 subjects with inactive chronic HBV infections. Data from the four HBeAg-positive subjects are shown in blue. Each red vertical branch leads to a clade of clones that came from one HBeAg-negative subject.
Sixty of the clones came from the sera of 4 HBeAg-positive subjects (identified in Fig. 1), and 214 of the clones came from 17 HBeAg-negative subjects (identified in Fig. 1). Fifteen of the 17 HBeAg-negative subjects had a single, distinct clade of HBV sequences. In one HBeAg-negative subject (N5), there were a single dominant clade and a smaller, separate clade of two sequences (each from a different extraction), suggesting that the subject had been infected by more than one virus. Only the sequences from the dominant N5 clade were included in the analysis. Subject N10 had two closely related clades, one of which was similar to that of subject N2. The existence of both N10 clades was confirmed by sequencing clones from a second extraction of HBV from a different tube of serum.
Selection pressure on the HBV core gene.
Maximum-likelihood analysis of the whole core gene alignment in PAML predicted that 68.1% of the 183 amino acid sites encoded by the HBV core gene were under negative selection (average ω = 0.06), 21.0% under neutral selection (ω = 1.00), and 10.8% under strong positive selection (ω = 4.24). Site-by-site analysis identified 19 amino acids that were under positive selection, although only 13 of these reached statistical significance (Table 2). The number of subjects whose virus contained one or more clones with a nonsynonymous mutation at each of these sites ranged from 4 to 12 of the 21 subjects. These observations suggest that there may be a limited number of sites in the core gene that are susceptible to positive selection pressure and that the molecular mechanisms that lead to positive selection pressure are common to many of the subjects. However, there was considerable variation between the subjects in both the number and the repertoire of sites that were under positive selection pressure (Table 3).
TABLE 2.
Amino acids of the HBV core gene that were under statistically significant positive selection pressure
| Amino acid position | Wild-type amino acid | Posterior probability | ω (±SEM) | No. of subjects positive at each sitea |
|
|---|---|---|---|---|---|
| HBeAg+ | HBeAg− | ||||
| 13 | V | 1.000 | 4.4 ± 0.5 | 0 | 6 |
| 14 | E | 1.000 | 4.4 ± 0.5 | 1 | 4 |
| 21 | S | 1.000 | 4.4 ± 0.5 | 0 | 8 |
| 77 | E | 1.000 | 4.4 ± 0.5 | 0 | 8 |
| 79 | P | 0.998 | 4.3 ± 0.5 | 0 | 7 |
| 87 | S | 0.955 | 4.2 ± 0.8 | 2 | 10 |
| 97 | I | 0.999 | 4.4 ± 0.5 | 0 | 10 |
| 105 | I | 0.998 | 4.3 ± 0.5 | 0 | 8 |
| 113 | E | 1.000 | 4.4 ± 0.5 | 1 | 8 |
| 130 | P | 1.000 | 4.4 ± 0.5 | 3 | 8 |
| 151 | R | 0.965 | 4.2 ± 0.8 | 0 | 4 |
| 155 | S | 0.973 | 4.3 ± 0.7 | 2 | 7 |
| 180 | E | 0.999 | 4.3 ± 0.5 | 0 | 8 |
The number of subjects with at least one clone with a nonsynonymous mutation at each site.
TABLE 3.
Number of clones with a nonsynonymous mutation at each of the positively selected sites in each subject
| Subject identifier | No. of clones | No. of clones with a mutation at amino acid site: |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 13 | 14 | 21 | 77 | 79 | 87 | 97 | 105 | 113 | 130 | 151 | 155 | 180 | ||
| P1 | 9 | 3 | 5 | |||||||||||
| P2 | 22 | 2 | 1 | |||||||||||
| P3 | 17 | 1 | 9 | |||||||||||
| P4 | 12 | 1 | 1 | 1 | ||||||||||
| N1 | 11 | 9 | 9 | 11 | 11 | 10 | 2 | 10 | ||||||
| N2 | 11 | 10 | 4 | |||||||||||
| N3 | 16 | 16 | 16 | 11 | 4 | 12 | 4 | 16 | ||||||
| N4 | 15 | 13 | 5 | 14 | 2 | 11 | 15 | 5 | 11 | |||||
| N5 | 28 | 26 | 28 | 28 | 28 | 28 | 2 | 28 | ||||||
| N6 | 12 | 2 | 2 | 3 | 12 | 12 | 12 | |||||||
| N7 | 16 | 5 | 7 | 11 | 7 | 11 | 7 | 11 | 4 | |||||
| N8 | 13 | 13 | 10 | 1 | 12 | |||||||||
| N9 | 7 | |||||||||||||
| N10 | 12 | 5 | 1 | 1 | 12 | 12 | ||||||||
| N11 | 8 | 8 | 8 | 7 | 8 | 8 | ||||||||
| N12 | 15 | 13 | 1 | 13 | 2 | 13 | ||||||||
| N13 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | |||
| N14 | 11 | 6 | 1 | 6 | 1 | 1 | 10 | 1 | ||||||
| N15 | 7 | 7 | 7 | 7 | 7 | 6 | 4 | |||||||
| N16 | 15 | 15 | 15 | 15 | 15 | 8 | 15 | |||||||
| N17 | 7 | 7 | 7 | 1 | ||||||||||
Ten of these positively selected sites (amino acids 13 to 130) were encoded by the nonoverlapping region of the C ORF. However, there was positive selection pressure on the amino acids at positions 151, 155, and 180, which are encoded in a region of the HBV genome that overlaps the P ORF, encoding amino acids 14/15, 18/19, and 43/44, respectively. None of these P ORF amino acids was associated with an ω of >1 when the HBV polymerase gene was analyzed (see below), and thus, mutations at these sites are likely to be due to positive selection pressure on the core gene.
The positively selected (ω > 2.0) amino acids of the core gene that did not reach statistical significance are shown in Table 4.
TABLE 4.
Amino acids in the HBV core gene that were under positive selection pressure (ω > 2.0) but did not reach statistical significance
| Amino acid position | Wild-type amino acid | Posterior probability | ω (±SEM) | No. of subjects positive at each sitea |
|
|---|---|---|---|---|---|
| HBeAg+ | HBeAg− | ||||
| 26 | S | 0.595 | 2.9 ± 1.6 | 0 | 4 |
| 35 | S | 0.743 | 3.5 ± 1.5 | 1 | 5 |
| 55 | L | 0.878 | 3.9 ± 1.2 | 0 | 8 |
| 84 | L | 0.506 | 2.6 ± 1.7 | 0 | 3 |
| 91 | V | 0.853 | 3.8 ± 1.2 | 0 | 10 |
| 177 | Q | 0.553 | 2.8 ± 1.6 | 0 | 4 |
The number of subjects with at least one clone with a nonsynonymous mutation at each site.
Influence of HLA class I alleles on selection pressure within the HBV core gene.
The primary purpose of this study was to determine whether there was a difference in the repertoires of amino acid sites under positive selection pressure between groups of HBeAg-negative subjects who were homozygous for either HLA-B*4001 (subjects N1 to N10) or HLA-B*5602 (subjects N11 to N17). Table 5 shows that amino acid mutations at 3 of the 13 positively selected sites were associated with these alleles. The serine at position 21 (P = 0.02) had a higher frequency of mutations in subjects who were homozygous for HLA-B*5602. The glutamic acids at both position 77 (P = 0.05) and position 113 (P = 0.002) had a higher frequency of mutations in subjects who were homozygous for HLA-B*4001.
TABLE 5.
Significance (Fisher's exact test) of associations between HLA-B*4001 (subjects N1 to N10) and HLA-B*5602 (subjects N11 to N17) genotypes and the number of HBeAg-negative subjects with at least one clone containing a nonsynonymous mutation at amino acid sites known to exhibit positive selection
| Site | No. of subjects |
P | |||
|---|---|---|---|---|---|
| HLA-B* 4001/4001 |
HLA-B* 5602/5602 |
||||
| NSa present | NS absent | NS present | NS absent | ||
| S 21 | 2 | 8 | 6 | 1 | 0.02 |
| E 77 | 7 | 3 | 1 | 6 | 0.05 |
| E 113 | 8 | 2 | 0 | 7 | 0.002 |
NS, nonsynonymous mutations.
In addition, there were three sites (Table 6) at which amino acid mutations were associated with the presence of an HLA-A*02 allele (subjects N1 to N7). These were the valine at position 13 (P = 0.03), the glutamic acid at position 14 (P = 0.01), and the glutamic acid at position 113 (P = 0.0004). Since the HLA-A*02 and HLA-B*4001 alleles were strongly associated in this population sample (Table 1), it is difficult to determine which HLA class I allele is primarily associated with the positive selection pressure on E113.
TABLE 6.
Significance (Fisher's exact test) of associations between the presence (subjects N1 to N7) or absence (subjects N8 to N17) of HLA-A*02 alleles and the number of subjects with at least one clone containing a nonsynonymous mutation at amino acid sites known to exhibit positive selection
| Site | No. of subjects |
P | |||
|---|---|---|---|---|---|
| HLA-A*02 present |
HLA-A*02 absent |
||||
| NSa present | NS absent | NS present | NS absent | ||
| V 13 | 5 | 2 | 1 | 9 | 0.03 |
| E 14 | 4 | 3 | 0 | 10 | 0.01 |
| E 113 | 7 | 0 | 1 | 9 | 0.0004 |
NS, nonsynonymous mutations.
Predictions of the strength of peptide binding to HLA class I alleles.
Artificial neural network predictions of the binding affinities of peptides containing positively selected wild-type and mutated amino acids to HLA class I alleles are shown in Table 7. Most peptides presented by HLA class I molecules to CD8+ T cells have a 50% inhibitory concentration (IC50) of less than 500 nM (14). Thus, the wild-type glutamic acid residues at positions 77 and 113 of the HBV core gene may both be contained in peptides that could bind to HLA-B*4001. All four of the amino acid mutations we found at these sites would reduce the binding affinities of these peptides to HLA-B*4001 to a level (<5,000 nM) that is unlikely to result in peptide presentation to CD8+ T cells. These data are also consistent with observations that peptides binding to HLA-B*4001 require an acidic amino acid at anchoring-peptide position 2 to bind to the basic lysine residue in the B pocket of HLA-B*4001 (29).
TABLE 7.
Artificial neural network predictions of the IC50s for binding of wild-type and mutated peptides containing positively selected amino acids to HLA class I alleles
| Best peptidea | HLA | Wild type |
Mutation 1 |
Mutation 2 |
|||
|---|---|---|---|---|---|---|---|
| Amino acid | IC50 (nM) | Amino acid | IC50 (nM) | Amino acid | IC50 (nM) | ||
| LEDPASREL | B*4001 | E77 | 98 | Q77 | 5,470 | D77 | 16,657 |
| RETVLEYLV | B*4001 | E113 | 107 | D113 | 14,886 | A113 | 18,688 |
| ASVELLSFL | A*0201 | V13, E14 | 3,203 | Various | 1,107-8,711 | ||
| ASVELLSFL | A*0206 | V13, E14 | 41 | Various | 42-629 | ||
| SCLTFGRETV | A*0201 | E113 | 3,485 | D113 | 6,570 | A113 | 2,751 |
| CLTFGRETV | A*0206 | E113 | 1,167 | D113 | 1,492 | A113 | 1,144 |
Positively selected amino acids are in boldface.
It is possible that peptides containing the valine and glutamic acid residues at positions 13 and 14 of the core gene can bind both the HLA-A*02 family alleles that were present in our sample, but neither of these amino acids is likely to be responsible for peptide anchoring to the B pocket. We found 15 different combinations of amino acid mutations at these two sites, only two of which reduced the binding affinity to greater than 5,000 nM. Similarly, the binding algorithm predicts that peptides containing the glutamic acid at position 113 have only weak binding to HLA-A*02 family alleles, and neither of the amino acid mutations found at position 113 had a significant influence on the predicted binding of these peptides.
Selection pressure on the HBV X gene.
Analysis of the X gene required the removal of 17 of the 274 clones from 3 of the 4 HBeAg-positive and 3 of the 17 HBeAg-negative subjects because of indels in the core promoter region. Maximum-likelihood analysis of the alignment of the terminal 67 amino acids of the X gene estimated that 98.0% were under negative selection (ω = 0.41), 0.5% under neutral selection (average ω = 1.00), and 1.5% under strong positive selection (average ω = 6.2). Site-by-site analysis identified the isoleucine at position 40 (position 127 of the full X gene) as being under positive selection pressure (ω = 6.45 ± 1.9), with a posterior probability of 1.000. There was no influence of the HLA class I genotype on the frequency of subjects with a nonsynonymous mutation at this site.
Selection pressure on the HBV polymerase gene.
Maximum-likelihood analysis of the alignment of the initial 150 amino acids of the P gene predicted that 92.3% were likely to be under negative selection (average ω = 0.13) and 7.3% under neutral selection (average ω = 1.00). There were no sites under positive selection pressure in this segment of the polymerase gene.
Influence of HBeAg status on positive selection pressure.
The number of positively selected core gene sites for which at least one nonsynonymous mutation was found was calculated for each subject. For example, subject N1 had seven sites and subject P1 had two sites with a nonsynonymous mutation (Table 3). The number of sites with nonsynonymous mutations per subject was higher (Kruskal-Wallis test; P = 0.01) in the HBeAg-negative (N1 to N17; median, 6; range, 0 to 10 out of 13 sites per subject) than in the HBeAg-positive (P1 to P4; median, 2; range, 2 to 3 out of 13 sites per subject) group. This was not due to a difference in the number of clones sequenced per subject, as this was higher in the HBeAg-positive (mean, 15.0 per subject) than in the HBeAg-negative (mean, 12.6 per subject) group (Table 3, column 2).
None of the 4 HBeAg-positive subjects and 11 of the 17 HBeAg-negative subjects had at least one clone containing a nonsynonymous mutation at amino acid 127 of the X gene (P = 0.04).
DISCUSSION
HBeAg seroconversion is an important stage in the evolution of a chronic HBV infection, because it coincides with the development of the HBV-specific CD8+ T-cell repertoire that is believed to suppress replication of the virus and reduce the risk of chronic hepatitis B, liver cirrhosis, and liver cancer (11, 13). HBeAg seroconversion is also associated with an increase in the number of mutations in core polypeptide amino acids (2, 9). Positive selection pressure from the host immune system, which drives the selection of escape mutations (3, 18), is responsible for some of these changes (12). However, others may be due to genetic drift or to changes in the amino acid sequence that compensate for any loss of core polypeptide function associated with the escape mutations (7, 23). The full repertoire of common escape mutations in HBV polypeptides has not been defined, partly because the strong influences of both the viral genotype and host genetics on the immune response (26) make specific virus-host interactions difficult to identify in complex populations. For this reason, we have chosen to study the influence of HLA class I on the repertoire of positively selected amino acid mutations in the HBV core polypeptide in the Tongan population, where there is limited diversity both in the prevalent HBV strains (unpublished data) and in HLA class I haplotypes (1).
We identified 13 amino acids of the HBV core polypeptide that were commonly under significant positive selection pressure. We also identified six codons in the core gene where a high ratio of nonsynonymous to synonymous nucleotide changes was suggestive of positive selection. The failure to reach statistical significance at these six codons may have been due to having insufficient statistical power to detect weak biological effects.
Our data showed a significantly higher frequency of nonsynonymous mutations at the 13 positively selected sites in HBVs from 17 HBeAg-negative relative to 4 HBeAg-positive subjects. This is consistent with data from a longitudinal study of patients undergoing HBeAg seroconversion, in which HBV from postseroconversion samples showed increased evidence of positive selection pressure (12). This raises the possibility that some of the selection pressure might be due to the CD8+ T cells that are known to be activated at the time of HBeAg seroconversion (22). Mutation of the amino acids responsible for the binding and positioning of peptides within the groove of HLA class I molecules in response to selection pressure from CD8+ T cells has been described in human immunodeficiency virus and HBV infections (3, 18), and disruption of T-cell recognition sites in these peptides is also possible (18). These escape mutations allow the virus to escape detection by CD8+ T cells, albeit at some cost to the function of the protein. In our data, HLA class I alleles were found to be significantly associated with the presence of amino acid mutations at 5 of the 13 sites that were under positive selection pressure. The wild-type amino acids at these sites could be responsible for peptide binding and/or positioning in the peptide groove of these class I alleles or could be part of the T-cell receptor recognition site in the peptide. Other possibilities are that these amino acid mutations could compensate for any loss of core gene function that was associated with other HLA class I-associated escape mutations, and type I statistical error.
The results of the peptide prediction algorithm analyses are consistent with the possibility that the glutamic acid residues at positions 77 and 113 of the core gene are responsible for peptide anchoring to the B pocket of HLA-B*4001 (29). The amino acid mutations that occurred at these sites were both predicted to decrease peptide binding to a level that would prevent presentation to CD8+ T cells. Thus, the amino acid mutations at E77 and E113 fit the pattern expected of escape mutations that allow infected hepatocytes to avoid detection by the immune system. CD8+ T-cell assays will be needed to confirm this.
In contrast, there was little evidence from the predicted peptide-binding analyses to support a role for V13, E14, or E113 in anchoring peptides to HLA-A*02 family alleles, although this possibility cannot be completely excluded. None of these three amino acids were at the peptide-anchoring position 2 of any peptides that could theoretically bind to HLA-A*02, and the mutations we detected at these sites did not consistently affect predicted peptide binding. Thus, if the statistical associations between nonsynonymous mutations at these sites and HLA-A*02 are not due to type I statistical error, they may either represent disruptions of the site recognized by T-cell receptors or be responsible for restoring function to the core gene that was lost because of other mutations. The possibility that there is disruption of the T-cell receptor recognition site could be tested using in vitro CD8+ T-cell assays.
Eight of the 13 positively selected amino acid sites were not associated with HLA class I alleles. It is possible that some of these amino acids are contained within peptides that bind the HLA-A*1101, HLA-A*2402, and HLA-A*3401 alleles but the associations were missed because of insufficient statistical power. It is also possible that some of these amino acids are contained in “promiscuous” peptides or overlapping peptides that bind more than one HLA class I molecule. The possibility that immune mechanisms other than CD8+ T cells contribute to the development of positive selection pressure is also of interest. These mechanisms could be upregulated by the adjuvant component of a therapeutic vaccine. For example, intracellular pattern recognition receptors or RNA interference might be upregulated by either CD8+ T cells or the innate immune system. Mutations in viral proteins that impair peptide processing have also been reported (27).
In addition to following up these data with studies of peptide-specific CD8+ T cells, it will also be valuable to determine which of these mutations are selected because they restore replicative capacity that is lost as a result of other immunity-driven escape mutations in the same protein (23). Escape mutations that arise from CD8+ T-cell responses driven by wild-type peptides may have no overall effect on viral replication if they are immediately counteracted by compensatory mutations. Such wild-type peptides may have little therapeutic benefit if they are included in a therapeutic vaccine. Alternatively, it may be worthwhile to develop strategies whereby a therapeutic vaccine targets common compensatory mutations before they emerge. For example, the amino acids at sites 87 and 97 had a high frequency of non-HLA-associated, nonsynonymous mutations both in our HBeAg-negative subjects (Table 2) and in a group of subjects from Singapore with genotype B infections (12). It is possible that these two amino acids are sensitive to positive selection pressure from a mechanism that is independent of CD8+ T cells, such as a common compensatory mechanism or escape from innate immunity.
It is probably not possible to predict which of our 13 amino acid sites might include compensatory mutations. Statistical correlations between the existence of immune-driven and compensatory mutations within subjects are not always strong (24). Tracking stable combinations of mutations through transmission pairs (24) is also unlikely to be successful with hepatitis B in New Zealand, as most people are unaware of the source of their original exposure. Even finding evidence that mutations at an amino acid site disrupt peptide binding to class I HLA does not exclude the possibility that the same amino acid cannot also undergo compensatory changes. Thus, in vitro studies of viral replication will probably be necessary to resolve this issue.
A study of a larger number of Tongan subjects should result in the identification of all the associations between common HLA class I alleles and positively selected amino acids in the core polypeptide. Identification of all the associations between HLA class I and positively selected amino acids would also be valuable because, by exclusion, it would allow identification of the amino acids that were escaping selection pressure from other mechanisms. Our incidental finding of associations with the HLA-A*02 family alleles, which were heterozygous in most subjects, suggests that it will not be necessary to restrict future studies to people who are homozygous for each HLA allele. This will greatly simplify study design. The disadvantage of studying Tongan subjects is that there is strong linkage disequilibrium between the HLA-A, HLA-B, and HLA-C genes. Thus, it is difficult to exclude the possibility that the association between HLA-B*5602 and S21 is due to the HLA-C*0102 allele. A follow-up study in a population with a more diverse repertoire of HLA haplotypes needs to be done.
Our data also provided an opportunity to determine whether the first 150 amino acids of the polymerase gene and the last 67 amino acids of the X gene include any sites that are susceptible to positive selection pressure. There was no evidence for positive selection pressure on any of these polymerase gene amino acids. CD8+ T cells responding to polymerase peptides have been described (21), but none were contained in the first 150 amino acids of the polypeptide. The possibility that X gene peptides stimulate CD8+ T cells in humans is less clear. X gene peptides have been shown to stimulate CD8+ T cells in HLA-A*0201 transgenic mice (16), but there is currently no supportive data for humans. Although I127 of the X gene was under selection pressure in our subjects, there was no evidence that this was related to the effect of any HLA class I allele.
In summary, we have developed an assay that has identified 13 amino acid sites in the HBV core gene that are susceptible to positive selection pressure in subjects with an inactive, HBeAg-negative chronic HBV infection. A significant association between an HLA class I allele and the presence of nonsynonymous mutations was found at five of these sites. This suggests that some of these amino acids are contained within peptides that are recognized by antiviral CD8+ T cells with clinically significant effects on viral replication. If follow-up CD8+ T-cell assays indicate that this is true, it should be possible to use this assay to define the full repertoire of viral peptides that bind to the HLA class I alleles that are common in areas where there is a high prevalence of chronic hepatitis B. These peptides may be useful components of a therapeutic vaccine for chronic hepatitis B and may be useful when designing assays for use as correlates of immunity in treatment trials.
Acknowledgments
This work was supported by the Maurice and Phyllis Paykel Trust; The Lloyd Morgan Lions Clubs Charitable Trust, New Zealand; and the Auckland Medical Research Foundation.
We thank Ika Vea, Lavili Ahokavi, Likuone Latu, Mele Vaka, Mele Taufa, Pauline Lolohea, and Janette Medforth for recruiting the volunteers, and the staff of the Labplus Sequencing Unit and Kristine Boxen of the School of Biological Sciences for the sequencing analyses. We are particularly grateful to the study subjects.
Footnotes
Published ahead of print on 21 October 2009.
REFERENCES
- 1.Abbott, W. G., C. F. Tukuitonga, M. Ofanoa, S. R. Munn, and E. J. Gane. 2006. Low-cost, simultaneous, single-sequence genotyping of the HLA-A, HLA-B and HLA-C loci. Tissue Antigens 68:28-37. [DOI] [PubMed] [Google Scholar]
- 2.Akarca, U. S., and A. S. Lok. 1995. Naturally occurring hepatitis B virus core gene mutations. Hepatology 22:50-60. [PubMed] [Google Scholar]
- 3.Bertoletti, A., A. Costanzo, F. V. Chisari, M. Levrero, M. Artini, A. Sette, A. Penna, T. Giuberti, F. Fiaccadori, and C. Ferrari. 1994. Cytotoxic T lymphocyte response to a wild type hepatitis B virus epitope in patients chronically infected by variant viruses carrying substitutions within the epitope. J. Exp. Med. 180:933-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boni, C., P. Fisicaro, C. Valdatta, B. Amadei, P. Di Vincenzo, T. Giuberti, D. Laccabue, A. Zerbini, A. Cavalli, G. Missale, A. Bertoletti, and C. Ferrari. 2007. Characterization of hepatitis B virus (HBV)-specific T-cell dysfunction in chronic HBV infection. J. Virol. 81:4215-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, T. T., R. G. Gish, R. de Man, A. Gadano, J. Sollano, Y. C. Chao, A. S. Lok, K. H. Han, Z. Goodman, J. Zhu, A. Cross, D. DeHertogh, R. Wilber, R. Colonno, and D. Apelian. 2006. A comparison of entecavir and lamivudine for HBeAg-positive chronic hepatitis B. N. Engl. J. Med. 354:1001-1010. [DOI] [PubMed] [Google Scholar]
- 6.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford, H., J. G. Prado, A. Leslie, S. Hue, I. Honeyborne, S. Reddy, M. van der Stok, Z. Mncube, C. Brander, C. Rousseau, J. I. Mullins, R. Kaslow, P. Goepfert, S. Allen, E. Hunter, J. Mulenga, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2007. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant HLA-B*5703-restricted Gag epitope in chronic human immunodeficiency virus type 1 infection. J. Virol. 81:8346-8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guindon, S., A. G. Rodrigo, K. A. Dyer, and J. P. Huelsenbeck. 2004. Modeling the site-specific variation of selection patterns along lineages. Proc. Natl. Acad. Sci. U. S. A. 101:12957-12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gunther, S. 2006. Genetic variation in HBV infection: genotypes and mutants. J. Clin. Virol. 36(Suppl. 1):S3-S11. [DOI] [PubMed] [Google Scholar]
- 10.Lai, C. L., D. Shouval, A. S. Lok, T. T. Chang, H. Cheinquer, Z. Goodman, D. DeHertogh, R. Wilber, R. C. Zink, A. Cross, R. Colonno, and L. Fernandes. 2006. Entecavir versus lamivudine for patients with HBeAg-negative chronic hepatitis B. N. Engl. J. Med. 354:1011-1020. [DOI] [PubMed] [Google Scholar]
- 11.Liaw, Y. F. 2009. Natural history of chronic hepatitis B virus infection and long-term outcome under treatment. Liver Int. 29(Suppl. 1):100-107. [DOI] [PubMed] [Google Scholar]
- 12.Lim, S. G., Y. Cheng, S. Guindon, B. L. Seet, L. Y. Lee, P. Hu, S. Wasser, F. J. Peter, T. Tan, M. Goode, and A. G. Rodrigo. 2007. Viral quasi-species evolution during hepatitis Be antigen seroconversion. Gastroenterology 133:951-958. [DOI] [PubMed] [Google Scholar]
- 13.Lok, A. S., E. J. Heathcote, and J. H. Hoofnagle. 2001. Management of hepatitis B: 2000—summary of a workshop. Gastroenterology 120:1828-1853. [DOI] [PubMed] [Google Scholar]
- 14.Lundegaard, C., K. Lamberth, M. Harndahl, S. Buus, O. Lund, and M. Nielsen. 2008. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8-11. Nucleic Acids Res. 36:W509-W512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maini, M. K., C. Boni, C. K. Lee, J. R. Larrubia, S. Reignat, G. S. Ogg, A. S. King, J. Herberg, R. Gilson, A. Alisa, R. Williams, D. Vergani, N. V. Naoumov, C. Ferrari, and A. Bertoletti. 2000. The role of virus-specific CD8(+) cells in liver damage and viral control during persistent hepatitis B virus infection. J. Exp. Med. 191:1269-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malmassari, S., Y. C. Lone, M. Zhang, C. Transy, and M. L. Michel. 2005. In vivo hierarchy of immunodominant and subdominant HLA-A*0201-restricted T-cell epitopes of HBx antigen of hepatitis B virus. Microbes Infect. 7:626-634. [DOI] [PubMed] [Google Scholar]
- 17.Marinos, G., N. V. Naoumov, and R. Williams. 1996. Impact of complete inhibition of viral replication on the cellular immune response in chronic hepatitis B virus infection. Hepatology 24:991-995. [DOI] [PubMed] [Google Scholar]
- 18.McMichael, A. J., and R. E. Phillips. 1997. Escape of human immunodeficiency virus from immune control. Annu. Rev. Immunol. 15:271-296. [DOI] [PubMed] [Google Scholar]
- 19.Missale, G., A. Redeker, J. Person, P. Fowler, S. Guilhot, H. J. Schlicht, C. Ferrari, and F. V. Chisari. 1993. HLA-A31- and HLA-Aw68-restricted cytotoxic T cell responses to a single hepatitis B virus nucleocapsid epitope during acute viral hepatitis. J. Exp. Med. 177:751-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norder, H., A. M. Courouce, P. Coursaget, J. M. Echevarria, S. D. Lee, I. K. Mushahwar, B. H. Robertson, S. Locarnini, and L. O. Magnius. 2004. Genetic diversity of hepatitis B virus strains derived worldwide: genotypes, subgenotypes, and HBsAg subtypes. Intervirology 47:289-309. [DOI] [PubMed] [Google Scholar]
- 21.Rehermann, B., P. Fowler, J. Sidney, J. Person, A. Redeker, M. Brown, B. Moss, A. Sette, and F. V. Chisari. 1995. The cytotoxic T lymphocyte response to multiple hepatitis B virus polymerase epitopes during and after acute viral hepatitis. J. Exp. Med. 181:1047-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rehermann, B., D. Lau, J. H. Hoofnagle, and F. V. Chisari. 1996. Cytotoxic T lymphocyte responsiveness after resolution of chronic hepatitis B virus infection. J. Clin. Invest. 97:1655-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneidewind, A., M. A. Brockman, J. Sidney, Y. E. Wang, H. Chen, T. J. Suscovich, B. Li, R. I. Adam, R. L. Allgaier, B. R. Mothe, T. Kuntzen, C. Oniangue-Ndza, A. Trocha, X. G. Yu, C. Brander, A. Sette, B. D. Walker, and T. M. Allen. 2008. Structural and functional constraints limit options for cytotoxic T-lymphocyte escape in the immunodominant HLA-B27-restricted epitope in human immunodeficiency virus type 1 capsid. J. Virol. 82:5594-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneidewind, A., Z. L. Brumme, C. J. Brumme, K. A. Power, L. L. Reyor, K. O'Sullivan, A. Gladden, U. Hempel, T. Kuntzen, Y. E. Wang, C. Oniangue-Ndza, H. Jessen, M. Markowitz, E. S. Rosenberg, R. P. Sekaly, A. D. Kelleher, B. D. Walker, and T. M. Allen. 2009. Transmission and long-term stability of compensated CD8 escape mutations. J. Virol. 83:3993-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sobao, Y., K. Sugi, H. Tomiyama, S. Saito, S. Fujiyama, M. Morimoto, S. Hasuike, H. Tsubouchi, K. Tanaka, and M. Takiguch. 2001. Identification of hepatitis B virus-specific CTL epitopes presented by HLA-A*2402, the most common HLA class I allele in East Asia. J. Hepatol. 34:922-929. [DOI] [PubMed] [Google Scholar]
- 26.Tan, A. T., E. Loggi, C. Boni, A. Chia, A. J. Gehring, K. S. Sastry, V. Goh, P. Fisicaro, P. Andreone, C. Brander, S. G. Lim, C. Ferrari, F. Bihl, and A. Bertoletti. 2008. Host ethnicity and virus genotype shape the hepatitis B virus-specific T-cell repertoire. J. Virol. 82:10986-10997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tenzer, S., E. Wee, A. Burgevin, G. Stewart-Jones, L. Friis, K. Lamberth, C. H. Chang, M. Harndahl, M. Weimershaus, J. Gerstoft, N. Akkad, P. Klenerman, L. Fugger, E. Y. Jones, A. J. McMichael, S. Buus, H. Schild, P. van Endert, and A. K. Iversen. 2009. Antigen processing influences HIV-specific cytotoxic T lymphocyte immunodominance. Nat. Immunol. 10:636-646. [DOI] [PubMed] [Google Scholar]
- 28.Tsai, S. L., M. H. Chen, C. T. Yeh, C. M. Chu, A. N. Lin, F. H. Chiou, T. H. Chang, and Y. F. Liaw. 1996. Purification and characterization of a naturally processed hepatitis B virus peptide recognized by CD8+ cytotoxic T lymphocytes. J. Clin. Invest. 97:577-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vasmatzis, G., C. Zhang, J. L. Cornette, and C. DeLisi. 1996. Computational determination of side chain specificity for pockets in class I MHC molecules. Mol. Immunol. 33:1231-1239. [DOI] [PubMed] [Google Scholar]
- 30.Webster, G. J., S. Reignat, D. Brown, G. S. Ogg, L. Jones, S. L. Seneviratne, R. Williams, G. Dusheiko, and A. Bertoletti. 2004. Longitudinal analysis of CD8+ T cells specific for structural and nonstructural hepatitis B virus proteins in patients with chronic hepatitis B: implications for immunotherapy. J. Virol. 78:5707-5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang, Z., W. S. Wong, and R. Nielsen. 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22:1107-1118. [DOI] [PubMed] [Google Scholar]
- 32.Yewdell, J. W. 2006. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity 25:533-543. [DOI] [PubMed] [Google Scholar]