Abstract
The human papillomavirus type 18 (HPV-18) E2 gene is inactivated in cervical carcinoma after integration of the viral DNA into the host cellular genome. Since E2 represses the transcription of the two viral oncogenes E6 and E7, integration which allows their strong expression is considered a major step in transformation by HPV. We show here that E2 is specifically degraded at the end of the G1 phase in a Brd4-independent manner, implying that its regulatory functions are cell cycle dependent. Degradation of E2 occurs via the Skp1/Cullin1/F-box Skp2 (SCFSkp2) ubiquitin ligase, since silencing of Skp2 induces stabilization of E2. In addition, the amino-terminal domain of E2 can interact with Skp2 as shown by coimmunoprecipitation experiments. We previously showed that E2 inhibits the anaphase-promoting complex/cyclosome (APC/C) ubiquitin ligase, leading to accumulation of several of its substrates. We demonstrate here that Skp2, which is a known APC/C substrate in G1, is also stabilized by E2. Therefore, by negative feedback, SCFSkp2 activation could lead to E2 degradation and E6/E7 expression specifically in the late G1 phase. Moreover, since the SCFSkp2 can trigger S-phase entry and Skp2 itself is a known oncogene, we believe that E2-mediated accumulation of Skp2, together with E2 degradation leading to putative release of E6 and E7 inhibition, could induce premature S-phase entry in HPV-infected cells, pointing to a direct role of E2 in the early steps of HPV-mediated transformation.
Human papillomaviruses (HPVs) are small DNA viruses that infect stratified epithelia in differentiation. They induce benign lesions, as well as cervical carcinomas, depending on the HPV type. These viruses encode two viral oncogenes, E6 and E7, which, among other transforming functions, can, respectively, inhibit p53 and pRb, thus deregulating cell proliferation (9, 18, 23, 24, 29). The transcription of E6 and E7 is negatively regulated by the viral E2 protein which binds directly to their promoter (8, 27). In 100% of cervical carcinomas associated with HPV-18, the viral DNA is integrated into the host cell genome, leading to disruption of the E2 open reading frame and uncontrolled E6 and E7 expression (28).
We and others have already shown that the E2 proteins from both low- and high-risk HPV types exhibit short half-life times due to degradation by the ubiquitin/proteasome pathway (4, 5, 11, 20), and Cullin-3 has recently been identified as a potential E2 ubiquitin ligase, although the involvement of other cullins has not been excluded (31). We have previously shown that, despite strong interactions of high-risk HPV E2 with Cdc20 and Cdh1, the activators of the anaphase-promoting complex/cyclosome (APC/C) ubiquitin ligase, E2 is not an APC/C substrate. In contrast, these interactions mediate stabilization of several APC/C substrates, presumably responsible for both G2/M arrest and chromosomal instability (3). Since these phenotypes are specific to high-risk HPV E2 proteins compared to the low-risk ones, we postulated that the E2-mediated chromosomal instability could participate in the transformation process by high-risk HPVs.
SCF (Skp1/Cullin1/F box) is a ring-finger ubiquitin ligase composed of four subunits including three invariable components (Rbx1, Cullin1, and Skp1) and 1 variable component, namely, F-box protein, such as Skp2 or βTrCP, that binds directly to the substrates (previous phosphorylation of the substrate is generally required for degradation via the SCFSkp2 pathway). Skp2 is degraded in G1 and G0 via the APC/C. At the end of G1, SCFSkp2 is activated by Skp2 accumulation after APC/C inhibition (1, 2) and promotes S-phase entry by degrading the cyclin/Cdk inhibitors p27 and p21 (6, 7, 19, 26, 30). This leads to activation of cyclin/Cdk complexes, which in turn phosphorylate pRb and release functional E2F that activates S-phase genes. Consequently, ectopic expression of stable Skp2 promotes S-phase entry, and overexpression of Skp2 is a common event in cancer (10, 12, 14).
We show here that levels of HPV-18 E2 vary during the cell cycle and demonstrate that E2 is degraded at the end of G1 while being accumulated from G2 to mid-G1. HPV-18 E2 interacts with Skp2, and the silencing of Skp2, in asynchronous cells or at the G1/S transition, induces E2 stabilization in such a way that the SCFSkp2 appears to be a major ubiquitin ligase involved in E2 degradation.
MATERIALS AND METHODS
Cell cultures, infections, transfections, and live microscopy.
HaCaT cells were grown in 10-cm petri dishes in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum.
Infections with recombinant adenoviruses expressing the green fluorescent protein (GFP) fusion proteins were done at a multiplicity of infection of 200 as previously described (3).
Cotransfections of the Skp2 expression vector with GFP-E2, GFP-ΔTAD, GFP, or untagged E2 expression vectors were performed by Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions, with 15 μg of each plasmid.
Transfections of the small interfering RNA (siRNA) against Skp2 (5′-AUUCUCUGAAUUUGCCCUGCAGAdTdT-3′) as previously described by Jiang et al. (13) or control siRNA (5′-GAGCUGCAAACAACUAUACdTdT) were performed by DharmaFECT1 (Thermo Scientific) immediately after infection, according to the manufacturer's instructions. When needed, cells were treated with cycloheximide (50 μg/ml) for 3 h.
For live microscopy, images were captured every 30 min in phase contrast and in fluorescence with a ×63 objective lens. The images were then processed and deconvolved with the softWoRx software and converted to Adobe Photoshop.
Synchronization and flow cytometry analyzes.
For G1/S synchronization, the cells were treated by thymidine (2.5 mM), immediately after infection, for 24 or 48 h depending on the experiments. When needed, the Skp2 siRNA was transfected 1 h after infection. For synchronization in metaphase, cells were first treated by thymidine for 24 h and then infected and simultaneously released for 15 h in the presence of nocodazole (100 ng/ml). After mitotic shake-off, the cells were released in normal medium for 5, 9, or 13 h.
Flow cytometry analyses were performed as previously described (3), and quantification of the green fluorescence was performed with the LSRII (Becton Dickinson) flow cytometer and BD FACSDiva software.
Immunoprecipitation and Western blot experiments.
For E2 coimmunoprecipitation with endogenous Skp2, HaCaT cells were synchronized at the G1/S transition for 24 h, infected under thymidine treatment, and grown in the presence of thymidine for 24 additional hours. Proteins were extracted in an extraction buffer (300 mM NaCl, 50 mM Tris-HCl [pH 8], 0.5% Nonidet P-40 [NP-40], 1 mM EDTA, protease, and phosphatase inhibitors) for 30 min at 4°C, followed by centrifugation. Extracts containing 1 mg of total proteins were immunoprecipitated with the rabbit anti-Skp2 antibody (51-1900; Zymed Laboratories) overnight at 4°C or GFP-Trap_A (ChromoTek) according to the manufacturer's instructions. Samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed in Western blot experiments.
For Western blot experiments, anti-GFP (TP401; Torrey Pines Biolabs), anti-β-tubulin (T4026; Sigma), anti-actin (A2066; Sigma), or anti-Skp2 (32-3300; Zymed Laboratories) were used.
In vitro Skp2 interactions with GST-E2 or GFP-E2 proteins.
Glutathione S-transferase (GST)-fused proteins produced in Escherichia coli BL21 were purified by affinity chromatography on glutathione-agarose beads. The binding reactions were carried out by incubating 500 ng of each GST fusion protein on beads with 10 μl of rabbit reticulocyte lysate containing 35S-radiolabeled in vitro-translated Skp2 in a final volume of 60 μl of binding buffer (10 mM HEPES [pH 7.7], 200 mM NaCl, 1 mM MgCl2, 2 mM dithiothreitol [DTT], 50 mM sucrose, 0.1% Triton X-100) for 1 h at 4°C on a rotating wheel. After centrifugation, beads were washed four times with 0.5 ml of binding buffer containing 1% Triton X-100, and proteins from the beads and input (1/10 of total proteins) were analyzed by SDS-PAGE, followed by autoradiography.
For interaction of in vitro-synthesized Skp2 with GFP fusion proteins, total proteins were extracted from infected cells in P300 buffer (20 mM NaH2PO4, 250 mM NaCl, 30 mM NaPPi, 0.1% NP-40, 5 mM EDTA, 5 mM DTT, and protease inhibitors). Extracts containing 500 μg of total proteins were immunoprecipitated by using anti-GFP antibody (11 814 460 001; Roche). Beads were washed in interaction buffer (20 mM Tris, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.1% NP-40, and protease inhibitors) and incubated in 25 μl of the same buffer together with 10 μl of in vitro-translated radiolabeled Skp2. Proteins eluted from beads and input (1/10 of total proteins) were separated by SDS-PAGE and revealed by autoradiography.
RESULTS
Degradation of the HPV-18 E2 protein is cell cycle dependent.
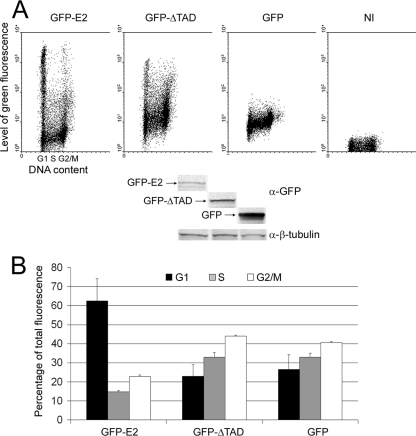
We have previously shown that HPV-18 E2 is degraded by the ubiquitin/proteasome pathway through its amino-terminal transactivation domain (4). The half-life of the full-length E2 protein in HeLa cells is about 50 min, whereas the protein deleted of the N-terminal transactivation domain (ΔTAD) is stable, with a half-life of about 6 h. We have previously checked that fusion of the GFP to the amino terminus of E2 does not affect its functions or its stability since both the fusion and the native proteins present the same short half-life times (4); thus, the study of the stability of GFP-E2 is an accurate measure of wild-type E2 behavior. To study the behavior of E2 during the cell cycle, we had to overexpress the protein in cells, since there is no natural cell line expressing E2. HaCaT cells (HPV-negative keratinocytes) were infected by recombinant adenoviruses expressing GFP-E2 (AdGFP-E2), GFP-ΔTAD (AdGFP-ΔTAD), or Flag-GFP (AdGFP). In these conditions, after 40 h of overexpression, the levels of expression of E2 were low, due to the protein instability compared to the deletant of the transactivation domain ΔTAD or the GFP (Fig. 1A, lower panel). In an asynchronous population of HaCaT cells infected with adenoviruses expressing the full-length GFP-E2, flow cytometry analyses demonstrated that cells in G1 displayed a wide range of green fluorescence, including two distinct populations of either very low levels (under 10) or very high levels (above 103) of GFP-E2 expression (Fig. 1A, upper panel, first dot plot). In contrast, G1 cells expressing GFP-ΔTAD or Flag-GFP showed more-homogeneous levels of fluorescence (Fig. 1A, upper panel, second and third dot plots). This indicates that modulation in quantities of the full-length E2 occurs during the G1 phase compared to ΔTAD. The means of GFP-E2 levels in S phase dramatically decreased compared to the G1 phase, whereas the ΔTAD levels slightly increased (see Fig. 1A, upper panel, and 1B for quantifications calculated from three independent experiments). Then, the levels of GFP-E2 increased in G2/M cells, although they could not reach the mean level of fluorescence observed in G1 cells. The TAD domain alone, fused to GFP, exhibited a behavior comparable to that of the full-length E2 protein (data not shown), thus confirming its major role in regulating E2 stability. This disparity of expression levels in G1 cells expressing E2 could be explained by stabilization of E2 from G2 to the end of G1, followed by a rapid degradation of E2 at the G1/S transition, or by a progressive degradation during the G1 phase which would persist during the S phase.
FIG. 1.
Degradation of the HPV-18 E2 protein is cell cycle-dependent. HaCaT cells were infected with AdGFP-E2, AdGFP-ΔTAD (E2 deleted of its amino-terminal domain), or Ad-GFP or not infected (NI). At 40 h after infection, flow cytometry analyses and Western blot analyses were performed. (A) In the upper panel, cells were analyzed for propidium iodide incorporation into DNA (x axis), giving the phases of the cell cycle as indicated under the first panel, and green fluorescence (y axis), indicating the amount of protein expressed in a logarithmic scale. In the lower panel, the results of a Western blot performed with equal amounts of proteins are shown. (B) Means of green fluorescence in G1, S, and G2/M cells represented as a percentage of total fluorescence and calculated from three independent infection experiments.
E2 is degraded at the end of the G1 phase.
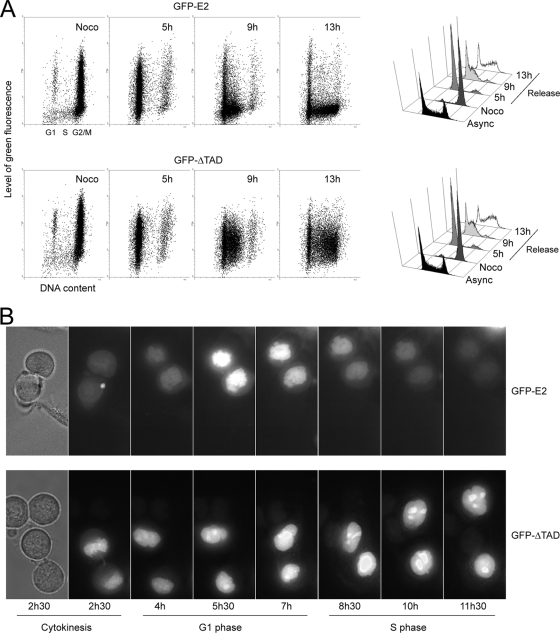
To further check whether the drop of GFP-E2 fluorescence observed in S phase resulted from sudden degradation of the protein at the end of G1 or from a progressive degradation during G1, cells infected with AdGFP-E2 or AdGFP-ΔTAD were synchronized in metaphase by nocodazole treatment and released for 13 h. Flow cytometry analyses showed that while the cells were progressing from G1 (5-h release) to early S (9-h release) and late S (13-h release), as shown on the right panels of Fig. 2A, the levels of GFP-E2 fluorescence dropped dramatically (Fig. 2A, dot plots of the upper panel), whereas at the same time points the levels of the amino-terminal-deleted protein GFP-ΔTAD remained comparable (Fig. 2A, dot plots of the lower panel). In time-lapse microscopy performed on the same cells released from the nocodazole block, the levels of GFP-E2 fluorescence increased during the G1 phase and then dropped dramatically at the end of G1 and the beginning of the S phase (Fig. 2B, upper panel). In sharp contrast, the levels of green fluorescence in AdGFP-ΔTAD-infected cells increased after cytokinesis but then remained constant in the late G1 and S phases (Fig. 2B, lower panel). The increase of the levels of the two proteins during G1 could be explained by two concomitant mechanisms. First, as the global quantity of proteins present in each daughter cell is divided by two after mitosis, we expect the levels of GFP-E2 and GFP-ΔTAD to be also divided by two at the beginning of G1. Second, we expect the global quantity of proteins to increase gradually during G1 because of ongoing transcription and translation after mitosis. Thus, since the levels of GFP-E2 first increased and then decreased rapidly in late G1, whereas the levels of GFP-ΔTAD first increased similarly but did not show any drop after mid-G1, these experiments strongly suggested that the amino-terminal domain of E2 could be a specific target for a G1/S ubiquitin ligase such as the SCFSkp2.
FIG. 2.
E2 is degraded at the end of the G1 phase. HaCaT cells infected by AdGFP-E2 or AdGFP-ΔTAD were synchronized in metaphase by nocodazole treatment and released. (A) In the left panel, cells were analyzed as in Fig. 1 after 0 (Noco), 5, 9, or 13 h of release. The right panel shows a classical representation of the phases of the cell cycle showing the G2/M arrest induced by nocodazole (Noco) and release of the metaphasic block after 5, 9, or 13 h. (B) Cells from the same experiment were subjected to live microscopy after nocodazole release. The first pictures show the phase contrast at 2.5 h of release corresponding to the end of cytokinesis.
Silencing of Skp2 induces stabilization of E2 in asynchronous cells.
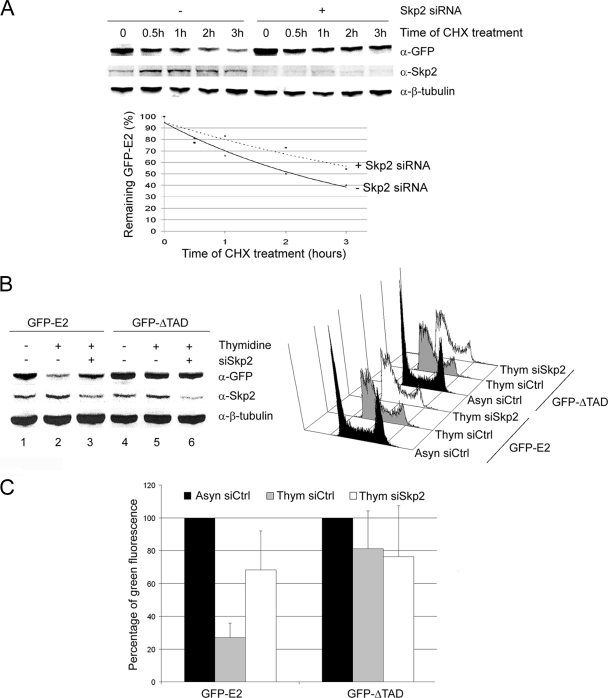
To check whether the SCFSkp2 could be involved in E2 degradation, we decided to test whether silencing of Skp2 by siRNA could stabilize E2 in asynchronous HaCaT cells subjected to cycloheximide treatment. Since Skp2 is necessary for the G1/S transition, we found, as expected, that treatment of cells with the Skp2 siRNA led to G1 arrest 40 h after transfection (data not shown). To avoid this cell cycle block, which could influence our results, we decided to study the behavior of E2 as early as 24 h after transfection of the Skp2 siRNA. In these conditions, the Skp2 knockdown was efficient (>60% of inhibition, Fig. 3A, upper panel) but did not yet affect the cell cycle (data not shown). Western blot analyzes showed that E2 was stabilized by about two times after expression of the Skp2 siRNA compared to cells treated by the control siRNA (Fig. 3A, upper panel, and lower panel for quantification). These results indicate that the SCFSkp2 is a major ubiquitin ligase leading to E2 degradation but do not allow us to exclude the involvement of other ubiquitin ligases.
FIG. 3.
The SCFSkp2 ubiquitin ligase mediates E2 degradation at the end of G1. (A) Western blot experiments performed with equal amounts of proteins from asynchronous HaCaT cells infected with AdGFP-E2 and transfected with Control (Ctrl) or Skp2 siRNA. After 24 h, cycloheximide was added to the medium, and cells were harvested after 0, 0.5, 1, 2, or 3 h of treatment and analyzed by Western blotting with the indicated antibodies. (B) HaCaT cells infected by AdGFP-E2 or AdGFP-ΔTAD were transfected by Control (Ctrl) or Skp2 siRNA and were either treated or not by thymidine as indicated. The left panel shows results of Western blot experiments performed with equal amounts of proteins. In the right panel, flow cytometry analyses of the same cells show the G1/S arrest of thymidine-treated cells (Thym) compared to asynchronous cells (Asyn) for both Ctrl and Skp2 siRNA transfections. (C) The graph shows the mean of green fluorescence as a percentage of G1 fluorescence in each condition, calculated from four independent infection experiments.
The SCFSkp2 ubiquitin ligase mediates E2 degradation at the end of G1.
To confirm the role of Skp2 in the specific degradation of E2 at the end of G1, we performed comparable Skp2 silencing experiments in cells synchronized at the G1/S transition by thymidine. Thymidine treatment induced a strong G1/S block (Fig. 3B, right panel), as well as a slight stabilization of Skp2 as expected (Fig. 3B, left panel, lanes 2 and 5), whereas the levels of full-length E2 concomitantly dropped by ∼4-fold compared to asynchronous cells (Fig. 3B, left panel, compare lanes 1 and 2, and Fig. 3C for quantification from four independent experiments). Silencing of Skp2 after siRNA transfection induced a 2- to 3-fold stabilization of E2 in cells synchronized at the G1/S transition (Fig. 3B, left panel, compare lanes 2 and 3, and Fig. 3C), whereas no change was observed for the ΔTAD protein in all conditions (Fig. 3B, left panel, compare lanes 4, 5, and 6, and Fig. 3C). These results indicate an involvement of the SCF complex in the specific degradation of E2 at the G1/S transition through the Skp2 F-box protein adaptor.
E2 binding to Brd4 does not interfere with E2 degradation in S phase.
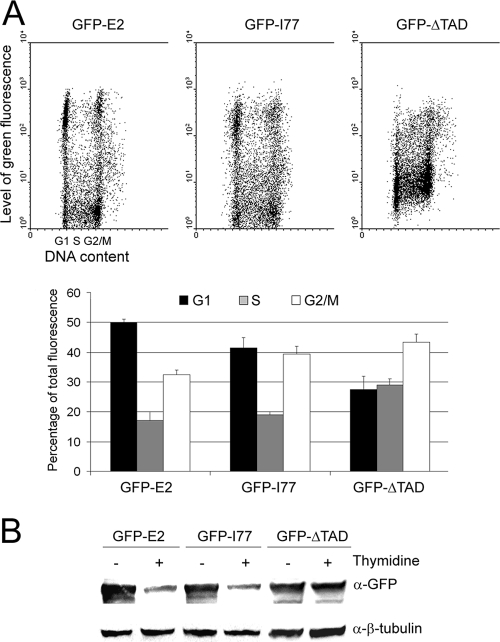
Since Brd4 binding has been implicated in stabilization of E2 (11, 15, 31), we decided to check whether the HPV-18 E2 I77A (I77) mutant, equivalent to the BPV-1, HPV-11, HPV-31, and HPV-16 E2 I73A mutants, unable to activate transcription and unable to bind Brd4 (11; H. Senechal and J. Archambault, unpublished data), showed the same behavior as the wild-type E2 regarding the fluorescence levels during the cell cycle. HaCaT cells were infected by AdGFP-E2, AdGFP-I77, or AdGFP-ΔTAD, and asynchronous cells were analyzed by flow cytometry as in Fig. 1A. As shown in Fig. 4A, the I77 mutant showed a fluorescence profile similar to that of the wild-type E2 protein, with a heterogeneous population in G1 and low levels of fluorescence in the S phase. However, after quantification, the levels of I77 in the S phase appeared to be only about half of the G1 level, whereas we used to obtain a 3- to 4-fold decrease with the wild-type E2 protein (Fig. 4A, lower panel). This could be explained by a less efficient degradation of the I77 mutant in S phase or by a decreased stabilization of I77 in G1 compared to the wild-type E2 due to the lack of Brd4 binding. We checked the G1/S levels of the I77 mutant compared to the wild-type E2 by Western blotting with or without thymidine treatment and found that the levels of both proteins were strongly decreased at the G1/S transition (Fig. 4B). We conclude from these experiments that Brd4 is not involved in the G1/S degradation of E2 by the SCFSkp2 but could be involved in stabilizing E2 in G1, although this effect appears to be quite marginal.
FIG. 4.
E2 binding to Brd4 does not interfere with E2 degradation in S phase. (A) For the upper panel, HaCaT cells were infected with AdGFP-E2, AdGFP-Ι77 (equivalent to HPV-16 E2 I73, unable to bind Brd4), or Ad-ΔTAD, and flow cytometry analyses of the cells were performed. Cells were analyzed for propidium iodide incorporation into DNA (x axis) and green fluorescence (y axis). The lower panel shows the mean green fluorescence in G1, S, and G2/M cells represented as a percentage of total fluorescence and calculated from two independent infection experiments. (B) Western blot experiments were performed with equal amounts of proteins from HaCaT cells infected by AdGFP-E2, AdGFP-I77, or AdGFP-ΔTAD and treated or not treated with thymidine as indicated.
Skp2 interacts with the amino-terminal domain of E2 synthesized in vivo but not in vitro.
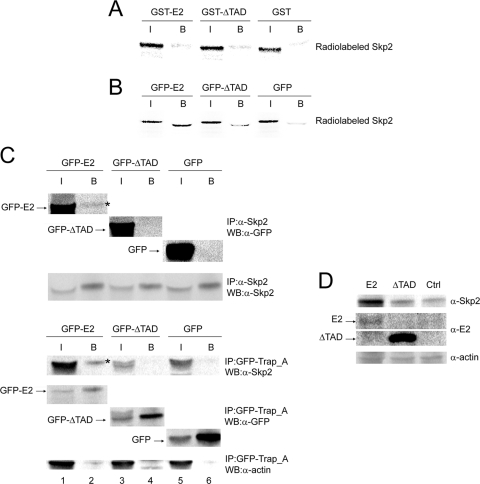
Since substrates of SCF directly interact with the F-box proteins, we decided to check whether E2 could interact with the Skp2 F-box protein. We first performed GST pulldown experiments with purified GST-E2 and in vitro-translated radiolabeled Skp2 or β-TrCP. We were unable to detect any specific interaction between E2 and β-TrCP (data not shown). At first, we did not detect the interaction between E2 and Skp2 in vitro either presumably because the bacterial E2 was not phosphorylated (Fig. 5A). Indeed, when the E2 protein was produced in cells instead of bacteria, we did detect a specific interaction between GFP-E2 and Skp2 (Fig. 5B). This result was confirmed by coimmunoprecipitation experiments between endogenous Skp2 and E2 in infected HaCaT cells treated by thymidine. For the last 5 h, lactacystin (10 μM final) was added to the medium to inhibit E2 degradation and to further stabilize the interaction with Skp2. In these conditions, we could detect coimmunoprecipitated GFP-E2 (but not GFP-ΔTAD and GFP controls) with the endogenous Skp2 (Fig. 5C, upper panel). In the reverse coimmunoprecipitation experiment using cotransfected GFP-E2 and Skp2, we could also detect Skp2 coimmunoprecipitated with GFP-E2 but not with GFP-ΔTAD or GFP (Fig. 5C, bottom panel). We also found interaction of E2 with two other members of the SCF complex, Skp1 and Cullin1, in TAP-TAG, followed by mass spectrometry experiments (data not shown). We deduced from these experiments that E2 and the SCFSkp2 complex interact but that a posttranslational modification of E2 such as phosphorylation is required, as expected for SCFSkp2 targets.
FIG. 5.
Skp2 interacts with the amino-terminal domain of E2 synthesized in vivo but not in vitro and is stabilized by E2. (A) GST fusion proteins bound to glutathione-agarose beads were incubated with 35S-radiolabeled in vitro-translated Skp2 as indicated and analyzed by SDS-PAGE, followed by autoradiography. I, input (1/10); B, beads. (B) Equal amounts of proteins extracted from AdGFP-E2-, AdGFP-ΔTAD-, or AdGFP-infected cells were immunoprecipitated with anti-GFP antibody. Beads were incubated with 35S-radiolabeled in vitro-translated Skp2. The radiolabeled proteins retained on the beads were analyzed by SDS-PAGE and revealed by autoradiography. I, input (1/10); B, beads. (C) For the top panel, equal amounts of proteins extracted from AdGFP-E2-, AdGFP-ΔTAD-, or AdGFP-infected HaCaT cells synchronized at the G1/S transition were immunoprecipitated with anti-Skp2 antibodies. Immunoprecipitates were analyzed by SDS-PAGE and revealed by anti-GFP or anti-Skp2 antibody as indicated. The coimmunoprecipitated E2 is indicated by an asterisk. I, input (1/50); B, beads. For the bottom panel, equal amounts of proteins extracted from C33 cells cotransfected with the indicated GFP fusion plasmids and pCS2 Skp2 were immunoprecipitated with the GFP-Trap_A beads. Immunoprecipitates were analyzed by SDS-PAGE and revealed by anti-GFP or anti-Skp2 antibody as indicated. The coimmunoprecipitated Skp2 is shown by an asterisk. I, input (1/50); B, beads. (D) C33 cells were cotransfected by untagged E2 (or untagged ΔTAD) and Skp2. Accumulation of cotransfected Skp2 in untagged E2-expressing cells is shown by Western blotting.
Expression of HPV-18 E2 leads to stabilization of Skp2.
Interestingly, Skp2 appeared stabilized by GFP-E2 in cotransfection experiments (Fig. 5C, bottom panel, compare lane 1 to lanes 3 and 5). This result was expected since we have previously shown that E2 is a repressor of the APC/C ubiquitin ligase (3), which is known to degrade Skp2 in G1. We verified that the native E2 protein could also stabilize Skp2 by performing similar experiments with the HPV-18 E2 protein with no GFP tag. In these conditions, we could also detect a strong stabilization of Skp2, although the quantity of E2 was quite low in these experiments (Fig. 5D), thus confirming that low levels of wild-type E2 are sufficient to inhibit APC/C-mediated degradation, as previously described (3). In addition, these experiments indicate that accumulation of Skp2 is concomitant with a decreased E2 expression, as expected from the data presented above.
DISCUSSION
We showed here that the levels of expression of the HPV-18 E2 protein are dependent on the cell cycle, with E2 being much more expressed in the G1 phase than in the S and G2/M phases. We could show that these differences of quantities could be explained by E2 degradation (through its amino-terminal transactivation domain) via the SCFSkp2 ubiquitin ligase at the end of the G1 phase.
Since differentiated cells do not express Skp2 in contrast to proliferating cells, the subsequent stabilization of E2 could explain why E2 is found accumulated in the granular layers of HPV-associated CIN lesions, while it is undetectable in the proliferating cells of the basal layers (17, 25), although E2 should be expressed since it is necessary for initiation of viral DNA replication. Thus, even if the SCFSkp2 prevents E2 from accumulating in the basal cell layers, we believe that the remaining quantity should be sufficient to allow viral DNA replication. A possibility is that chromatin-associated E2 involved in replication could be stabilized, as suggested by inhibition of its degradation through interaction with Brd4 (11, 15, 31), although Brd4 binding does not prevent E2 from the SCFSkp2-promoted degradation in our experiments.
An important consequence of the G1/S degradation of E2 could be the release of E6 and E7 repression by E2 in late G1 that could occur even in the presence of nonintegrated viral DNA and therefore of intact E2 open reading frame and putative E2 expression, as shown in CINII and CINIII (22, 28). Since the main oncogenic function of E7 is to induce S-phase entry, it is fair to believe that its peak of activity has to occur at the G1/S transition, which correlates with E2 degradation. We could show here that overexpression of E2 stabilizes Skp2, which is a known oncogene. Indeed, high levels of Skp2 accelerate the G0/G1 and G1/S transitions and overexpression of Skp2 has been described in many cancers (10). Stabilization of Skp2 by E2 could lead to increased levels of Skp2 before the cells reach the G1/S transition. Through negative feedback, accumulation of Skp2 by E2 would lead to E2 degradation via the SCFSkp2, allowing high E6 and E7 expression in G1 and activation of S-phase entry.
We have already published that high-risk HPV E2 proteins, through inhibition of APC/C, stabilize cyclin B2 and induce chromosomal instability, as well as centrosome overduplication, probably via stabilization of other APC/C substrates such as Aurora A (reference 3 and results not shown). APC/C inhibition and thus Skp2 stabilization are induced at very low levels of E2, whereas the E2-induced apoptosis published by us and others necessitates higher levels of expression. Thus, the APC/C inhibition could occur in the proliferating basal cells where the E2 levels are low, whereas the proapoptotic activity of E2 might rather play a role in the superficial layers for the release of the new viruses for example. The chromosomal instability caused by E2-mediated APC/C inhibition led us to postulate for the first time that high-risk HPV E2 proteins might have an oncogenic potential (3). A recent study suggests that chromosomal instability induced by high-risk HPVs could be a cause rather than a consequence of viral DNA integration (28). This indicates that HPV proteins other than E6 and E7 could be involved in high-risk HPV-induced chromosomal instability and transformation. Furthermore, it has more recently been shown that E2 of cutaneous high-risk HPV-8 can induce tumors in mice in the absence of other HPV proteins (21). Interestingly, similar experiments involving the low-risk HPV-11 E2, which has been shown to be unable to induce chromosomal instability in tissue culture (3), do not lead to the proliferative phenotype in mice (16). Taken together, these new data, including the results presented here, lead us to reassess the role of high-risk HPV E2 proteins as potential oncogenes in the early steps of HPV-induced cancers.
Acknowledgments
We thank Catherine Bonne-Andrea for the gift of the Skp2 expression vector, Sebastien Teissier for help with analyses of the microscopy images, and Alice Quek for technical help in experiments.
Footnotes
Published ahead of print on 14 October 2009.
REFERENCES
- 1.Bashir, T., N. V. Dorrello, V. Amador, D. Guardavaccaro, and M. Pagano. 2004. Control of the SCFSkp2-Cks1 ubiquitin ligase by the APC/CCdh1 ubiquitin ligase. Nature 428:190-193. [DOI] [PubMed] [Google Scholar]
- 2.Bashir, T., and M. Pagano. 2004. Don't skip the G1 phase: how APC/CCdh1 keeps SCFSKP2 in check. Cell Cycle 3:850-852. [PubMed] [Google Scholar]
- 3.Bellanger, S., S. Blachon, F. Mechali, C. Bonne-Andrea, and F. Thierry. 2005. High-risk but not low-risk HPV E2 proteins bind to the APC activators Cdh1 and Cdc20 and cause genomic instability. Cell Cycle 4:1608-1615. [DOI] [PubMed] [Google Scholar]
- 4.Bellanger, S., C. Demeret, S. Goyat, and F. Thierry. 2001. Stability of the human papillomavirus type 18 E2 protein is regulated by a proteasome degradation pathway through its amino-terminal transactivation domain. J. Virol. 75:7244-7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blachon, S., S. Bellanger, C. Demeret, and F. Thierry. 2005. Nucleo-cytoplasmic shuttling of high-risk human papillomavirus E2 proteins induces apoptosis. J. Biol. Chem. 280:36088-36098. [DOI] [PubMed] [Google Scholar]
- 6.Bornstein, G., J. Bloom, D. Sitry-Shevah, K. Nakayama, M. Pagano, and A. Hershko. 2003. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J. Biol. Chem. 278:25752-25757. [DOI] [PubMed] [Google Scholar]
- 7.Carrano, A. C., E. Eytan, A. Hershko, and M. Pagano. 1999. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat. Cell Biol. 1:193-199. [DOI] [PubMed] [Google Scholar]
- 8.Demeret, C., C. Desaintes, M. Yaniv, and F. Thierry. 1997. Different mechanisms contribute to the E2-mediated transcriptional repression of human papillomavirus type 18 viral oncogenes. J. Virol. 71:9343-9349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyson, N., P. M. Howley, K. Münger, and E. Harlow. 1989. The human papillomavirus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934-937. [DOI] [PubMed] [Google Scholar]
- 10.Frescas, D., and M. Pagano. 2008. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat. Rev. Cancer 8:438-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gagnon, D., S. Joubert, H. Senechal, A. Fradet-Turcotte, S. Torre, and J. Archambault. 2009. Proteasomal degradation of the papillomavirus E2 protein is inhibited by overexpression of the bromodomain-containing protein 4 (Brd4). J. Virol. 83:4127-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gstaiger, M., R. Jordan, M. Lim, C. Catzavelos, J. Mestan, J. Slingerland, and W. Krek. 2001. Skp2 is oncogenic and overexpressed in human cancers. Proc. Natl. Acad. Sci. USA 98:5043-5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang, F., N. P. Caraway, R. Li, and R. L. Katz. 2005. RNA silencing of S-phase kinase-interacting protein 2 inhibits proliferation and centrosome amplification in lung cancer cells. Oncogene 24:3409-3418. [DOI] [PubMed] [Google Scholar]
- 14.Kudo, Y., S. Kitajima, S. Sato, M. Miyauchi, I. Ogawa, and T. Takata. 2001. High expression of S-phase kinase-interacting protein 2, human F-box protein, correlates with poor prognosis in oral squamous cell carcinomas. Cancer Res. 61:7044-7047. [PubMed] [Google Scholar]
- 15.Lee, A. Y., and C. M. Chiang. 2009. Chromatin adaptor Brd4 modulates E2 transcription activity and protein stability. J. Biol. Chem. 284:2778-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leykauf, K., K. Kabsch, N. Gassler, L. Gissmann, A. Alonso, and J. Schenkel. 2008. Expression of the HPV11 E2 gene in transgenic mice does not result in alterations of the phenotypic pattern. Transgenic Res. 17:1-8. [DOI] [PubMed] [Google Scholar]
- 17.Maitland, N. J., S. Conway, N. S. Wilkinson, J. Ramsdale, J. R. Morris, C. M. Sanders, J. E. Burns, P. L. Stern, and M. Wells. 1998. Expression patterns of the human papillomavirus type 16 transcription factor E2 in low- and high-grade cervical intraepithelial neoplasia. J. Pathol. 186:275-280. [DOI] [PubMed] [Google Scholar]
- 18.Münger, K., B. A. Werness, N. Dyson, W. C. Phelps, E. Harlow, and P. M. Howley. 1989. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 8:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pagano, M., S. W. Tam, A. M. Theodoras, P. Beer-Romero, G. Del Sal, V. Chau, P. R. Yew, G. F. Draetta, and M. Rolfe. 1995. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science 269:682-685. [DOI] [PubMed] [Google Scholar]
- 20.Penrose, K., and A. McBride. 2000. Proteasome-mediated degradation of the papillomavirus E2-TA protein is regulated by phosphorylation and can modulate viral genome copy number. J. Virol. 74:6031-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfefferle, R., G. P. Marcuzzi, B. Akgul, H. U. Kasper, F. Schulze, I. Haase, C. Wickenhauser, and H. Pfister. 2008. The human papillomavirus type 8 E2 protein induces skin tumors in transgenic mice. J. Investig. Dermatol. 128:2310-2315. [DOI] [PubMed] [Google Scholar]
- 22.Sathish, N., P. Abraham, A. Peedicayil, G. Sridharan, S. John, and G. Chandy. 2004. Human papillomavirus 16 E6/E7 transcript and E2 gene status in patients with cervical neoplasia. Mol. Diagn. 8:57-64. [DOI] [PubMed] [Google Scholar]
- 23.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV16 E6 and E6-AP complex functions as ubiquitin-protein ligase in the ubiquitination of p53. Cell 75:495-505. [DOI] [PubMed] [Google Scholar]
- 24.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 25.Stevenson, M., L. C. Hudson, J. E. Burns, R. L. Stewart, M. Wells, and N. J. Maitland. 2000. Inverse relationship between the expression of the human papillomavirus type 16 transcription factor E2 and virus DNA copy number during the progression of cervical intraneoplasia. J. Gen. Virol. 81:1825-1832. [DOI] [PubMed] [Google Scholar]
- 26.Sutterluty, H., E. Chatelain, A. Marti, C. Wirbelauer, M. Senften, U. Muller, and W. Krek. 1999. p45SKP2 promotes p27Kip1 degradation and induces S phase in quiescent cells. Nat. Cell Biol. 1:207-214. [DOI] [PubMed] [Google Scholar]
- 27.Thierry, F., and M. Yaniv. 1987. The BPV1-E2 trans-acting protein can be either an activator or a repressor of the HPV18 regulatory region. EMBO J. 6:3391-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vinokurova, S., N. Wentzensen, I. Kraus, R. Klaes, C. Driesch, P. Melsheimer, F. Kisseljov, M. Durst, A. Schneider, and M. von Knebel Doeberitz. 2008. Type-dependent integration frequency of human papillomavirus genomes in cervical lesions. Cancer Res. 68:307-313. [DOI] [PubMed] [Google Scholar]
- 29.Werness, B. A., A. J. Levine, and P. M. Howley. 1990. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 248:76-79. [DOI] [PubMed] [Google Scholar]
- 30.Yu, Z. K., J. L. Gervais, and H. Zhang. 1998. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21CIP1/WAF1 and cyclin D proteins. Proc. Natl. Acad. Sci. USA 95:11324-11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng, G., M. R. Schweiger, G. Martinez-Noel, L. Zheng, J. A. Smith, J. W. Harper, and P. M. Howley. 2009. Brd4 regulation of papillomavirus protein E2 stability. J. Virol. 83:8683-8692. [DOI] [PMC free article] [PubMed] [Google Scholar]