Abstract
Recent genome-wide screens have highlighted an important role for transportin 3 in human immunodeficiency virus type 1 (HIV-1) infection and preintegration complex (PIC) nuclear import. Moreover, HIV-1 integrase interacted with recombinant transportin 3 protein under conditions whereby Moloney murine leukemia virus (MLV) integrase failed to do so, suggesting that integrase-transportin 3 interactions might underscore active retroviral PIC nuclear import. Here we correlate infectivity defects in transportin 3 knockdown cells with in vitro protein binding affinities for an expanded set of retroviruses that include simian immunodeficiency virus (SIV), bovine immunodeficiency virus (BIV), equine infectious anemia virus (EIAV), feline immunodeficiency virus (FIV), and Rous sarcoma virus (RSV) to critically address the role of integrase-transportin 3 interactions in viral infection. Lentiviruses, with the exception of FIV, display a requirement for transportin 3 in comparison to MLV and RSV, yielding an infection-based dependency ranking of SIV > HIV-1 > BIV and EIAV > MLV, RSV, and FIV. In vitro pulldown and surface plasmon resonance assays, in contrast, define a notably different integrase-transportin 3 binding hierarchy: FIV, HIV-1, and BIV > SIV and MLV > EIAV. Our results therefore fail to support a critical role for integrase binding in dictating transportin 3 dependency during retrovirus infection. In addition to integrase, capsid has been highlighted as a retroviral nuclear import determinant. Accordingly, MLV/HIV-1 chimera viruses pinpoint the genetic determinant of sensitization to transportin 3 knockdown to the HIV-1 capsid protein. We therefore conclude that capsid, not integrase, is the dominant viral factor that dictates transportin 3 dependency during HIV-1 infection.
The early events in the retroviral life cycle, typified by reverse transcription and subsequent cDNA integration, occur within the context of high-molecular-weight nucleoprotein complexes that are derived from the core of the infecting virus particle (4, 20, 21, 36). This presents a particular challenge for the viruses, as the chromosomal targets of integration reside within the nucleus, which is separated from the cytoplasm by a double lipid bilayer. The nuclear membrane houses nuclear pore complexes that permit the passive diffusion of macromolecules with diameters of less than ∼9 nm (34). At 56 nm (36), the human immunodeficiency virus type 1 (HIV-1) preintegration complex (PIC), derived from a prototypical retrovirus, grossly exceeds the diffusion limit. Different retroviruses have evolved different mechanisms to deal with the nuclear membrane barrier. The gammaretrovirus Moloney murine leukemia virus (MLV) relies on cell division for productive infection (35), apparently subverting the membrane when it dissolves during the M phase of the cell cycle (32, 42). HIV-1, in contrast, is actively transported during interphase (6), reflecting its ability to efficiently infect terminally differentiated and growth-arrested cells in the absence of cell division (31, 48).
A number of HIV-1 PIC components, including matrix (MA), Vpr, integrase (IN), and the central DNA flap that forms during reverse transcription, have been implicated in nuclear import. However, this aspect of HIV-1 biology is relatively muddled, with multiple reports both supporting and refuting important roles for each of these components (see references 15, 19, and 51 for recent reviews). These apparent inconsistencies may be due to any number of reasons. HIV-1 might employ numerous overlapping mechanisms such that the alteration of one leads to minimal or partial phenotypes. Many viral mutations can exert pleiotropic affects, often complicating data interpretation. Moreover, the identification of the HIV-1 capsid (CA) protein as a dominant infectivity determinant in growth-arrested cells suggested that virus uncoating compared to active nuclear transport may be the rate-limiting step in this phase of the life cycle (51). IN also continues to attract particular interest, as it is the one protein component of the PIC that must remain associated with the reverse transcript after nuclear entry.
Although numerous host cell factors have been implicated in HIV-1 nuclear transport, the field has yet to reach a consensus on the salient player(s). Various import proteins, including the importin α/importin β heterodimer (22, 23, 25), importin 7 (2, 22, 54), NUP153 (49), and transportin 3 (TNPO3 or TRN-SR2) (13), have been shown to interact with HIV-1 IN. TNPO3, a member of the karyopherin β protein family that imports serine/arginine-rich splicing factors (SR proteins) into the nucleus (27), has generated particular interest as of late. In addition to its identification as an HIV-1 IN binding protein (13), TNPO3 was identified in two independent genome-wide short interfering RNA (siRNA) screens for host factors required by HIV-1 for infection (5, 30). TNPO3 depletion yielded significant HIV-1 (5, 13, 30) and HIV-2 (13) but much less dramatic MLV (5, 13, 30) infection defects, indicating a potential role as a lentivirus-specific host cofactor. Moreover, the block to HIV-1 infection was pinpointed at PIC nuclear import (13). Furthermore, recombinant TNPO3 protein was shown to bind HIV-1 IN but not MLV IN in vitro, suggesting that the TNPO3-IN interaction might underlie lentivirus-specific PIC nuclear import (13).
In this study we critically addressed the hypothesis that TNPO3 is indeed an IN-dependent cofactor of lentiviral infection. We correlated the infectivities of an expanded set of retroviral vectors in cells depleted for TNPO3 expression with relative IN-TNPO3 binding affinities. Our findings recapitulate the previously reported in vitro interaction between TNPO3 and HIV-1 IN (13). However, we find TNPO3 to be a relatively prolific IN binding protein, displaying affinity for the majority of tested IN proteins. Moreover, protein binding profiles did not correlate with the requirement for the host factor during virus infection. Furthermore, analyses of MLV/HIV-1 (MHIV) chimera viruses that harbor parts of MLV Gag and/or IN for the corresponding HIV-1 determinants highlight a role for CA in mediating the requirement for TNPO3 during HIV-1 infection.
MATERIALS AND METHODS
TNPO3 sequence analysis.
Whole-genome shotgun (WGS) sequences and expressed sequence tags (ESTs) of TNPO3 orthologs available in the GenBank database were identified by searching the NCBI sequence database with Basic Local Alignment Search Tool (BLAST) (www.ncbi.nlm.nih.gov/BLAST) using cross-species megaBLAST. TNPO3 sequences of human (GenBank accession number NM_012470.2), rodent (Mus musculus) (accession number NM_177296.4), rhesus macaque (Macaca mulatta) (accession number XM_001092689.1), and bovine (Bos taurus) (accession number NM_001098090.1) origins were previously annotated. While Felis catus TNPO3 was unknown, equine (Equus caballus) and chicken (Gallus gallus) orthologs were incomplete and/or incorrect. The feline sequence was assembled by using overlapping reads from WGS contigs ACBE01084855.1, ACBE01084851.1, ACBE01084858.1, AANG01550855.1, ACBE01084853.1, AANG01536946.1, ACBE01084860.1, AANG01398020.1, ACBE01084852.1, AANG01398022.1, AANG01536945.1, ACBE01084854.1, AANG01536947.1, ACBE01084859.1, ACBE01084842.1, AANG01537011.1, ACBE01084856.1, ACBE01084862.1, ACBE01084855.1, AANG01797569.1, ACBE01084863.1, AANG01398018.1, AANG01398021.1, and AC114336.2 of the raw organism WGS traces. The Equus caballus sequence was confirmed and corrected: (i) residues corresponding to human TNPO3 amino acids 759 and 760 were annotated using EST NW_001867413.1, and (ii) the N-terminal region was annotated using WGS trace sequence ti_1270106774. The subsequent correction of 11 presumptive sequencing errors yielded an N terminus identical to the human N-terminal 40 residues (data not shown). ESTs BU260361.1, BU382083.1, BU213567.1, and BU324095.1 were used to assemble the internal chicken TNPO3 sequence corresponding to human residues 688 to 759, while EST BU324095.1 was used to complete the C terminus of the avian protein.
Cells, viruses, and infections.
HeLa, 293T, and HeLa-T4 cells were grown in Dulbecco's modified Eagle's medium supplemented to contain 10% fetal bovine serum, 100 IU/ml penicillin, and 100 μg/ml streptomycin. HeLa cells stably transduced with empty lentiviral vector pLKO.1 or pLKO.1 expressing TNPO3-specific short hairpin RNA (shRNA) (GGCGCACAGAAATTATAGAA) (Sigma-Aldrich, St. Louis, MO) were additionally grown in 1 μg/ml puromycin. HeLa-T4 cells were transfected with 100 nM TNPO3-directed (CGACAUUGCAGCUCGUGUA) or mismatched control (CGCCUAUGUAGCUCGUGUA [changes are underlined]) (Thermo Scientific Dharmacon Products, Lafayette, CO) siRNA by using RNAiMax (Invitrogen Corporation, Carlsbad, CA).
All viral inocula in this study were pseudotyped with the vesicular stomatitis virus G envelope (Env) glycoprotein (VSV-G). The virus-specific molecular clones used were as follows: pBH2 and pBS-BC4MGppt for bovine immunodeficiency virus (BIV) (3) (obtained from Michael Kaleko, Advanced Vision Therapies, Inc.), pEV53 and an equine infectious anemia virus (EIAV)-green fluorescence protein (GFP) transfer vector for EIAV (38, 39) (gifts from John Olsen, University of North Carolina), pNL4-3env-GFP (24) for HIV-1 (kindly provided by Dana Gabuzda, Dana-Farber Cancer Institute), pRCAS-GFP for Rous sarcoma virus (RSV) (10) (Addgene plasmid 13878, obtained from Constance Cepko via Addgene, Cambridge, MA), pFP93 and pGINSIN for feline immunodeficiency virus (FIV) (29, 33) (gifts from Eric Poeschla, Mayo Clinic), and pFB-hrGFP (40) (obtained from Joseph Sodroski, DFCI) and pCG-GagPol (45, 46) (provided by Richard Mulligan, Harvard Medical School) for MLV. Simian immunodeficiency virus (SIV)-based pSIVmac239ΔnefΔenvEGFP (donated by J. Sodroski) was derived from pSIVΔnefEGFP (1) (a kind gift from Ronald Desrosiers, Harvard Medical School). Env sequences were deleted by first changing the ATG start codon to TCC to create a BspE1 site. The 1.1-kb fragment between this and the native BspE1 site at position 8013 (GenBank accession number M33262.1) was excised, leaving Tat, Rev, and the Rev-responsive element intact.
Vector particles were generated by transfecting 293T cells in 10-cm plates with 10 μg total of various ratios of virus production plasmids together with VSV-G expression vector pMD.G (37) using Fugene 6 (Roche Molecular Biochemicals, Indianapolis, IN). Cells were washed 16 h after transfection, and supernatants collected 28 to 72 h thereafter were clarified by centrifugation, filtered through 0.45 μm filters, and concentrated by ultracentrifugation at 53,000 × g for 2 h at 4°C. GFP reporter virus infections were conducted in triplicate in 48-well plates. Cells were replenished with fresh medium following 16 h of virus absorption, and the percentage of positive cells was determined 48 h postinfection by using a FACsCanto flow cytometer (BD, Franklin Lakes, NJ) equipped with FACSDIVA software. Inocula were adjusted to yield ∼30 to 40% GFP-positive cells in control samples, with all infections scoring between 5 and 60% positive.
Env-deleted MHIV chimera plasmids MHIV-mMA12CA-Luc, MHIV-mMA12-Luc, MHIV-mIN-Luc, and MHIV-mMA12CA/mIN-Luc as well as parental HIV-1LAI-based pLaiΔenvLuc2 were generous gifts from Michael Emerman and Masahiro Yamashita (50, 52). Viruses were generated by cotransfecting 293T cells with pCG-VSV-G (45, 46) using Fugene 6. Supernatants collected at 36 and 60 h posttransfection were pooled and spin concentrated as described above. Single-round MLV-Luc was produced as described previously (45). Cells infected in duplicate in 12-well plates for 12 h were rinsed with phosphate-buffered saline and replenished with fresh medium. At 2 days postinfection, resulting levels of luciferase activity in cell extracts were normalized to corresponding levels of total protein as described previously (45). Inocula were adjusted such that control cells yielded 1 × 103 to 1.2 × 104 relative light units/μg.
Western blotting.
Cells were lysed with radioimmunoprecipitation assay buffer (20 mM HEPES [pH 7.5], 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 0.5 M EDTA, Complete protease inhibitor [Roche Molecular Biochemicals]) at the time of infection. Following protein concentration determinations by Bradford assay (Bio-Rad, Hercules, CA), 50 μg or various dilutions were fractionated through 10 to 20% Tris-glycine gels (Invitrogen Corporation). Proteins transferred onto a polyvinylidene fluoride membrane were blotted with a 1:100 dilution of mouse anti-TNPO3 antibody (Abcam Inc., Cambridge, MA), followed by a 1:10,000 dilution of rabbit anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibody (Dako North America Inc., Carpinteria, CA). HRP-conjugated anti-β-actin antibody (1:10,000 dilution; Abcam Inc.) was used to control for gel loading.
DNA constructs for protein expression.
TNPO3 gene fragments amplified from Thermo Scientific Open Biosystems (Huntsville, AL) clone BC009923 using primers AE3385 (5′-GACGGATCCATTATGGAAGGAGCAAAGCCG)/AE3422 (5′-GATCTCCCATGCATGAACCGAACGCTGCAGCTCCC) and AE3421 (5′-GGGAGCTGCAGCGTTCGGTTCATGCATGGGAGATC)/AE3356 (5′-GCAGAATTCCTATCGAAACAACCT) were combined, reamplified using AE3385/AE3356, and digested with BamHI and EcoRI. The resulting fragment ligated into BamHI/EcoRI-digested pGEX-6P-3 vector DNA (GE Healthcare, Piscataway, NJ) yielded pGEX6P3-TNPO3, which directed the expression of the glutathione S-transferase (GST)-TNPO3 fusion protein in Escherichia coli.
The RSV IN coding region amplified from pATV-8 (28) using AE175 (5′-CAACATATGCCCTTGAGAGAGGCTAA) and AE3494 (5′-GCGCGCTCGAGCTATGCAAAAAGAGGGC) was digested with NdeI and XhoI and ligated into NdeI/XhoI-digested pET-15b DNA (EMD Biosciences, Madison, WI) to yield pET15b-RSVIN. The SIV IN expression vector pCP-SIV-IN-6H was generously donated by Peter Cherepanov, Imperial College London. Expression plasmids for the following His6-tagged INs were previously described: pETINH1 for MLV (26) (kindly provided by Monica Roth, Rutgers University), pHT-EIAV (16), pCP-BIV-IN-6H for BIV, pCP-HIV2-IN-6H for HIV-2, pCP-MPMV-IN-6H for Mason-Pfizer monkey virus (MPMV) (11) (gifts from P. Cherepanov), pT7-T(His)-FIVIN (44) (obtained from Samson Chow, University of California at Los Angeles), and pINSD.His for HIV-1 (7, 14).
Recombinant protein purification.
All INs were studied as His6-tagged fusion proteins; expression and purification were performed essentially as previously described (11). Briefly, shaker flasks of E. coli strain BL21 or its PC2 derivative (11) were grown in LB medium at 28°C to an A600 of 0.6 to 0.8 prior to induction with 0.4 mM isopropyl-thio-β-d-galactopyranoside (IPTG) for 4 h, after which cells were harvested and lysed by sonication in buffer A {1 M NaCl, 7.5 mM 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate (CHAPS), 0.5 mM phenylmethanesulfonyl fluoride, 25 mM Tris-HCl (pH 7.4)}. Lysate clarified by centrifugation at 40,000 × g for 30 min at 4°C was incubated with 2 ml Ni-nitrilioacetate (NTA) agarose beads (Qiagen, Valencia, CA) for 3 h at 4°C in buffer A containing 25 mM imidazole. After extensive washing, protein was eluted with buffer A-200 mM imidazole. IN-containing fractions diluted with 4 volumes of 7.5 mM CHAPS-50 mM Tris-HCl (pH 7.4) were injected into a 1-ml HiTrap heparin column (GE Healthcare), and bound proteins were eluted with a linear gradient of 0.25 to 1 M NaCl in 7.5 mM CHAPS-50 mM Tris-HCl (pH 7.4). MLV IN was further purified by gel filtration on a Superdex-200 column in buffer A, whereas MPMV IN was purified using just Ni-NTA beads. IN proteins concentrated by ultrafiltration using Amicon ultracentrifugal filters with a 10,000-molecular-weight cutoff (Millipore, Billerica, MA) were dialyzed against buffer A-10% (wt/vol) glycerol, flash-frozen in liquid N2, and stored at −70°C.
BL21 cells transformed with pGEX6P3-TNPO3 were grown at 37°C to an A600 of 0.35 to 0.45 prior to 4 h of induction with 0.4 mM IPTG. Bacteria were harvested and lysed by sonication in cold buffer B (150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol [DTT], 50 mM Tris-HCl [pH 7.4]). The crude extract, incubated with 25 U/ml Benzonase nuclease (EMD Biosciences) for 30 min at 25°C to degrade bulk E. coli DNA, was clarified by centrifugation at 40,000 × g for 30 min at 4°C, and the supernatant was incubated with 2 ml glutathione-Sepharose beads (GE Healthcare) for 3 h at 4°C. After washing in excess buffer B, a 50% gel slurry was incubated with 20 U PreScission protease (GE Healthcare) at 4°C for 16 to 24 h to remove the GST tag. Flowthrough and wash fractions containing tagless TNPO3 were pooled, concentrated, flash-frozen in liquid N2, and stored at −70°C. Recombinant GST (12) and lens epithelium-derived growth factor p75 (LEDGFp75) (47) proteins were purified following bacterial expression as previously described.
GST pulldown assays.
GST-TNPO3 protein concentrations were determined by densitometric scans of Coomassie-R250-stained gel images (FluorChem FC2; Alpha Innotech, San Leandro, CA) relative to bovine serum albumin (BSA) standard curves. GST-TNPO3 (12 μg, which corresponded to 0.46 μM during binding) or control GST (12 μg) absorbed onto glutathione-Sepharose beads (30-μl settled volume) was resuspended in 200 μl of PD buffer (150 mM NaCl, 5 mM MgCl2, 5 mM DTT, 0.1% Nonidet P40, 25 mM Tris-HCl [pH 7.4]) containing 10 μg BSA (12). IN (0.4 μM) was added, and the mixture was gently rocked for 3 h at 4°C. After aspiration of the supernatant, the settled beads resuspended in 600 μl PD buffer were allowed to sediment without centrifugation. The wash was repeated twice, and bound proteins were eluted in Laemmli sample buffer containing 50 mM EDTA-50 mM DTT. IN pulldown was confirmed in some experiments by blotting with a 1:1,000 dilution of HRP-conjugated anti-His6 antibody (Clontech, Mountain View, CA).
Ni-NTA pulldown assays.
INs were adsorbed onto Ni-NTA agarose beads in NTA-PD buffer (150 mM NaCl, 2 mM MgCl2, 25 mM imidazole, 0.1% Nonidet P40, 50 mM Tris-HCl [pH 7.4]). Beads (25-μl settled volume containing 0.4 μM IN) resuspended in 300 μl NTA-PD buffer containing 10 μg of BSA were incubated without (input controls) or with 0.4 μM TNPO3 for 3 h at 4°C. Following supernatant aspiration, beads washed three times in ice-cold NTA-PD buffer were boiled in Laemmli sample buffer as described above. IN proteins were detected by staining with Coomassie-R250, whereas TNPO3 was detected by Western blotting.
SPR.
A BIAcore 3000 biosensor system (GE Healthcare) instrument was used for surface plasmon resonance (SPR). Protein (ca. 1,000 resonance units [RU], corresponding to 1 ng/mm2) was immobilized onto carboxymethyl-5 chips using an equal mixture of N-ethyl-N-(dimethylaminopropyl)carbodiimide-N-hydroxysuccinimide in 10 mM sodium acetate buffer (pH 5.0). Unreacted activated sites were blocked with 1 M ethanolamine. Reference surfaces were treated the same way except that protein ligand was omitted.
Binding studies were conducted with HBS-EP buffer (150 mM NaCl, 3 mM EDTA, 0.005% surfactant P20, 10 mM HEPES [pH 7.4]) at 25°C. For association rate constant (Kass) (M−1 s−1) determinations, various concentrations of IN in binding buffer were allowed to bind to immobilized tagless TNPO3 at a flow rate of 20 μl/min. The dissociation rate constant (Kdiss) (s−1) was measured following replacement with binding buffer. Surface regeneration was conducted with 20 mM NaOH.
Sensograms corrected for residual binding to reference surfaces were analyzed by using BIAevaluation 3.2 software. Data were globally fit using the simultaneous Kass/Kdiss menu, which first generated rate equations and then determined values for the parameters in the equation that best fit the experimental data. The data, which were fit to the 1:1 model of interaction, were used to derive the equilibrium dissociation constant (KD = Kdiss/Kass). The half-life (t1/2) of the TNPO3-IN complex was calculated as ln2/Kdiss. Calculated maximum response values (Rmax) were in the range of 150 to 250 RU, with χ2 and residuals of the fit of the experimental data to the model of 0.3 to 2.0 and ±2 to ±5, respectively.
Affinity measurements carried out in the reverse orientation, with retroviral INs immobilized onto biosensor chips washed with multiple tagless TNPO3 concentrations, yielded results in agreement with the TNPO3 on-chip orientation (data not shown).
RESULTS
Experimental strategy.
Retroviral PICs must access cellular chromosomes to effect cDNA integration, and lentiviruses are thought to interact with specific karyopherin and/or nucleoporin proteins to effect energy-dependent transport through intact nuclear pore complexes (see references 15, 19, and 51 for recent reviews). Recent studies have highlighted a role for the karyopherin β protein TNPO3 in lentiviral nuclear import (5, 13, 30). MLV, in contrast, efficiently infected TNPO3 knockdown cells (5, 13, 30), and recombinant TNPO3 and MLV IN proteins failed to interact under conditions where an HIV-1 IN-TNPO3 interaction was detected. Based on these observations, it was previously proposed that IN-TNPO3 interactions might underscore active retroviral PIC nuclear import (13). To critically test this model, we expanded the strategy to include additional viral vectors and corresponding IN proteins derived from the lentiviruses SIV, BIV, EIAV, and FIV as well as the alpharetrovirus RSV. Although we did not have access to analogous vectors, recombinant HIV-2 and betaretroviral MPMV proteins were included in some IN binding assays.
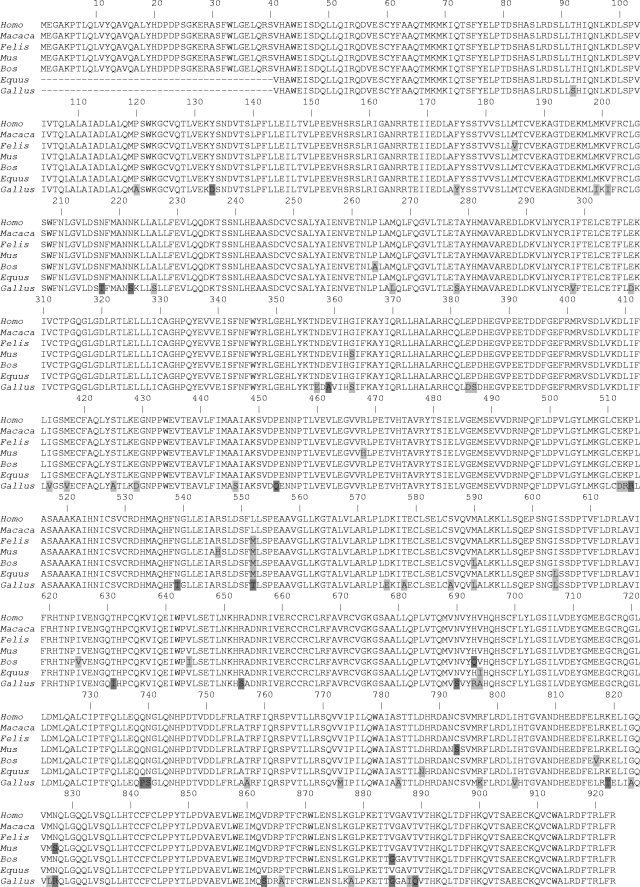
Similar to previously reported approaches (5, 13, 30), we wished to knock down TNPO3 in readily transfectable human cells. As challenge vectors were based on infectious agents derived from various animal hosts, we initially compared the sequences of the corresponding TNPO3 proteins to assess the validity of this strategy. Several fully annotated TNPO3 sequences, including those from human, rhesus macaque, mouse, and cow, were available in the NCBI database. The feline sequence, which was unknown, along with partial/incorrect horse and chicken sequences were annotated by using WGS and EST contigs (see Materials and Methods). Although this yielded the full-length cat sequence, we were unable to unambiguously complete the horse and chicken sequences. Each of these sequences lacks ca. 40 N-terminal residues that correspond to those encoded by human TNPO3 exon 1 (Fig. 1).
FIG. 1.
Amino acid sequence alignment of TNPO3 orthologs from Homo sapiens, rhesus macaque (Macaca mulatta), domestic cat (Felis catus), mouse (Mus musculus), cow (Bos taurus), horse (Equus caballus), and chicken (Gallus gallus) reveals the indicated nonhuman amino acid positions replaced by conserved (light gray) or nonhomologous (dark gray) residues as defined by previously reported chemical groupings (43). Numbers mark human sequence positions.
The alignment revealed a high degree of evolutionary conservation among orthologs: human and macaque TNPO3 are identical, whereas the other species are ≥98% homologous to the human protein. The cat sequence harbors only two conservative amino acid substitutions (Fig. 1). Chicken, the most divergent from humans, still retained 93.7% identity over their common 883-residue stretch. These results validate previously reported assessments of MLV infectivity in human cells knocked down for TNPO3 expression (5, 13, 30) and support the current strategy to expand the analysis to viruses derived from a range of host species.
Retroviral dependencies on cellular TNPO3.
Single-round GFP reporter viruses were produced by cotransfecting either one or two virus-based plasmids with the pMD.G VSV-G expression construct. Most GFP transcripts were initiated from natural viral long terminal repeat (LTR) promoters, with the exceptions of BIV, which utilized a modified myeloid proliferative sarcoma LTR, and EIAV and FIV, which employed internal cytomegalovirus (CMV) promoters. The transcripts were fully spliced and likely exported using similar cellular mRNA export mechanisms, with the exception of the FIV vector, which additionally harbored the woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) (55).
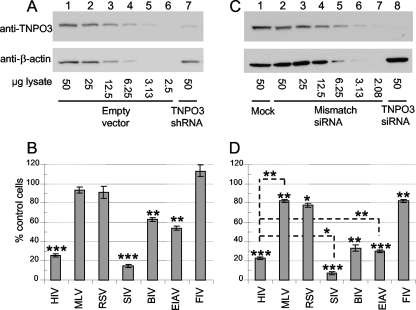
Titers of viruses were initially measured to determine the level of each stock required to transduce ∼30 to 40% of HeLa cells, corresponding to theoretical multiplicities of infection of ∼0.35 to 0.5. TNPO3 levels were then knocked down by using RNA interference. The stable transduction of HeLa cells with a pLKO.1-shRNA vector yielded approximate 7- to 10-fold reductions in the level of TNPO3 protein compared to that for empty vector-transduced cells (Fig. 2A). Numbers of GFP-positive cells were determined 2 days after virus challenge, and results were expressed as the percent knockdown-cell infection normalized to control cells (Fig. 2B). The various viruses exhibited a relatively wide range of sensitivity to TNPO3 knockdown. Consistent with data from previous reports, HIV-1 infectivity was significantly (P < 0.001) impaired by the knockdown, while the MLV titer was basically unimpaired (5, 13). RSV, an alpharetrovirus, behaved similarly to MLV. SIV, BIV, and EIAV exhibited considerable ∼6.9-fold (P < 0.001), 1.6-fold, and 1.9-fold (P < 0.01 each) infection defects, respectively, whereas FIV was surprisingly innocuous to the knockdown (Fig. 2B).
FIG. 2.
Retroviral infectivity profiles of TNPO3 knockdown cells. (A) Diluted lysate of empty vector control cells (from neat to 1:20) were loaded into lanes 1 to 6 to define a TNPO3 standard curve. Scanning densitometry revealed approximate 7- to 10-fold protein reductions in shRNA-expressing cells (lane 7); β-actin served as a gel loading control. (B) Results of five independent experiments (means ± standard errors of the means [SEM]) expressed as percent control-cell infection. (C) Western blots of mock-transfected HeLa-T4 cells (lane 1) or cells transfected with mismatched control (lanes 2 to 7) or TNPO3-targeting (lane 8) siRNA. (D) Infectivities (means ± SEM) (n = 4 experiments) of indicated retroviral vectors, expressed as percent infectivity in TNPO3 knockdown cells compared to mismatch siRNA-transfected controls. Results in A and C are representative data from duplicate Western blot experiments. Statistical significance values shown in B and D were calculated by using a Student's two-sided t test. Asterisks directly above bars indicate differences between knockdown and control cells for a given virus, whereas asterisks above lines indicate differences between indicated viruses. ***, P < 0.0001; **, P < 0.001; *, P < 0.01.
To address the possibility that residual TNPO3 levels might contribute to the unexpected FIV phenotype, TNPO3 was subsequently knocked down by transient siRNA transfection. Transfected HeLa-T4 cells indeed revealed a stronger knockdown phenotype, yielding approximate 14- to 20-fold reductions in protein levels at 72 h posttransfection, commensurate with the time of virus challenge (Fig. 2C). The further reduction in TNPO3 levels yielded reductions in relative viral titers across the board, although the rank in virus infectivity did not change from the prior condition (compare Fig. 2D to B). Thus, SIV remained the most affected by TNPO3 knockdown. The lower TNPO3 levels did reduce the relative FIV titer (P < 0.01), but this effect was quite similar to the marginal defects observed with MLV and RSV (Fig. 2D). In summary, shRNA- or siRNA-mediated knockdown of TNPO3 in HeLa cells yielded the following viral pattern of sensitivity to infection: SIV > HIV-1 > BIV and EIAV > MLV, RSV, and FIV.
TNPO3 is a prolific IN binding protein.
The results of the above-described experiments demonstrated clear differences in TNPO3 dependencies during retroviral infection. This established an ideal situation to test if IN binding affinity indeed correlated with the observed virological phenotypes. Recombinant His6-tagged IN proteins expressed in E. coli cells were purified to near homogeneity by using standard methods. TNPO3 was expressed and purified as a GST fusion protein, and the GST tag was removed for some experiments by subsequent proteolysis.
(i) In vitro pulldown assays.
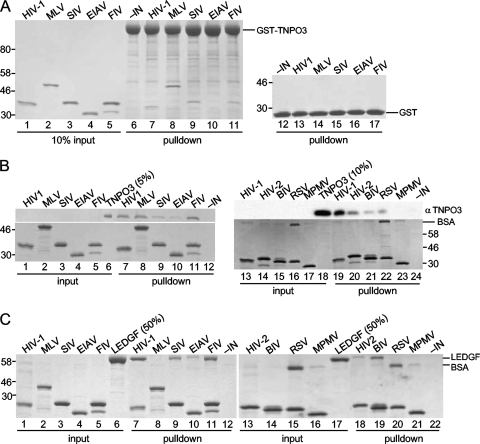
We initially interrogated potential IN-TNPO3 interactions by using reciprocal pulldown assays. GST-TNPO3 immobilized onto beads was first tested versus the control GST protein for its ability to bind soluble IN. In agreement with previously reported results (13), HIV-1 IN was pulled down by GST-TNPO3 (Fig. 3A, compare lane 7 to lane 6). Contrary to data from the previous report, MLV IN also readily bound GST-TNPO3 (Fig. 3A, lane 8). GST-TNPO3 recovered SIV (Fig. 3A, lane 9) and FIV (lane 11) IN proteins similarly as HIV-1, with EIAV IN binding not detected under these conditions (lane 10). Increasing the sensitivity of assay detection by Western blotting revealed low-level EIAV IN binding to GST-TNPO3 (data not shown). GST alone failed to recover any of the IN proteins, confirming the specificity of the observed TNPO3-IN interactions (Fig. 3A, lanes 12 to 17).
FIG. 3.
Pulldown analyses of IN-TNPO3 binding. (A) GST pulldowns. Lanes 1 to 5 contained 10% of input IN protein. Proteins bound to GST-TNPO3 (lanes 6 to 11)- and GST (lanes 12 to 17)-conjugated beads were released by boiling; gels were developed by Coomassie blue staining. IN was omitted from the reaction mixtures loaded into lanes 6 and 12. (B) Mock (lanes 1 to 5 and 13 to 17) or TNPO3-containing (lanes 7 to 12 and 19 to 24) Ni-NTA pulldowns were conducted with the indicated bead-bound INs. Eluted proteins were detected by Western blotting (top) or Coomassie blue staining (bottom). Input levels of TNPO3 were analyzed in lanes 6 and 18; IN was omitted from the reaction mixtures loaded into lanes 12 and 24. (C) Same as B except that LEDGFp75 was tested in place of TNPO3 and all proteins were detected with Coomassie blue. RSV IN and, to a lesser extent, MPMV displayed nonspecific binding to BSA (B, lanes 16, 17, 22, and 23, and C, lanes 15, 16, 20, and 21), which was included at 10 μg in all pulldown assays. The lack of RSV IN-LEDGFp75 binding (C, lane 20) indicated a specific RSV IN-TNPO3 interaction (B, lane 22). Migration positions of molecular mass standards in kDa are marked adjacent to gel panels.
In the reverse Ni-NTA pulldown format, bead-bound INs were assayed for their abilities to capture tagless TNPO3 protein (Fig. 3B). The results in large part mirrored those obtained via GST-TNPO3 pulldowns: SIV and EIAV recovered less TNPO3 than either HIV-1 or MLV IN, while FIV IN appeared to bind TNPO3 better in the Ni-NTA assay than in the GST pulldown assay (compare top lanes 7 to 11 in Fig. 3B to the same lanes in Fig. 3A). The lack of TNPO3 binding to Ni-NTA beads in the absence of IN protein established the specificity of the interactions (Fig. 3B, lane 12). IN proteins derived from additional viruses, HIV-2, BIV, RSV, and MPMV, were also tested in the Ni-NTA pulldown assay. With the exception of MPMV, each protein demonstrated some TNPO3 binding affinity (Fig. 3B, lanes 19 to 24). These data indicate that TNPO3 possesses affinity for IN proteins derived from gamma- and alpharetroviruses in addition to lentiviruses. LEDGFp75, a critical lentivirus-specific DNA integration cofactor (reviewed in references 17 and 41), displays lentiviral specificity in IN binding assays (8, 11). Ni-NTA pulldowns were therefore repeated with LEDGFp75 to ascertain the binding specificity of our IN preparations. As expected, each lentiviral IN protein pulled down LEDGFp75 under conditions where MLV, RSV, and MPMV IN failed to do so (Fig. 3C, lanes 7 to 12 and 18 to 22).
(ii) Affinity measurement by SPR.
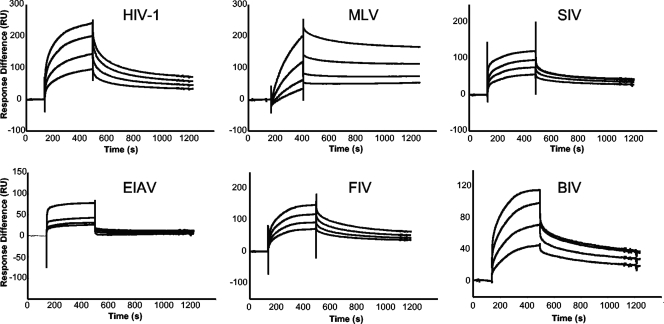
The results of the above-described experiments confirmed TNPO3 as an HIV-1 IN binding protein and additionally revealed binding to INs derived from viruses outside of the genus Lentivirus. SPR was next used to develop acute quantitative pictures of TNPO3-IN binding interactions. Akin to the pulldown experiments, SPR was conducted by use of two experimental formats. In one, chip-bound TNPO3 was exposed to various IN proteins, whereas in the other format, IN proteins bound to different chip surfaces were washed with TNPO3. Both platforms gave overall similar data, although the results generated with immobilized TNPO3 fit better to theoretical binding models. Accordingly, Kass and Kdiss values were calculated from sensograms generated by flowing various concentrations of HIV-1, MLV, SIV, EIAV, FIV, and BIV INs over immobilized TNPO3 (Fig. 4 and Table 1).
FIG. 4.
SPR analysis of IN-TNPO3 interactions. Sensograms revealing association and dissociation binding phases were generated by using the following IN concentration ranges: 156.25 nM to 1.25 μM for HIV-1, 444.4 nM to 3.5 μM for MLV, 78.13 nM to 1.25 μM for SIV, 312.5 nM to 5 μM for EIAV, 78.13 nM to 625 nM for FIV, and 312.5 nM to 2.5 μM for BIV. RSV IN behaved as a relatively sticky analyte in this format and was therefore omitted from the SPR analysis.
TABLE 1.
SPR-based rate and equilibrium constants
| Analyte | Mean Kass ± SD (M−1 s−1) | Mean Kdiss ± SD (s−1) | t1/2 (s) | KD (nM) |
|---|---|---|---|---|
| HIV-1 | 1.21 × 104 ± 28.9 | 3.16 × 10−3 ± 8.9 × 10−6 | 219.3 | 261 |
| MLV | 5.23 × 103 ± 2.4 | 2.53 × 10−3 ± 3.7 × 10−6 | 395.2 | 484 |
| SIV | 1.03 × 104 ± 5.5 | 3.67 × 10−3 ± 4.2 × 10−5 | 188.8 | 356 |
| EIAV | 7.71 × 103 ± 25.4 | 9.67 × 10−3 ± 5.7 × 10−4 | 71.7 | 1,254 |
| FIV | 9.47 × 103 ± 37.3 | 1.79 × 10−3 ± 7.1 × 10−6 | 387.2 | 189 |
| BIV | 7.08 × 103 ± 20.8 | 1.98 × 10−3 ± 1.0 × 10−5 | 350.0 | 280 |
Consistent with the pulldown studies, the different INs bound TNPO3 with a range of affinities. The KD for the TNPO3-HIV-1 IN interaction was 261 nM (Table 1). Despite marginally slower association and dissociation phases, FIV and BIV INs exhibited similar binding affinities, yielding KD values of 189 nM and 280 nM, respectively (Table 1). SIV IN bound TNPO3 about 1.4-fold more weakly than HIV-1 IN. As observed in the GST pulldown assay, SPR scored the EIAV IN-TNPO3 interaction as relatively weak. The comparatively slow association during binding and the fast dissociation from TNPO3 contributed to an almost fivefold loss in EIAV IN affinity and a threefold loss in complex stability (t1/2) compared to HIV-1 (Fig. 4 and Table 1). In contrast, MLV IN bound TNPO3 with an intermediate affinity (KD = 484 nM) (Table 1). SPR-based affinities of IN for TNPO3, ranked from strongest to weakest, were as follows: FIV, HIV-1, and BIV > SIV and MLV > EIAV.
MHIV chimera viruses highlight a role for HIV-1 CA in TNPO3 dependency.
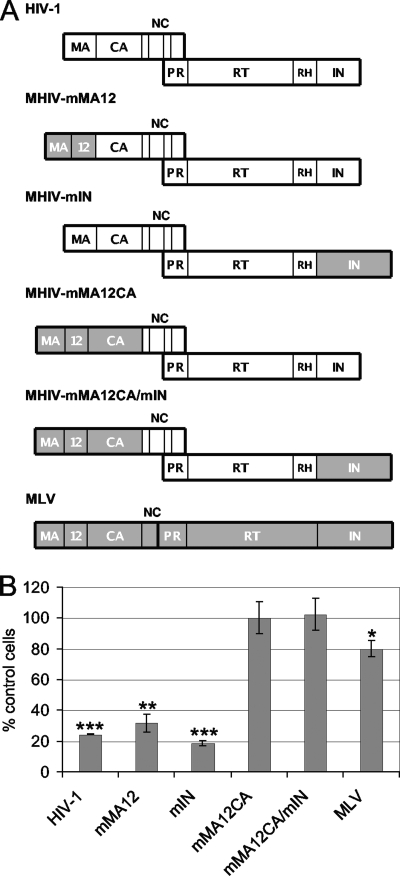
Our findings indicated that IN-TNPO3 binding affinities do not underscore the respective viral dependencies on TNPO3 during infection. We therefore sought to determine other viral factors that might contribute to the phenotype. As TNPO3 knockdown preferentially inhibited infection by HIV-1 over MLV (5, 13, 30) (Fig. 2), previously described MHIV chimera viruses, which swap various parts of MLV for the corresponding HIV-1 proteins (50, 52), were investigated. Indeed, these hybrids were previously used to highlight a dominant role for HIV-1 CA in conferring infectivity on growth-arrested cells (50, 52). The following chimera viruses were utilized here: MHIV-mMA12CA, where MLV MA, p12, and CA replaced HIV-1 MA and CA (p12 is specific to MLV); MHIV-mMA12, harboring MLV MA and p12 in place of HIV-1 MA; MHIV-mIN, where MLV IN replaced the HIV-1 protein; and MHIV-mMA12CA/mIN, where MLV Gag and IN determinants were combined in the same construct (Fig. 5A). Parental luciferase reporter viruses were VSV-G pseudotyped alongside the chimerae, and the resulting infectivities in stably knocked down HeLa cells were expressed as percentages of control cell infection levels (Fig. 5B).
FIG. 5.
Infectivity profiles of MHIV chimera viruses versus HIV-1 and MLV. (A) Schematics of HIV-1 (white), MLV (gray), and chimera viral Gag and Pol proteins. NC, nucleocapsid; PR, protease; RT, reverse transcriptase; RH, RNase H. (B) Results of two independent experiments (means ± SEM) expressed as the percent infectivity on TNPO3-depleted compared to control cells. Student's two-sided t test comparisons of these values revealed significance parameters (***, P < 0.001; **, P < 0.01; *, P < 0.05). In contrast, relative MHIV-mMA12 and MHIV-mIN titers did not significantly differ (P > 0.05) from the HIV-1 value.
Replacing HIV-1 MA and CA with the corresponding MLV determinants strikingly rendered the MHIV-mMA12CA chimera insensitive to TNPO3 knockdown. Swapping the CA of this virus back to the HIV-1 protein, however, resensitized MHIV-mMA12 to the knockdown, highlighting a role for CA in determining TNPO3 dependency during HIV-1 infection (Fig. 5B). The MHIV-mIN chimera, which harbored MLV IN alongside HIV-1 Gag determinants, was accordingly sensitive to the knockdown. Moreover, the combined MHIV-mMA12CA/mIN chimera virus behaved in a TNPO3-independent manner (Fig. 5B). Thus, all viruses that harbored HIV-1 CA depended on TNPO3 for optimal infectivity regardless of whether they carried HIV-1 or MLV IN.
DISCUSSION
Recent genome-wide siRNA screens (5, 30) for host proteins necessary during HIV-1 infection identified TNPO3, a member of the karyopherin β family that imports spliceosomal SR proteins into the nucleus (27). TNPO3 knockdown inhibited HIV-1 infectivity without substantially affecting MLV or adeno-associated viral vectors. The block to HIV-1 infection was further pinpointed after reverse transcription but prior to or concomitant with integration, suggesting that HIV-1 might require TNPO3 as a PIC nuclear import cofactor. Indeed, data reported previously by Christ et al. (13) subsequently supported this interpretation, as a decreased level of formation of 2-LTR-containing circles, a signature of nuclear import, as well as fewer GFP-tagged HIV-1 PICs were identified in the nuclei of TNPO3 knockdown cells. Moreover, those authors determined that TNPO3 bound HIV-1 IN by reciprocal yeast two-hybrid and recombinant protein pulldown assays. As MLV IN failed to interact with TNPO3, these results suggested that an IN-TNPO3 interaction might underscore the biology of lentiviral PIC nuclear import. The expanded study described here indicates that this interpretation may have been premature.
Differential requirements for TNPO3 during retroviral infection.
We investigated the role of TNPO3 in retroviral biology by biochemical and genetic analyses using an expanded set of lentiviruses alongside RSV and MLV. Viral infectivities were compared in knockdown HeLa cells that exhibited up to a 20-fold reduction in TNPO3 protein levels. HIV-1, SIV, BIV, and EIAV exhibited wide-ranging but significant infectivity defects under these conditions, unlike FIV, which behaved virtually indistinguishably from MLV and RSV (Fig. 2). Given that the sequence of feline TNPO3 is more similar to the human protein than is either the equine or bovine ortholog (Fig. 1), the phenotype is unlikely confounded by the study of a cat virus in human cells. We therefore considered FIV-specific vector elements that might have complicated the interpretation of results. The internal CMV promoter played no apparent role, as data from both Brass et al. (5) and our own studies (data not shown) revealed LTR- and CMV-driven HIV-1 vectors that were equally affected by TNPO3 knockdown. FIV was the only vector that harbored the WPRE mRNA transport element, but follow-up studies with a WPRE-containing HIV-1 vector again showed a clear sensitivity to TNPO3 knockdown (data not shown). FIV IN revealed the greatest affinity for TNPO3 binding among the tested IN proteins (Table 1 and Fig. 4), which could contribute to its relative insensitivity to TNPO3 knockdown during infection. However, the infectivity profiles of other viruses have led us to conclude that IN is not the predominant retroviral factor that determines TNPO3 dependency (see below). Although future experiments with additional cell lines and alternative FIV vectors will be necessary to make a definitive conclusion, our results indicate that FIV is, at best, minimally dependent on TNPO3 and thus may rely on other karyopherin β family members for its nuclear import.
IN binding affinities do not correlate with the requirement for TNPO3 during retroviral infection.
In light of our viral infectivity analysis and the suggested role of IN in TNPO3-mediated nuclear transport (13), we investigated the profiles of binding of various IN proteins to recombinant TNPO3 using qualitative (reciprocal pulldown) and quantitative (SPR) approaches. GST and Ni-NTA pulldown assays not only confirmed the HIV-1 IN-TNPO3 interaction but also extended the specificity of binding to all lentiviral IN proteins tested (Fig. 3 and data not shown). Interestingly, MLV IN also bound TNPO3 in both pulldown assays, revealing that the interaction with TNPO3, although conserved among the Lentiviridae, is not limited to this genus.
Quantitative SPR measurements support TNPO3 as a prolific IN binding protein. Differences in the KD values for the IN-TNPO3 interaction were within twofold for HIV-1, SIV, FIV, and BIV INs; however, EIAV IN bound human TNPO3 approximately fivefold less efficiently than did HIV-1 IN (Table 1). Considering that equine TNPO3 is 95.6% identical and 100% homologous to human TNPO3 (Fig. 1), we speculate that the difference in binding affinities is due to comparatively unique EIAV IN sequence determinants (9, 18). Taken alongside the results of virus infection, these data refute IN as a critical viral factor that determines TNPO3 dependency: based on relatively weak EIAV IN-TNPO3 binding, one would predict a corresponding acute viral infection defect upon TNPO3 knockdown if IN was the sole determinant dictating host factor dependency. Instead, HIV-1 and SIV displayed greater TNPO3 dependencies than did EIAV (Fig. 2). Moreover, the affinity of MLV IN for TNPO3 was comparable to that of HIV-1 IN (less than a twofold difference) (Table 1), yet MLV infectivity was in large part independent of cellular TNPO3 levels (5, 13, 30) (Fig. 2).
CA determines TNPO3 dependency during HIV-1 infection.
The CA protein plays a dominant role in determining HIV-1 infection of growth-arrested cells (50, 52, 53). We utilized previously described MHIV chimera viruses wherein different MLV proteins, including IN, MA, and/or CA, were swapped for their HIV-1 counterparts to further investigate viral genetic determinants of TNPO3 dependency. Replacing HIV-1 MA-CA with MLV MA-p12-CA remarkably rendered the resulting MHIV-mMA12CA chimera insensitive to TNPO3 knockdown (Fig. 5). As this virus retained wild-type HIV-1 IN, these results additionally discount a dominant role for IN in mediating TNPO3 dependency during HIV-1 infection. Accordingly, the MHIV-mIN chimera, which harbored MLV IN in place of the HIV-1 protein, was sensitive to TNPO3 knockdown. Therefore, IN does not dictate the requirement for TNPO3 during HIV-1 infection. Moreover, because MHIV-mMA12 relied on TNPO3 for its infection, we conclude that CA is a dominant viral determinant of TNPO3 dependency during HIV-1 infection. In support of this view, we have, through a related series of experiments, identified CA point mutations that render HIV-1 insensitive to TNPO3 knockdown (K. Lee and V. N. KewalRamani, unpublished data). Taken together, our results discount an important role for IN in determining the requirement for TNPO3 during infection and instead highlight a dominant role for CA in this aspect of HIV-1 biology. Ongoing work in the laboratory is aimed at determining the underpinning mechanisms.
Acknowledgments
This study relied on the contributions of reagents from the following generous colleagues: Peter Cherepanov, Samson Chow, Ronald Desrosiers, Michael Emerman, Dana Gabuzda, Michael Kaleko, Richard Mulligan, John Olsen, Eric Poeschla, Monica Roth, Joseph Sodroski, Didier Trono, and Masahiro Yamashita.
This work was supported by the HIV Drug Resistance Program (V.N.K.), NIH grant AI052014 (A.E.), and the Harvard University Center for AIDS Research, an NIH-funded program (grant P30AI060354), which is supported by the following NIH institutes and centers: NIAID, NCI, NIMH, NIDA, NICHD, NHLBI, and NCCAM.
The contents of the manuscript do not necessarily reflect the views of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Published ahead of print on 21 October 2009.
REFERENCES
- 1.Alexander, L., R. S. Veazey, S. Czajak, M. DeMaria, M. Rosenzweig, A. A. Lackner, R. C. Desrosiers, and V. G. Sasseville. 1999. Recombinant simian immunodeficiency virus expressing green fluorescent protein identifies infected cells in rhesus monkeys. AIDS Res. Hum. Retroviruses 15:11-21. [DOI] [PubMed] [Google Scholar]
- 2.Ao, Z., G. Huang, H. Yao, Z. Xu, M. Labine, A. W. Cochrane, and X. Yao. 2007. Interaction of human immunodeficiency virus type 1 integrase with cellular nuclear import receptor importin 7 and its impact on viral replication. J. Biol. Chem. 282:13456-13467. [DOI] [PubMed] [Google Scholar]
- 3.Berkowitz, R., H. Ilves, W. Y. Lin, K. Eckert, A. Coward, S. Tamaki, G. Veres, and I. Plavec. 2001. Construction and molecular analysis of gene transfer systems derived from bovine immunodeficiency virus. J. Virol. 75:3371-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowerman, B., P. O. Brown, J. M. Bishop, and H. E. Varmus. 1989. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 3:469-478. [DOI] [PubMed] [Google Scholar]
- 5.Brass, A. L., D. M. Dykxhoorn, Y. Benita, N. Yan, A. Engelman, R. J. Xavier, J. Lieberman, and S. J. Elledge. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921-926. [DOI] [PubMed] [Google Scholar]
- 6.Bukrinsky, M., N. Sharova, M. Dempsey, T. Stanwick, A. Bukrinskaya, S. Haggerty, and M. Stevenson. 1992. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc. Natl. Acad. Sci. U. S. A. 89:6580-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bushman, F. D., A. Engelman, I. Palmer, P. Wingfield, and R. Craigie. 1993. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc. Natl. Acad. Sci. U. S. A. 90:3428-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Busschots, K., J. Vercammen, S. Emiliani, R. Benarous, Y. Engelborghs, F. Christ, and Z. Debyser. 2005. The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J. Biol. Chem. 280:17841-17847. [DOI] [PubMed] [Google Scholar]
- 9.Cannon, P. M., E. D. Byles, S. M. Kingsman, and A. J. Kingsman. 1996. Conserved sequences in the carboxyl terminus of integrase that are essential for human immunodeficiency virus type 1 replication. J. Virol. 70:651-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, C. M., D. M. Smith, M. A. Peters, M. E. Samson, J. Zitz, C. J. Tabin, and C. L. Cepko. 1999. Production and design of more effective avian replication-incompetent retroviral vectors. Dev. Biol. 214:370-384. [DOI] [PubMed] [Google Scholar]
- 11.Cherepanov, P. 2007. LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res. 35:113-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cherepanov, P., E. Devroe, P. A. Silver, and A. Engelman. 2004. Identification of an evolutionarily-conserved domain in LEDGF/p75 that binds HIV-1 integrase. J. Biol. Chem. 279:48883-48892. [DOI] [PubMed] [Google Scholar]
- 13.Christ, F., W. Thys, J. De Rijck, R. Gijsbers, A. Albanese, D. Arosio, S. Emiliani, J. C. Rain, R. Benarous, A. Cereseto, and Z. Debyser. 2008. Transportin-SR2 imports HIV into the nucleus. Curr. Biol. 18:1192-1202. [DOI] [PubMed] [Google Scholar]
- 14.Craigie, R., A. B. Hickman, and A. Engelman. 1995. Integrase, p. 53-71. In J. Karn (ed.), HIV volume 2: biochemistry, molecular biology, and drug discovery. IRL Press, New York, NY.
- 15.De Rijck, J., L. Vandekerckhove, F. Christ, and Z. Debyser. 2007. Lentiviral nuclear import: a complex interplay between virus and host. Bioessays 29:441-451. [DOI] [PubMed] [Google Scholar]
- 16.Engelman, A. 1996. Biochemical characterization of recombinant equine infectious anemia virus integrase. Protein Expr. Purif. 8:299-304. [DOI] [PubMed] [Google Scholar]
- 17.Engelman, A., and P. Cherepanov. 2008. The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication. PLoS Pathog. 4:e1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engelman, A., and R. Craigie. 1992. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J. Virol. 66:6361-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fassati, A. 2006. HIV infection of non-dividing cells: a divisive problem. Retrovirology 3:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fassati, A., and S. P. Goff. 2001. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J. Virol. 75:3626-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fassati, A., and S. P. Goff. 1999. Characterization of intracellular reverse transcription complexes of Moloney murine leukemia virus. J. Virol. 73:8919-8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fassati, A., D. Gorlich, I. Harrison, L. Zaytseva, and J. M. Mingot. 2003. Nuclear import of HIV-1 intracellular reverse transcription complexes is mediated by importin 7. EMBO J. 22:3675-3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. U. S. A. 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He, J., Y. Chen, M. Farzan, H. Choe, A. Ohagen, S. Gartner, J. Busciglio, X. Yang, W. Hofmann, W. Newman, C. R. Mackay, J. Sodroski, and D. Gabuzda. 1997. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature 385:645-649. [DOI] [PubMed] [Google Scholar]
- 25.Hearps, A. C., and D. A. Jans. 2006. HIV-1 integrase is capable of targeting DNA to the nucleus via an importin alpha/beta-dependent mechanism. Biochem. J. 398:475-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jonsson, C. B., G. A. Donzella, and M. J. Roth. 1993. Characterization of the forward and reverse integration reactions of the Moloney murine leukemia virus integrase protein purified from Escherichia coli. J. Biol. Chem. 268:1462-1469. [PubMed] [Google Scholar]
- 27.Kataoka, N., J. L. Bachorik, and G. Dreyfuss. 1999. Transportin-SR, a nuclear import receptor for SR proteins. J. Cell Biol. 145:1145-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katz, R. A., C. A. Omer, J. H. Weis, S. A. Mitsialis, A. J. Faras, and R. V. Guntaka. 1982. Restriction endonuclease and nucleotide sequence analyses of molecularly cloned unintegrated avian tumor virus DNA: structure of large terminal repeats in circle junctions. J. Virol. 42:346-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khare, P. D., N. Loewen, W. Teo, R. A. Barraza, D. T. Saenz, D. H. Johnson, and E. M. Poeschla. 2008. Durable, safe, multi-gene lentiviral vector expression in feline trabecular meshwork. Mol. Ther. 16:97-106. [DOI] [PubMed] [Google Scholar]
- 30.Konig, R., Y. Zhou, D. Elleder, T. L. Diamond, G. M. Bonamy, J. T. Irelan, C. Y. Chiang, B. P. Tu, P. D. De Jesus, C. E. Lilley, S. Seidel, A. M. Opaluch, J. S. Caldwell, M. D. Weitzman, K. L. Kuhen, S. Bandyopadhyay, T. Ideker, A. P. Orth, L. J. Miraglia, F. D. Bushman, J. A. Young, and S. K. Chanda. 2008. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135:49-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis, P., M. Hensel, and M. Emerman. 1992. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 11:3053-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis, P. F., and M. Emerman. 1994. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J. Virol. 68:510-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loewen, N., R. Barraza, T. Whitwam, D. T. Saenz, I. Kemler, and E. M. Poeschla. 2003. FIV vectors. Methods Mol. Biol. 229:251-271. [DOI] [PubMed] [Google Scholar]
- 34.Mattaj, I. W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67:265-306. [DOI] [PubMed] [Google Scholar]
- 35.Miller, D. G., M. A. Adam, and A. D. Miller. 1990. Gene transfer by retrovirus vectors occurs only in cells that are actively replicating at the time of infection. Mol. Cell. Biol. 10:4239-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, M., C. Farnet, and F. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naldini, L., U. Blömer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 38.Olsen, J. C. 1998. Gene transfer vectors derived from equine infectious anemia virus. Gene Ther. 5:1481-1487. [DOI] [PubMed] [Google Scholar]
- 39.O'Rourke, J. P., G. C. Newbound, D. B. Kohn, J. C. Olsen, and B. A. Bunnell. 2002. Comparison of gene transfer efficiencies and gene expression levels achieved with equine infectious anemia virus- and human immunodeficiency virus type 1-derived lentivirus vectors. J. Virol. 76:1510-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perron, M. J., M. Stremlau, B. Song, W. Ulm, R. C. Mulligan, and J. Sodroski. 2004. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. U. S. A. 101:11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poeschla, E. M. 2008. Integrase, LEDGF/p75 and HIV replication. Cell. Mol. Life Sci. 65:1403-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roe, T., T. C. Reynolds, G. Yu, and P. O. Brown. 1993. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 12:2099-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwartz, R. M., and M. O. Dayhoff. 1978. Matrices for detecting distance relationships, p. 353-358. In M. O. Dayhoff (ed.), Atlas of protein sequence and structure, vol. 5, suppl. 3. National Biochemical Research Foundation, Washington, DC. [Google Scholar]
- 44.Shibagaki, Y., M. L. Holmes, R. S. Appa, and S. A. Chow. 1997. Characterization of feline immunodeficiency virus integrase and analysis of functional domains. Virology 230:1-10. [DOI] [PubMed] [Google Scholar]
- 45.Shun, M. C., J. E. Daigle, N. Vandegraaff, and A. Engelman. 2007. Wild-type levels of human immunodeficiency virus type 1 infectivity in the absence of cellular emerin protein. J. Virol. 81:166-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ulm, J. W., M. Perron, J. Sodroski, and R. C. Mulligan. 2007. Complex determinants within the Moloney murine leukemia virus capsid modulate susceptibility of the virus to Fv1 and Ref1-mediated restriction. Virology 363:245-255. [DOI] [PubMed] [Google Scholar]
- 47.Vandegraaff, N., E. Devroe, F. Turlure, P. A. Silver, and A. Engelman. 2006. Biochemical and genetic analyses of integrase-interacting proteins lens epithelium-derived growth factor (LEDGF)/p75 and hepatoma-derived growth factor related protein 2 (HRP2) in preintegration complex function and HIV-1 replication. Virology 346:415-426. [DOI] [PubMed] [Google Scholar]
- 48.Weinberg, J. B., T. J. Matthews, B. R. Cullen, and M. H. Malim. 1991. Productive human immunodeficiency virus type 1 (HIV-1) infection of nonproliferating human monocytes. J. Exp. Med. 174:1477-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woodward, C. L., S. Prakobwanakit, S. Mosessian, and S. A. Chow. 2009. Integrase interacts with nucleoporin NUP153 to mediate the nuclear import of human immunodeficiency virus type 1. J. Virol. 83:6522-6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamashita, M., and M. Emerman. 2004. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J. Virol. 78:5670-5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamashita, M., and M. Emerman. 2006. Retroviral infection of non-dividing cells: old and new perspectives. Virology 344:88-93. [DOI] [PubMed] [Google Scholar]
- 52.Yamashita, M., and M. Emerman. 2005. The cell cycle independence of HIV infections is not determined by known karyophilic viral elements. PLoS Pathog. 1:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamashita, M., O. Perez, T. J. Hope, and M. Emerman. 2007. Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS Pathog. 3:1502-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zaitseva, L., P. Cherepanov, L. Leyens, S. J. Wilson, J. Rasaiyaah, and A. Fassati. 2009. HIV-1 exploits importin 7 to maximize nuclear import of its DNA genome. Retrovirology 6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zufferey, R., J. E. Donello, D. Trono, and T. J. Hope. 1999. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J. Virol. 73:2886-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]