Abstract
MicroRNA 122 (miR-122) promotes hepatitis C virus (HCV) RNA abundance through a direct interaction with the viral RNA and stimulates the mevalonate pathway in the animal liver. We found that overexpression of miR-122 enhanced viral RNA accumulation without affecting genes in the mevalonate pathway, such as the 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR) gene. However, inhibition of miR-122 decreased both HCV RNA and HMGCR RNA with little effects on the rates of HCV and HMGCR RNA synthesis. Loss of HCV RNA could not be restored by isoprenoid intermediate metabolites. Overall, these findings suggest that miR-122 modulates viral RNA abundance independently of its effect on isoprenoid metabolism.
Hepatitis C virus (HCV) is a positive-stranded, hepatotropic RNA virus belonging to the Flaviviridae. HCV has been estimated to infect approximately 170 million people worldwide, leading to chronic liver disease and hepatocellular carcinoma (14, 25). With sustained virological response rates in current therapies limited to 50%, a greater understanding of HCV virology and pathogenesis is needed to treat this disease more effectively (24).
MicroRNAs are small, approximately 22-nucleotide RNAs that normally repress cellular gene expression in a sequence-specific manner, either through RNA degradation or through translational inhibition that can be accompanied by targeted RNA degradation (3, 5, 18). We have recently found that microRNA 122 (miR-122), which is expressed at a high abundance in the liver (4), acts in an unusual manner to stimulate accumulation of HCV RNA by interacting with the 5′ noncoding region of the viral genome (Fig. 1) (19, 20). Functional sequestration of miR-122 resulted in a large reduction in viral RNA abundance, suggesting that miR-122 may be a potential target for antiviral therapeutics. Clues to normal functions for miR-122 in the liver came from studies in which miR-122 was sequestered by modified antisense oligomers in the livers of mice and nonhuman primates (9-11, 23). In each case, sequestration of miR-122 resulted in reduced mevalonate pathway activity, lowered liver and plasma cholesterol levels, and reduced fat accumulation in the liver (9-11, 23). The mevalonate pathway functions to convert mevalonate to cholesterol and isoprenoid intermediates (Fig. 1). The physiological effects of miR-122 sequestration were accompanied by decreased expression of transcripts encoding proteins of this pathway, such as 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR) and squalene epoxidase (Fig. 1) (9-11, 23). Because these transcripts do not contain miR-122 binding sites, miR-122 is thought to regulate their expression by downregulation of an inhibitor for these genes.
FIG. 1.
Predicted interactions between miR-122 and the mevalonate pathway and HCV. See the text for details of how miR-122 promotes HCV RNA abundance and enhances accumulation of mevalonate pathway enzyme transcripts and accumulation of cholesterol. The compound lovastatin that blocks the pathway's rate-limiting enzyme, HMGCR, is shown. The interaction of viral protein NS5A with geranylgeranylated cellular protein FBL2 is indicated. CoA, coenzyme A; PP, pyrophosphate.
Interestingly, HCV infection also alters lipid metabolism through an incompletely characterized mechanism that involves cleavage and activation of the regulatory transcription factor sterol regulatory element binding protein (SREBP) (28, 30). Cholesterol, synthesized in the sterol branch of the mevalonate pathway (Fig. 1), is needed for virus entry and exit from infected cells (15, 21). A second, nonsterol branch of the mevalonate pathway produces geranylgeranyl and farnesylpyrophosphate isoprenoids, which are often used in posttranslational modification of proteins (Fig. 1). The geranylgeranylated protein FBL2 is also an important host factor for HCV replication (29). Given the importance of mevalonate pathway end products for HCV, it is not surprising that inhibition of this pathway with HMGCR inhibitors, such as lovastatin, inhibits HCV replication. This inhibition can be reversed by supplementation with the downstream biosynthetic intermediates mevalonate and geranylgeraniol.
Because miR-122 plays key roles in both the HCV life cycle and the regulation of the mevalonate pathway, we examined whether miR-122 modulates HCV RNA expression through stimulation of the mevalonate pathway.
Because miR-122 regulates the mevalonate pathway in mouse and nonhuman primate livers (9-11, 23), we examined whether overexpression of miR-122 could modulate HCV and the mevalonate pathway in cultured human liver NNeo/C-5B cells that continuously replicate genotype 1b HCV RNA (17). Briefly, cells were transfected with 50 nM of distinct, duplexed microRNA mimics (20). Total RNA was prepared at 48 h after transfection, and the abundances of specific RNAs were determined by Northern blotting and Storm PhosphorImager analyses (Molecular Dynamics, Sunnyvale, CA) (20) (Fig. 2). As was observed previously (20), overexpression of miR-122 mimics stimulated HCV RNA accumulation relative to the Lipofectamine 2000 control treatment (no RNA) results (Fig. 2A), while transfection of a miR-122 mimic with a mutation at position 3 (122-p3) or an unrelated microRNA let-7 mimic did not enhance HCV RNA abundance (Fig. 2A). In contrast, the ectopic expression of miR-122 did not elicit an appreciable change in the abundance of the mevalonate pathway transcript HMGCR (Fig. 2A). These findings demonstrate that upregulation of HCV RNA abundance by ectopic expression of miR-122 can be uncoupled from effects on HMGCR mRNA abundance in cultured liver cells.
FIG. 2.
Effects of miR-122 on HCV and mevalonate pathway gene transcript abundance. (A) HCV and HMGCR RNA abundance after overexpression of microRNAs in Huh7-5B replicon cells. (B) HCV and HMGCR RNA abundance after overexpression of microRNAs and treatment with lovastatin. (C) HCV and HMGCR RNA abundance after inhibition of miR-122 with 2′O-methylated antisense miR-122 oligonucleotide or a randomized oligonucleotide control. In each panel, RNA abundance was determined in Northern blots and normalized with respect to actin RNA and expressed as the change relative to the result for the Lipofectamine 2000 (no RNA) or Let-7 control. Error bars display the standard error of the mean. 122, wild-type miR-122 duplex RNA; no RNA, Lipofectamine 2000 addition; 122-p3, point mutation at position 3 of the seed sequence in miR-122; 122-p356, point mutations at positions 3, 5, and 6 of the seed sequence in miR-122; 122-p2-8, point mutation from positions 2 through 8 of the seed sequence in miR-122; let7, Let-7a duplex RNA; Random, 2′O-methylated random oligonucleotide; 122as, antisense miR-122 oligonucleotide. Experiments were performed three to seven times in each case. *, P < 0.05; ***, P < 0.001 (Student's t test).
Because HMGCR RNA abundance was not affected by miR-122 overexpression under normal growth conditions, we overexpressed miR-122 in cells that were depleted of isoprenoids by treatment with an HMGCR inhibitor, lovastatin, for 24 h (Fig. 2B). Under these conditions, the miR-122 mimic stimulated HCV RNA accumulation to an extent similar to that observed without lovastatin (compare HCV graphs in Fig. 2A and B). Again, transfection of 122-p3 microRNA mimics did not enhance viral RNA abundance. On the other hand, transfection of miR-122 mimics with mutations at positions 3, 5, and 6 (122-p356) or positions 2 through 8 (122-p2-8) slightly inhibited viral RNA accumulation (Fig. 2B). At 48 h, miR-122 stimulation of HCV was much less pronounced (data not shown), likely a consequence of downregulation on HCV RNA abundance after prolonged lovastatin treatment (Fig. 3A) (22, 31). In contrast to results for non-lovastatin-treated cells (Fig. 2A), cells treated with lovastatin showed a 45% increase in HMGCR RNA abundance in response to ectopic expression of miR-122 compared to the let-7 control duplex (compare HMGCR graphs in Fig. 2A and B). Interestingly, miR-122 duplexes mutated in the seed region also had a stimulatory effect on HMGCR, but to a lesser extent than wild-type miR-122 (Fig. 2B). It is therefore possible that the targeted mRNA in the mevalonate pathway mRNA that is modulated by miR-122 does not depend on an intact microRNA seed sequence-target seed match interaction, as has been observed for microRNA-mRNA interactions (8).
FIG. 3.
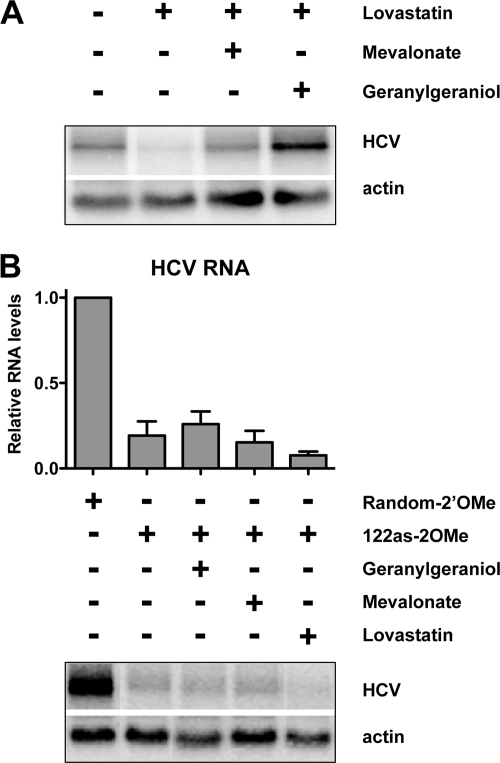
Effects of mevalonate pathway intermediates and miR-122 on HCV RNA abundance in lovastatin-treated cells. (A) Effects of mevalonate pathway intermediates on lovastatin-induced inhibition of HCV. A representative Northern blot is shown. (B) Lack of rescue of HCV RNA abundance by mevalonate pathway intermediates during sequestration of miR-122. The graph shows data from three independent experiments. Error bars display the standard error of the mean. Representative Northern blots for HCV and actin RNAs are shown below. Random-2OMe, 2′O methylated random sequence oligonucleotide; 122as-2OMe, 2′O methylated antisense miR-122 oligonucleotide.
We further tested whether miR-122 could affect mevalonate pathway gene expression by sequestration of miR-122 with antisense oligonucleotides (20). Indeed, a loss of HCV RNA and a moderate but significant decrease of HMGCR mRNA abundance were observed when miR-122 was inhibited (P = 0.026) (Fig. 2C). The approximately twenty percent decrease in HMGCR RNA mirrored what was observed with mouse liver in which miR-122 was sequestered by antisense RNAs (11). Overall, these results indicate that miR-122-modulated HMGCR RNA and HCV RNA accumulation can occur in independent ways in cultured cells.
Because miR-122 inhibition by antisense oligonucleotides led to a decrease in both HCV and HMGCR RNA abundance (Fig. 2C), we tested whether mevalonate pathway inhibition by antisense miR-122 contributed to the decrease in HCV abundance. It is known that HCV RNA abundance is dramatically diminished when the mevalonate pathway is inhibited with lovastatin (16, 22, 31) and that this repression can be rescued by replenishing cells with biosynthetic intermediates of the mevalonate pathway (22, 31). Indeed, the addition of 50 mM lovastatin to NNeo/C-5B cells reduced HCV RNA abundance, and this reduction could be rescued by supplementation of the culture medium with 10 mM mevalonate or 10 μM geranylgeraniol (Fig. 3A).
We next asked whether antisense miR-122 inhibition of HCV could also be rescued with mevalonate pathway intermediates. Untreated and lovastatin-treated cells were transfected with 100 nM concentrations of oligonucleotides complementary to miR-122 (122as-2′OMe), or random sequence 2′O-methylated oligonucleotides (Random-2′OMe), using the Lipofectamine 2000 transfection reagent. Total RNA was prepared at 72 h after transfection, and RNA abundance was examined by Northern blotting. As expected (20), inhibition of miR-122 with antisense oligonucleotides specifically diminished HCV RNA abundance fivefold (Fig. 3B). However, in contrast to what was observed for lovastatin-treated cells, the addition of mevalonate and geranylgeraniol did not rescue HCV RNA abundance to the level observed for the Random-2′OMe-treated control. Furthermore, lovastatin and antisense miR-122 had a combined inhibitory effect on HCV RNA abundance.
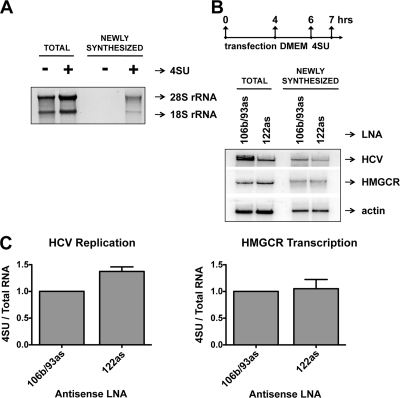
Finally, we examined whether miR-122 upregulated HCV and HMGCR RNA abundance by modulating the rates of HCV RNA synthesis or HMGCR transcription. To simultaneously measure rates of viral and host cell RNA synthesis, we modified a protocol initially described by Cleary and colleagues (6). Briefly, cultured cells were incubated with 4-thiouridine (4SU) for a short period of time. The modified nucleoside is then converted to 4SU-triphosphate, which is incorporated into newly synthesized RNA by both HCV RNA-dependent RNA polymerase NS5B and host DNA-dependent RNA polymerase II. Total RNA was purified, 4SU-labeled RNA was biotinylated, and 4SU-containing RNAs were isolated on streptavidin beads. Total and newly synthesized RNAs were then examined by Northern blotting (Fig. 4A). Figure 4A demonstrates that 4SU incorporation allows for the specific enrichment of newly synthesized rRNA. To test the effects of miR-122 on rates of HCV replication and HMGCR transcription, cells were transfected with 25 nM antisense, locked nucleic acid-modified miR-122 (Antisense LNA) or control antisense LNA that binds miR-106b and miR-93 (Fig. 4B) (10). Cells were incubated in transfection mix for 4 h and then incubated in complete media for 2 h. Pulse labeling was done from 6 to 7 h posttransfection, and newly synthesized RNA was analyzed as described above (Fig. 4B). Northern blotting, which detects both plus- and minus-stranded HCV RNA (Fig. 4B and 4C), showed a decrease in total HCV RNA of approximately 40% after treatment with LNA miR-122 compared to levels for antisense-treated controls. Inspection of newly synthesized HCV RNA after LNA miR-122 treatment revealed that the rate of viral RNA replication was slightly increased at this time (Fig. 4C). These results suggest that effects of miR-122 sequestration on rates of HCV RNA synthesis are moderate and that other posttranscriptional steps that affect HCV RNA turnover may determine the dramatic loss of HCV seen after miR-122 sequestration (Fig. 2C). Such steps may include decreased viral mRNA translation in the absence of miR-122 (13), which could increase the number of replication-competent HCV RNAs, leading to a temporary increase in RNA synthesis. Such an increase in RNA synthesis would not lead to sustained viral RNA abundance if accompanied by other, as-yet-uncharacterized effects of miR-122 sequestration, which could involve mislocalization or decreased RNA protection from innate immune responses. Notably, no significant changes in HMGCR transcription rates were observed in this experimental protocol (Fig. 4B and C), further confirming that miR-122's effects on HCV happen rapidly and independently of its effects on genes that regulate cholesterol synthesis.
FIG. 4.
Effects of miR-122 on rates of HCV RNA replication and HMGCR transcription. (A) Methylene blue stain of a Northern blot of total and streptavidin-enriched (newly synthesized) rRNA isolated from cells incubated in the presence or absence of 4SU. (B) Transfection and pulse-labeling strategy (top). Total RNA abundance and newly synthesized RNAs of HCV, HMGCR, and actin RNA during inhibition of miR-106b/93 or miR-122 by antisense locked nucleic acids (106b/93as or 122as, respectively). A Northern blot is shown (bottom). (C) Quantitation of the Northern blot analyses (Fig. 4B) from three independent experiments. Data are represented as the fraction of newly synthesized RNA (4SU) per amount of template (total RNA) relative to the miR-106b/93 control. Error bars display the standard error of the mean. LNA, antisense, locked nucleic acid oligonucleotide.
Our findings suggest that miR-122 does not affect HCV RNA abundance directly through the mevalonate pathway. It remains possible that miR-122 could regulate HCV RNA abundance in the intact liver through other effects on lipid metabolism. For example, a reduced response rate to antiviral therapy is associated with hepatosteatosis, and antisense miR-122 has been shown to reduce steatosis in mice (1, 11, 26). Therefore, HCV therapy with antisense miR-122 could have a two-pronged effect, acting on underlying host steatosis, in parallel with causing a direct reduction in HCV RNA abundance. Clinical trials are currently under way to test the feasibility of antisense miR-122, as well as statins, for HCV therapy (2, 27). Our results indicate that on a cellular level, miR-122 inhibition and statin treatment act on HCV RNA abundance through independent mechanisms. Notably, the combination of the two inhibitors yields an even greater decrease in viral RNA abundance. This finding could have important consequences for HCV therapy; recent work has revealed that a combination therapy of statins with other antivirals also promotes HCV replicon clearance and, moreover, prevents the emergence of drug-resistant virus, which is a significant obstacle for HCV therapy (7). It is thus encouraging that combination therapy of antisense miR-122 with drugs targeting isoprenoid biosynthesis could give rise to even greater therapeutic effects.
Acknowledgments
We thank members of the Sarnow laboratory and Gusti Zeiner for stimulating discussions and Karla Kirkegaard for helpful comments. We also thank John Boothroyd for miR-106b/93 LNA and Santaris, Inc., for SPC3649 miR-122 LNA.
K.L.N. is supported by a Fellowship from the Alberta Heritage Foundation for Medical Research. P.S. is funded by the National Institutes of Health.
Footnotes
Published ahead of print on 21 October 2009.
REFERENCES
- 1.Asselah, T., L. Rubbia-Brandt, P. Marcellin, and F. Negro. 2006. Steatosis in chronic hepatitis C: why does it really matter? Gut 55:123-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bader, T., J. Fazili, M. Madhoun, C. Aston, D. Hughes, S. Rizvi, K. Seres, and M. Hasan. 2008. Fluvastatin inhibits hepatitis C replication in humans. Am. J. Gastroenterol. 103:1383-1389. [DOI] [PubMed] [Google Scholar]
- 3.Brennecke, J., A. Stark, R. B. Russell, and S. M. Cohen. 2005. Principles of microRNA-target recognition. PLoS Biol. 3:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, J., E. Nicolas, D. Marks, C. Sander, A. Lerro, M. A. Buendia, C. Xu, W. S. Mason, T. Moloshok, R. Bort, K. S. Zaret, and J. M. Taylor. 2004. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter. RNA Biol. 1:106-113. [DOI] [PubMed] [Google Scholar]
- 5.Chekulaeva, M., and W. Filipowicz. 2009. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol. 21:452-460. [DOI] [PubMed] [Google Scholar]
- 6.Cleary, M. D., C. D. Meiering, E. Jan, R. Guymon, and J. C. Boothroyd. 2005. Biosynthetic labeling of RNA with uracil phosphoribosyltransferase allows cell-specific microarray analysis of mRNA synthesis and decay. Nat. Biotechnol. 23:232-237. [DOI] [PubMed] [Google Scholar]
- 7.Delang, L., J. Paeshuyse, I. Vliegen, P. Leyssen, S. Obeid, D. Durantel, F. Zoulim, A. Op de Beeck, and J. Neyts. 2009. Statins potentiate the in vitro anti-hepatitis C virus activity of selective hepatitis C virus inhibitors and delay or prevent resistance development. Hepatology 50:6-16. [DOI] [PubMed] [Google Scholar]
- 8.Doench, J. G., and P. A. Sharp. 2004. Specificity of microRNA target selection in translational repression. Genes Dev. 18:504-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elmén, J., M. Lindow, S. Schutz, M. Lawrence, A. Petri, S. Obad, M. Lindholm, M. Hedtjarn, H. F. Hansen, U. Berger, S. Gullans, P. Kearney, P. Sarnow, E. M. Straarup, and S. Kauppinen. 2008. LNA-mediated microRNA silencing in non-human primates. Nature 452:896-899. [DOI] [PubMed] [Google Scholar]
- 10.Elmén, J., M. Lindow, A. Silahtaroglu, M. Bak, M. Christensen, A. Lind-Thomsen, M. Hedtjarn, J. B. Hansen, H. F. Hansen, E. M. Straarup, K. McCullagh, P. Kearney, and S. Kauppinen. 2008. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 36:1153-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esau, C., S. Davis, S. F. Murray, X. X. Yu, S. K. Pandey, M. Pear, L. Watts, S. L. Booten, M. Graham, R. McKay, A. Subramaniam, S. Propp, B. A. Lollo, S. Freier, C. F. Bennett, S. Bhanot, and B. P. Monia. 2006. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 3:87-98. [DOI] [PubMed] [Google Scholar]
- 12.Reference deleted.
- 13.Henke, J. I., D. Goergen, J. Zheng, Y. Song, C. G. Schuttler, C. Fehr, C. Junemann, and M. Niepmann. 2008. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 27:3300-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoofnagle, J. H. 2002. Course and outcome of hepatitis C. Hepatology 36:S21-29. [DOI] [PubMed] [Google Scholar]
- 15.Huang, H., F. Sun, D. M. Owen, W. Li, Y. Chen, M. Gale, and J. Ye. 2007. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc. Natl. Acad. Sci. U. S. A. 104:5848-5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikeda, M., K.-I. Abe, M. Yamada, H. Dansako, K. Naka, and N. Kato. 2006. Different anti-HCV profiles of statins and their potential for combination therapy with interferon. Hepatology 44:117-125. [DOI] [PubMed] [Google Scholar]
- 17.Ikeda, M., M. Yi, K. Li, and S. M. Lemon. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson, R. J., and N. Standart. 2007. How do microRNAs regulate gene expression? Sci. STKE 2007:re1. [DOI] [PubMed] [Google Scholar]
- 19.Jopling, C. L., S. Schutz, and P. Sarnow. 2008. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe 4:77-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jopling, C. L., M. Yi, A. M. Lancaster, S. M. Lemon, and P. Sarnow. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309:1577-1581. [DOI] [PubMed] [Google Scholar]
- 21.Kapadia, S. B., H. Barth, T. Baumert, J. A. McKeating, and F. V. Chisari. 2007. Initiation of hepatitis C virus infection is dependent on cholesterol and cooperativity between CD81 and scavenger receptor B type I. J. Virol. 81:374-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapadia, S. B., and F. V. Chisari. 2005. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc. Natl. Acad. Sci. U. S. A. 102:2561-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krutzfeldt, J., N. Rajewsky, R. Braich, K. G. Rajeev, T. Tuschl, M. Manoharan, and M. Stoffel. 2005. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438:685-689. [DOI] [PubMed] [Google Scholar]
- 24.Modi, A. A., and J. H. Hoofnagle. 2007. New therapies for hepatitis C. Hepatology 46:615-617. [DOI] [PubMed] [Google Scholar]
- 25.Moradpour, D., F. Penin, and C. M. Rice. 2007. Replication of hepatitis C virus. Nat. Rev. Microbiol. 5:453-463. [DOI] [PubMed] [Google Scholar]
- 26.Poynard, T., V. Ratziu, J. McHutchison, M. Manns, Z. Goodman, S. Zeuzem, Z. Younossi, and J. Albrecht. 2003. Effect of treatment with peginterferon or interferon alfa-2b and ribavirin on steatosis in patients infected with hepatitis C. Hepatology 38:75-85. [DOI] [PubMed] [Google Scholar]
- 27.Santaris Pharma. 2009. Targeting microRNAs using LNA. Santaris Pharma, Hoersholm, Denmark. http://www.santaris.com/Technology,microRNA/Default.aspx.
- 28.Su, A. I., P. G. Schultz, F. V. Chisari, J. P. Pezacki, L. Wodicka, A. D. Brideau, L. Supekova, R. Thimme, S. Wieland, J. Bukh, and R. H. Purcell. 2002. Genomic analysis of the host response to hepatitis C virus infection. Proc. Natl. Acad. Sci. U. S. A. 99:15669-15674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, C., M. Gale, B. C. Keller, H. Huang, M. S. Brown, J. L. Goldstein, and J. Ye. 2005. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol. Cell 18:425-434. [DOI] [PubMed] [Google Scholar]
- 30.Waris, G., D. J. Felmlee, F. Negro, and A. Siddiqui. 2007. Hepatitis C virus induces proteolytic cleavage of sterol regulatory element binding proteins and stimulates their phosphorylation via oxidative stress. J. Virol. 81:8122-8130. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Ye, J., C. Wang, R. Sumpter, M. S. Brown, J. L. Goldstein, and M. Gale. 2003. Disruption of hepatitis C virus RNA replication through inhibition of host protein geranylgeranylation. Proc. Natl. Acad. Sci. U. S. A. 100:15865-15870. [DOI] [PMC free article] [PubMed] [Google Scholar]