Abstract
One of the most characteristic features of African swine fever virus gene expression is its use of two polyproteins, pp220 and pp62, to produce several structural proteins that account for approximately 32% of the total protein virion mass. Equimolecular amounts of these proteins are the major components of the core shell, a thick protein layer that lies beneath the inner envelope, surrounding the viral nucleoid. Polyprotein pp220, which is located immediately underneath the internal envelope, is essential for the encapsidation of the core of the viral particle. In its absence, the infection produces essentially coreless particles. In this study we analyzed, by means of an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible virus, the role of polyprotein pp62 in virus assembly. Polyprotein pp62 is indispensable for viral replication. The repression of polyprotein pp62 expression does not alter late gene expression or the proteolytic processing of the polyprotein pp220. However, it has a profound impact on the subcellular localization of polyprotein pp220. Electron microscopy studies revealed that polyprotein pp62 is necessary for the correct assembly and maturation of the core of the viral particle. Its repression leads to the appearance of a significant fraction of empty particles, to an increase in the number of immature-like particles, and to the accumulation of defective particles. Immunoelectron microscopy analysis showed a clear correlation between the amount of polyprotein pp62, the quantity of polyprotein pp220, and the state of development of the core, suggesting that the complete absence of polyprotein pp62 during morphogenesis would produce a homogenous population of empty particles.
African swine fever virus (ASFV), (39, 44), a large enveloped DNA-containing arbovirus, is the only member of the Asfarviridae family (18). The viral genome is a double-stranded DNA molecule of 170 to 190 kbp in length with hairpin loops and terminal inverted repeats. It carries more than 150 open reading frames (ORFs). These include genes encoding structural proteins; a variety of enzymes involved in DNA replication and repair, gene transcription, and protein modification; proteins potentially involved in the modulation of virus-host interaction; and proteins involved in viral virulence and host range (17, 45). However, approximately 50% of the genes lack any known or predictable function.
Comparative genome analysis suggests that ASFV shares a monophyletic origin with the members of the proposed nucleocytoplasmic large DNA viruses, the four virus families Asfarviridae, Iridoviridae, Phycodnaviridae, and Poxviridae and the yet-unnamed family containing the virus Acanthamoeba polyphaga mimivirus (26, 27). Interestingly, nucleocytoplasmic large DNA viruses are part of the structure-based PRD1-adenovirus viral lineage hypothesis that unites double-stranded-DNA tailless viruses that use similar architectural principles, a trimeric major capsid protein with a double β-barrel structure (32). Members of the PRD1-adenovirus lineage infect cells in all three domains of cellular life.
In its natural hosts, the wild swine warthogs and bushpigs and the argasid ticks of the genus Ornithodoros, ASFV infection is asymptomatic with low-viremia titers, often resulting in persistence (3, 30, 43). In domestic pigs, however, ASFV produces a highly lethal hemorrhagic fever for which animal slaughter and area quarantine are the only methods of disease control (34).
The virus particle has an overall icosahedral shape, with an average diameter of 200 nm (11). It contains more than 50 proteins (22) and consists of several concentric layers enclosing an electron-dense nucleoid (6, 11, 15). The viral core is composed of two domains: the central DNA-containing nucleoid and a surrounding thick protein coat referred to as core shell. The core is enwrapped by an inner lipid envelope (8, 24) that lies beneath the icosahedral capsid (20, 21, 23). Extracellular particles possess an additional envelope derived from the plasma membrane (11).
ASFV particles are formed within perinuclear cytoplasmic viral factories (6, 13, 33) that in several respects resemble aggresomes, perinuclear inclusions containing accumulations of cellular protein aggregates (31). Thus, both ASFV factories and aggresomes are located close to the microtubule-organizing center, exclude cellular organelles, recruit chaperones and mitochondria, and collapse the vimentin network into distinct cage structures (16, 25, 37). The first morphological evidence of viral assembly is the appearance of precursor membranous structures within the viral factories. These viral membranes, which are thought to be derived from the endoplasmic reticulum (8, 38), become icosahedral particles by the progressive edification of the capsid layer at the convex face of the precursor membranes, while simultaneously the developing particles acquire the core material at the concave surface (6, 12, 23). The resulting intracellular mature virions are infectious (9), although their relevance for in vivo infection has not yet been established. A fraction of the intracellular particles reaches the plasma membrane by a microtubule-mediated transport (2, 16, 28) and are released by budding (11) to give rise to the infectious extracellular enveloped virions or are projected out at the tip of long filopodium-like protrusions to enhance cell-to-cell spread (29).
One unusual characteristic of ASFV gene expression is that several of its major structural proteins are synthesized as two large polyprotein precursors. Thus, gene CP2745L encodes polyprotein pp220, which after proteolytic processing gives rise to proteins p150, p37, p34, and p14, and gene CP530R encodes polyprotein pp62, which is processed into the two major core proteins p35 and p15 (41, 42). Equimolecular amounts of the products of these polyproteins are found in the core shell of the viral particle and account for one-third of the total protein content of the ASFV virion (5).
Polyprotein pp220 is a key component of the core shell. When polyprotein pp220 is expressed in transfected cells, it autoassembles to form long (tens of micrometers) and thick (24-nm) coats that are bound, through the N-terminal myristic moiety of polyprotein pp220, to the membranes of diverse cellular compartments (7). The encapsidation of the components of the core, including the viral DNA, depends on the expression of the polyprotein pp220. Thus, when the expression of this protein is repressed by means of an inducible virus, coreless particles accumulate in the viral factory (7). In the current model for virus assembly, core formation begins with the insertion of the N-terminal myristic moiety of polyprotein pp220 at the concave face of the internal envelope, whereas the assembly of the capsid proteins p72 and pB438L occurs simultaneously at the convex face of the envelope.
The role of polyprotein pp62 in the construction of the core shell is less well understood. When a myristoylable polyprotein pp220 and polyprotein pp62 are coexpressed in transfected cells, they assemble into elongated zipper-like structures that resemble the core shell of the viral particle bound to a lipid membrane at each side (5). Polyprotein pp62 is believed to be located at the centers of these structures, interacting with the two opposite layers of membrane-bound polyprotein pp220. This layout is supported by the detection in the middle of these structures of a thin electron-dense line that is also found in the core shell of the ASFV particle, where it is strongly labeled by antibodies against polyprotein pp62 (5).
This report analyzes the role of polyprotein pp62 in ASFV morphogenesis. We describe the construction and properties of a conditional lethal mutant of ASFV with an inducible copy of gene CP530R. The results presented indicate that polyprotein pp62 is essential for the correct assembly of the core of the viral particle. Repression of polyprotein pp62 expression leads to the delocalization of polyprotein pp220 products and to the accumulation in the viral factory of abnormal particles with different levels of core development.
MATERIALS AND METHODS
Cells and viruses.
Vero, Vero C1008, and COS-1 cells were obtained from the American Type Culture Collection and grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum (FCS). The ASFV strain BA71V and the recombinant vGUSREP have been described previously (19, 23).
Antibodies.
The monospecific rabbit polyclonal sera against the polyprotein pp220 products p150, p37/p14, and p34, the polyprotein pp62 product p35, and the major capsid protein p72; the mouse monoclonal antibodies 18HH7 and 24AG4 against polyprotein pp220 product p150 and 17LD3 against protein p72; and the rat polyclonal antibody against protein p54 have been described previously (4, 9, 40, 41).
Plasmid construction.
The intermediate transfer vectors pIND3 and pIND4, designed to allow the inducible expression of a target gene after homologous recombination with virus vGUSREP, have been described previously (20). These plasmids contain a cassette formed by the viral inducible promoter p72.I*, consisting of the strong late promoter p72.4 separated by 2 bp from the core sequence of the Escherichia coli lac operator O1 (10), the lacZ gene under the control of the strong late promoter p72, and two multiple-cloning sites to allow the cloning of the target gene and the corresponding upstream and downstream flanking sequences (23). The plasmids were constructed as follows. A 2.5-kb XbaI-HindIII DNA fragment containing the complete CP530R ORF was excised from pKS-CP530R (4) and inserted into XbaI/HindIII-digested pIND1 to obtain the plasmid pIND1.CP530R.ORF. A synthetic DNA fragment of 987 bp, which contains the nucleotide sequence from position −979 to −15 relative to the translation initiation codon of the CP530R gene, was obtained by PCR using BA71V genomic DNA as a template and the oligonucleotides 5′-GTTGGTACCGAATAATTTTAAAATTGAATGG and 5′-CGAGCGGCCGCTTTGTGTCTACTTTGAAC, which contain KpnI and NotI restriction sites (underlined) at their respective 5′ ends. Plasmid pIND1.pp62 was generated by inserting the KpnI/NotI-digested PCR fragment into the KpnI/NotI-linearized plasmid pIND1.CP530R.ORF. Next, the cassette containing the β-galactosidase marker and the p72.I inducible promoter in this pIND1-derived plasmid was replaced by a similar cassette from pIND3 and pIND4, which contains the more repressive p72.I* inducible promoter. For this, the XbaI/KpnI restriction fragment was excised from pIND1.pp62 and replaced by the XbaI/KpnI fragments from pIND3 and pIND4 to obtain the final transfer vectors pIND3.pp62 and pIND4.pp62, respectively. These vectors differ in the direction of transcription of the lacZ gene with respect to the inducible promoter.
Generation of recombinant virus v62i.
Recombinant viruses were generated essentially as previously described (36) with minor modifications. Briefly, COS cells were infected with virus vGUSREP and transfected with plasmids pIND3.pp62 and pIND4.pp62 in the presence of different concentrations of IPTG (isopropyl-β-d-thiogalactopyranoside). At 48 h postinfection (hpi), the cells were harvested and the recombinant viruses were isolated by sequential rounds of plaque purification in Vero C1008 cells in the presence of IPTG. The inducer concentration at which virus production was maximal, 0.5 mM, was chosen for the growth and purification of the recombinant virus. Similar results were obtained with the two plasmids used, and one virus clone, v62i, obtained from the pIND3.pp62-transfected cells was selected for further characterization.
Plaque assays.
Vero C1008 cell monolayers in six-well plates were infected with 100 PFU of recombinant v62i or parental BA71V. After 1 h, the inoculum was removed and the cells were overlaid with DMEM containing 0.6% Noble agar and 2% FCS in the presence or absence of 0.5 mM IPTG. Seven days later, the medium was removed and the monolayers were stained with 1% crystal violet.
One-step virus growth curves.
COS cell monolayers in 24-well plates were infected with 5 PFU of recombinant v62i or parental BA71V per cell. After a 1-h adsorption, the cells were incubated in DMEM supplemented with 2% FCS. IPTG (0.5 mM) was added immediately after the adsorption period or at 12 or 18 hpi. Infected cells with their culture supernatants were harvested at different times postinfection, sonicated, and titrated by plaque assay in the presence of 0.5 mM IPTG.
Metabolic labeling.
COS cells were mock infected or infected with 5 PFU of BA71V or v62i virus per cell in the absence or in the presence of 0.5 mM IPTG. The cells were pulse-labeled from 16 to 18 hpi with 500 μCi of [35S]methionine-[35S]cysteine (EXPRE35S35S protein labeling mix; Perkin-Elmer Inc.) per ml. Proteins were resolved by sodium dodecyl sulfate (SDS)-12% polyacrylamide gel electrophoresis (PAGE) and detected by autoradiography. Quantification of protein bands was performed with a Bio-Rad GS710 densitometer and Quantity One software (Bio-Rad Laboratories).
Indirect immunofluorescence.
Vero cells grown in coverslips were infected with a multiplicity of infection (MOI) of 1 PFU per cell of recombinant v62i or parental BA71V and maintained in the presence or absence of 0.5 mM IPTG. At the indicated times after infection, the infected cells were fixed for 45 min with 4% paraformaldehyde in 1× phosphate-buffered saline (PBS). Fixed cells were permeabilized for 15 min with 0.1% Triton X-100 in 1× PBS and incubated twice for 10 min in 1× PBS containing 50 mM NH4Cl. Samples were then sequentially blocked for 30 min with blocking buffer (1% cold fish skin gelatin, 0.1% Triton X-100, 10% normal goat serum, 1× PBS) and incubated for 1 h with primary and corresponding secondary antibodies diluted with blocking buffer. The cells were finally mounted with Mowiol/Dabco on glass slides. Preparations were examined using a Zeiss LSM 520 laser scanning confocal microscope. Images were processed using Adobe Photoshop software.
The monoclonal antibodies were used at a dilution of 1/100, and the polyclonal antibodies were used at a dilution of 1/300. The secondary antibodies used were goat anti-rat, goat anti-rabbit, and goat anti-mouse immunoglobulin G antibodies coupled to Alexa 488 or Alexa 555 (1/500; Molecular Probes).
Purification and analysis of viral particles.
The purification of extracellular virus particles from the supernatant of infected cells was done following the procedure described by Carrascosa et al. (14) with minor modifications. COS cells cultured in 150-mm plates (50 plates per virus and condition) were infected with BA71V or with the recombinant v62i or v220i at an MOI of at least 2 PFU per cell. At 39 hpi, culture supernatants were centrifuged at 2,000 rpm for 10 min in a GS3 Sorvall rotor to remove cell debris, and the extracellular virions were concentrated by centrifugation in a GS3 Sorvall rotor at 8500 rpm for 6.5 h at 4°C. Virus pellets were resuspended in 1× PBS and further purified by centrifugation through a 25% sucrose cushion in a Sorvall AH-627 rotor at 25,000 rpm for 1.5 h. The pelleted virus was resuspended in PBS, mixed with Percoll to a final concentration of 45% in PBS, and subjected to equilibrium gradient sedimentation in a TFT 56.13 Sorvall rotor at 20,000 rpm for 30 min at 4°C. Aliquots of the fractions were analyzed for protein content by Western immunoblotting with anti-p150, anti-p35, and anti-p72 antibodies.
Immunoblotting.
Protein samples were electrophoresed in 12% SDS-polyacrylamide gels, transferred to nitrocellulose, and probed with the indicated antibodies against ASFV structural proteins. Protein detection was performed using peroxidase-conjugated antibodies and the ECL system (Amersham Pharmacia Biotech).
Electron microscopy.
For conventional Epon section analysis, COS cells were infected with 10 PFU per cell of BA71V or recombinant v62i in the absence or in the presence of 0.5 mM IPTG and fixed at the indicated times with 2% glutaraldehyde in 200 mM HEPES (pH 7.4) for 1 h at room temperature. Postfixation was carried out with 1% OsO4 and 1.5% K3Fe(CN)6 in H2O at 4°C for 30 min. Samples were dehydrated with acetone and embedded in Epon according to standard procedures.
For freeze-substitution, cells infected as described above were fixed at 24 hpi for 1 h with 4% formaldehyde and 0.1% glutaraldehyde in 200 mM HEPES, pH 7.2, on ice. The specimens were cryoprotected with 30% glycerol for 30 min, rapidly frozen in liquid propane (−180°C), and stored in liquid nitrogen. Freeze-substitution and embedding in Lowicryl K4M were carried out as described previously (6). Ultrathin sections were collected on nickel grids coated with Formvar and carbon and processed for immunogold labeling of ASFV proteins as described previously (6). Specimens were examined at 80 kV in a Jeol 1010 electron microscope.
RESULTS
Construction of a conditionally lethal ASFV mutant with an inducible CP530R gene.
To study the role of polyprotein pp62 in virus replication, we constructed an ASFV recombinant, v62i, in which the expression of the polyprotein-encoding gene CP530R is controlled by the E. coli lac operator/repressor system (Fig. 1A). The starting virus for the construction was vGUSREP, a recombinant virus derived from the BA71V strain that constitutively expresses the E. coli lac repressor (23). To allow the inducible expression of polyprotein pp62, the promoter of gene CP530R was replaced in vGUSREP by the inducible ASFV promoter p72.I*, composed of the strong late viral promoter p72.4 separated by 2 bp from the operator sequence O1. This reduced distance between operator and promoter results in a strong repression of gene expression (23).
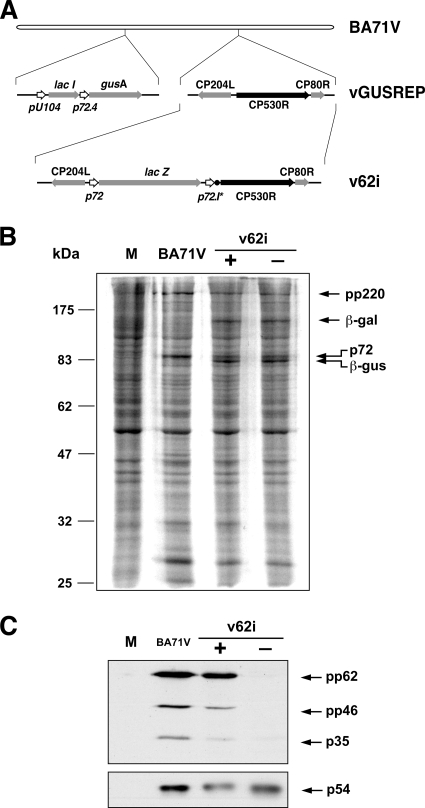
FIG. 1.
Construction and characterization of v62i virus. (A) Genomic structure of the ASFV recombinant v62i. The inducible virus v62i derives from the intermediate virus vGUSREP, which constitutively expresses the lacI gene encoding the Lac repressor. In v62i, the gene CP530R is under the control of the inducible promoter p72.I*, consisting of the synthetic late promoter (p72.4) followed by the lac operator sequence (•). The reporter genes lacZ and gusA, used for the selection of the recombinants, are also shown. (B) Protein profile of cells infected with the recombinant v62i. COS cells were mock infected (lane M) or infected with the parental BA71V or with the recombinant v62i in the presence (+) or absence (−) of 0.5 mM IPTG. The cells were pulse-labeled with [35S]methionine-[35S]cysteine from 16 to 18 hpi, lysed, resolved by SDS-PAGE, and detected by fluorography. The positions of the molecular weight markers, the polyprotein pp220, the major capsid protein p72, and the reporter proteins β-galactosidase (β-gal) and β-glucoronidase (β-gus) are indicated. (C) IPTG-dependent expression of polyprotein pp62. COS cells were mock infected (lane M) or infected with the parental BA71V or with the recombinant v62i in the presence (+) or absence (−) of 0.5 mM IPTG. At 12 hpi, the cells were lysed, resolved by SDS-PAGE, and analyzed by Western blotting with antibodies against the polyprotein pp62-derived product p35 and the protein p54 as a loading control. Arrows indicate the positions of protein p54, polyprotein pp62, the intermediate processing product polyprotein pp46, and the final product p35.
Inducible expression of polyprotein pp62.
To confirm that the expression of polyprotein pp62 by v62i can be regulated by the inducer IPTG, cells infected with the recombinant virus under permissive or nonpermissive conditions were labeled with [35S]methionine-[35S]cysteine from 16 to 18 hpi, a period in which late gene expression is in progress. Mock- and BA71V-infected cells labeled under the same conditions were used as negative and positive controls, respectively. As shown in Fig. 1B, similar overall protein profiles were observed when parental BA71V and recombinant v62i infections were compared, with the exception that extra bands corresponding to the reporters β-glucuronidase and β-galactosidase were detected in the cells infected with the inducible recombinant. Since polyprotein pp62 and its proteolytic products are not clearly detectable in total cell extracts, its expression was analyzed by Western blotting using antibodies against protein p35, one of the end products of the processing of this polyprotein. As shown in Fig. 1C, polyprotein pp62 synthesis was drastically abrogated under nonpermissive conditions. Densitometric quantification revealed that under permissive conditions, the expression level of polyprotein pp62 was similar to that observed in control BA71V infections, whereas in the absence of IPTG, it was reduced to 2.7% of the control value.
Inducer dependence of recombinant v62i.
The effect of the repression of polyprotein pp62 expression on the ability of the recombinant virus to form lysis plaques was demonstrated by plaque assay (Fig. 2A). Under permissive conditions, the numbers of lysis plaques were similar for both parental and recombinant viruses, although the plaque size was slightly smaller in v62i infections. The omission of IPTG led to a dramatic decrease in the number of plaques formed by the recombinant virus compared with control BA71V virus.
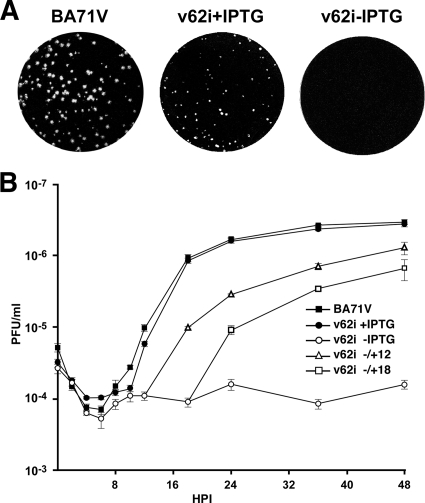
FIG. 2.
v62i is a lethal conditional mutant. (A) Effect of IPTG on plaque formation. Vero C1008 cell monolayers were infected with the parental BA71V or v62i in the presence or absence of 0.5 mM IPTG. After 7 days, plaques were visualized using crystal violet. (B) One-step growth curves of v62i. COS cells were infected with 5 PFU per cell of BA71V or v62i. At the indicated times after infection, the total virus titer of each sample was determined by plaque assay. The recombinant virus was grown in the presence (+IPTG) or absence (−IPTG) of 0.5 mM IPTG or was grown under restrictive conditions for 12 (−/+12) or 18 (−/+18) hpi and then induced with 0.5 mM IPTG. The results are represented as the means from three independent experiments. Error bars indicate the standard errors of the means.
The reduction in the number of lysis plaques observed under restrictive conditions could indicate a role for polyprotein pp62 in the dissemination of the virus or in viral replication. To clarify this, one-step growth curves were performed by infecting cells at a MOI of 5 PFU/cell with BA71V or with v62i in the presence or absence of 0.5 mM IPTG and analyzing the total virus production. Figure 2B shows that under permissive conditions, the growth curve of the recombinant virus is slightly delayed compared with that of the parental virus, although the finals titer, obtained at 48 hpi, are very similar in both viruses. Without the inducer, however, the total virus yield obtained in v62i infections did not increase significantly with time, with a difference in total virus production of 2 log units from 18 hpi onwards and a maximal difference of ca. 2.5 log units at 36 hpi.
This result indicates that the phenotype observed is not due to an effect on the capability of the virus to spread from cell to cell; instead, the absence of the inducer produces a strong inhibition of virus replication.
We next analyzed the ability of cells infected with v62i and grown for different times under nonpermissive conditions to produce infectious particles upon IPTG addition. The reduction in the final titer observed in these postinduction experiments was proportional to the length of the incubation period under restrictive conditions (Fig. 2B). Thus, when IPTG was added at 12 hpi, an early time for virus assembly (6, 13), the maximal virus yield, reached at 48 hpi, was about 46% of that observed for v62i grown under permissive conditions throughout the infection, and when the inducer was added at 18 hpi, the maximal yield at 48 hpi was 24% of that obtained in the control.
Altogether, the plaque assay and one-step growth experiments indicate that polyprotein pp62 is essential for viral replication and that the recombinant v62i is an IPTG-dependent lethal conditional mutant.
Processing of polyprotein pp220 is independent of the expression of polyprotein pp62.
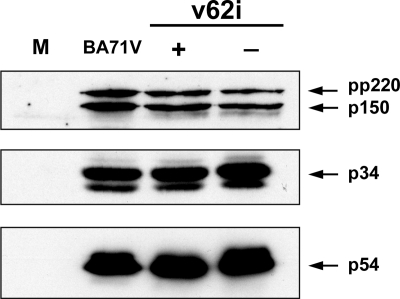
Using the inducible virus v220i, in which the expression of polyprotein pp220 is regulated by IPTG, we have previously shown that the proteolytic processing of polyprotein pp62 requires the expression of polyprotein pp220 (5). We were interested in determining in detail the effect that the repression of polyprotein pp62 has on the expression and proteolytic processing of polyprotein pp220. Polyprotein processing was monitored by Western immunoblotting of extracts of v62i-infected cells maintained under permissive or restrictive conditions for 12 h using antibodies specific for polyprotein pp220 and its products p150 and p34 (Fig. 3). As a control, uninfected cells or cells infected with the parental BA71V were analyzed as described above. Also, the expression of protein p54 was analyzed for reference. As we can observe in Fig. 3, the repression of polyprotein pp62 expression has no detectable effect on the expression or processing of polyprotein pp220.
FIG. 3.
Proteolytic processing of polyprotein pp220 in v62i-infected cells. COS cells were mock infected (lane M) or infected with the parental BA71V or with the recombinant v62i in the presence (+) or absence (−) of 0.5 mM IPTG. At 12 hpi, the cells were lysed, resolved by SDS-PAGE, and analyzed by Western blotting using antibodies against the polyprotein pp220-derived products p150 and p34 and the protein p54 as a loading control. Arrows indicate the positions of protein p54, polyprotein pp220, and the products p150 and p34.
Repression of polyprotein pp62 alters the subcellular localization of polyprotein pp220.
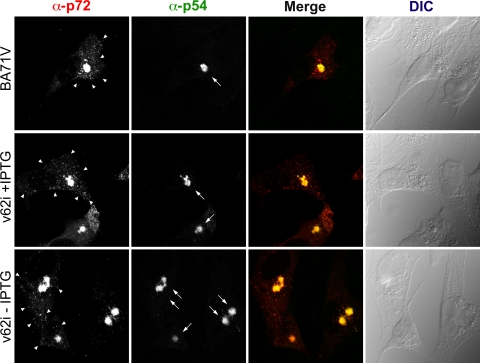
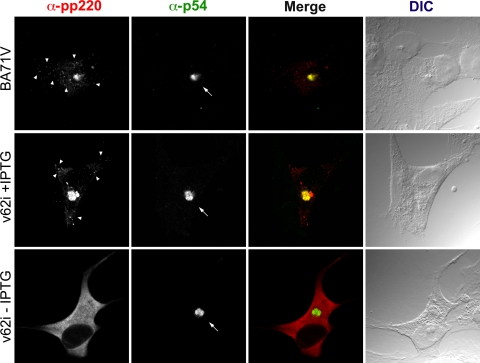
Since polyproteins pp62 and pp220 assemble into the core shell, it seems reasonable to think that although the repression of polyprotein pp62 does not affect the expression and processing of polyprotein pp220, it could have an important impact on its localization. To evaluate this hypothesis, we studied by confocal immunofluorescence the effect of the repression of polyprotein pp62 on the localization of polyprotein pp220. As can be seen in Fig. 4, very similar immunofluorescence profiles were observed when cells infected with the parental BA71V or with the recombinant virus and maintained during 12 h under permissive or restrictive conditions were analyzed by confocal microscopy using antibodies specific for p72, the major component of the icosahedral capsid, or p54, a transmembrane protein that localizes at the inner envelope of the viral particle. Thus, under both conditions, the fluorescence associated with these two antibodies strongly labeled the viral factory, which appears to be normal in size and location. Furthermore, a punctate pattern, characteristic of viral particles, can be detected with the anti-p72 antibody independently of the presence of the inducer. However, when the monoclonal antibody 18HH7, specific for polyprotein pp220, was used to analyze the distribution of this protein in infected cells (Fig. 5), it was evident that the repression of polyprotein pp62 has a strong influence on the localization of polyprotein pp220. Thus, the strong fluorescent signal that under permissive conditions is associated mainly with the viral factory and faintly with viral particles is located, under restrictive conditions, dispersed uniformly throughout the cytoplasm of the infected cell, excluding the nucleus. This unexpected behavior was verified using a different monoclonal antibody specific for polyprotein pp220 (24AG4) and a rabbit polyclonal antibody against polyprotein pp220/p150 (not shown).
FIG. 4.
Immunofluorescence microscopy analysis of the subcellular localization of the capsid protein p72 in v62i-infected cells. Vero cells were fixed at 12 hpi with the parental BA71V or with v62i in the presence or absence of 0.5 mM IPTG. Samples were incubated with antibodies against protein p72 or the inner envelope protein p54, which was used as a marker for the viral factory. Antibodies were detected by using secondary antibodies coupled to Alexa 555 and Alexa 488, respectively. Differential interference contrast (DIC) microscopy of the samples is shown at the right. Arrows and arrowheads indicate the positions of the viral factories and viral particles, respectively.
FIG. 5.
Immunofluorescence microscopy analysis of the subcellular localization of polyprotein pp220 in v62i-infected cells. Vero cells were fixed at 12 hpi with the parental BA71V or v62i in the presence or absence of 0.5 mM IPTG. Samples were incubated with the monoclonal antibody 18HH against the polyprotein pp220/p150 or with an antibody against the inner envelope protein p54, which was used as a marker for the viral factory. Antibodies were detected by using secondary antibodies coupled to Alexa 555 and Alexa 488, respectively. Differential interference contrast (DIC) microscopy of the samples is shown at right. Arrows and arrowheads indicate the positions of the viral factories and viral particles, respectively.
Morphogenesis of v62i virus under nonpermissive conditions.
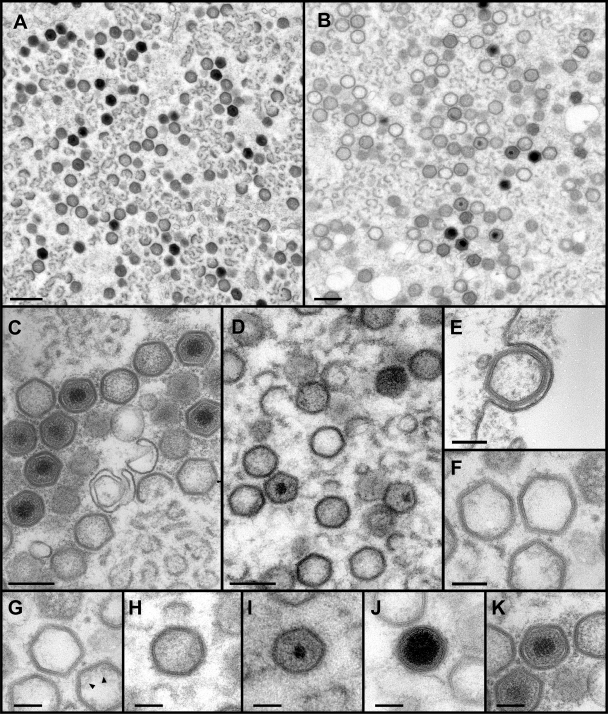
We have previously mentioned that when polyprotein pp220 is repressed, the viral morphogenesis results in the production of particles that lack the core. The abnormal localization of this polyprotein in cells infected with v62i under restrictive conditions and the presence of a punctate pattern in the immunofluorescence using antibodies against the capsid protein p72 suggest a similar phenotype when the expression of polyprotein pp62 is repressed. To investigate in detail the effect of polyprotein pp62 repression on virus morphogenesis, v62i-infected cells maintained for 24 h under permissive or nonpermissive conditions were analyzed by electron microscopy along with cells infected by the parental virus BA71V (Fig. 6). When factories produced by the infection with BA71V (Fig. 6A) or with v62i under permissive conditions (Fig. 6C) are compared with those produced by v62i under restrictive conditions (Fig. 6B), we can observe that the numbers of viral membrane precursors, icosahedral intermediates, and viral particles in different stages of maturation are very similar in both cases, indicating that the repression of polyprotein pp62 synthesis did not apparently affect the assembly of the inner viral envelope and the outer capsid. However, the relative abundance of the particles in each of the different stages of maturation is clearly very different. We wanted to obtain a quantitative evaluation of the different types of particles observed in the factories under the different conditions. For this, we first needed to establish a nomenclature for the different types of particles observed. To ensure that we were comparing similar sections of complete particles, we analyzed particles displaying a hexagonal outline. Although it is impossible to distinguish precisely among the continuum of electron densities displayed by the ASFV viral particles as they progress toward the final stages of assembly, we can clearly differentiate between two extremes of electron density: (i) empty particles (Fig. 6G), which are virtually devoid of core material or, in some cases, display small amounts of core material apparently bound to the internal envelope and are very similar to the particles produced when the expression of polyprotein pp220 is repressed (Fig. 6F), and (ii) particles that contain an electron-dense nucleoid that can be mature, containing a well-centered nucleoid surrounded by a dense core shell of uniform thickness (Fig. 6K), or defective, with an overdeveloped (Fig. 6J) or underdeveloped (Fig. 6I) nucleoid, usually noncentered, surrounded by an abnormal core shell. Particles that are not empty and do not contain a nucleoid are designated immature (Fig. 6H). Table 1 shows a quantification of the relative abundance of each type of particle in factories of cells infected with BA71V or with the recombinant v62i and maintained for 24 hpi under permissive or restrictive conditions. The presence of similar numbers of mature and immature icosahedral particles is characteristic of the viral factories of cells infected with BA71V or with the recombinant virus under permissive conditions. Empty or defective particles are rarely detected. In contrast, when the infection with v62i under restrictive conditions is examined under similar conditions, empty particles account for approximately 18% of the virus particles detected, and nearly all the particles with nucleoids are defective particles (17%). These defective particles are very similar to the particles produced when the expression of the viral proteinase is blocked (1); however, the defective particles formed when the expression of polyprotein pp62 is repressed show a high variability in the size of the nucleoid and a less developed core shell domain, with a reduced electron density. The remaining 65% of the particles analyzed were classified as immature-like particles, in contrast with the 52% found in factories of cells infected with BA71V or the 51% found in factories of cells infected with v62i under permissive conditions.
FIG. 6.
Effect of polyprotein pp62 repression on virus assembly. Electron microscopy of COS cells infected with BA71V (A), v62i (B to E and G to K), and v220i (F) and maintained in the presence (C and K) or absence (A, B, D, E, F, and G) of 0.5 mM IPTG is shown. The samples were processed at 24 hpi by conventional Epon embedding. Whereas factories of parental virus (A) or v62i under permissive conditions (C) show essentially mature (K) and immature (H) viral particles, factories of v62i grown in the absence of the inducer (B and D) display a heterogeneous population of immature (H), empty (G), and defective (I and J) icosahedral particles. Empty particles appear to be devoid of core material or occasionally display some electron density close to the inner envelope (arrowheads in panel G). They are very similar to the particles produced when the expression of polyprotein pp220 is repressed (F). All types of icosahedral particles, including empty particles (E), are released from the infected cell. Bars, 1 μm (A and B), 200 nm (C and D), and 100 nm (E, F, G, H, I, J, and K).
TABLE 1.
Quantification of the different types of particles within factories of cells infected with the indicated virusesa
| Virus | % (mean ± SEM) of particles |
|||
|---|---|---|---|---|
| Empty | Immature | Defective | Mature | |
| BA71V | 0.08 ± 0.09 | 52.39 ± 3.16 | 2.61 ± 0.57 | 45.14 ± 3.09 |
| v62i | ||||
| With IPTG | 1.33 ± 0.89 | 51.78 ± 3.48 | 3.18 ± 1.00 | 43.51 ± 3.86 |
| Without IPTG | 18.23 ± 1.03 | 64.85 ± 1.78 | 16.62 ± 1.14 | 0.64 ± 0.15 |
Thin sections of COS cells infected with BA71V or with v62i, maintained for 24 h in the presence or absence of IPTG, and fixed at 24 hpi were examined under the electron microscope. For each factory the percentages of empty, immature, defective, and mature particles were estimated. More than 1,000 particles were counted for each virus and condition in at least 25 different factories.
Altogether, these observations indicate that polyprotein pp62 is essential for the correct assembly and maturation of the core of the viral particle. The repression of this polyprotein leads to the appearance of a significant fraction of empty particles, to an increase in the number of immature-like particles, and to the accumulation of defective particles.
Immunoelectron microscopy of intracellular defective v62i particles.
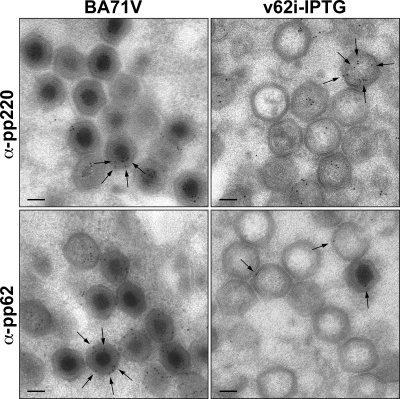
The presence of a heterogeneous population of unusual particles (empty and defective) when the expression of polyprotein pp62 is repressed raises the question of which type of structure is the real phenotype due to the lack, during its assembly, of polyprotein pp62. One possibility is that empty particles are formed when no or very small amounts of polyprotein pp62 are available and that immature-like and defective particles were the result of the presence of small quantities of polyprotein pp62 during assembly. Moreover, when polyprotein pp220 expression is repressed, empty particles, structurally very similar to those found under restrictive conditions in v62i infections, accumulate in the viral factory. It seems clear, then, that some amount of polyprotein pp220 must be present in the immature particles and in the defective particles found in cells infected with v62i in the absence of inducer. It is also likely that the presence of polyprotein pp220 in the empty particles will be minimal, if it is present at all. To verify these hypotheses, we quantified the amounts of polyproteins pp62 and pp220 present in the different types of particles by immunoelectron microscopy. For this, cells infected with BA71V or with the recombinant v62i under restrictive conditions were processed at 24 hpi by freeze-substitution followed by Lowicryl embedding. Ultrathin sections from the samples were then incubated with antibodies against polyproteins pp220 and pp62, followed by protein A-gold. Table 2 shows a quantification of the labeling density obtained for each antibody on the mature, defective, immature, and empty icosahedral particles produced by the different viruses under the different conditions. Examples of the labeling patterns for these proteins are illustrated in Fig. 7. From the study of the data presented in Table 2, we can conclude that there is a direct correlation for each type of particle between the amount of polyprotein pp62, the quantity of polyprotein pp220, and the state of development of the core of the particle. Thus, empty particles contain trace amounts of polyprotein pp62 (0.7% of the label corresponding to polyprotein pp62 detected by immunoelectron microscopy in a BA71V mature particle [Table 2]) and very little polyprotein pp220 (6.6%) and appear to be essentially devoid of core material, occasionally displaying some electron density close to the inner envelope. Immature particles, which display a uniformly electron-dense core, contain small amounts of polyprotein pp62 (3.8%) (considerably less than a normal immature particle [59.5%]) and contain 40.1% of the polyprotein pp220 (compared with 58.5% for a normal immature particle). Finally, defective particles contain the highest levels of both polyprotein pp62 (17.0%) and polyprotein pp220 (60.9%) and consequently display a more developed core, although both the core shell and the nucleoid of these defective particles are abnormal: the nucleoid appears over- or underdeveloped, and the core shell is irregular and displays a lower electron density than a mature particle. It is important to note that these highest levels are considerably lower than the levels found in the BA71V mature particles.
TABLE 2.
Quantification of polyproteins pp62 and pp220 within the different types of virus particles by immunoelectron microscopya
| Antibody and virus | Mean no. ± SEM/% of gold grains in particles |
|||
|---|---|---|---|---|
| Empty | Immature | Defective | Mature | |
| Anti-pp62 | ||||
| BA71V | ND | 3.37 ± 0.28/59.5 | ND | 5.67 ± 0.36/100 |
| v62i (without IPTG) | 0.04 ± 0.02/0.7 | 0.22 ± 0.03/3.8 | 0.96 ± 0.16/17.0 | ND |
| Anti-pp220 | ||||
| BA71V | ND | 3.82 ± 0.34/58,5 | ND | 6.53 ± 0.40/100 |
| v62i (without IPTG) | 0.43 ± 0.08/6.6 | 2.62 ± 0.19/40.1 | 3.98 ± 0.36/60.9 | ND |
Immunogold labeling was performed as described in the legend to Fig. 7. Quantification was performed for closed hexagonal profiles at the assembly sites. For each condition and antibody, gold grains within at least 100 particles of each type were counted. Percentages are with respect to the mean number of gold grains detected in the mature BA71V particles. ND, not determined.
FIG. 7.
Immunoelectron microscopy of v62i virus particles. COS cells were infected with BA71V and with the recombinant v62i in the absence of IPTG. At 24 hpi, the cells were fixed and processed by freeze-substitution. Ultrathin Lowicryl sections were incubated with antibodies against the polyprotein pp220-derived protein p150 (pp220) and polyprotein pp62-derived protein p35 (pp62), followed by incubation with protein A-gold (10-nm diameter). Gold grains are indicated by arrows. Bars, 100 nm.
These observations indicate that the presence of polyprotein pp62 is necessary for the recruitment or the retention of polyprotein pp220 in the assembling particle. Also, the data show a positive relationship between the amount of polyprotein pp62 present in the particle and the degree of core development found, suggesting that the complete absence of polyprotein pp62 during morphogenesis would produce a homogenous population of empty particles.
Purification and analysis of extracellular v62i aberrant particles.
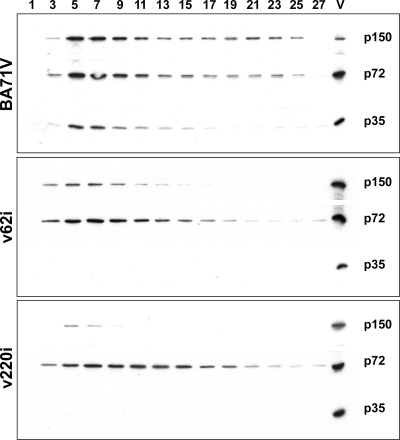
To further characterize the aberrant particles produced by the infection of v62i virus under restrictive conditions, we purified them from the supernatants of cells infected with the recombinant virus and maintained in the absence of IPTG for 39 h, a time when extracellular virus production has not reached the maximum but there is a small amount of cellular debris in the supernatant of the cultures. As a reference for the content in the proteolytic products of the polyprotein pp220, we prepared under similar conditions particles from the supernatants of cells infected with the recombinant v220i and maintained in the absence of IPTG (negative control) or infected with the parental BA71V (positive reference). Extracellular virions collected from the culture media were sedimented into an isopycnic Percoll gradient as previously established (14). The gradients were then fractionated from the bottom, and each fraction was evaluated by Western immunoblotting with antibodies against protein p72, the polyprotein pp220-derived protein p150, and the polyprotein pp62-derived protein p35. As shown in Fig. 8, the amount of p150 detected in the particles corresponding to the v62i infections is smaller than that detected in the parental virus but significantly larger that the amount found in the particles purified from v220i infections, which correspond mainly to empty particles. This result supports the previous results obtained by immunoelectron microscopy indicating the presence of a heterogeneous population of particles, none of which contains a normal amount of polyprotein pp220, and also agrees with the immunofluorescence microscopy results indicating that a significant proportion of the polyprotein products is out of the normal morphogenetic pathway.
FIG. 8.
Analysis of extracellular v62i virus particles. Extracellular virions collected from clarified culture supernatants of infections with BA71V or with v62i and v220i under restrictive conditions were semipurified by using a sucrose cushion, adjusted to the same p72 content, and purified by equilibrium sedimentation in a Percoll gradient. Aliquots of the gradient fractions were analyzed by Western immunoblotting with antibodies against protein p72, polyprotein pp220-derived protein p150, and polyprotein pp62-derived protein p35. The positions of proteins p150, p72, and p35 are indicated.
On the other hand, the fact that the aberrant particles are released to the extracellular medium indicates that this mutant phenotype is irreversible and that in postinduction experiments the newly synthesized polyprotein pp62 has no access to the previously assembled particles. We therefore interpret the partial restoration of infectivity observed after IPTG addition (Fig. 2B) as a consequence of the de novo assembly of mature virions from precursor membranes.
DISCUSSION
The results presented in this work indicate that polyprotein pp62 has an essential role in the assembly of the core of the ASFV particle. Using a conditional lethal mutant of ASFV with an IPTG-inducible copy of gene CP530R, we show that polyprotein pp62 is essential for viral replication. The repression of polyprotein pp62 expression does not have a significant impact on the synthesis of viral structural proteins such as p72, p54, or polyprotein pp220 and does not alter the size or location of the viral factory. These results indicate that DNA replication and late gene expression are not significantly affected under restrictive conditions. However, the subcellular location of polyprotein pp220 strongly depends on the expression of polyprotein pp62. Thus, the repression of polyprotein pp62 leads to an even distribution of polyprotein pp220 in the cytoplasm of the infected cell, in contrast with its accumulation at the viral factory observed under permissive conditions. Surprisingly, despite this abnormal localization, proteolytic processing of polyprotein pp220 by pS273R, the viral protease, appears to occur normally.
When studied by electron microscopy, the number of membrane precursors, viral icosahedral intermediates, and icosahedral particles found at the viral factories of cells infected by v62i were very similar independently of the conditions studied. However, the repression of polyprotein pp62 expression produces a profound effect on the development state of the core of the icosahedral particles. Under restrictive conditions, very few normal mature particles are found, in agreement with the observed reduction in the number of infectious particles produced. There is an accumulation of immature-like particles compared with the case for the viral factories produced under permissive conditions or by infection with BA71V. Empty and defective particles, which are rarely detected under permissive conditions, accumulate in the absence of the inducer. These are abnormal particles that show profound defects in the development of the viral core. When the contents of these particles in polyproteins pp62 and pp220 were analyzed by immunoelectron microscopy, a clear correlation was found between the amount of polyprotein pp62, the quantity of polyprotein pp220, and the state of development of the core of the particle. The lowest levels of polyproteins pp62 and pp220 are found in the empty particles, which are virtually devoid of core material. An increase in the label of both polyproteins is found in immature particles, which show a core of homogeneous electron density. The highest level of label for both polyproteins is associated with defective particles, which display an aberrant nucleoid and whose core shell is irregular, with an electron density lower than that of a mature particle. These results indicate that the expression of polyprotein pp62 is necessary for the recruitment or the retention of polyprotein pp220 in the assembling particle. The strong correlation found between the amount of polyprotein pp62 present in a particle and its degree of core development suggests that the complete absence of polyprotein pp62 during morphogenesis would produce a homogenous population of empty particles. We interpret the presence of the other types of particles detected as the result of repression system leakage, since they contain various amounts of polyprotein pp62. These results agree with the proposed role for polyprotein pp62 as an organizer of the interactions between the polyprotein pp220 layers that compose the core shell of the viral particle and indicate that although equimolecular amounts of polyprotein pp62- and polyprotein pp220-derived products are required for the correct assembly of a mature ASFV particle, very small amounts of polyprotein pp62 suffice for the initiation of core formation.
In our current model for the assembly of the viral particle, core formation begins with the interaction of the N-terminal myristic moiety of polyprotein pp220 with the internal face of the inner envelope. The fact that polyproteins pp220 and pp62 autoassemble in transfected cells in a structure very similar to the authentic core shell suggests that further core encapsidation is driven by protein-protein interactions between polyproteins pp220 and pp62 to construct the core shell, the matrix-like protein layer that surrounds the nucleoid of the viral particle. Polyprotein pp62 is believed to be located at the center of the core shell, interacting with two opposite layers of polyprotein pp220. This layout is supported by the detection in the middle of the core shell of a thin electron-dense line that is strongly labeled by antibodies against polyprotein pp62 (5). Further core encapsidation will be mediated by the interactions of the inner layer of polyprotein pp220 with other core material. In this model, the absence of polyprotein pp62 will block the core encapsidation after the assembly of the first layer of polyprotein pp220 under the inner envelope, and since the assembly of the external domains of the particle occurs independently of core encapsidation, this will result in particles containing only a layer of polyprotein pp220 bound to the inner envelope of the particle.
However, the results presented here show a discrepancy with the model in the amount of polyprotein pp220 present in the empty particles. We have found that empty particles do not contain significant amounts of polyprotein pp220, whereas the model suggests that the incorporation of the first layer of polyprotein pp220 would be independent of the presence of polyprotein pp62.
This discrepancy may be explained if we examine the proteolytic processing of polyprotein pp220. Surprisingly, we have found that the proteolytic processing of polyprotein pp220 is not affected by the repression of polyprotein pp62 expression, although under these conditions polyprotein pp220 is mislocalized and accumulates at the cytoplasm of the infected cell. One interesting question, which is germane to the explanation of the discrepancy in the amount of polyprotein pp220 found in the viral particles, is whether the delocalization of polyprotein pp220 to the cytoplasm occurs after the protein has been previously localized in an assembling particle. The fact that polyprotein pp220 is proteolytically processed at a normal extent strongly argues in favor of this hypothesis, because the processing of ASFV polyproteins is tightly associated with virus assembly. Experiments involving pulse-chase followed by subcellular fractionation indicate that the proteolytic processing of polyproteins pp220 and pp62 occurs after the association of the viral polyproteins with membranes, which in the case of polyprotein pp62 probably occurs by its association with polyprotein pp220 (5). This dependence on the encapsidation for the processing of polyproteins is further supported by the finding that when the assembly of icosahedral particles is blocked by repressing the expression of some structural or nonstructural proteins (p72, p54, or pB602L), the two viral polyproteins, which are localized into the so-called zipper-like structures or inclusion bodies, are not processed (20, 23, 35). Furthermore, when polyprotein pp220 is repressed, polyprotein pp62, which is not incorporated in the coreless particles produced, remains unprocessed (7). The surprising finding that the repression of polyprotein pp62 does not affect the processing of polyprotein pp220 argues in favor of the idea that the delocalization of the protein is posterior to its encapsidation, suggesting that polyprotein pp62 is required neither for the targeting of polyprotein pp220 to the precursor membranes nor for its processing during assembly but that it is essential to mediate the interactions that allow the stabilization of the processed products during assembly. Thus, we propose that during the formation of the empty particles, polyprotein pp220 interacts with the inner envelope through its N-terminal myristic acid and is processed by the pS273R protease, but the near absence of polyprotein pp62 blocks any further assembly, leaving polyprotein pp220 products as the only core components associated with the precursor developing particle; eventually, these proteins will move out of the assembling particle and will disperse throughout the cytoplasm of the infected cell.
Although the results presented in this report have been interpreted within the model for the core shell proposed by Andrés et al. (5), in which the structure of the core shell is seen as two layers of polyprotein pp220 bound by a central sheet of polyprotein pp62, our results do not directly support or negate this model. Equally well suited for the interpretation of our results could have been a model in which the layer of polyprotein pp220 bound to the inner envelope interacts with polyprotein pp62, which in turn directly recruits further core components. Further experimental evidence is necessary to understand the true architecture of the core shell domain of the ASFV viral particle.
Acknowledgments
We thank E. M. Forero, M. Guerra, and M. Rejas for technical assistance.
This work was supported by grants from the Wellcome Trust (075813/C/04/Z) and the Spanish Ministerio de Ciencia e Innovación (BFU2007-61647) and by an institutional grant from Fundación Ramón Areces. J. M. Rodríguez was supported by the Ramón y Cajal program of the Ministerio de Ciencia e Innovación.
Footnotes
Published ahead of print on 21 October 2009.
REFERENCES
- 1.Alejo, A., G. Andrés, and M. L. Salas. 2003. African swine fever virus proteinase is essential for core maturation and infectivity. J. Virol. 77:5571-5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alves de Matos, A. P., and Z. G. Carvalho. 1993. African swine fever virus interaction with microtubules. Biol. Cell 78:229-234. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, E. C., G. H. Hutchings, N. Mukarati, and P. J. Wilkinson. 1998. African swine fever virus infection of the bushpig (Potamochoerus porcus) and its significance in the epidemiology of the disease. Vet. Microbiol. 62:1-15. [DOI] [PubMed] [Google Scholar]
- 4.Andrés, G., A. Alejo, C. Simón-Mateo, and M. L. Salas. 2001. African swine fever virus protease: a new viral member of the SUMO-1-specific protease family. J. Biol. Chem. 276:780-787. [DOI] [PubMed] [Google Scholar]
- 5.Andrés, G., A. Alejo, J. Salas, and M. L. Salas. 2002. African swine fever virus polyproteins pp220 and pp62 assemble into the core shell. J. Virol. 76:12473-12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrés, G., C. Simón-Mateo, and E. Viñuela. 1997. Assembly of African swine fever virus: role of polyprotein pp220. J. Virol. 71:2331-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrés, G., R. García-Escudero, M. L. Salas, and J. M. Rodríguez. 2002. Repression of African swine fever virus polyprotein pp220-encoding gene leads to the assembly of icosahedral core-less particles. J. Virol. 76:2654-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrés, G., R. García-Escudero, C. Simón-Mateo, and E. Viñuela. 1998. African swine fever virus is enveloped by a two-membraned collapsed cisterna derived from the endoplasmic reticulum. J. Virol. 72:8988-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrés, G., R. García-Escudero, E. Viñuela, M. L. Salas, and J. M. Rodríguez. 2001. African swine fever virus structural protein pE120R is essential for virus transport from the assembly sites to the plasma membrane but not for infectivity. J. Virol. 75:6758-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Betz, J. L., and J. R. Sadler. 1981. Variants of a cloned synthetic operator. I. A palindromic dimer lactose operator derived from one strand of the cloned 40-base pair operator. Gene 13:1-12. [DOI] [PubMed] [Google Scholar]
- 11.Breese, S. S. Jr., and C. J. DeBoer. 1966. Electron microscope observation of African swine fever virus in tissue culture cells. Virology 28:420-428. [DOI] [PubMed] [Google Scholar]
- 12.Brookes, S. M., A. D. Hyatt, T. Wise, and R. M. E. Parkhouse. 1998. Intracellular virus DNA distribution and the acquisition of the nucleoprotein core during African swine fever virus particle assembly: ultrastructural in situ hybridisation and DNase-gold labelling. Virology 249:175-188. [DOI] [PubMed] [Google Scholar]
- 13.Brookes, S. M., L. K. Dixon, and R. M. E. Parkhouse. 1996. Assembly of African swine fever virus: quantitative ultrastructural analysis in vitro and in vivo. Virology 224:84-92. [DOI] [PubMed] [Google Scholar]
- 14.Carrascosa, A. L., M. del Val, J. F. Santarén, and E. Viñuela. 1985. Purification and properties of African swine fever virus. J. Virol. 54:337-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carrascosa, J. L., J. M. Carazo, A. L. Carrascosa, N. García, A. Santisteban, and E. Viñuela. 1984. General morphology and capsid fine structure of African swine fever virus particles. Virology 132:160-172. [DOI] [PubMed] [Google Scholar]
- 16.Carvalho, Z. G., A. P. Alves de Matos, and C. Rodrigues-Pousada. 1988. Association of African swine fever virus with the cytoskeleton. Virus Res. 11:175-192. [DOI] [PubMed] [Google Scholar]
- 17.Chapman, D. G., V. Tcherepanov, C. Upton, and L. K. Dixon. 2008. Comparison of the genome sequences of non-pathogenic and pathogenic African swine fever virus isolates. J. Gen. Virol. 89:397-408. [DOI] [PubMed] [Google Scholar]
- 18.Dixon, L. K., J. M. Escribano, C. Martins, D. L. Rock, M. L. Salas, and P. J. Wilkinson. 2005. The Asfarviridae, p. 135-143. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy. Eigth Report of the International Committee for the Taxonomy of Viruses. Elsevier/Academic Press, London, United Kingdom.
- 19.Enjuanes, L., A. L. Carrascosa, M. A. Moreno, and E. Viñuela. 1976. Titration of African swine fever virus. J. Gen. Virol. 32:471-477. [DOI] [PubMed] [Google Scholar]
- 20.Epifano, C., J. Krijnse-Locker, M. L. Salas, J. M. Rodríguez, and J. Salas. 2006. The African swine fever virus nonstructural protein pB602L is required for formation of the icosahedral capsid of the virus particle. J. Virol. 80:12260-12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Epifano, C., J. Krijnse-Locker, M. L. Salas, J. Salas, and J. M. Rodríguez. 2006. Generation of filamentous instead of icosahedral particles by repression of African swine fever virus structural protein pB438L. J. Virol. 80:11456-11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esteves, A., M. I. Marques, and J. V. Costa. 1986. Two-dimensional analysis of African swine fever virus proteins and proteins induced in infected cells. Virology 152:192-206. [DOI] [PubMed] [Google Scholar]
- 23.García-Escudero, R., G. Andrés, F. Almazán, and E. Viñuela. 1998. Inducible gene expression from African swine fever virus recombinants: analysis of the major capsid protein p72. J. Virol. 72:3185-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hawes, P. C., C. L. Netherton, T. Wileman, and P. Monaghan. 2008. The envelope of intracellular African swine fever virus is composed of a single lipid bilayer. J. Virol. 82:7905-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heath, C. M., M. Windsor, and T. Wileman. 2001. Aggresomes resemble sites specialized for virus assembly. J. Cell Biol. 153:449-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iyer, L. M., L. Aravind, and E. V. Koonin. 2001. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 75:11720-11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iyer, L. M., S. Balaji, E. V. Koonin, and L. Aravind. 2006. Evolutionary genomics of nucleocytoplasmic large DNA viruses. Virus Res. 117:156-184. [DOI] [PubMed] [Google Scholar]
- 28.Jouvenet, N., P. Monaghan, M. Way, and T. Wileman. 2004. Transport of African swine fever virus from assembly sites to the plasma membrane is dependent on microtubules and conventional kinesin. J. Virol. 78:7990-8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jouvenet, N., M. Windsor, J. Rietdorf, P. Hawes, P. Monaghan, M. Way, and T. Wileman. 2006. African swine fever virus induces filopodia-like projections at the plasma membrane. Cell. Microbiol. 8:1803-1811. [DOI] [PubMed] [Google Scholar]
- 30.Kleiboeker, S. B., T. G. Burrage, G. A. Scoles, D. Fish, and D. L. Rock. 1998. African swine fever virus infection in the argasid host, Ornithodoros porcinus porcinus. J. Virol. 72:1711-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kopito, R. R. 2000. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10:524-530. [DOI] [PubMed] [Google Scholar]
- 32.Krupovic, M., and D. H. Bamford. 2008. Virus evolution: how far does the double beta-barrel viral lineage extend? Nat. Rev. Microbiol. 6:941-948. [DOI] [PubMed] [Google Scholar]
- 33.Moura Nunes, J. F., J. D. Vigario, and A. M. Terrinha. 1975. Ultrastructural study of African swine fever virus replication in cultures of swine bone marrow cells. Arch. Virol. 49:59-66. [DOI] [PubMed] [Google Scholar]
- 34.Plowright, W., G. R. Thomson, and J. A. Neser. 1994. African swine fever, p. 568-599. In J. A. W. Coetzer, G. R. Thomson, and R. C. Tustin (ed.), Infectious diseases of livestock, 1st ed., vol. 1. Oxford University Press Southern Africa, Cape Town, South Africa. [Google Scholar]
- 35.Rodríguez, J. M., R. García-Escudero, M. L. Salas, and G. Andrés. 2004. African swine fever virus structural protein p54 is essential for the recruitment of envelope precursors to assembly sites. J. Virol. 78:4299-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodríguez, J. M., F. Almazán, E. Viñuela, and J. F. Rodríguez. 1992. Genetic manipulation of African swine fever virus: construction of recombinant viruses expressing the β-galactosidase gene. Virology 188:239-250. [DOI] [PubMed] [Google Scholar]
- 37.Rojo, G., M. Chamorro, M. L. Salas, E. Viñuela, J. M. Cuezva, and J. Salas. 1998. Migration of mitochondria to viral assembly sites in African swine fever virus-infected cells. J. Virol. 72:7583-7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rouiller, I., S. M. Brookes, A. D. Hyatt, M. Windsor, and T. Wileman. 1998. African swine fever virus is wrapped by the endoplasmic reticulum. J. Virol. 72:2373-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salas, J., M. L. Salas, and E. Viñuela. 1999. African swine fever virus: a missing link between poxviruses and iridoviruses?, p. 467-480. In E. Domingo, R. G. Webster, and J. J. Holland (ed.), Origin and evolution of viruses. Academic Press, London, United Kingdom.
- 40.Sanz, A., B. García-Barreno, M. L. Nogal, E. Viñuela, and L. Enjuanes. 1985. Monoclonal antibodies specific for African swine fever virus proteins. J. Virol. 54:199-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simón-Mateo, C., G. Andrés, and E. Viñuela. 1993. Polyprotein processing in African swine fever virus: a novel gene expression strategy for a DNA virus. EMBO J. 12:2977-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simón-Mateo, C., G. Andrés, F. Almazán, and E. Viñuela. 1997. Proteolytic processing in African swine fever virus: evidence for a new structural polyprotein, pp62. J. Virol. 71:5799-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomson, G. R., M. D. Gainaru, and A. F. van Dellen. 1980. Experimental infection of warthogs (Phacochoerus aethiopicus) with African swine fever virus. Onderstepoort J. Vet. Res. 47:19-22. [PubMed] [Google Scholar]
- 44.Tulman, E. R., G. A. Delhon, B. K. Ku, and D. L. Rock. 2009. African swine fever virus. Curr. Top. Microbiol. Immunol. 328:43-87. [DOI] [PubMed] [Google Scholar]
- 45.Yáñez, R. J., J. M. Rodríguez, M. L. Nogal, L. Yuste, C. Enriquez, J. F. Rodríguez, and E. Viñuela. 1995. Analysis of the complete nucleotide sequence of African swine fever virus. Virology 208:249-278. [DOI] [PubMed] [Google Scholar]