Abstract
Using a cell-based replicon screen, we identified a class of compounds with a thiazolidinone core structure as inhibitors of hepatitis C virus (HCV) replication. The concentration of one such compound, BMS-824, that resulted in a 50% inhibition of HCV replicon replication was ∼5 nM, with a therapeutic index of >10,000. The compound showed good specificity for HCV, as it was not active against several other RNA and DNA viruses. Replicon cells resistant to BMS-824 were isolated, and mutations were identified. A combination of amino acid substitutions of leucine to valine at residue 31 (L31V) and glutamine to leucine at residue 54 (Q54L) in NS5A conferred resistance to this chemotype, as did a single substitution of tyrosine to histidine at amino acid 93 (Y93H) in NS5A. To further explore the region(s) of NS5A involved in inhibitor sensitivity, genotype-specific NS5A inhibitors were used to evaluate a series of genotype 1a/1b hybrid replicons. Our results showed that, consistent with resistance mapping, the inhibitor sensitivity domain also mapped to the N terminus of NS5A, but it could be distinguished from the key resistance sites. In addition, we demonstrated that NS5A inhibitors, as well as an active-site inhibitor that specifically binds NS3 protease, could block the hyperphosphorylation of NS5A, which is believed to play an essential role in the viral life cycle. Clinical proof of concept has recently been achieved with derivatives of these NS5A inhibitors, indicating that small molecules targeting a nontraditional viral protein like NS5A, without any known enzymatic activity, can also have profound antiviral effects on HCV-infected subjects.
Hepatitis C virus (HCV) is the major causative agent for non-A, non-B hepatitis worldwide, which affects more than 3% of the world population. HCV establishes chronic infections in a large percentage of infected individuals, increasing the risk for developing liver cirrhosis and, in some cases, hepatocellular carcinoma. Although the current standard of care for HCV infection involves the use of PEGylated interferon and ribavirin, a large proportion of patients fail to respond to this therapy, and treatment is associated with frequent and sometimes serious side effects (9). Given the limited efficacy of the current therapy, the development of safer and more effective therapies is of tremendous importance.
HCV is a positive-strand RNA virus belonging to the family Flaviviridae. The HCV genome consists of a ∼9.6-kb RNA with a large open reading frame encoding a polyprotein of ∼3,010 amino acids. The polyprotein is cleaved co- and posttranslationally by both cellular and viral proteases into at least 10 different products (10, 11). The viral proteins required for RNA replication include NS3, NS4A, NS4B, NS5A, and NS5B (4, 19). NS3 consists of an amino-terminal protease domain required for the cleavage of the remaining nonstructural proteins and a carboxyl-terminal helicase/NTPase domain (8, 11, 30). NS4A serves as a cofactor for NS3 protease and helicase activities (8). NS4B is an integral membrane protein involved in the formation of the membranous web, where HCV replication complexes are thought to assemble (7). NS5A is a membrane-associated phosphoprotein present in basally phosphorylated (p56) and hyperphosphorylated (p58) forms (15, 31). It was previously reported that only p58-defective mutants could be complemented in trans (1), and NS5A is involved in HCV virion production (22, 34), suggesting that different forms of NS5A exert multiple functions at various stages of the viral life cycle. The N terminus of NS5A (domain I) has been crystallized in alternative dimer forms and contains zinc- and RNA-binding domains (20, 33). The ability of NS5A to bind to zinc (32) and RNA (14) has been demonstrated in vitro. NS5A has been shown to interact with a number of host proteins, is implicated in interferon resistance in vivo, and has been the subject of several reviews (13, 21). NS5B functions as the viral RNA-dependent RNA polymerase (2). Previous studies have shown that the NS3-NS5B proteins are all essential for HCV replication and are believed to form the HCV replicase complex (4, 18, 19).
The development of the cell-based HCV replicon system provides a means for the large-scale screening of HCV inhibitors against multiple viral targets. The use of a cell-based replication assay likely includes essential functions that previously could not be evaluated with in vitro enzyme assays. The disadvantages for the advancement of HCV inhibitors targeting nonenzymatic proteins are (i) the potential for structure-activity relationships (SAR) to be difficult to interpret based on the complexity of cell-based systems, (ii) the lack of a system for validation, and (iii) difficulty in predicting if in vitro potency can translate into in vivo effect. Therefore, during the process of developing HCV NS5A inhibitors, we established a series of assays and checkpoints prior to entering the clinic. This is the first report in a series of articles detailing the development of HCV NS5A inhibitors that has culminated in the demonstration of clinical efficacy for this novel mechanistic class of HCV inhibitor (25).
In this report, we have used a previously described cell-based approach (26) to identify a novel compound that specifically inhibits HCV RNA replication. Through the use of resistance selection, we have demonstrated that the inhibitor targets the HCV NS5A protein, thereby establishing that the function of NS5A in replication can be inhibited by small molecules. In addition, using genotype-specific inhibitors, we have further shown that the N terminus of NS5A plays an essential role in compound activity by both 50% effective concentration (EC50) determinations as well as a functional assay to evaluate NS5A hyperphosphorylation.
MATERIALS AND METHODS
Cell culture and compounds.
Huh-7 cells were grown in Dulbecco's modified Eagle medium (DMEM) with 100 U/ml penicillin-streptomycin and 10% fetal bovine serum (FBS). Both bovine viral diarrhea virus (BVDV) and HCV replicon cell lines were isolated as previously described (26) and maintained in medium that also contained 0.3 to 0.5 mg/ml Geneticin (G418). Huh-7 cells cured of a Con1 replicon were generated as previously described (17) and propagated in DMEM with penicillin-streptomycin and 10% FBS. Compounds used in this study were synthesized at Bristol-Myers Squibb.
FRET assay.
A fluorescence resonance energy transfer (FRET) assay was performed as previously described (26, 28). Briefly, after 72 h at 37°C, replicon cell plates were washed with phosphate-buffered saline and then used for FRET assay by the addition of 30 μl of the FRET peptide assay reagent per well. The assay reagent consisted of 1× luciferase cell culture lysis buffer, 150 mM NaCl, and 20 μM FRET peptide. The plate containing assay reagent was then read in a kinetic mode in a Cytofluor 4000 instrument, which had been set to a 340-nm excitation/490-nm emission automatic mode for 20 cycles. EC50s were calculated as the concentration of compound which caused a 50% reduction in HCV FRET activity.
Isolation of resistant replicons.
The selection of resistant replicon cells was performed by growing genotype 1b replicon cells in medium containing a concentration of either 5 μM BMS-858 or 2 μM BMS-824. Medium containing the compound was added to monolayers of HCV 1b-377-neo replicon cells at ∼25% confluence in the presence of 0.5 mg/ml G418. Replicon cells maintained in the presence of dimethyl sulfoxide (DMSO) were used as a control. After 5 to 6 weeks, control DMSO-selected replicon cells and compound-selected cells were tested for compound sensitivity by using the HCV replicon FRET assay. In addition, individual colonies derived from the selections were isolated, expanded into homogeneous cell lines, and tested for compound sensitivity.
cDNA cloning and rescue of the resistant phenotype.
Total RNA was isolated from both control replicon cells and homogeneous cell lines selected with compound using Trizol (Gibco-BRL) according to the manufacturer's protocol. Total RNA was treated extensively with DNase, cleaned up by phenol-chloroform extraction, and ethanol precipitated.
To test for resistance, 14 μg total RNA containing 2 ng HCV RNA was electroporated into 4 × 106 naive Huh-7 cells as previously described (17). A portion of these cells (2.5 × 105 cells) was plated onto 35-mm dishes and incubated at 37°C with or without 2 μM BMS-858 or 5 μM BMS-182, an HCV protease inhibitor (PI). At various times postelectroporation, total RNA was isolated using the RNeasy kit (Qiagen), and HCV RNA levels were determined using quantitative reverse transcription (qRT)-PCR (17).
To select for a cell line, the remaining 1.75 × 106 electroporated cells were placed into a 100-mm dish and selected for 4 weeks with 0.5 mg/ml G418, changing the medium every 3 to 5 days. After 4 weeks, selected cells were trypsinized for expansion, and a portion was plated in medium containing either DMSO or 2 μM BMS-858. Forty-eight hours later, total RNA was isolated by using the RNeasy kit (Qiagen), and HCV RNA levels were determined by qRT-PCR.
Total RNA from selected cells was also used to generate HCV cDNAs. The entire HCV open reading frame was generated and amplified in a single fragment by using SuperScript One-Step reverse transcriptase PCR for long templates (Gibco-BRL) and primers targeting the encephalomyocarditis virus (EMC) internal ribosome entry site and 3′ untranslated region. Reaction products were gel purified and cloned directly into pCR2.1-TOPO by using a TOPO TA cloning kit (Invitrogen). The DNA sequence of the entire HCV nonstructural coding region was determined for multiple clones.
To put substitutions back into the HCV 1b-377-neo replicon, cDNAs containing changes were digested with EcoRI and HpaI, and the correct-sized fragments were gel purified and ligated into similarly digested HCV 1b-377-neo DNA. Clones containing the correct sequence were identified by restriction digestion and confirmed by sequence analysis. To perform transient assays, the EcoRI-to-HindIII fragment containing the Y93H substitution was cloned into a Blast/Luc 1-377 1b replicon containing a blasticidin/luciferase fusion gene in place of the neomycin phosphotransferase gene (17). Alternatively, the L31V and Q54L mutants were generated in the Blast/Luc 1-377 1b replicon by using the Stratagene QuikChange mutagenesis kit according to the manufacturer's directions.
Transient replication assays and generation of cell lines.
RNA transcripts were synthesized in vitro by using ScaI-digested DNAs and the T7 MegaScript transcription kit (Ambion) according to the manufacturer's directions. For transient replication assays, subconfluent cured Huh-7 cells in a 35-mm dish were transfected with 2.5 μg of RNA transcript using DMRIE-C (Invitrogen) according to the manufacturer's directions. Four hours later, the transfectant was removed, replaced with DMEM plus 10% FBS with or without compound, and then incubated at 37°C. At various time points, cells were harvested, and luciferase activity was determined by using the Renilla luciferase assay kit (Promega). To generate cell lines, 4 × 106 cured Huh-7 cells were electroporated with 10 μg of RNA transcript and plated into 100-mm dishes. After 24 h, selective medium containing 0.5 mg/ml G418 was added, and the medium was changed every 3 to 5 days. After ∼4 weeks, small colonies were visible, which were isolated and expanded for further analysis.
Generation of hybrid replicons.
Hybrid replicons were generated based on the genotype 1b Con1 sequence (19) and the genotype 1a H77 sequence (36). The genotype 1b replicon was made exactly as described previously by Lohmann et al. (19). The genotype 1a replicon was made as previously described (17). To generate hybrid replicons, regions of the NS5A gene of the genotype 1a H77 strain or the genotype 1b Con1 strain were cloned into the genotype 1b or 1a replicon by using standard molecular biology techniques, and the correct clones were confirmed by sequence analysis.
Transient expression assays.
Mammalian transient expression assays using the vaccinia virus-T7 hybrid system were performed as previously described, with minor modifications (10). Briefly, monolayers of BHK-21 cells were infected with vTF7-3 at a multiplicity of infection of 1 PFU per cell for 1 h at room temperature. After the removal of the inoculum, cells were transfected with a mixture of plasmid DNA, Plus reagent, and Lipofectamine (Invitrogen) according to the manufacturer's directions and incubated in the absence or presence of compound for various times. For [33P]orthophosphate labeling, 3.5 h after transfection, cells were washed three times with phosphate-free DMEM and labeled for 4 h with phosphate-free DMEM containing 3% dialyzed FBS, 200 μCi of [33P]orthophosphate per ml, and either DMSO or compound. Cells were harvested, resuspended in 150 μl lysis buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1% NP-40, 1 mM EDTA), and incubated for 30 min on ice, and supernatant was harvested after pelleting. Extracts were immunoprecipitated as described below and treated with calf intestinal alkaline phosphatase (CIP). For pulse-chase experiments, cells at 4 h posttransfection were starved for 45 min in minimal essential medium without methionine and labeled for 30 min with 100 μCi of 35S-labeled methionine per ml. Cells were either harvested (time zero) or washed two times with phosphate-buffered saline and further incubated for 4 h with DMEM containing 5% FBS and either DMSO or compound.
Protein analysis.
For Western analysis, transfected cells were lysed at 7 h postinfection by using cell dissociation buffer, and material from equal numbers of cells was separated on an 8% acrylamide gel by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). After electrophoretic transfer, the HCV NS5A protein was detected with rabbit antiserum specific for NS5A and secondary goat anti-rabbit horseradish peroxidase-conjugated antibody, followed by the ECL detection system (Amersham Biosciences).
For immunoprecipitation, 25 μl of extract was heated at 95°C for 4 min in the presence of 0.5% SDS. Three microliters of NS5A-specific antiserum was added with 160 μl dilution buffer (190 mM NaCl, 60 mM Tris [pH 7.4], 6 mM EDTA, 1.25% Triton X-100) and incubated at 4°C overnight. Samples were incubated at 4°C for 1 h with 40 μl Ultralink immobilized protein G beads and were washed three times with wash solution (0.1% Triton X-100, 0.02% SDS, 150 mM NaCl, 50 mM Tris [pH 7.5], 5 mM EDTA) and one time with wash solution lacking Triton X-100 and SDS. Immunoprecipitated samples were boiled in SDS sample dye and separated by SDS-PAGE. Gels were treated for fluorography, dried, and exposed to X-ray film.
Phosphatase treatment.
Immunoprecipitated samples were washed with phosphatase buffer (50 mM Tris [pH 7.5], 10 mM MgCl2, 1 mM dithiothreitol, 100 mM NaCl) and incubated with 15 U of CIP for 1 h at 37°C in 100 μl phosphatase buffer. Samples were washed with wash solution lacking detergent, boiled in SDS sample dye, and analyzed by SDS-PAGE.
RESULTS
Identification of HCV-specific inhibitors.
The availability of HCV replicon cell lines provides a system for the study of viral RNA replication and the screening of antiviral compounds. We have used a cell-based high-throughput HCV/BVDV replicon screen (26) to identify specific inhibitors of HCV. Briefly, this assay uses a mixture of HCV and BVDV replicon cell lines that allows one to determine the potency and specificity of a compound by measuring cytotoxicity, the inhibition of HCV replication, and activity against the closely related BVDV in a single well (26). The reduction of HCV replication is measured indirectly by monitoring HCV NS3 protease activity using a FRET assay. Cytotoxicity is determined by alamar blue staining, and BVDV replication is monitored by measuring luciferase activity, since this replicon contains a luciferase reporter gene as part of its genome. Using this approach, we screened a portion of the Bristol-Myers Squibb chemical deck and identified a class of compounds with a thiazolidinone core as inhibitors of HCV (29). One such compound, BMS-858 (Fig. 1), gave 100% inhibition of HCV at 15 μM with no detectable toxicity. An initial titration of BMS-858 on the HCV replicon cells yielded an EC50 of 0.57 to 1.0 μM with a 50% cytotoxic concentration (CC50) of >50 μM and no detectable activity on the related BVDV replicon cell line (EC50 of >50 μM).
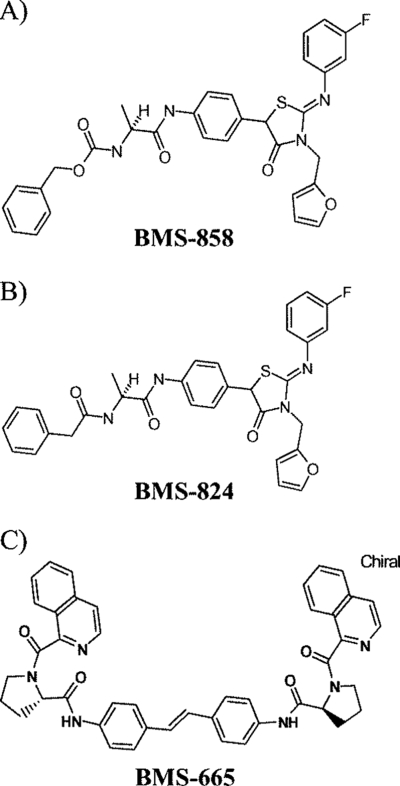
FIG. 1.
Chemical structures of compounds used in this study. (A) Structure of screen hit BMS-858. (B) Structure of library compound BMS-824. (C) Structure of BMS-665.
To determine if SAR were coherent for BMS-858, a series of related compounds were prepared and examined with the HCV replicon assay. These efforts yielded the more potent inhibitor BMS-824 (Fig. 1). The structure of BMS-824 differs from that of BMS-858 only by the replacement of the benzyl carbamate element by a phenylacetamide moiety that is 1 atom shorter. The EC50 for BMS-824 against HCV was initially determined to be ∼5 nM, which reflects an increase in potency of >100-fold compared to that of BMS-858. To be more confident that this compound series was specific for HCV, BMS-824 was tested against several RNA and DNA viruses, including the closely related pestivirus BVDV (Table 1). As can be seen, BMS-824 did not demonstrate significant activity against human immunodeficiency virus (HIV), human rhinovirus, respiratory syncytial virus, or BVDV. The BVDV replicon was inhibited with an EC50 of 1.7 μM. Since the BVDV replicon is contained in the same cell background as that of the HCV replicon (Huh-7 cells), this inhibition may reflect some cellular toxicity rather than a specific antiviral effect. This was demonstrated for other unrelated compounds showing BVDV activity and was attributed to the increased sensitivity of a virus-based luciferase reporter assay for BVDV versus an alamar blue-based toxicity measurement (data not shown; 26). The therapeutic window of BMS-824 on HCV was >10,000, demonstrating the increased specificity of BMS-824 for HCV compared to that for BVDV.
TABLE 1.
Antiviral selectivity of BMS-824
| Replicon or virusa | Cell line | EC50 (μM) | CC50 (μM) |
|---|---|---|---|
| HCV replicon | Huh-7 | 0.005 | >50 |
| BVDV replicon | Huh-7 | 1.7 | >50 |
| BVDV | MDBK | 5 | >150 |
| HIV | MT2 | 10 | 23 |
| HRV | MRC-5 | >100 | >100 |
| RSV | Hep-2 | 14 | 14 |
HRV, human rhinovirus; RSV, respiratory syncytial virus.
To explore which viral proteins these compounds might be targeting, BMS-858 was first tested in in vitro enzyme assays for the HCV NS3 protease and NS5B polymerase. BMS-858 did not inhibit HCV protease or polymerase in these assays, yielding 50% inhibitory concentrations (IC50s) of >100 μM (data not shown).
Isolation of resistant replicons.
In an effort to determine the target of the BMS-858/BMS-824 series, BMS-858 was used to select for resistance on HCV replicon cells. Medium containing 5 μM of BMS-858 was added to subconfluent monolayers of replicon cells as described in Materials and Methods. Replicon cells incubated with DMSO-containing medium in parallel were used as a control. After individual colonies were visible, single colonies were picked, expanded, and tested for sensitivity to compounds by using the FRET assay. The selected cells (858R) were sensitive to an unrelated compound inhibiting the HCV NS3 protease but demonstrated >5-fold resistance to BMS-858 versus control DMSO-selected HCV replicon cells (Table 2). Likewise, BMS-824 demonstrated >600-fold resistance on BMS-858-selected cells, going from an EC50 of ∼8 nM on wild-type cells to an EC50 >5 μM on 858R cells (Table 2), thus confirming cross-resistance and compound relatedness.
TABLE 2.
Compound sensitivities of replicon cell lines
| Compound | EC50 (μM) |
Fold resistance | EC50 (μM) |
Fold resistance | CC50 (μM) | ||
|---|---|---|---|---|---|---|---|
| Wild typea | 858R | Wild typeb | Y93H | ||||
| BMS-858 | 1.0 | >5 | >5 | 0.5 | >5 | >10 | >5 |
| BMS-824 | 0.008 | >5 | >625 | 0.005 | >5 | >1,000 | >5 |
| PI | 0.098 | 0.069 | 0.7 | 0.086 | 0.092 | 1.1 | >5 |
DMSO-selected control cells.
Cell line derived from transfection of a wild-type replicon.
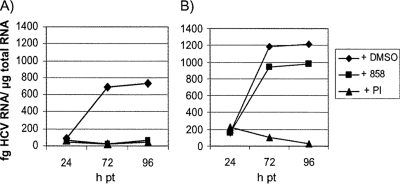
We used several approaches to demonstrate that the resistant phenotype was due to a viral and not a cellular mutation. First, we determined whether HCV viral RNA from the resistant cells could transfer the resistant phenotype to naive Huh-7 cells. If the observed resistance is virally based, the viral RNA from 858R cells should transfer the resistant phenotype, while cell-based resistance would not be transferred. Total RNA was extracted from both wild-type and resistant cells (858R) and transfected into naive Huh-7 cells. A portion of the transfected cells was plated in the presence or absence of HCV inhibitors, and HCV RNA levels were determined at various time posttransfection by using qRT-PCR. The compounds used included 5 μM of an HCV NS3 PI as well as 4 μM of BMS-858. As shown in Fig. 2, by 96 h posttransfection, wild-type levels of HCV RNA were reduced more than 90% by both the NS3 PI and BMS-858. In contrast, for cells transfected with RNA from the 858R cell line, levels of HCV RNA were reduced more than 95% by the PI but only 20% in the presence of BMS-858.
FIG. 2.
Resistance to BMS-858 in cells transfected with RNA from wild-type or 858R cells. Naive Huh-7 cells were electroporated with total cellular RNA containing 2 ng HCV RNA isolated from wild-type (A) or 858R (B) cell lines and incubated with or without compound. HCV RNA levels were determined at various times posttransfection (pt).
To further ensure that resistance was due to the viral RNA and not the transfected cellular RNA, the remaining cells from the initial transfection were plated and grown in the presence of G418 to select for cells containing transfected HCV RNA. After 4 weeks of passaging, cells were plated in the presence or absence of 4 μM BMS-858 or 5 μM PI, and HCV RNA levels were determined 48 h later. The cell line derived from wild-type transfected RNA showed more than an 85% reduction in HCV RNA levels in the presence of either BMS-858 or PI (data not shown). The cell line derived from transfection of 858R RNA showed no reduction in HCV RNA levels when incubated with BMS-858, while RNA levels were reduced more than 85% by the NS3 PI. Taken together, these results demonstrate that viral RNA could transfer the resistant phenotype, suggesting that it contained a mutation responsible for BMS-858 resistance.
Identification of the BMS-858 target.
Further confirmation of virus-based resistance was provided by mapping the causal mutation(s) for resistance. To determine the target of BMS-858, eight different cDNA clones encompassing the entire HCV nonstructural coding region were generated from two independently isolated resistant cell lines (cell lines B and C), and one was generated from a wild-type cell line. Sequence analysis of the HCV nonstructural coding region identified a single predominant mutation site. All three clones from cell line B had a T-to-C substitution at nucleotide 4943, resulting in an amino acid substitution of Tyr93 to His in NS5A. Likewise, all four clones from cell line C had an A-to-G substitution at nucleotide 4944, resulting in an amino acid substitution of Tyr93 to Cys.
To determine if the Y93H change was necessary and sufficient to confer resistance to BMS-858, the single mutation was placed into the HCV 1b-377-neo replicon. RNA transcripts of this clone, in parallel with the wild-type replicon clone, were transfected into Huh-7 cells and subjected to G418 selection. After 3 weeks of selection, colonies were isolated and expanded, and the sensitivity of these cells to BMS-858 was examined by using a FRET assay. On the wild-type cells, BMS-858 had an EC50 of 0.5 μΜ (Table 2), while the Y93H cell line showed no inhibition up to 5 μΜ, the highest concentration tested. Further testing with the more potent compound BMS-824 showed that there was more than a 1,000-fold window of resistance in Y93H cells compared to wild-type cells (Table 2), confirming that the histidine substitution in NS5A was responsible for the resistant phenotype against this compound series.
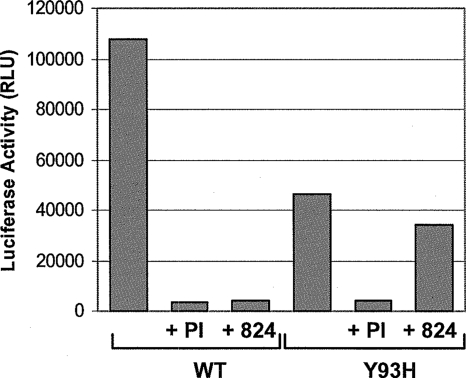
This resistant phenotype was further validated by engineering the Y93H mutation into a genotype 1b replicon containing a luciferase reporter gene, which can be used to monitor replication in a transient reporter assay. The replication of the wild-type and Y93H mutant replicons was monitored over time in the presence or absence of either 1 μM BMS-824 or 2 μM NS3 PI. The time of maximum replication efficiency for both the wild-type and mutant RNAs was determined to be 72 h posttransfection (data not shown). As shown in Fig. 3, the replication efficiency of the Y93H replicon was ∼30 to 43% of the level of the wild type at 72 h, indicating that the resistant mutant had reduced fitness. Both replicons were inhibited by PI with similar efficiencies. In contrast, 1 μM of BMS-824 inhibited the wild-type replicon by more than 90%, but the Y93H mutant was inhibited only 15%, indicating that this mutation resulted in resistance to the NS5A compound. This was further confirmed by titrating BMS-824 against the replicons. This experiment yielded EC50s of 22 nM and 4.8 μM for the wild type and mutant, respectively, indicating that the Y93H substitution conferred >200-fold resistance to BMS-824. In contrast, the potency of compounds targeting NS3 protease or NS5B polymerase was not affected by the Y93H mutation (data not shown). Transient replication studies using a Y93C mutant indicated that this substitution also conferred resistance to BMS-858 and BMS-824, similar to the Y93H mutant (data not shown).
FIG. 3.
Transient replication of wild-type and Y93H HCV genomes. Huh-7 cells were transfected with wild-type (WT) or Y93H replicon RNA and incubated in the presence or absence of 2 μM PI or 1 μM BMS-824. Luciferase activities (relative light units [RLU]) were determined with lysates of cells harvested 72 h after transfection. The 4-h value was used to correct for differences in transfection efficiencies. This graph represents data from one of two experiments giving similar results.
To see if alternative substitutions were chosen with a more potent inhibitor, 2 μM BMS-824 was also used to select for resistance on genotype 1b replicon cells. Mapping of the BMS-824-resistant cell lines revealed two new consensus mutations in NS5A, L31V and Q54L. By using site-directed mutagenesis, these substitutions were made singly and in combination in the luciferase-containing genotype 1b replicon. When tested in a transient replication assay, none of the mutants conferred resistance to a control HCV PI (Table 3). In contrast, the Q54L mutant conferred low-level resistance to BMS-824, while the L31V change yielded somewhat greater resistance (Table 3). However, when both the L31V and Q54L mutations were present in combination, BMS-824 resistance increased significantly, suggesting that both changes were required to maximally affect compound potency. A similar resistance profile was observed for these mutants with BMS-858 (Table 3), even though selection with this compound generated resistance at amino acid 93.
TABLE 3.
Transient testing of selected mutations
| Compound | Fold resistance relative to wild typea |
|||
|---|---|---|---|---|
| Wild type | L31V | Q54L | L31V/Q54L | |
| BMS-824 | 1 | 60 | 9 | 354 |
| BMS-858 | 1 | 18 | 7 | 100 |
| PI | 1 | 2 | 2 | 2 |
The relative change in resistance is determined by dividing the EC50 obtained with mutant HCV replicons by the EC50 obtained with the wild-type replicon.
Adaptive mutations are required to allow efficient replication of the HCV genotype 1b replicon, and the most prominent mutations are located in NS5A (4, 16, 18). To determine whether the potency of BMS-824 was affected by these mutations, we evaluated BMS-824 against replicon cell lines with various adaptive changes. This included cell lines with a deletion of serine 2197 in NS5A, one with an S2204I substitution in NS5A (4), a cell line with a K1846T substitution in NS4B (12), and one with the K1846T change plus E1202G and T1280I substitutions in NS3. BMS-824 showed similar potencies against all four cell lines (Table 4), suggesting that the adaptive mutations do not play a role in BMS-824 sensitivity.
TABLE 4.
Effect of adaptive mutations on compound potencya
| Adaptive mutation(s) | Mutation site(s) | BMS-824 EC50 (μM) | BMS-824 CC50 (μM) |
|---|---|---|---|
| S2197 deletion | NS5A | 0.011 | >50 |
| S2204I | NS5A | 0.008 | >50 |
| K1846T | NS4B | 0.007 | >50 |
| E1202G, T1280I, K1846T | NS3 and NS4B | 0.010 | >50 |
Adaptive mutations are listed first as the wild-type residue and then as its position in the HCV polyprotein, followed by the residue to which it was changed.
The N terminus of NS5A is important for genotype 1a coverage.
All three resistance mutations (Y93H, L31V, and Q54L) map to the N terminus of NS5A, a region believed to be important for dimer formation, RNA binding, and membrane association of the replication complex. To further define the region of NS5A involved in inhibitor sensitivity, we took advantage of the fact that this compound series showed a loss of potency against genotype 1a. For instance, while the EC50 of BMS-824 was ∼5 to 20 nM against the genotype 1b replicon, the EC50 against the genotype 1a replicon was >30 μM, indicating a >1,500-fold loss in potency.
To investigate the region responsible for this loss of genotype 1a coverage and thereby further define the region of NS5A important for inhibitor activity, a series of genotype 1a/1b chimeric replicons was made. Previously, we showed that it was possible to replace the NS5A gene in the genotype 1b replicon with the NS5A gene from genotype 1a and still have a viable replicon (17). The HCV coding sequence of this clone (termed genotype 1a NS5A) is from the Con1 genotype 1b sequence except that the NS5A gene has been replaced with the NS5A sequence from the H77 genotype 1a clone, and there is an adaptive mutation of K1846T in NS4B to (i) promote efficient replication and (ii) ensure that inhibitor sensitivity is not affected by adaptive mutations in the NS5A protein (Fig. 4A). The Con1-derived genotype 1b NS5A sequence and the genotype 1a NS5A sequence from H77 share ∼80% amino acid identity overall, and both sequences contain a tyrosine at residue 93 and a leucine at residue 31 (Fig. 4B). We first compared the EC50 values of BMS-824 on the genotype 1b, 1a, and 1a NS5A replicon cell lines to see if the transfer of the NS5A gene was sufficient to confer the loss of genotype 1a coverage. As shown in Fig. 4A, the potency of BMS-824 on the genotype 1a NS5A cell line was greatly reduced (EC50 of >10 μM) compared to its activity against the replicon that was entirely genotype 1b. This is similar to what was observed with the genotype 1a cell line, demonstrating that the sequence within the NS5A gene was responsible for the loss of genotype 1a coverage. To further define the region responsible for this, additional hybrids were made by replacing smaller regions of the NS5A gene (Fig. 4A). Initially, a genotype 1b chimera containing the last 218 amino acids of genotype 1a H77 NS5A was generated (termed 5′1b/3′1a 5A) and tested in a transient replication assay. On the 5′1b/3′1a 5A clone, BMS-824 gave EC50s similar to those of the control genotype 1b replicon, suggesting that the 5′ half of NS5A was important for inhibitor activity. We then focused on the N-terminal region of NS5A, constructing hybrids that contained only the first 76 amino acids of the other genotype 1a or 1b subtype. Within these 76 amino acids, there are 15 residues that differ between genotypes 1a and 1b (Fig. 4B), yielding ∼80% identity within this region. Interestingly, a replicon with a chimeric genotype 1a NS5A molecule with only the first 76 amino acids from genotype 1b NS5A (76-1b on 1a 5A) was now completely sensitive to compound BMS-824. Conversely, the chimeric genotype 1b replicon containing the N-terminal 76 residues of genotype 1a NS5A was not inhibited by BMS-824, showing more than a 550-fold loss in sensitivity to BMS-824. Results from these hybrid experiments are summarized in Fig. 4A and demonstrate that the lack of genotype 1a coverage with BMS-824 is due to the first 76 amino acids of NS5A. Since the hybrid replicon 76-1a on genotype 1b 5A contains leucine at amino acid 31 and tyrosine at amino acid 93, the wild-type residues for genotype 1b NS5A, yet exhibits reduced sensitivity to BMS-824, this suggests that the region involved in genotype 1a/1b coverage can be separated from primary resistance sites at L31 and Y93.
FIG. 4.
Schematic depicting the nonstructural region of HCV replicon RNAs and alignment of the N-terminal 100 amino acids of genotype 1a and 1b NS5A. (A) Genotype 1a and 1b sequences are shown, with the genotype 1a sequence being represented by a hatched fill pattern, and the positions of the nonstructural proteins are indicated. The nomenclature used for each construct is shown on the left, and the EC50 of BMS-824 or BMS-665 for each construct is indicated at the right. nd, not determined. (B) The genotype 1a sequence is derived from the H77 strain, and genotype 1b is Con1. Two dots indicate identical amino acid residues between genotypes 1a and 1b.
Further work with this compound series yielded inhibitors that now exhibited activity against genotype 1a, although their genotype 1a potency was reduced compared to their activity against genotype 1b. One such inhibitor (BMS-665) (Fig. 1C) had an EC50 of ∼10 nM against the genotype 1b replicon and an EC50 of 200 to 400 nM against the genotype 1a replicon. As shown in Fig. 4A, BMS-665 displayed similar potencies against both the genotype 1a full-length and genotype 1a NS5A hybrid replicons. When tested against the genotype 1b and 1a NS5A replicons containing the L31V substitution, more-than-50-fold resistance was observed for BMS-665, further demonstrating that the resistance and inhibitor sensitivity regions are distinct.
Given the expanded genotype coverage of BMS-665, this inhibitor was also tested in the more biologically relevant JFH-1 genotype 2a infectious virus assay. Parallel testing against the genotype 1b replicon yielded EC50s of 5 nM for the genotype 1b replicon and 20 nM against the virus, suggesting only a slight loss of potency against genotype 2a virus.
Inhibition of p58 production.
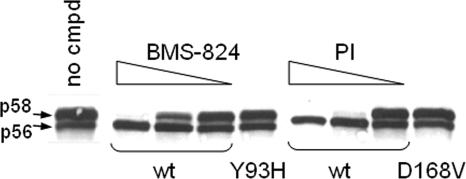
NS5A is known to be a phosphoprotein present in basally phosphorylated (p56) and hyperphosphorylated (p58) forms (15, 31). Previously reported studies suggested that hyperphosphorylation is important for the HCV life cycle, especially for the transition from RNA replication to packaging (22, 34). Since hyperphosphorylation requires the expression of the HCV polyprotein NS3-NS5A (23), we decided to examine inhibitors of NS3 protease, NS5B polymerase, and NS5A to assess (i) whether these inhibitors, when they bind to their targets, would alter p58 formation and (ii) to which stage(s) of polyprotein processing these inhibitors would bind. A vaccinia virus transient replication system was used to determine the effect of these inhibitors on NS5A hyperphosphorylation. DNA encoding a genotype 1b HCV subgenomic replicon under the control of a T7 promoter was transfected into T7-expressing vaccinia virus-infected cells and treated with DMSO, BMS-824, or an NS3 protease or NS5B polymerase allosteric-site inhibitor, and levels of p58 were then measured by Western blot analysis. As shown in Fig. 5, both p56 and p58 were present in DMSO-treated cells, while p58 formation was blocked during treatment with BMS-824 or NS3 PI in a dose-dependent manner. In contrast, treatment with an HCV NS5B polymerase inhibitor did not affect p58 levels (data not shown). In addition, BMS-824 and NS3 PI did not affect the level of p58 expressed from replicons containing the Y93H (NS5A) and D186V (NS3 protease) resistance mutations, respectively. Titration of the NS5A and NS3 inhibitors revealed that these compounds block the hyperphosphorylation of NS5A with IC50s similar to their EC50s obtained in the genotype 1b replicon cell line (Fig. 5), suggesting that there is a correlation between the blockage of p58 formation and the inhibition of replicon replication.
FIG. 5.
Effect of compound on p58 production. DNAs encoding the wild-type (wt)-, Y93H-, or D168V-containing 1-377 1b HCV replicons were expressed in a vaccinia virus transient expression system treated with either DMSO (no cmpd) or a titration of BMS-824 (0.05 to 0.002 μM) or NS3 PI (0.1 to 0.01 μM). NS5A (Y93H) and NS3 (D168V) mutants were treated with 0.05 μM BMS-824 and 0.1 μM PI, respectively. Cell lysates were separated by SDS-PAGE on 8% gels, and NS5A proteins were identified by Western immunoblotting using an anti-NS5A antibody. NS5A-specific bands, both p56 and p58, were quantified by phosphorimaging.
Loss of genotype coverage and p58 effect correlate.
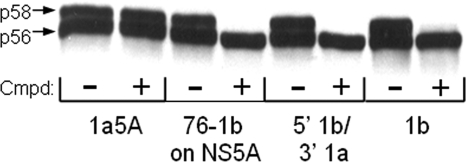
The transient expression system was used to determine whether the amino acid requirements for genotype 1a coverage with BMS-824 also correlated with its effect on NS5A hyperphosphorylation. The effect of the compound on p58 production was determined for the following replicons: genotype 1b, genotype 1a NS5A, 5′1b/3′1a, and 76-1b on 1a 5A (Fig. 4A). Similar to its lack of potency on the genotype 1a replicon or the genotype 1a NS5A hybrid replicon, BMS-824 did not alter the hyperphosphorylation of genotype 1a NS5A (Fig. 6). However, treatment of the hybrid NS5A molecule containing the 5′ 230 amino acids from genotype 1b and the 3′ 218 amino acids from genotype 1a (5′1b/3′1a) with BMS-824 did block p58 production. In fact, the transfer of just the first 76 amino acids of genotype 1b NS5A into the genotype 1a replicon (76-1b on 1a 5A) resulted in a protein whose hyperphosphorylation was blocked by BMS-824. Therefore, the ability of BMS-824 to affect p58 production correlated with the presence of the first 76 amino acids of genotype 1b NS5A, similar to the requirement for BMS-824 efficacy.
FIG. 6.
Inhibitor effect on genotype 1a/1b NS5A hybrids. DNAs encoding various HCV replicons were expressed in a vaccinia virus transient expression system treated with either DMSO (−) or 0.1 μM BMS-824 (+). Cell lysates were separated by SDS-PAGE on 8% gels, and NS5A proteins were identified by Western immunoblotting using an anti-NS5A antibody. Cmpd, compound.
Both NS5A and PIs block NS5A hyperphosphorylation.
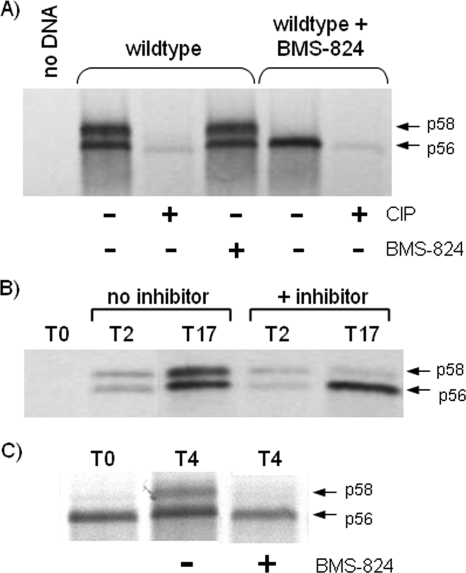
To confirm that our compounds were actually altering the hyperphosphorylation of NS5A, [33P]orthophosphate labeling was performed during the transient expression of the wild-type HCV genotype 1b replicon, and samples immunoprecipitated with NS5A antibody were treated with or without CIP. In cells expressing the HCV replicon in the absence of BMS-824 (wild type), both p56 and p58 [33P]orthophosphate-labeled bands were detected, and treatment with CIP resulted in their disappearance, confirming that they were the phosphorylated forms of NS5A (Fig. 7A). Cells treated with BMS-824 (wild type plus BMS-824) expressed only the faster-migrating p56 form of NS5A, which also disappeared upon CIP treatment, indicating that the compound is indeed altering the hyperphosphorylation of NS5A. Once p56 and p58 were formed, incubation with compound had no effect on hyperphosphorylated NS5A (Fig. 7A), suggesting that the inhibitor does not affect the stability of p58. This can be seen further in a time course experiment where cells transiently expressing HCV were harvested at 2 and 17 h after DNA transfection. In the absence of the compound, levels of p56 and p58 increased similarly over time (Fig. 7B). When the compound was added 30 min after DNA transfection, p56 levels clearly increased over time, while p58 levels remained unchanged (Fig. 7B), further suggesting that the compound does not affect p58 once it is already made but rather blocks its production.
FIG. 7.
Blocking NS5A hyperphosphorylation. (A) Cells transiently expressing the HCV replicon were [33P]orthophosphate labeled in the presence or absence of BMS-824, and extracts were immunoprecipitated with antibody to NS5A. Precipitated samples were then incubated in phosphatase buffer with or without CIP or BMS-824. A lysate from mock-transfected cells (no DNA) that had been infected only with vaccinia virus was included as a control. Samples treated with BMS-824 during labeling are indicated at the top of the gel. (B) Cells transiently expressing the HCV replicon were treated with or without BMS-824 and harvested at various times posttransfection. Western immunoblotting was performed on cell extracts by using an NS5A-specific antibody. The times of harvest are indicated above the gel. (C) Cells transiently expressing the HCV replicon were labeled with [35S]methionine for 30 min and chased in the presence (+) or absence (−) of either BMS-824 or PI (PI not shown). Cells were harvested at 4 h postlabeling and immunoprecipitated with NS5A-specific antibody.
To further explore how p58 production may be blocked, transfected cells were pulse-labeled for 30 min with [35S]methionine and chased in the presence or absence of either BMS-824 or a PI. Immunoprecipitation with NS5A-specific antibody was used to monitor p58 production. At the end of the labeling period (time zero), p56 is predominantly seen, a portion of which was chased into p58 by 4 h (Fig. 7C). When either BMS-824 or PI was included during the chase, p58 was not detected (Fig. 7C) (protease not shown). Given that both BMS-824 and the PI blocked p58 production, these results suggest that p58 is made after p56 either from a polyprotein or during complex formation, which can be disrupted by the binding of NS3 by the PI or NS5A by BMS-824.
DISCUSSION
The use of a cell-based HCV replicon assay to screen a diverse collection of compounds resulted in the identification of a specific inhibitor of HCV. The initial lead, BMS-858, demonstrated a modest potency of ∼1 μM versus the HCV replicon but could be used to generate a resistant HCV replicon cell line. SAR exploration led to the discovery of a related compound (BMS-824) that was cross-resistant to the BMS-858-selected cells and was >100-fold more potent against HCV. Resistance mapping of both BMS-858- and BMS-824-selected cells suggested that NS5A is the target of these compounds. Since NS5A is a novel target with no known enzymatic activity, we built our confidence in these inhibitors by using various viral and enzymatic counterscreens to show specificity for HCV and to show that adaptive mutations do not affect potency and multiple assays to demonstrate that resistance is virus based.
Resistance selection with BMS-858 and BMS-824 yielded substitutions of Y93H and Y93C, and L31V/Q54L, respectively, in NS5A. Replicons containing these mutations showed substantially reduced susceptibility to both BMS-858 and BMS-824. Two structurally related compounds induced different resistance substitutions but did exhibit cross-resistance, providing useful information for SAR development to improve potency and specificity as well as further define the binding pocket and reveal additional contact residues involved in inhibitor activity. Another group also reported NS5A inhibitors that select for Y93H as well as additional resistance mutations A92V and R157W, since their chemotype is different (6). The Y93H mutant replicated at only 30 to 40% of the level of the wild type, indicating that its fitness is greatly reduced. Although the L31V/Q54L double mutant had a minimal impact on replication efficiency, replicating at 90 to 100% of the level of the wild type, the requirement for two linked mutations to achieve high-level resistance suggests a higher genetic barrier for this resistance to emerge.
Studies with the chimeric NS5A replicons demonstrated that the lack of genotype 1a coverage with BMS-824 can be attributed to the first 76 amino acids of NS5A since the transfer of just these residues is sufficient to transfer sensitivity to the inhibitor. Amino acids 1 to 76 of genotype 1a and 1b NS5A proteins show 80% identity, similar to the overall level of homology for the entire NS5A protein, with just 15 amino acid differences between the two genotypes. Although both the resistance mutations and the inhibitor sensitivity region map to the first 100 amino acids of NS5A, they are distinct. Residues 31 and 93, key resistance residues, are the same in genotypes 1a and 1b, yet BMS-858 and BMS-824 are not active against the genotype 1a strain, suggesting that the region involved in genotype 1a/1b coverage can be separated from the primary resistance sites. This is further supported by the fact that the L31V substitution in a genotype 1a replicon does confer resistance to a related compound with genotype 1a activity. This suggests that the primary conformation of NS5A, or of NS5A in the replication complex, is the major determinant for inhibitor sensitivity, while L31 is one of the determinants for resistance selection. Although amino acid 54 differs between the two genotypes, initial mutagenesis studies suggested that this change does not account for the different sensitivities of genotypes 1a and 1b (data not shown). In the course of determining the key N-terminal residues responsible for inhibitor sensitivity, most of the 15 amino acids that differ between genotype 1a and 1b NS5A have been changed to the other subtype. The genotypic and phenotypic analysis of these mutants and their sensitivity to NS5A inhibitors will be presented elsewhere. BMS-665 was also active against the genotype 2a infectious virus, showing only a slight loss in potency compared to the genotype 1b replicon, suggesting that this class of compounds may have the potential for broad genotype coverage.
Both the resistance mutations and the region affecting genotype 1a/1b coverage map to the N terminus of NS5A. This region is conserved among HCV genotypes, suggesting that it plays an important role in HCV replication. The NS5A protein consists of three putative domains (domains I, II, and III), with the resistance mutations residing in domain I. Domain I consists of the first 213 amino acids of the protein and contains a membrane-anchoring helix in the N-terminal 30 amino acids. An 18-mer peptide derived from this region (amino acids 3 to 20) was previously shown to be a potent inhibitor of HCV as well as other members of the Flaviviridae and HIV (5). Inhibition of the HCV replicon by BMS-858 and BMS-824 occurs by a mechanism distinct from the inhibitory effect of the peptide. Downstream of the alpha-helix, there is a zinc-binding motif within the N terminus of domain I, although the role of zinc binding in NS5A function is unclear. The structure of domain I was previously determined (20, 33), and it was shown to form a dimer via contacts near the N-terminal ends of the molecules, which can adopt different conformations. It is possible that our compounds interfere with NS5A dimer formation, as residue 93 forms part of the interface between two NS5A molecules. We have tried to explore this with pulldown experiments using replicons expressing His- and Flag-tagged NS5A proteins. However, to date, the data are not conclusive, and it is unclear whether this is due to a technical issue or because dimers are forming in cis and not in trans. Whether BMS-824 inhibition is due to its targeting a small region of the interface critical to the dimer interaction needs to be determined since small nonpeptidic molecules that interfere with protein-protein interactions have been identified (3). NS5A has also been shown to interact with RNA, and it is possible that the BMS-858/BMS-824 series affects its ability to bind RNA, which is a subject for future study. Domain I has been shown to interact with a number of host cell proteins, including cellular kinases, and an effect on a kinase pathway could account for the significant potency of BMS-824. It seems unlikely that our compounds are targeting a kinase, however, since a similar potency was observed previously for cell lines expressing different levels of hyperphosphorylated NS5A due to their distinct adaptive mutations or an A2199T substitution (24), which greatly increases levels of p58 production (data not shown). A kinase inhibitor would most likely exhibit an inverse correlation between the level of NS5A hyperphosphorylation and replication inhibition, as was previously shown for inhibitors of protein kinase CKI-α which is involved in the hyperphosphorylation of HCV NS5A (27).
Consistent with the requirement for expression of the HCV NS3-NS5A polyprotein for NS5A hyperphosphorylation (23), we have shown that NS5A and NS3 PIs, but not an NS5B polymerase inhibitor, altered NS5A p58 formation. However, collectively, information from this study and other previously reported data raise several interesting questions: does the alteration of NS5A p58 formation by NS3 or NS5A inhibitors occur when NS3 and/or NS5A is still part of a polyprotein or fully processed proteins? If they are in the fully processed form, are NS3 and NS5A in a replication complex? Why does the blockage of p58 formation and inhibition of replicon replication correlate? While it is easy to envision how an NS5A inhibitor can prevent p58 formation, it is not clear how the PI, which binds to NS3, would be having an effect. As shown in Fig. 5, the blockage of p58 formation by the NS3 PI occurred in a dose-dependent manner. This indicates that the blockage of p58 formation is due to the inhibitor binding either NS3 in the polyprotein form or processed NS3, either alone or as part of a replication complex. Although the binding of the PI to NS3 in the polyprotein form could explain the blockage of downstream NS5A hyperphosphorylation, we, and others, do not have any direct evidence that NS3 PIs bind NS3 in the polyprotein form. Given that (i) the NS3 PI used in this study is an active-site inhibitor and the cocrystallization of NS3 and inhibitors of this chemotype has been achieved (35) and (ii) data suggest that p56 is the precursor to p58 (15, 23), this would strongly support the hypothesis that the NS3 PI binds to NS3 and alters NS5A p58 formation within the same replication complex. This is consistent with results from the pulse-chase experiment showing that p58 production is still blocked by the addition of PI even when p56 is already made, suggesting that p58 is being formed from p56 in the context of a multiprotein complex, and this conversion can be disrupted by the binding of the NS3 inhibitor. Similarly, the alteration of NS5A hyperphosphorylation by adaptive mutations outside NS5A is most likely due to a conformational change of the kinase substrate NS5A, but studies with the NS3 inhibitor clearly demonstrate that a small, non-kinase inhibitor, specifically targeting a defined viral target, can also alter the hyperphosphorylation of NS5A, further building our confidence in the NS5A inhibitors. The fact that our NS5A inhibitors do affect hyperphosphorylation provides a phenotype that is linked with our compounds and an assay to assess the activity of our NS5A inhibitors. Furthermore, the suppression of p58 formation by NS5A inhibitors is suggestive of a conformational change in NS5A. Since the EC50 (concentration of NS5A inhibitor required to inhibit 50% of replicon replication) and IC50 (concentration of NS5A inhibitor required to block 50% of p58 formation) of our inhibitors correlate, it clearly demonstrates that very small amounts of NS5A inhibitors can have a profound effect on the NS5A conformation. Our current working model, based on these compounds and additional more potent analogs, is that NS5A forms an oligomeric complex consisting of multiple replication sites and that a single inhibitor molecule binds to an NS5A dimer and induces a conformational change that is communicated to adjacent unbound NS5A oligomers. A cooperative conformational change in the NS5A polymer chain could block the entire viral RNA replication network.
There is a great need for additional HCV antivirals to provide more effective, better-tolerated treatment options and for use in combination therapy, which will be necessary for building successful treatment regimens. In general, the development of antivirals has focused on viral proteins with known enzymatic activities, such as protease or polymerase. This is the first paper in a planned series showing that the NS5A protein of HCV can be targeted by small molecules, resulting in specific and potent inhibition. Although essential for HCV replication, the function of NS5A is still poorly understood, and no enzymatic activity has been ascribed to it. The exploration and optimization of the BMS-858/BMS-824 chemotype have yielded a number of interesting NS5A inhibitors displaying different phenotypes, which provide a powerful set of tools to study NS5A function. Further work with this compound series should allow us to determine the exact mechanism of action of BMS-824 and enhance our understanding of the function of the NS5A protein. Additional work in this series, which will be reported elsewhere, has also led to the development of more potent inhibitors, which have recently shown excellent effects on HCV-infected subjects (25), demonstrating a proof of concept that inhibitors targeting a nontraditional target like NS5A can have profound antiviral effects in vivo and that in vitro potency can translate into in vivo effect.
Acknowledgments
We thank Ralf Bartenschlager for providing the native Huh-7 cell line, and were are grateful to our Bristol-Myers Squibb (BMS) colleagues Jin-hua Sun, Karen Rigat, Brian Terry, Cheryl Ferraro, Chris Cianci, Steve Levine, Ronald Rose, Betsy Eggers, and Michael Wichroski for their contribution to the screening of the BMS deck or performing counterscreens. In addition, we thank our BMS colleagues for helpful discussions during the course of this work and Mark Cockett for his continued support.
Footnotes
Published ahead of print on 7 October 2009.
REFERENCES
- 1.Appel, N., U. Herian, and R. Bartenschlager. 2005. Efficient rescue of hepatitis C virus RNA replication by trans-complementation with nonstructural protein 5A. J. Virol. 79:896-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behrens, S. E., L. Tomei, and R. DeFrancesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 3.Berg, T., S. B. Cohen, J. Desharnais, C. Sonderegger, D. J. Maslyar, J. Goldberg, D. L. Boger, and P. K. Vogt. 2002. Small-molecule antagonists of Myc/Max dimerization inhibit Myc-induced transformation of chicken embryo fibroblasts. Proc. Natl. Acad. Sci. U. S. A. 99:3830-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, G., A. Montero, P. Gastaminza, C. Whitten-Bauer, S. F. Wieland, M. Isogawa, B. Fredericksen, S. Selvarajah, P. A. Gallay, M. R. Ghadiri, and F. V. Chisari. 2008. A virocidal amphipathic α-helical peptide that inhibits hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 105:3088-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conte, I., C. Giuliano, C. Ercolani, F. Narjes, U. Koch, M. Rowley, S. Altamura, R. De Francesco, P. Neddermann, G. Migliaccio, and I. Stansfield. 2009. Synthesis and SAR of piperazinyl-N-phenylbenzamides as inhibitors of hepatitis C virus RNA replication in cell culture. Bioorg. Med. Chem. Lett. 19:1779-1783. [DOI] [PubMed] [Google Scholar]
- 7.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Failla, C., L. Tomei, and R. DeFrancesco. 1994. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J. Virol. 68:3753-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foster, G., and P. Mathurin. 2008. Hepatitis C virus therapy to date. Antivir. Ther. 13(Suppl. 1):3-8. [PubMed] [Google Scholar]
- 10.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. A second hepatitis C virus-encoded proteinase. Proc. Natl. Acad. Sci. U. S. A. 90:10583-10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grakoui, A., C. Wychowski, C. Lin, S. M. Feinstone, and C. M. Rice. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67:1385-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo, J., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holler, T. P., T. Parkinson, and D. C. Pryde. 2009. Targeting the non-structural proteins of hepatitis C virus: beyond hepatitis C virus protease and polymerase. Expert Opin. Drug Discov. 4:3. [DOI] [PubMed] [Google Scholar]
- 14.Huang, L., J. Hwang, S. D. Sharma, M. R. Hargittai, Y. Chen, J. J. Arnold, K. D. Raney, and C. E. Cameron. 2005. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J. Biol. Chem. 280:36417-36428. [DOI] [PubMed] [Google Scholar]
- 15.Kaneko, T., Y. Tanji, S. Satoh, M. Hijikata, S. Shinichi, K. Kimura, and K. Shimotohno. 1994. Production of two phosphoproteins from the NS5A region of the hepatitis C viral genome. Biochem. Biophys. Res. Commun. 205:320-326. [DOI] [PubMed] [Google Scholar]
- 16.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemm, J. A., M. Liu, R. E. Rose, R. Fridell, D. R. O'Boyle II, R. Colonno, and M. Gao. 2005. Replication-competent chimeric hepatitis C virus subgenomic replicons. Intervirology 48:183-191. [DOI] [PubMed] [Google Scholar]
- 18.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lohmann, V., F. Korner, J.-O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNA in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 20.Love, R. A., O. Brodsky, M. J. Hickey, P. A. Wells, and C. N. Cronin. 2009. Crystal structure of a novel dimeric form of NS5A domain I protein from hepatitis C virus. J. Virol. 83:4395-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacDonald, A., and M. Harris. 2004. Hepatitis C virus NS5A: tales of a promiscuous protein. J. Gen. Virol. 85:2485-2502. [DOI] [PubMed] [Google Scholar]
- 22.Miyanari, Y., K. Atsuzawa, N. Usuda, K. Watashi, T. Hishiki, M. Zayas, R. Bartenschlager, T. Wakita, M. Hijikata, and K. Shimotohno. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089-1097. [DOI] [PubMed] [Google Scholar]
- 23.Neddermann, P., A. Clementi, and R. De Francesco. 1999. Hyperphosphorylation of the hepatitis C virus NS5A protein requires an active NS3 protease, NS4A, NS4B, and NS5A encoded on the same polyprotein. J. Virol. 73:9984-9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neddermann, P., M. Quintavalle, C. Di Pietro, A. Clementi, M. Cerretani, S. Altamura, L. Bartholomew, and R. De Francesco. 2004. Reduction of hepatitis C virus NS5A hyperphosphorylation by selective inhibition of cellular kinases activates viral RNA replication in cell culture. J. Virol. 78:13306-13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nettles, R., C. Chien, E. Chung, A. Persson, M. Gao, M. Belema, N. A. Meanwell, M. P. DeMicco, T. C. Marbury, R. Goldwater, P. Northup, J. Coumbis, W. K. Kraft, M. R. Charlton, J. C. Lopez-Talavera, and D. Grasela. 2008. BMS-790052 is a first-in-class potent hepatitis C virus (HCV) NS5A inhibitor for patients with chronic HCV infection: results from a proof-of-concept study, abstr. LB12. Abstr. 59th Annu. Meet. Am. Assoc. Study Liver Dis., San Francisco, CA.
- 26.O'Boyle, D. R., II, P. T. Nower, J. A. Lemm, L. Valera, J.-H. Sun, K. Rigat, R. Colonno, and M. Gao. 2005. Development of a cell-based high-throughput specificity screen using a hepatitis C virus-bovine viral diarrhea virus dual replicon assay. Antimicrob. Agents Chemother. 49:1346-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quintavalle, M., S. Sambucini, C. Di Pietro, R. De Francesco, and P. Neddermann. 2006. The α isoform of protein kinase CKI is responsible for hepatitis C virus NS5A hyperphosphorylation. J. Virol. 80:11305-11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ripka, A., J. A. Campbell, A. C. Good, P. M. Scola, N. Sin, and B. Venables. March 2004. Hepatitis C virus inhibitors. U.S. patent 20040048802 A1.
- 29.Romine, J. L., S. W. Martin, L. B. Snyder, M. Serrano-Wu, M. Deshpande, D. Whitehouse, J. Lemm, D. O'Boyle, M. Gao, and R. Colonno. February 2007. Iminothiazolidinones as inhibitors of HCV replication. U.S. patent 7,183,302 B2.
- 30.Tai, C.-L., W.-K. Chi, D.-S. Chen, and L.-H. Hwang. 1996. The helicase activity associated with hepatitis C virus nonstructural protein 3 (NS3). J. Virol. 70:8477-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanji, Y., T. Kaneko, S. Satoh, and K. Shimotohno. 1995. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J. Virol. 69:3980-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tellinghuisen, T. L., J. Marcotrigiano, A. E. Gorbalenya, and C. M. Rice. 2004. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J. Biol. Chem. 279:48576-48587. [DOI] [PubMed] [Google Scholar]
- 33.Tellinghuisen, T. L., J. Marcotrigiano, and C. M. Rice. 2005. Structure of the zinc-binding domain of an essential replicase component of hepatitis C virus reveals a novel fold. Nature 435:374-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tellinghuisen, T. L., K. L. Foss, and J. Treadaway. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4:e1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsantrizos, Y. S., G. Bolger, P. Bonneau, D. R. Cameron, N. Goudreau, G. Kukolj, S. R. LaPlante, M. Llinas-Brunet, H. Nar, and D. Lamarre. 2003. Macrocyclic inhibitors of the NS3 protease as potential therapeutic agents of hepatitis C virus infection. Angew. Chem. Int. Ed. Engl. 42:1355-1360. [DOI] [PubMed] [Google Scholar]
- 36.Yanagi, M., R. Purcell, S. Emerson, and J. Bukh. 1997. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. U. S. A. 94:8738-8743. [DOI] [PMC free article] [PubMed] [Google Scholar]