Abstract
Human cytomegalovirus (HCMV) is a member of the betaherpesvirus family that, unlike other herpesviruses, triggers a strong innate immune response in infected cells that includes transcription of the beta interferon gene via activation of interferon regulatory factor 3 (IRF3). IRF3 activation requires signaling from pattern recognition receptors that is initiated by their interaction with specific pathogen-associated molecules. However, while IRF3-activating pathways are increasingly well characterized, the cellular molecules involved in HCMV-mediated IRF3-dependent beta interferon transcription are virtually unknown. We undertook a systematic examination of new and established IRF3-terminal pathway components to identify those that are essential to HCMV-triggered IRF3 activation. We show here that IRF3 activation induced by HCMV infection involves the newly identified protein STING but, in contrast to infections with other herpesviruses, occurs independently of the adaptor molecule IPS-1. We also show that the protein DDX3 contributes to HCMV-triggered expression of beta interferon. Moreover, we identify Z-DNA binding protein 1 (ZBP1) as being essential for IRF3 activation and interferon beta expression triggered by HCMV, as well as being sufficient to enhance HCMV-stimulated beta interferon transcription and secretion. ZBP1 transcription was also found to be induced following exposure to HCMV in a JAK/STAT-dependent manner, thus perhaps also contributing to a positive feedback signal. Finally, we show that constitutive overexpression of ZBP1 inhibits HCMV replication. ZBP1 was recently identified as a cytosolic pattern recognition receptor of double-stranded DNA, and thus, we propose a model for HCMV-mediated IRF3 activation that involves HCMV-associated DNA as the principal innate immune-activating pathogen-associated molecular pattern.
Human cytomegalovirus (HCMV) is the archetypal member of the betaherpesvirus family, an ancient group of large enveloped double-stranded DNA (dsDNA) viruses. HCMV is a ubiquitous, opportunistic, and immunoevasive pathogen that persists indefinitely in immunocompetent hosts despite continuously inducing strong antiviral immune responses. HCMV infection is mostly asymptomatic in healthy individuals, but the virus is a significant cause of morbidity and mortality during artificial or human immunodeficiency virus (HIV)-induced immunosuppression (33, 43). HCMV is also the leading microbial cause of birth defects when acute infection occurs during pregnancy (3). In addition, persistent infection of the vasculature and subsequent chronic inflammatory stimulation have been strongly linked with the development of cardiovascular diseases, such as atherosclerosis, restenosis, and transplant vascular sclerosis (reviewed in reference 66).
The innate immune response against incoming pathogens plays a pivotal role during primary infection, especially in patients with defects in adaptive immunity. Indeed, HCMV infection of host cells rapidly triggers a strong induction of type I interferons (IFNs), IFN-stimulated genes (ISGs), and proinflammatory cytokines (1, 4, 8, 20, 38, 63, 84, 85), a process that serves to establish an antiviral state in infected and neighboring cells. Type I IFNs include IFN-β (secreted by epithelial cells and fibroblasts) and IFN-α subtypes 1 to 14 (macrophages and dendritic cells). IFN-α/β activate signaling pathways via the type I IFN receptor, involving phosphorylation of signal transducer and activator of transcription 1 (STAT1) and STAT2. STAT1 and STAT2 lead to the transcription of ISGs that encode proteins that act to impair intracellular molecular activities required for virus replication (56). Currently, it is not known how HCMV initially triggers the IFN response or how the virus manages to escape the antiviral activities of that response.
Significant progress has been made in recent years regarding our understanding of the pathways and molecules involved in the induction of IFN and ISGs, especially by RNA viruses (see references 25 and 45 for reviews). In response to activation signals initiated by pathogen-associated molecular patterns (PAMPs), an enhanceosome complex containing IFN regulatory factor 3 (IRF3), nuclear factor κB (NF-κB), activating transcription factor 2, CREB-binding protein (CBP), and p300 is formed in the IFN-β promoter region (29, 34, 75, 81). While evidence suggests that IFN-β induction can occur in the absence of NF-κB (73), IRF3 is necessary for IFN responses to viral, including HCMV (58), infection in fibroblasts (14).
IRF3 is a constitutively expressed protein that normally shuttles freely between the cytoplasm and the nucleus. However, PAMP-triggered phosphorylation of C-terminal serine residues results in IRF3 homodimerization, association with coactivators (CBP and p300), and nuclear accumulation (18, 29, 60, 62, 81). Activation of IRF3 occurs in response to infection with HCMV (4, 14, 20, 24, 37, 38, 42, 47), a betaherpesvirus, and an ICP0 deletion mutant of the alphaherpesvirus herpes simplex virus type 1 (HSV-1) (11, 30, 36, 47). Interestingly, HCMV-mediated IRF3 activation and IFN-β transcription are intensified in the absence of viral-gene expression (14), most likely due to the inactivation of HCMV-encoded inhibitory proteins that impair NF-κB function (70). However, there has been no demonstration of an HCMV-specific obstructive effect on IRF3 activity. Interestingly, the closely related CMV of rhesus macaques completely blocks IRF3 activation even in the absence of viral-gene expression (13). It is also important to note that activated IRF3 can itself induce the expression of a subset of ISGs (independently of IFN), which leads to generation of an antiviral cellular state (5, 8, 14, 42, 84, 85). This HCMV-triggered IFN-independent induction of ISGs appears to play a prominent role in the innate immune response to the virus (42), presumably because HCMV can interfere with IFN-dependent (JAK/STAT) signaling (44).
Phosphorylation of IRF3 occurs via TANK binding kinase 1 (TBK1) and IκB kinase ɛ (IKKɛ) (18, 29, 60, 62, 81). While IRF3 is thought to be a direct target of these kinases, recent work has identified another molecule that is required for TBK1/IKKɛ-mediated IRF3 activation, perhaps as an adaptor. Stimulator of IFN genes (STING) (23), or mediator of IRF3 activation (MITA) (83), is an endoplasmic reticulum (ER)- and mitochondrion-associated protein essential to IRF3 activation in response to Sendai virus (SeV), HSV-1, Listeria monocytogenes, and IFN-stimulatory dsDNA. Zhong et al. indicated that STING is a phosphorylation target of TBK1 (83), but the molecule's exact biochemical role with respect to IRF3 activation is not clear. Furthermore, DEAD box protein 3 (DDX3) was also found to be essential for IRF3-dependent transcription. DDX3 is a helicase and phosphorylation target of TBK1 that binds to the IFN-β promoter independently of IRF3 (59, 64). DDX3 contributes to IFN-β induction triggered by Listeria, transfected polyinosine-polycytosine [poly(I·C)], transfected poly(dA-dT), lipopolysaccharide (LPS), SeV, and vaccinia virus (59, 64). The importance of DDX3 for antiviral immune responses is underscored by the fact that it is a target of inhibition for the vaccinia virus protein K7 (59).
Signals terminating in IRF3 phosphorylation originate from pattern recognition receptors (PRRs), which are either cell surface, endosomal, or cytoplasmic proteins that detect specific PAMPs. IRF3-specific PRRs include Toll-like receptors (TLRs) and cytoplasmic helicases (see reference 72 for a review). Human TLRs include 12 known molecules (69), of which only TLR3 and TLR4 (reacting with extracellular dsRNA and LPS, respectively) lead to IRF3 activation. Retinoic acid-inducible gene I (RIG-I) (80) and melanoma differentiation-associated gene 5 (MDA5) (79) are RNA helicases that contain N-terminal caspase recruitment domains (CARDs) that recognize cytoplasmic poly(I·C) and virus-associated dsRNA (12, 21, 46, 52, 53, 79). Activation of IRF3-specific kinases by PRRs further involves adaptor molecules that act as points of integration for multiple upstream signaling receptors. These include Toll/interleukin-1 receptor domain-containing adaptor-inducing IFN-β (TRIF) (78) (also called TICAM1 [41]) and the CARD-containing molecule IFN-β promoter stimulator 1 (IPS-1; also called MAVS, VISA, and CARDIF [26, 35, 61, 77, 80]). As such, TLR3 and TLR4 require TRIF, while MDA5 and RIG-I employ IPS-1 for activation of IRF3-specific kinases. IRF3 phosphorylation in response to SeV and transfected poly(I·C) suggests that human fibroblasts (a common HCMV in vitro target) possess functional cytoplasmic RNA helicase PRRs and IPS-1. IPS-1 is essential for IRF3 activation, not only by diverse RNA viruses (see reference 28 for a review), but also by alphaherpesviruses (10, 48, 49) and vaccinia virus (82). The phylogenetic diversity of viruses triggering IRF3 activation through IPS-1 implies that the protein may in fact be an essential component of all innate antiviral cellular activation.
IRF3-dependent, but TLR-independent, transcription of IFN-β is also triggered by the presence of cytoplasmic dsDNA (22, 65). Recent work has indicated the existence of at least two separate IRF3-terminal pathways activated by cytoplasmic AT-rich dsDNA. Cytoplasmic murine Z-form DNA binding protein 1 (ZBP1) (also called DLM-1 [15, 19, 51]) was first shown to be involved in IFN-β induction by both cytosolic DNA and HSV-1 (68, 74). The protein was subsequently given the additional name DNA-dependent activator of IRFs (DAI) (68). However, induction of IFN-β by HSV or dsDNA can occur in the absence of ZBP1 (31, 68, 74), indicating the presence of an additional dsDNA-sensing pathway. RNA polymerase III (POL3) was recently shown to convert cytoplasmic AT-rich dsDNA into dsRNA containing a 5′ triphosphate moiety that then activates canonical RIG-I-dependent signaling (2, 10), a pathway to which IPS-1 is essential. In human cells, the POL3-dependent pathway is important for IFN-β activation by multiple DNA viruses, including HSV-1, adenovirus, and Epstein-Barr virus (2, 10), leading to speculation that IPS-1-dependent pathways might be central to all cytoplasmic virus detection (10).
While roles for TBK1 (20) and IRF3 (14) in HCMV-mediated IRF3 activation and IFN-β induction are established, the identities of additional upstream receptors and signaling molecules are unknown. Interestingly, HCMV particle entry is necessary for IRF3 activation and ISG induction, perhaps an indication of the importance of a cytoplasmic PRR (24, 38). It has also been speculated that membrane fusion events occurring during viral entry might activate IRF3 (24). Research using human fibroblasts indicates that TLR molecules are not involved (24, 42) but that an uncharacterized pathway may be (40). Since a systematic evaluation of key members of innate antiviral pathways has not been performed with HCMV, we decided to examine the roles of multiple components of the IRF3 activation pathways in this context. Our results indicate that, unlike other herpesviruses, IPS-1, the key adaptor protein for POL3 and RNA helicase-dependent signaling, is not involved in IFN induction by HCMV. In contrast, the dsDNA sensor ZBP1 is essential for HCMV-mediated IFN-β transcription and the generation of an antiviral response, thus suggesting that the HCMV-associated PAMP responsible for triggering IRF3 activation is dsDNA. In addition, STING also contributes to HCMV-mediated IRF3 activation and DDX3 plays a role in HCMV-mediated IFN-β transcription.
MATERIALS AND METHODS
Reagents.
The dsRNA mimic poly(I·C) was obtained from Amersham Biosciences (product no. 27-4729) and resuspended in Millipure water. LPS was obtained from Sigma (product no. L2654) and resuspended in Millipure water. Cycloheximide (CHX) was obtained from Sigma (product no. C 4859) and used at 200 μg ml−1 cell culture medium. Hygromycin B was obtained from InvivoGen (product no. ant-hg-1) and used at 300 μg ml−1 cell culture medium. Puromycin was obtained from Clontech (product no. 631305) and used at 2 μg ml−1 cell culture medium. Lipofectamine 2000 transfection reagent was obtained from Invitrogen (product no. 11668-019) and used according to the manufacturer's instructions. HiPerfect transfection reagent was obtained from Qiagen (product no. 301702). Human recombinant IFN-β was obtained from PBL (product no. 11415-1).
Virus and cell culture.
Human foreskin fibroblasts stably transfected with the human telomerase gene (THFs) to extend passage life were obtained from W. Bresnahan (University of Minnesota) (6). THFs were propagated in Dulbecco's minimal essential medium (DMEM) containing 10% fetal calf serum (FCS) and antibiotics at 37°C in 5% CO2. HCMV strain AD169 was obtained from the ATCC, propagated on primary human foreskin fibroblasts, and purified by centrifugation through a 30% sorbitol cushion for 1 h at 22,000 rpm in a Beckman SW32 rotor. The titers of virus stocks were determined using endpoint serial dilution assays on primary human fibroblasts (50). Unless otherwise indicated, HCMV infections were performed in duplicate in confluent 35-mm dishes with 0.5 ml DMEM at a multiplicity of infection (MOI) of 3 for 6 h. 1 ml DMEM with 10% FCS was added back to the infected cells after the initial 1 h. Inactivation of HCMV particles using UV irradiation was performed in a Stratalinker for a length of time sufficient to block expression of protein from the HCMV open reading frame UL123 (IE1) and to induce IRF3 nuclear localization as determined by an immunofluorescence assay, as described previously (14). SeV was obtained from Charles River Laboratories and used at a concentration of 160 hemagglutinin units ml−1 of cell culture medium.
RNA interference.
Cells were plated at 30 to 40% confluence in 35-mm dishes the day before transfection with small interfering RNA (siRNA). Five microliters of siRNA (20 μM stock) was mixed with 10 μl HiPerfect in 95 μl OptiMem (Gibco) and added to cells containing 2.3 ml OptiMem. The cells were transfected twice, 8 h apart, and incubated for 16 h, and the OptiMem was replaced with DMEM with 10% FCS. The cells were allowed to expand for 3 to 4 days to near confluence and transfected once more 16 h before treatment. siRNA sequences against IRF3 and STING were as described in references 14 and 23, respectively. Other siRNA sequences were as follows: NS, 5′-UCGUAAGUAAGCGCAACCC-3′; IPS-1 5′-GGGUUCUUCUGAGAUUGAA-3′; DDX3, 5′-CGAGAGAGUUGGCAGUACA-3′; ZBP1 no. 06, 5′-GGCCACCUUGAACAAAGAA-3′; ZBP1 no. 57, 5′-GGGCGGGACUGAUCCUGAA-3′.
Generation of stable cell lines.
THF lines either stably expressing ZBP1 or STAT1 Y701F or containing IRF3-dependent firefly luciferase were constructed using the pRevTRE retroviral plasmid system (Clontech) as described previously (9). Full-length ZBP1 was subcloned from a pcDNA3.1 expression vector (described in reference 15) into pRevTRE to make THF-ZBP1 cells. STAT1 Y701F was similarly subcloned from a pRc/CMV vector (described in reference 76) into pRevTRE. IRF3 reporter THFs (THF-P55C1B) were generated by stably transfecting a luciferase open reading frame downstream of an IRF3-dependent promoter element containing repeated PRD1 sites (termed P55-C1B; originally described in reference 81). The P55-C1B-luciferase region was subcloned from the original pBL plasmid (a kind gift from T. Fujita, Kyoto University) into pRevTRE. Proper target insertion was verified by sequencing. Replication-defective recombinant retrovirus was produced by transfecting pRevTRE into Phoenix A cells using Lipofectamine 2000 (Invitrogen) and harvesting the supernatant after 48 h (27). Cell debris was removed from the supernatant by centrifugation (3,000 × g; 10 min), and the supernatant was filtered through two 0.45-μm filters. Subconfluent target cells were infected with retrovirus for 16 h in the presence of 4 μg/ml Polybrene. After the cells reached confluence, they were split into DMEM plus 10% FCS containing 100 μg/ml hygromycin. The transduced cells were passaged in the presence of increasing hygromycin (to 300 μg/ml) until the cultures were resistant to the compound. Four separately constructed cultures of hygromycin-resistant THF-P55C1B cells were assayed for their responsiveness to HCMV exposure in the form of luciferase expression, and the culture exhibiting the largest dynamic range of expression was used for all assays.
IFN-sensitive reporter THFs (THF-ISRE) were constructed using the pGreenFire ISRE reporter lentivector system according to the manufacturer's protocol (System Biosciences). Briefly, pGreenFire ISRE is a dual reporter vector containing an IFN-sensitive responsive element (ISRE) that controls the expression of both green fluorescent protein (GFP) and firefly luciferase. Vesicular stomatitis virus G protein-pseudotyped lentiviral particles were generated by cotransfecting 293T cells with pGreenFire-ISRE and packaging expression vectors encoding vesicular stomatitis virus G, HIV Gag, and HIV Rev (pPACK-H1 plasmid mix; System Biosciences). The virus was harvested, and target cells were infected as described above. The cell cultures were purified by retrieving GFP-positive cells by fluorescence-activated cell sorting following overnight treatment with 1,000 U/ml human IFN-β. Experimentally, THF-ISRE cells were used to quantify the synthesis and secretion of type I IFN in two ways: first, to measure the IFN secretion from other (non-THF-ISRE) cells by exposing them to media harvested from target cells, and second, to measure IFN secretion from THF-ISRE themselves following conditional exposure to stimuli as described previously.
Luciferase reporter assays.
Confluent THF-P55C1B or THF-ISRE cells were treated as indicated in six-well dishes. At the time of assay, the cells were washed twice with 1× phosphate-buffered saline (PBS), trypsinized, spun down at 2,000 × g for 10 min, and resuspended in 200 μl DMEM-FCS. In duplicate, 75 μl of the cell mixture was added to 75 μl One-Glo lysis/luciferin reagent (Promega; no. E6120) in a black 96-well plate. Luminescence was determined on a Veritas luminometer (Turner Biosystems).
When quantifying the secretion of type I IFN by target cells, THF-ISRE cells were grown to confluence in black 96-well plates. Confluent target cells were treated with HCMV virions rendered transcriptionally inactive by UV irradiation (UV-HCMV) or SeV as described above in six-well dishes. At 3 h postinfection, the culture medium was removed, the cells were washed three times with 1× PBS (to completely remove residual virus), and 1.5 ml fresh DMEM-FCS was added. At 7 h postinfection, 50 μl medium from target cells was transferred to each of four wells of confluent THF-ISRE cells and incubated at 37°C. At 16 h posttreatment, 50 μl One-Glo reagent was added to each well, and the luminescence was read as described above.
RNA isolation and semiquantitative RT-PCR.
Total RNA was isolated using the Mini RNA Isolation II kit according to the manufacturer's protocol (Zymo Research; product no. R1030) and quantified by UV spectrometry. RNA samples were treated with DNase using the DNA-Free RNA Kit according to the manufacturer's protocol (Zymo Research; product no. 1013). Single-stranded cDNA for use as a PCR template was made from total RNA using random hexamers to prime first-strand synthesis by Superscript III reverse transcriptase (Invitrogen; product no. 11754) as described in the manufacturer's protocol. Comparison of mRNA expression between samples (e.g., infected versus untreated) was performed using SYBR green-based semiquantitative real-time RT-PCR (qPCR) with the Applied Biosystems Sequence Detection System according to the method of Livak and Schmittgen (32). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as a housekeeping gene to establish a baseline against which target genes were compared between samples (described in reference 14). Other primer sequences used were as follows: IFN-β (F), 5′-AAACTCATGAGCAGTCTGCA-3′, and (R), 5′-AGGAGATCTTCAGTTTCGGAGG-3′; ZBP1 (F), 5′-TGGTCATCGCCCAAGCACTG-3′, and (R), 5′-GGCGGTAAATCGTCCATGCT-3′.
Immunoblotting.
Native IRF3 immunoblotting was performed as described previously (13). Denaturing (sodium dodecyl sulfate) immunoblotting was performed as follows. Following cell trypsinization and centrifugal pelleting at 2,000 × g for 10 min, whole-cell lysates were harvested in 2% sodium dodecyl sulfate lysis buffer (50 mM Tris-HCl, 20% glycerol). The lysates were electrophoresed in 10% polyacrylamide gels and transferred onto polyvinylidene difluoride membranes (Millipore) using semidry transfer at 400 mA for 1.5 h. The blots were blocked at room temperature for 2 h using 10% nonfat milk in 1× PBS containing 0.1% Tween 20. The blots were exposed to primary antibody in 5% nonfat milk in 1× PBS containing 0.1% Tween 20 for 16 h at room temperature. The blots were then washed in 1× PBS containing 0.1% Tween 20 for 20, 15, and 5 min, followed by deionized water for 5 min. One hour of exposure to horseradish peroxidase-conjugated secondary antibodies and subsequent washes were as described for the primary antibodies. The antibodies were visualized using enhanced chemiluminescence (Pierce). The antigens and the antibodies used against them were as follows: GAPDH, Santa Cruz no. SC-51906; IRF3, Santa Cruz no. SC-9082; DDX3, Bethyl no. A300-474A; IPS-1, Bethyl no. A300-782A; phospho-IRF3, Millipore no. 07-581. ZBP1 was described previously (31); STING was a kind gift from G. Barber (University of Miami) (23).
RESULTS
IPS-1 is not required for HCMV-mediated, IRF3-dependent transcription or IFN-β secretion.
Based on the diversity of stimuli (viral and otherwise) that trigger IPS-1-dependent signaling and the possibility that the unidentified sensor(s) of HCMV may also signal via IPS-1, we asked whether this protein was required for HCMV-mediated IRF3 activation in human fibroblasts. Since the focus of this investigation was the characterization of factors involved in the activation (rather than inhibition) of IRF3 and IFN-β, we used UV-HCMV, as described in Materials and Methods. These particles are incapable of exhibiting phenotypes (known or unknown) that may dampen innate immune activation and associated signal readouts. UV-HCMV particles have been shown to properly deliver virion-associated proteins subcellularly (85) and are otherwise biologically similar to live virus.
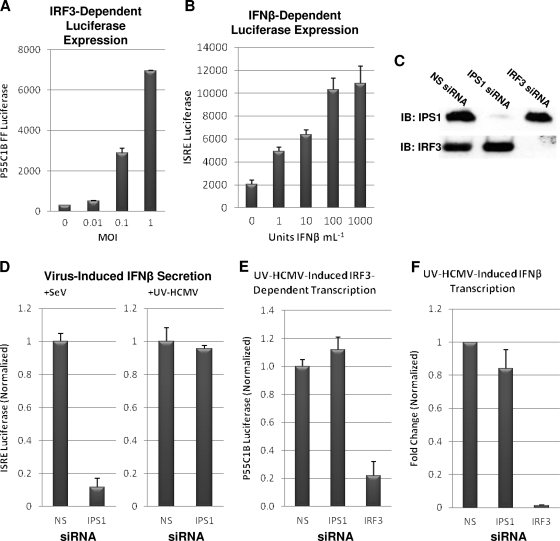
To examine the role of IPS-1, as well as other potential host genes, in HCMV-mediated activation of IRF3-dependent transcription and secretion, we constructed two THF reporter cell lines. Cell line THF-P55C1B contained firefly luciferase downstream of repeated PRD1 sites, which are DNA targets of phosphorylated IRF3 (81), and thus, luciferase is expressed in response to IRF3 activation by UV-HCMV. As shown in Fig. 1A, luciferase expression by THF-P55C1B cells occurs in response to exposure to UV-HCMV in a dose-dependent manner. The second cell line (THF-ISRE) contained an ISRE that controlled the expression of a GFP-plus-luciferase dual reporter in response to secreted or exogenously added type I IFN. As shown in Fig. 1B, luciferase expression by THF-ISRE cells occurred in response to IFN-β exposure in a dose-dependent manner. We next knocked down IPS-1 expression in these reporter cells using transfected siRNA. In contrast to siRNA containing a nonspecific (NS) sequence, siRNA directed against IPS-1 or IRF3 nearly eliminated their target protein (Fig. 1C). This degree of IPS-1 knockdown was sufficient to strongly reduce IFN-β secretion (as measured by ISRE-driven luciferase expression) triggered by SeV, a response requiring the protein (61) (Fig. 1D). However, unlike siRNA-mediated knockdown of IRF3, IPS-1 knockdown did not reduce IRF3- or IFN-dependent luciferase induction by HCMV relative to that observed in control cells (Fig. 1E).
FIG. 1.
IPS-1 does not contribute to UV-HCMV-mediated IRF3-dependent transcription, IFN-β transcription, or IFN-β secretion. (A) THF-P55C1B cells were treated in duplicate for 6 h with UV-HCMV at the indicated MOIs. The values presented are average levels of detected luciferase plus standard errors of the mean (SEM) from cell lysates; (B) THF-ISRE cells were treated in duplicate for 6 h with IFN-β at the indicated concentrations. The values presented are average levels of detected luciferase (plus SEM) from cell lysates. (C) Immunoblot (IB) showing diminishment of IPS-1 and IRF3 proteins from THF-ISRE cells treated with the indicated siRNAs. (D) IFN-β-dependent luciferase expression (plus SEM) from THF-ISRE cells following treatment with the indicated siRNAs and subsequent exposure to either SeV or UV-HCMV (MOI = 1). (E) IRF3-dependent luciferase expression (plus SEM) from THF-P55C1B cells following treatment with the indicated siRNAs and subsequent exposure to UV-HCMV (MOI = 1). (F) IFN-β gene transcription change relative to untreated cells following exposure of siRNA-treated THFs to UV-HCMV. The values are the changes of UV-HCMV-treated cells relative to untreated cells normalized to the changes obtained from THFs treated with NS siRNA (set to 1).
To examine the role of IPS-1 in HCMV-induced, IRF3-dependent transcription of endogenous host genes, we used qPCR to compare HCMV-triggered expression of the IFN-β gene in THFs transfected with IPS-1-specific or NS siRNA. While siRNA-mediated knockdown of IRF3 in THFs strongly inhibited IFN-β induction, IFN-β expression did not differ significantly between cells treated with NS and those treated with IPS-1-directed siRNA (Fig. 1F). Based on these results, we conclude that IPS-1 is not required for HCMV-mediated activation of IRF3 in human fibroblasts.
HCMV infection upregulates transcription of ZBP1 in a STAT1-dependent manner.
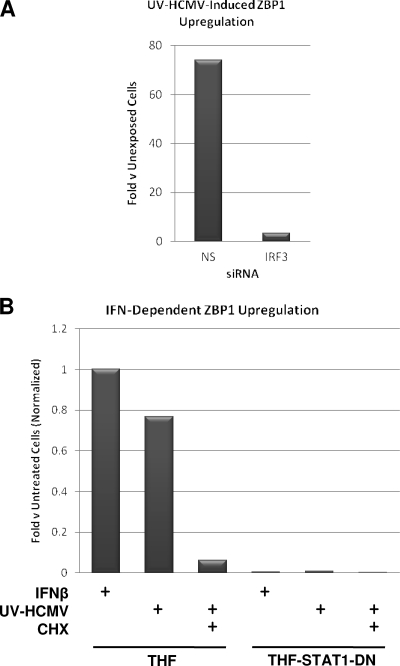
Recently, ZBP1 was identified as a sensor of cytoplasmic dsDNA in mouse embryonic fibroblasts, and upregulation of the gene following exposure to either transfected dsDNA or IFN-β was shown (68). We therefore decided to ask whether the same ZBP1 transcriptional dynamics are exhibited by human fibroblasts in response to treatment with HCMV. Exposure of THFs to UV-HCMV for 6 h resulted in ZBP1 transcriptional induction in control cells treated with NS siRNA relative to unexposed cells, as determined by qPCR (Fig. 2A). ZPB1 induction was inhibited in the presence of IRF3-directed siRNA (Fig. 2A), indicating that ZBP1 was either transcribed as a direct target of IRF3 or induced by IRF3-dependent IFN-β induction. We next examined whether IFN-β-stimulated JAK/STAT signaling was required for ZBP1 upregulation (as seen in murine cells) or whether ZBP1 induction by HCMV was IFN independent. To do this, we used parental THFs and THFs stably overexpressing a dominant-negative form of STAT1 containing a Y→F mutation at amino acid 701 (THF-STAT1-DN). First, both cell types were exposed to IFN-β (100 U/ml) for 6 h, and qPCR was used to compare ZBP1 expression to that in untreated cells. As shown in Fig. 2B, IFN-β triggered upregulation of ZBP1 relative to untreated cells in THF but not THF-STAT1-DN cells. Next, the cells were exposed to UV-HCMV (MOI = 3) in the presence or absence of CHX, a treatment done to block translation of all proteins, including IFN-β. In this case, UV-HCMV treatment induced ZBP1 transcription in THFs, but not in the presence of CHX (Fig. 2B). Furthermore, UV-HCMV failed to induce ZBP1 in THF-STAT1-DN cells under any conditions (Fig. 2B). Based on these observations, we conclude that upregulation of ZBP1 transcription following exposure to UV-HCMV is likely the result of JAK/STAT pathway stimulation by virus-induced, IRF3-dependent IFN-β secretion.
FIG. 2.
IRF3 activation and STAT1-dependent signaling contribute to UV-HCMV-induced upregulation of ZBP1 mRNA. (A) Change as determined by qPCR of ZBP1 mRNA in THFs exposed to UV-HCMV relative to that in unexposed cells following transfection of NS or IRF3-directed siRNA. (B) Change of ZBP1 mRNA relative to untreated cells in THFs and THF-STAT1-DN cells treated with IFN-β (100 U/ml) or UV-HCMV in the presence and absence of CHX (200 μg/ml; untreated control cells were also treated with CHX). The values are the changes in treated relative to untreated cells normalized to the change observed in IFN-β-treated THFs (set to 1).
ZBP1 is required for UV-HCMV-mediated IRF3-dependent transcription and IFN-β synthesis.
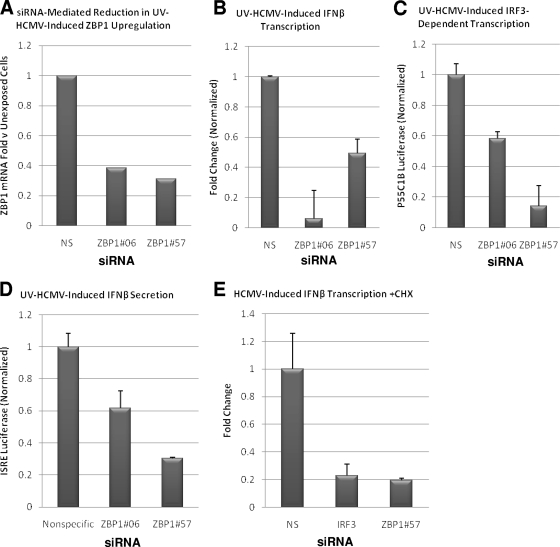
As mentioned above, ZBP1 was implicated in the induction of IFN-β by both cytosolic dsDNA and infection with HSV-1 (68, 74). To determine whether ZBP1 is involved in the IFN response triggered by HCMV, we examined the activation of innate immune signaling by UV-HCMV following knockdown of ZBP1. In untreated THFs, ZBP1 mRNA is expressed at low levels (not shown), but as discussed above (Fig. 2), transcription is strongly induced following exposure to UV-HCMV. Unfortunately, the reactivity of available anti-ZBP1 antibodies was too poor to observe endogenous cellular protein following HCMV-triggered expression. Nevertheless, two different siRNAs directed against ZBP1 were able to substantially reduce the upregulation of ZBP1 mRNA following exposure to UV-HCMV (Fig. 3A), as well as to knock down stably overexpressed ZBP1 protein in THFs (see below). As shown in Fig. 3B, treatment of THFs with ZBP1-targeted siRNA reduced HCMV-triggered IFN-β transcription relative to cells treated with NS siRNA. Similarly, IRF3- and IFN-β-dependent luciferase expression from THF-P55C1B and THF-ISRE cells, respectively, in response to HCMV was also reduced when the cells were transfected with ZBP1-directed siRNA prior to virus exposure (Fig. 3C and D).
FIG. 3.
ZBP1 contributes to UV-HCMV-triggered IRF3-dependent transcription and IFN-β secretion. (A) qPCR showing UV-HCMV-triggered induction of ZBP1 mRNA relative to untreated THFs in the presence of ZBP1-targeted or NS siRNA. The values represent ZBP1 mRNA change relative to that of unexposed cells normalized to the change observed in cells treated with NS siRNA (set to 1). (B) Change in IFN-β gene transcription relative to that in unexposed cells following exposure of siRNA-treated THFs to UV-HCMV. The values (plus standard errors of the mean) are the changes in UV-HCMV-treated cells relative to untreated cells normalized to the changes obtained from THFs treated with NS siRNA (set to 1). (C) Expression of IRF3-dependent luciferase following treatment of THF-P55C1B cells with UV-HCMV in the presence of NS or ZBP1-specific siRNAs. The values presented (plus standard errors of the mean) are normalized to NS siRNA-treated cells (set to 1). (D) Expression of IFN-β-dependent luciferase following treatment of THF-ISRE reporter cells with UV-HCMV in the presence of NS or ZBP1-specific siRNA. The values displayed (plus standard errors of the mean) are normalized to UV-HCMV-induced luciferase expression in NS siRNA-treated cells (set to 1). (E) IFN-β gene transcription change relative to unexposed cells following exposure of siRNA-treated THFs to HCMV and CHX (200 μg/ml). The values (plus standard errors of the mean) are the changes in HCMV- and CHX-treated cells relative to those in non-HCMV-treated, CHX-treated cells that were normalized to HCMV-induced changes obtained from THFs treated with NS siRNA and CHX (set to 1).
Given the induction of ZBP1 mRNA in response to UV-HCMV, we wondered whether de novo synthesis of ZBP1 protein is required for the receptor's detection of the virus. We thus examined HCMV-induced IFN-β transcription in cells treated with ZBP1-targeted siRNA in the presence of CHX. This was compared to HCMV-induced IFN-β transcription in cells transfected with NS siRNA in the presence of CHX. As such, siRNA treatment reduces ZBP1 protein existing prior to HCMV exposure and CHX prevents any new ZBP1 from being synthesized. Under these conditions, HCMV-induced IFN-β transcription was diminished to a level similar to that seen following knockdown of IRF3 (Fig. 3E). Based on these results, we conclude that ZBP1 is required during the initial detection of HCMV by the cell's innate antiviral signaling apparatus.
ZBP1 overexpression enhances UV-HCMV-mediated IFN-β transcription and secretion.
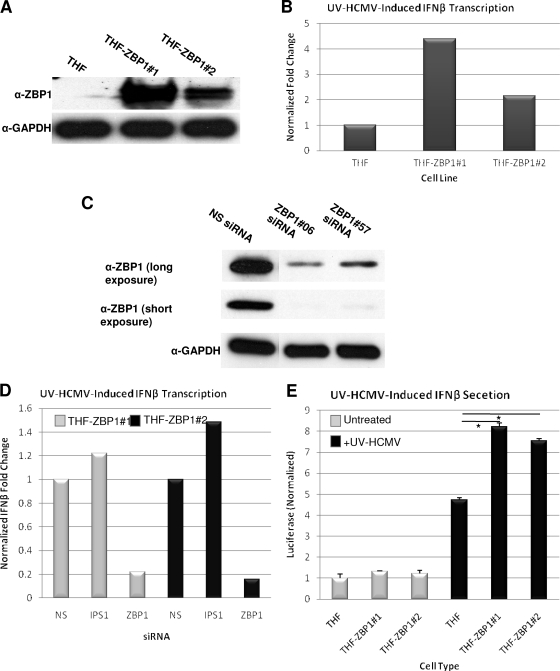
If ZBP1 participates in the initiation and amplification of the anti-HCMV innate reaction, we reasoned that overexpression of ZBP1 prior to treatment with UV-HCMV might increase the IRF3-dependent transcriptional response upon virus entry. We therefore created two THF lines that stably express ZBP1 (THF-ZBP1 no. 1 and no. 2) (Fig. 4A). While these cells did not exhibit an increase in constitutive IFN-β mRNA transcription (not shown), following treatment with UV-HCMV, an approximately two- to fourfold increase in IFN-β gene transcription relative to UV-HCMV-triggered IFN-β mRNA induction seen in parental cells was observed (Fig. 4B). ZBP1 expression did not lead to basal IFN-β secretion (Fig. 4E) but did result in significantly increased UV-HCMV-induced IFN-β secretion as measured by IFN-dependent luciferase expression following exposure of THF-ISRE cells to media collected from THF and THF-ZBP1 cells treated with UV-HCMV (Fig. 4D). The increased IFN-β transcription was nearly eliminated upon treatment of THF-ZBP1 cells with siRNA targeting ZBP1, thus demonstrating that ZBP1 is responsible for this effect (Fig. 4C and D). In contrast, upon treatment of THF-ZBP1 cells with siRNA directed against IPS-1, a slight increase in IFN-β induction was observed in both cell lines (Fig. 4C), in agreement with our data described above and further demonstrating that POL3 and IPS-1 are not necessary for UV-HCMV-triggered innate immune responses. Increased IFN-β production upon prior expression of ZBP1 is consistent with UV-HCMV-dependent upregulation of ZBP1, resulting in a positive feedback loop of IRF3 activation during HCMV replication.
FIG. 4.
Ectopic expression of ZBP1 increases UV-HCMV-induced IFN-β transcription independently of IPS-1. (A) Immunoblot showing overexpression of ZBP1 in two stable THF lines. (B) Induction of IFN-β mRNA as measured by qPCR in UV-HCMV-exposed relative to unexposed cells; the values shown are normalized to the IFN-β induction observed in THF parental cells (set to 1). (C) Immunoblot showing siRNA-mediated knockdown of stably expressed ZBP1 from THF-ZBP1 cells using two different ZBP1-targeted siRNA sequences. (D) Induction of IFN-β mRNA as measured by qPCR in UV-HCMV-exposed relative to unexposed THF-ZBP1 cells following treatment with the indicated siRNAs; the values shown are normalized to the IFN-β induction observed in THF-ZBP1 cells treated with NS siRNA (set to 1). (E) IFN-β secretion as measured by luciferase expression from THF-ISRE cells following exposure to media collected from THF or THF-ZBP1 cells that were either untreated or treated with UV-HCMV. The values (plus standard errors of the mean) are normalized to untreated THFs that were not exposed to UV-HCMV (set to 1). *, P < 0.05.
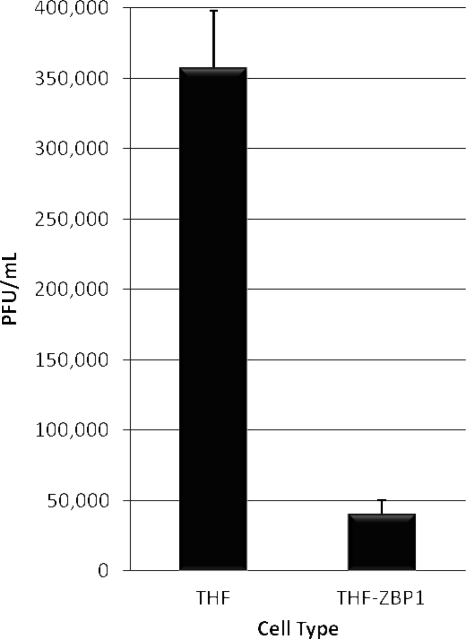
ZBP1 overexpression inhibits replication of HCMV.
Since ZBP1 appears to be important in HCMV-triggered IFN synthesis, as well as capable of enhancing this response, we decided to ask whether the physiological effects of the protein are antiviral, specifically with respect to HCMV. We therefore compared growth of the virus on both parental THF and THF-ZBP1 no. 1 cells. We infected both cell types with HCMV (MOI = 3) in duplicate and subsequently harvested cell culture media at day 3 postinfection. The titer of virus from this medium was next determined using an endpoint dilution assay on primary human fibroblasts. As shown in Fig. 5, the amount of progeny virus produced on THF-ZBP1 cells was approximately sevenfold lower than that produced on parental THFs, a result consistent with an anti-HCMV cellular response being conferred by the activity of ZBP1. It is worth noting that if the observed ZBP1-dependent anti-HCMV effect is ultimately IFN dependent, it may be dampened to some degree by the JAK/STAT inhibitory phenotype exhibited by HCMV (44).
FIG. 5.
Overexpression of ZBP1 inhibits replication of HCMV. In duplicate, THF or THF-ZBP1 no. 1 cells were infected with HCMV as described in the text. Virus from infected cell media was then quantified using endpoint dilution assays of primary human fibroblasts. The values presented are PFU/ml plus the standard error of the mean for each cell type.
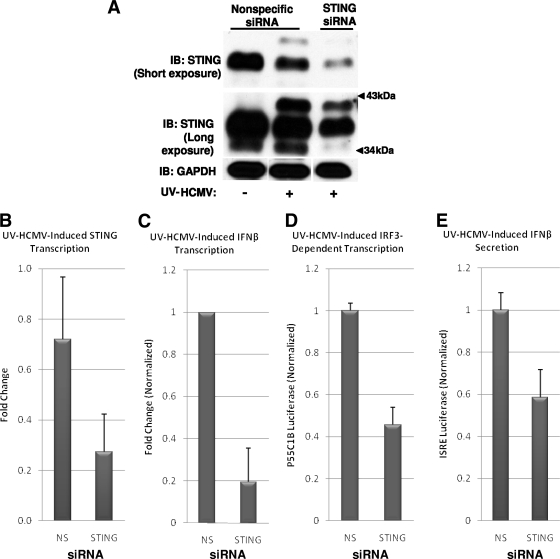
STING contributes to HCMV-mediated, IRF3-dependent IFN-β transcription and secretion.
Recently, a mitochondrion- and ER-associated protein termed STING was identified and found to be necessary for IRF3 activation and IFN-β induction by cytoplasmic dsDNA and infection with Listeria, SeV, and HSV-1 (23, 83). Since the protein is essential in IRF3 activation by diverse stimuli, including viral infection, we decided to examine its necessity for HCMV-mediated IRF3 activation.
Treatment of THFs with siRNA directed against STING was performed as described previously (23) and resulted in a decrease in the levels of STING mRNA and protein (Fig. 6A and B). Electrophoresed STING protein from whole-cell lysates of untreated THFs presented as a more quickly migrating, less voluminous band and a more slowly migrating, more abundant band (Fig. 6A). Interestingly, in lysates from THFs exposed to UV-HCMV, a more slowly migrating third band was also evident (Fig. 6A), perhaps representing a posttranslational modification, such as phosphorylation triggered by virus infection. As shown in Fig. 6, exposure of THFs transfected with STING-directed siRNA to UV-HCMV resulted in diminished IFN-β transcription (Fig. 6C) and secretion (Fig. 6E), as well as decreased IRF3-dependent transcription (Fig. 6D), relative to THFs treated with NS siRNA. Thus, consistent with its apparent role as an essential downstream phosphorylation target of TBK1 (83), STING appears to be involved in HCMV-mediated IRF3 activation.
FIG. 6.
STING contributes to UV-HCMV-induced IRF3-dependent transcription, IFN-β transcription, and IFN-β secretion. (A) Immunoblot (IB) showing siRNA-mediated depletion of STING protein from UV-HCMV-treated THFs treated with STING-specific, but not NS, siRNA. (B) Change of STING gene transcription in cells treated with UV-HCMV in the presence of either NS or STING-directed siRNA relative to that in cells treated with NS siRNA only. (C) Change of IFN-β gene transcription in UV-HCMV-exposed relative to unexposed THFs following treatment with the indicated siRNAs. The values displayed (plus standard errors of the mean) represent UV-HCMV-induced change with siRNA treatment normalized to the changes obtained from THFs treated with NS siRNA (set to 1). (D) Expression of IRF3-dependent luciferase following treatment of THF-P55C1B reporter cells with UV-HCMV in the presence of NS or STING-specific siRNA. The values displayed (plus standard errors of the mean) are normalized to UV-HCMV-induced luciferase expression in NS siRNA-treated cells (set to 1). (E) Expression of IFN-β-dependent luciferase following treatment of THF-ISRE reporter cells with UV-HCMV in the presence of NS or STING-specific siRNA. The values displayed (plus standard errors of the mean) are normalized to UV-HCMV-induced luciferase expression in NS siRNA-treated cells (set to 1).
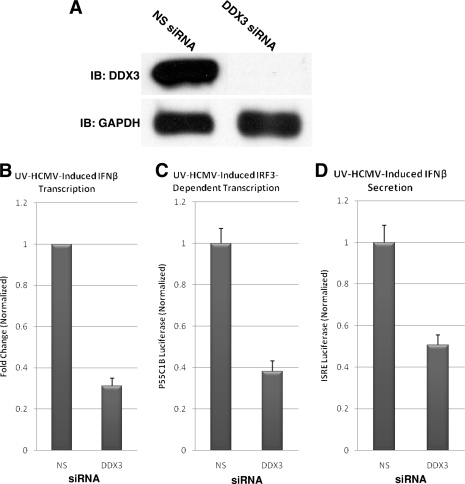
DDX3 contributes to HCMV-mediated, IRF3-dependent transcription and IFN-β secretion.
DDX3 was recently identified as a phosphorylation target of TBK1 and IKKɛ (64) that is necessary for complete IRF3-dependent activation of IFN-β by various stimuli (59, 64). DDX3 appears to function independently of IRF3 activation, however, in that it facilitates IRF3-mediated gene transcription but not IRF3 phosphorylation or DNA binding (64). A role for DDX3 in IFN-β induction by HCMV or other herpesviruses has not been investigated, however.
As shown in Fig. 7A, THFs express large amounts of DDX3 protein, and siRNA directed against DDX3 diminished endogenous protein to undetectable levels. Cells treated with siRNA directed against DDX3 displayed reduced UV-HCMV-mediated induction of IFN-β gene transcription relative to THFs transfected with NS siRNA (Fig. 7B). DDX3-targeted siRNA transfection also inhibited IRF3-dependent luciferase transcription (Fig. 7C) and IFN-β secretion (Fig. 7D) in response to exposure to UV-HCMV. It thus appears that, as with vaccinia viruses, poly(I·C), LPS, Listeria, and dsDNA, DDX3 is an important component of HCMV-mediated, IRF3-dependent IFN-β induction.
FIG. 7.
DDX3 is necessary for HCMV-induced IRF3-dependent transcription, IFN-β transcription, and IFN-β secretion. (A) Immunoblot (IB) showing siRNA-mediated depletion of DDX3 protein from THFs treated with DDX3-specific, but not NS, siRNA. (B) Change of IFN-β gene transcription in UV-HCMV-exposed relative to unexposed THFs following treatment with the indicated siRNAs. The values displayed (plus standard errors of the mean) represent the UV-HCMV-induced change with siRNA treatment normalized to the changes obtained from THFs treated with NS siRNA (set to 1). (C) Expression of IRF3-dependent luciferase following treatment of THF-P55C1B reporter cells with UV-HCMV in the presence of NS or DDX3-specific siRNA. The values displayed (plus standard errors of the mean) are normalized to UV-HCMV-induced luciferase expression in NS siRNA-treated cells (set to 1). (D) Expression of IFN-β-dependent luciferase following treatment of THF-ISRE reporter cells with UV-HCMV in the presence of NS or DDX3-specific siRNA. The values displayed (plus standard errors of the mean) are normalized to UV-HCMV-induced luciferase expression in NS siRNA-treated cells (set to 1).
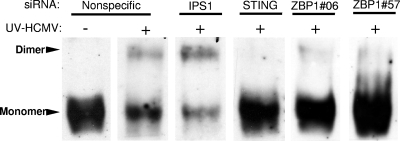
ZBP1 and STING, but not IPS-1, are required for HCMV-induced IRF3 dimerization.
Upstream signaling from PRRs and associated adaptor and kinase molecules results in IRF3 activation by phosphorylation, leading to its homodimerization and nuclear accumulation. However, recent data have questioned whether virus-triggered phosphorylation is an invariant phenomenon that is correlated with IRF3 activation (39). To verify that the upstream molecules investigated here are involved in IRF3 phosphorylation (as opposed to postactivation functions, e.g., nuclear accumulation and transcription), we examined UV-HCMV-induced IRF3 dimerization during siRNA-mediated protein knockdown. As shown in Fig. 8, monomeric IRF3 from unstimulated THFs migrates rapidly as a single large band. Following activation, IRF3 appears as both a rapidly migrating monomeric band and a more slowly migrating dimeric band, both of which are distinctly fainter than that from unstimulated cells. UV-HCMV-triggered IRF3 dimerization is apparent in THFs treated with NS and IPS-1-directed siRNAs, indicating that activation is not impaired in these cells. UV-HCMV-induced dimerization is not evident in cells treated with siRNA directed against ZBP1 or STING, indicating that IRF3 activation requires the full presence of these proteins. We thus conclude that the functions of ZBP1 and STING are contributory to HCMV-mediated IRF3 phosphorylation. Since IPS-1 was not required for IRF3 activation or IRF3-dependent transcription, these results further support our conclusion that HCMV activates the IRF3-terminal innate signaling pathway independently of IPS-1 and POL3.
FIG. 8.
STING, DDX3, and ZBP1, but not IPS-1, are required for UV-HCMV-stimulated IRF3 dimerization. Native immunoblots show the IRF3 dimerization status in THFs treated with the indicated siRNAs following mock (−) or actual (+) exposure to UV-HCMV.
DISCUSSION
Our results indicate that the cytoplasmic dsDNA sensor ZBP1 is important in HCMV-mediated activation of IRF3. This observation implicates virus-associated dsDNA as the major IRF3-activating PAMP of HCMV and is consistent with the fact that virus replication is not required for IRF3 activation, unlike what has been observed for other viruses, such as SeV (42). However, many questions remain regarding the nature and location of the DNA detection. For example, (i) whether the virus-associated DNA that triggers IRF3 activation is genomic or otherwise, (ii) the location of the stimulatory DNA within the virion (e.g., nucleocapsid or tegument), and (iii) where ZBP1 initially encounters the DNA subcellularly. While HCMV tegument-associated mRNA has been demonstrated (7), the existence of DNA outside the nucleocapsid is, to our knowledge, not known. Furthermore, the process of intracellular transport of HCMV genomes to the nucleus is poorly understood (16, 17, 67), and thus, the extent to which HCMV genomic DNA is exposed to the cytoplasm has not been examined in great detail. While the subcellular localization of ZBP1 is predominantly cytoplasmic, protein can be detected in the nucleus (15). It is thus possible that HCMV genomic DNA either is exposed to the cytoplasm during infection or is detected by ZBP1 in the nucleus.
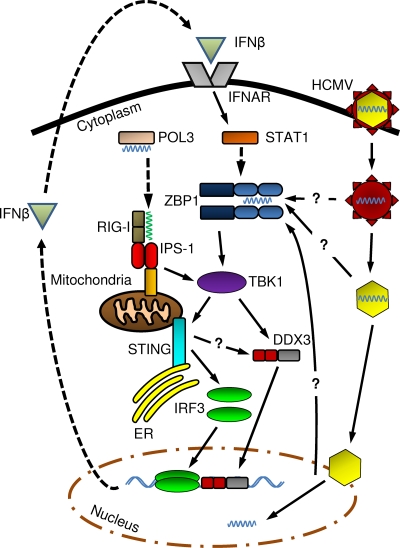
Our data further establish that ZBP1-mediated signaling is independent of IPS-1, thus indicating that this is perhaps a unique IRF3-terminal pathway. Based on this observation, involvement of the recently described dsDNA-triggered POL3-dependent pathway (2, 10) can be ruled out in the case of HCMV. We therefore propose the following model for HCMV-mediated IRF3 activation in human fibroblasts (illustrated in Fig. 9). HCMV particles enter the host cell following fusion with cellular membranes. ZBP1 in either the cytoplasm or nucleus is activated by dimerization (74) following contact with tegument- or nucleocapsid-associated DNA. ZBP1 triggers kinase activity of TBK1, resulting in the direct phosphorylation of DDX3 and the phosphorylation of IRF3 through uncharacterized mechanisms requiring STING (a role for STING in DDX3 phosphorylation is possible). Nuclear accumulation and DNA binding of phosphorylated DDX3 and IRF3 lead to the transcription of IFN-β, as well as ISGs. IFN-β synthesis/secretion stimulates JAK/STAT pathway-dependent expression of ISGs, including ZBP1. HCMV infection also triggers a posttranslational modification of STING by an unknown process that may require TBK1/IKKɛ or IRF3. HCMV-mediated induction of ZBP1 expression, and perhaps STING posttranslational modification, results in a positive feedback loop subsequently amplifying IRF3 activation and activity. ZBP1 appears to exhibit an antiviral effect on HCMV replication, but whether this results from subsequent IRF3- or IFN-dependent activity remains to be explored.
FIG. 9.
Proposed model of HCMV-triggered, IRF3-dependent transcriptional induction and ZBP1 expression. The schematic shows HCMV-triggered IRF3 activation and HCMV-mediated induction of ZBP1 expression in human fibroblasts. The dashed arrows indicate de novo transcription or translation, and the solid arrows indicate molecular interaction or signal transduction (e.g., phosphorylation).
With the exception of TBK1/IKKɛ (20), our study is the first to analyze cellular factors essential for IRF3 activation by HCMV. Moreover, while molecules necessary for IRF3 activation by other herpesviruses have been identified, the variety of species and cell types employed in previous studies renders it difficult to draw general conclusions about the components of innate cellular pathways activated by these viruses. For example, innate immune cells (e.g., dendritic cells and macrophages) express and employ assemblages of PRR molecules and associated adaptors that are distinct from nonimmune cells (e.g., fibroblasts and epithelial cells). The most informative comparisons are therefore likely to be those made between similar cell types, especially within the same species. Along these lines, TLR molecules have been shown to be important for IRF3- or IRF7-dependent induction of type I IFNs by numerous herpesviruses, although typically in innate immune cells and not fibroblasts (16, 67). Expression of type I IFNs following exposure to HSV-2 was nearly eliminated in murine fibroblasts obtained from IPS-1 knockout animals (48, 49). Furthermore, a role for RIG-I in HSV-2-mediated IRF3 activation in RAW264.7 cells is also established (48). Whether this response is dependent upon POL3-mediated conversion of dsDNA to dsRNA remains a likely but untested possibility (10). It is possible that despite their evolutionary closeness, HSV-1 and HSV-2 differentially activate innate signaling pathways. The gammaherpesvirus Epstein-Barr virus activates IRF3 through engagement of RIG-I by small dsRNA molecules expressed in infected cells (2, 54, 55), signaling that also requires IPS-1. These data, along with the involvement of IPS-1 in IRF3 activation by vaccinia virus (17, 82), another dsDNA virus, might suggest IPS-1 is an integral component of a general cytoplasmic antiviral signaling apparatus. However, our results show that IPS-1 is not required for HCMV-mediated or ZBP1-dependent IRF3 activation or IFN-β induction in fibroblasts, thus demonstrating the existence of a likely novel IRF3-activating mechanism. It is intriguing that despite their molecular similarities, even viruses of the same family (i.e., HCMV and HSV) trigger innate immune responses via distinct signaling pathways.
ZBP1 activity is triggered by association of its DNA binding domains with dsDNA. This interaction has been shown to induce homodimerization of the protein and the assembly of a signaling complex that includes TBK1 and leads to IRF3 phosphorylation (68, 74). Recently, Sanchez et al. observed IFN-α secretion from murine fibroblasts following transfection of herpesviral genomic DNA (57). The authors went on to identify homologous repetitive regions in the genomes of murine gammaherpesvirus 68 and Kaposi's sarcoma-associated herpesvirus that were capable of triggering IFN induction when exposed to cytoplasm; however, potential roles for ZBP1 or POL3 were not examined. We have also observed rapid and strong IRF3 nuclear accumulation in THFs following the transfection of genomic DNA of HCMV (unpublished observations). Whether a specific region of the HCMV genome is responsible for activating ZBP1 has not been investigated. Interestingly, HCMV and HSV may show similarity in at least partial involvement of ZBP1 during virus-mediated IFN induction. Takaoka et al. showed that siRNA-mediated knockdown of ZBP1 from murine L929 cells diminished (but did not eliminate) induction of IFN-β mRNA following HSV-1 treatment (68). Coupling this observation with data obtained for POL3 (10) indicates that HSV is actually capable of triggering multiple signaling pathways during infection but that HCMV is not. Many such phenomena regarding herpesvirus-triggered IFN induction in mammalian hosts require further examination. Hopefully the creation of ZBP1 and POL3 double-knockout mice will facilitate unraveling the relative importance of these innate DNA sensors for CMV and HSV.
HCMV may be unique among herpesviruses in that live wild-type virus does not inhibit IRF3 activation or IRF3-dependent transcriptional activity (4, 14, 20, 24, 37, 38, 42, 47). Moreover, seemingly contradictory observations have been made with regard to this phenomenon. For instance, complete HCMV particle entry into the host cell has been shown to be necessary for virus-triggered IFN and ISG induction (24, 38), yet mere exposure of host cells to a soluble form of HCMV-encoded, virion surface-associated glycoprotein B (gB) can induce IRF3 activation and ISG induction (4, 5, 63). However, since gB is essential for viral entry, it has so far not been possible to examine whether gB is necessary for IRF3 activation by HCMV. In fact, our data render it unlikely that gB or gB-mediated membrane fusion events are essential for IRF3 activation by HCMV, since activation was inhibited upon treatment with siRNA to ZBP1, a cytosolic sensor of DNA. Nevertheless, how gB triggers IFN induction remains to be clarified, and we are currently investigating the requirement for the molecules examined here in gB-mediated IRF3 activation.
We showed that the ER- and mitochondrion-associated protein STING is necessary for HCMV-mediated IRF3 activation. This finding is consistent with the essential role of the molecule in TBK1-mediated IRF3 activation by diverse stimuli (23, 83). Moreover, fibroblasts acquired from STING knockout mice failed to express IFN-β in response to exposure to HSV-1, in contrast to wild-type cells (23). Interestingly, an additional, more slowly migrating form of STING was observed following HCMV infection that was not observed following transfection of poly(I·C) (data not shown). Whether the change in migration represents a posttranslational modification that is specific to HCMV infection remains a possibility that requires exploration.
The importance of DDX3 in HCMV-triggered, IRF3-dependent transcription represents an additional novel finding of this study. The determination that DDX3 is involved in IRF3 transcriptional activity is a new discovery, and many questions surrounding this phenomenon still remain. What is known, however, is that DDX3 represents a kinase substrate of TBK1/IKKɛ that does not itself influence the phosphorylation of IRF3 (64). While we showed that STING is required for IRF3 dimerization, it is not known whether the protein is also necessary for TBK1-mediated DDX3 activation. Phosphorylated DDX3 binds to the IFN-β promoter and is required for full IRF3-dependent transcription, but no interaction between DDX3 and IRF3 was detected (64). Thus, although DDX3 is a virus- and PAMP-activated protein, its precise role in IRF3-dependent transcription remains to be elucidated. The function of DDX3 in HCMV-mediated IRF3-dependent transcription also requires additional exploration in order to be fully characterized.
In summary, our data indicate that host cells detect HCMV entry via the dsDNA sensor ZBP1, which then activates the innate IFN response via IRF3. In addition, HCMV separately induces proinflammatory genes via activation of the transcription factor NF-κB in a TLR2-dependent process that requires attachment but not entry (24, 42). The resulting release of proinflammatory and immune-stimulatory proteins likely contributes to the chronic inflammation and immune stimulation observed in HCMV-infected individuals. Future work will be required to understand the importance of HCMV-mediated IFN and proinflammatory molecule secretion to chronic infection.
Acknowledgments
We thank Wade Bresnahan for providing telomerized human fibroblasts, Glenn Barber for providing STING antibody, and Takahashi Fujita for providing the P55-C1B reporter plasmid.
This work was supported by American Heart Association grant 0730325N (V.R.D.), a grant from the Medical Research Foundation of Oregon (V.R.D.), and NIH grant R01AI070890 (K.J.F.).
Footnotes
Published ahead of print on 21 October 2009.
REFERENCES
- 1.Abate, D., S. Watanabe, and E. Mocarski. 2004. Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J. Virol. 78:10995-11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ablasser, A., F. Bauernfeind, G. Hartmann, E. Latz, K. A. Fitzgerald, and V. Hornung. 2009. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol. 10:1065-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alford, C. A., S. Stagno, R. F. Pass, and W. J. Britt. 1990. Congenital and perinatal cytomegalovirus infection. Rev. Infect. Dis. 12:745-753. [DOI] [PubMed] [Google Scholar]
- 4.Boehme, K. W., J. Singh, S. T. Perry, and T. Compton. 2004. Human cytomegalovirus elicits a coordinated cellular antiviral response via envelope glycoprotein B. J. Virol. 78:1202-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyle, K. A., R. L. Pietropaolo, and T. Compton. 1999. Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol. Cell. Biol. 19:3607-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bresnahan, W. A., G. E. Hultman, and T. Shenk. 2000. Replication of wild-type and mutant human cytomegalovirus in life-extended human diploid fibroblasts. J. Virol. 74:10816-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bresnahan, W. A., and T. Shenk. 2000. A subset of viral transcripts packaged within human cytomegalovirus particles. Science 288:2373-2376. [DOI] [PubMed] [Google Scholar]
- 8.Browne, E. P., B. Wing, D. Coleman, and T. Shenk. 2001. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 75:12319-12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cantrell, S. R., and W. A. Bresnahan. 2005. Interaction between the human cytomegalovirus UL82 gene product (pp71) and hDaxx regulates immediate-early gene expression and viral replication. J. Virol. 79:7792-7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiu, Y. H., J. B. Macmillan, and Z. J. Chen. 2009. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138:576-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins, S. E., R. S. Noyce, and K. L. Mossman. 2004. Innate cellular response to virus particle entry requires IRF3 but not virus replication. J. Virol. 78:1706-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui, S., K. Eisenacher, A. Kirchhofer, K. Brzozka, A. Lammens, K. Lammens, T. Fujita, K. K. Conzelmann, A. Krug, and K. P. Hopfner. 2008. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol. Cell 29:169-179. [DOI] [PubMed] [Google Scholar]
- 13.DeFilippis, V., and K. Fruh. 2005. Rhesus cytomegalovirus particles prevent activation of interferon regulatory factor 3. J. Virol. 79:6419-6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeFilippis, V. R., B. Robinson, T. M. Keck, S. G. Hansen, J. A. Nelson, and K. Früh. 2006. Interferon regulatory factor 3 is necessary for induction of antiviral genes during human cytomegalovirus infection. J. Virol. 80:1032-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deigendesch, N., F. Koch-Nolte, and S. Rothenburg. 2006. ZBP1 subcellular localization and association with stress granules is controlled by its Z-DNA binding domains. Nucleic Acids Res. 34:5007-5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delale, T., A. Paquin, C. Asselin-Paturel, M. Dalod, G. Brizard, E. E. Bates, P. Kastner, S. Chan, S. Akira, A. Vicari, C. A. Biron, G. Trinchieri, and F. Briere. 2005. MyD88-dependent and -independent murine cytomegalovirus sensing for IFN-alpha release and initiation of immune responses in vivo. J. Immunol. 175:6723-6732. [DOI] [PubMed] [Google Scholar]
- 17.Deng, L., P. Dai, T. Parikh, H. Cao, V. Bhoj, Q. Sun, Z. Chen, T. Merghoub, A. Houghton, and S. Shuman. 2008. Vaccinia virus subverts a mitochondrial antiviral signaling protein-dependent innate immune response in keratinocytes through its double-stranded RNA binding protein, E3. J. Virol. 82:10735-10746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491-496. [DOI] [PubMed] [Google Scholar]
- 19.Fu, Y., N. Comella, K. Tognazzi, L. F. Brown, H. F. Dvorak, and O. Kocher. 1999. Cloning of DLM-1, a novel gene that is up-regulated in activated macrophages, using RNA differential display. Gene 240:157-163. [DOI] [PubMed] [Google Scholar]
- 20.Gravel, S. P., and M. J. Servant. 2005. Roles of an IkappaB kinase-related pathway in human cytomegalovirus-infected vascular smooth muscle cells: a molecular link in pathogen-induced proatherosclerotic conditions. J. Biol. Chem. 280:7477-7486. [DOI] [PubMed] [Google Scholar]
- 21.Hornung, V., J. Ellegast, S. Kim, K. Brzozka, A. Jung, H. Kato, H. Poeck, S. Akira, K. K. Conzelmann, M. Schlee, S. Endres, and G. Hartmann. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994-997. [DOI] [PubMed] [Google Scholar]
- 22.Ishii, K. J., C. Coban, H. Kato, K. Takahashi, Y. Torii, F. Takeshita, H. Ludwig, G. Sutter, K. Suzuki, H. Hemmi, S. Sato, M. Yamamoto, S. Uematsu, T. Kawai, O. Takeuchi, and S. Akira. 2006. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat. Immunol. 7:40-48. [DOI] [PubMed] [Google Scholar]
- 23.Ishikawa, H., and G. N. Barber. 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455:674-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juckem, L. K., K. W. Boehme, A. L. Feire, and T. Compton. 2008. Differential initiation of innate immune responses induced by human cytomegalovirus entry into fibroblast cells. J. Immunol. 180:4965-4977. [DOI] [PubMed] [Google Scholar]
- 25.Kawai, T., and S. Akira. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7:131-137. [DOI] [PubMed] [Google Scholar]
- 26.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981-988. [DOI] [PubMed] [Google Scholar]
- 27.Kinsella, T. M., and G. P. Nolan. 1996. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 7:1405-1413. [DOI] [PubMed] [Google Scholar]
- 28.Lee, M. S., and Y. J. Kim. 2007. Signaling pathways downstream of pattern-recognition receptors and their cross talk. Annu. Rev. Biochem. 76:447-480. [DOI] [PubMed] [Google Scholar]
- 29.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, R., R. S. Noyce, S. E. Collins, R. D. Everett, and K. L. Mossman. 2004. The herpes simplex virus ICP0 RING finger domain inhibits IRF3- and IRF7-mediated activation of interferon-stimulated genes. J. Virol. 78:1675-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lippmann, J., S. Rothenburg, N. Deigendesch, J. Eitel, K. Meixenberger, V. van Laak, H. Slevogt, D. P. N′Guessan, S. Hippenstiel, T. Chakraborty, A. Flieger, N. Suttorp, and B. Opitz. 2008. IFNbeta responses induced by intracellular bacteria or cytosolic DNA in different human cells do not require ZBP1 (DLM-1/DAI). Cell. Microbiol. 10:2579-2588. [DOI] [PubMed] [Google Scholar]
- 32.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 33.Ljungman, P. 1996. Cytomegalovirus infections in transplant patients. Scand. J. Infect. Dis. Suppl. 100:59-63. [PubMed] [Google Scholar]
- 34.Merika, M., A. J. Williams, G. Chen, T. Collins, and D. Thanos. 1998. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol. Cell 1:277-287. [DOI] [PubMed] [Google Scholar]
- 35.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167-1172. [DOI] [PubMed] [Google Scholar]
- 36.Mossman, K. L., P. F. Macgregor, J. J. Rozmus, A. B. Goryachev, A. M. Edwards, and J. R. Smiley. 2001. Herpes simplex virus triggers and then disarms a host antiviral response. J. Virol. 75:750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Navarro, L., K. Mowen, S. Rodems, B. Weaver, N. Reich, D. Spector, and M. David. 1998. Cytomegalovirus activates interferon immediate-early response gene expression and an interferon regulatory factor 3-containing interferon-stimulated response element-binding complex. Mol. Cell. Biol. 18:3796-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Netterwald, J. R., T. R. Jones, W. J. Britt, S. J. Yang, I. P. McCrone, and H. Zhu. 2004. Postattachment events associated with viral entry are necessary for induction of interferon-stimulated genes by human cytomegalovirus. J. Virol. 78:6688-6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noyce, R. S., S. E. Collins, and K. L. Mossman. 2009. Differential modification of interferon regulatory factor 3 following virus particle entry. J. Virol. 83:4013-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noyce, R. S., S. E. Collins, and K. L. Mossman. 2006. Identification of a novel pathway essential for the immediate-early, interferon-independent antiviral response to enveloped virions. J. Virol. 80:226-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oshiumi, H., M. Matsumoto, K. Funami, T. Akazawa, and T. Seya. 2003. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat. Immunol. 4:161-167. [DOI] [PubMed] [Google Scholar]
- 42.Paladino, P., D. T. Cummings, R. S. Noyce, and K. L. Mossman. 2006. The IFN-independent response to virus particle entry provides a first line of antiviral defense that is independent of TLRs and retinoic acid-inducible gene I. J. Immunol. 177:8008-8016. [DOI] [PubMed] [Google Scholar]
- 43.Pass, R. F. 2001. Cytomegalovirus, p. 2675-2705. In P. M. Howley, David M. Knipe, Diane E. Griffin, Robert A. Lamb, Malcolm A. Martin, Bernard Roizman, and Stephen E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 44.Paulus, C., S. Krauss, and M. Nevels. 2006. A human cytomegalovirus antagonist of type I IFN-dependent signal transducer and activator of transcription signaling. Proc. Natl. Acad. Sci. U. S. A. 103:3840-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perry, A. K., G. Chen, D. Zheng, H. Tang, and G. Cheng. 2005. The host type I interferon response to viral and bacterial infections. Cell Res. 15:407-422. [DOI] [PubMed] [Google Scholar]
- 46.Pichlmair, A., O. Schulz, C. P. Tan, T. I. Naslund, P. Liljestrom, F. Weber, and C. Reis e Sousa. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997-1001. [DOI] [PubMed] [Google Scholar]
- 47.Preston, C. M., A. N. Harman, and M. J. Nicholl. 2001. Activation of interferon response factor-3 in human cells infected with herpes simplex virus type 1 or human cytomegalovirus. J. Virol. 75:8909-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rasmussen, S. B., S. B. Jensen, C. Nielsen, E. Quartin, H. Kato, Z. J. Chen, R. H. Silverman, S. Akira, and S. R. Paludan. 2009. Herpes simplex virus infection is sensed by both Toll-like receptors and retinoic acid-inducible gene-like receptors, which synergize to induce type I interferon production. J. Gen. Virol. 90:74-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rasmussen, S. B., L. N. Sorensen, L. Malmgaard, N. Ank, J. D. Baines, Z. J. Chen, and S. R. Paludan. 2007. Type I interferon production during herpes simplex virus infection is controlled by cell-type-specific viral recognition through Toll-like receptor 9, the mitochondrial antiviral signaling protein pathway, and novel recognition systems. J. Virol. 81:13315-13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reed, L., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 51.Rothenburg, S., T. Schwartz, F. Koch-Nolte, and F. Haag. 2002. Complex regulation of the human gene for the Z-DNA binding protein DLM-1. Nucleic Acids Res. 30:993-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saito, T., and M. Gale, Jr. 2008. Differential recognition of double-stranded RNA by RIG-I-like receptors in antiviral immunity. J. Exp. Med. 205:1523-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saito, T., D. M. Owen, F. Jiang, J. Marcotrigiano, and M. Gale, Jr. 2008. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature 454:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Samanta, M., D. Iwakiri, T. Kanda, T. Imaizumi, and K. Takada. 2006. EB virus-encoded RNAs are recognized by RIG-I and activate signaling to induce type I IFN. EMBO J. 25:4207-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Samanta, M., D. Iwakiri, and K. Takada. 2008. Epstein-Barr virus-encoded small RNA induces IL-10 through RIG-I-mediated IRF-3 signaling. Oncogene 27:4150-4160. [DOI] [PubMed] [Google Scholar]
- 56.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanchez, D. J., D. Miranda, Jr., V. Arumugaswami, S. Hwang, A. E. Singer, A. Senaati, A. Shahangian, M. J. Song, R. Sun, and G. Cheng. 2008. A repetitive region of gammaherpesvirus genomic DNA is a ligand for induction of type I interferon. J. Virol. 82:2208-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 59.Schroder, M., M. Baran, and A. G. Bowie. 2008. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J. 27:2147-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Servant, M. J., B. ten Oever, C. LePage, L. Conti, S. Gessani, I. Julkunen, R. Lin, and J. Hiscott. 2001. Identification of distinct signaling pathways leading to the phosphorylation of interferon regulatory factor 3. J. Biol. Chem. 276:355-363. [DOI] [PubMed] [Google Scholar]
- 61.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669-682. [DOI] [PubMed] [Google Scholar]
- 62.Sharma, S., B. R. tenOever, N. Grandvaux, G. P. Zhou, R. Lin, and J. Hiscott. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300:1148-1151. [DOI] [PubMed] [Google Scholar]
- 63.Simmen, K. A., J. Singh, B. G. Luukkonen, M. Lopper, A. Bittner, N. E. Miller, M. R. Jackson, T. Compton, and K. Fruh. 2001. Global modulation of cellular transcription by human cytomegalovirus is initiated by viral glycoprotein B. Proc. Natl. Acad. Sci. U. S. A. 98:7140-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Soulat, D., T. Burckstummer, S. Westermayer, A. Goncalves, A. Bauch, A. Stefanovic, O. Hantschel, K. L. Bennett, T. Decker, and G. Superti-Furga. 2008. The DEAD-box helicase DDX3X is a critical component of the TANK-binding kinase 1-dependent innate immune response. EMBO J. 27:2135-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stetson, D. B., and R. Medzhitov. 2006. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity 24:93-103. [DOI] [PubMed] [Google Scholar]
- 66.Streblow, D. N., S. L. Orloff, and J. A. Nelson. 2001. Do pathogens accelerate atherosclerosis? J. Nutr. 131:2798S-2804S. [DOI] [PubMed] [Google Scholar]
- 67.Tabeta, K., P. Georgel, E. Janssen, X. Du, K. Hoebe, K. Crozat, S. Mudd, L. Shamel, S. Sovath, J. Goode, L. Alexopoulou, R. A. Flavell, and B. Beutler. 2004. Toll-like receptors 9 and 3 as essential components of innate immune defense against mouse cytomegalovirus infection. Proc. Natl. Acad. Sci. U. S. A. 101:3516-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takaoka, A., Z. Wang, M. K. Choi, H. Yanai, H. Negishi, T. Ban, Y. Lu, M. Miyagishi, T. Kodama, K. Honda, Y. Ohba, and T. Taniguchi. 2007. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448:501-505. [DOI] [PubMed] [Google Scholar]
- 69.Takeda, K., and S. Akira. 2003. Toll receptors and pathogen resistance. Cell Microbiol. 5:143-153. [DOI] [PubMed] [Google Scholar]
- 70.Taylor, R. T., and W. A. Bresnahan. 2006. Human cytomegalovirus IE86 attenuates virus- and tumor necrosis factor alpha-induced NFkappaB-dependent gene expression. J. Virol. 80:10763-10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taylor, R. T., and W. A. Bresnahan. 2005. Human cytomegalovirus immediate-early 2 gene expression blocks virus-induced beta interferon production. J. Virol. 79:3873-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thompson, A. J., and S. A. Locarnini. 2007. Toll-like receptors, RIG-I-like RNA helicases and the antiviral innate immune response. Immunol. Cell Biol. 85:435-445. [DOI] [PubMed] [Google Scholar]
- 73.Wang, X., S. Hussain, E. J. Wang, X. Wang, M. O. Li, A. Garcia-Sastre, and A. A. Beg. 2007. Lack of essential role of NF-kappa B p50, RelA, and cRel subunits in virus-induced type 1 IFN expression. J. Immunol. 178:6770-6776. [DOI] [PubMed] [Google Scholar]
- 74.Wang, Z., M. K. Choi, T. Ban, H. Yanai, H. Negishi, Y. Lu, T. Tamura, A. Takaoka, K. Nishikura, and T. Taniguchi. 2008. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc. Natl. Acad. Sci. U. S. A. 105:5477-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 76.Wen, Z., Z. Zhong, and J. E. Darnell, Jr. 1995. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 82:241-250. [DOI] [PubMed] [Google Scholar]
- 77.Xu, L. G., Y. Y. Wang, K. J. Han, L. Y. Li, Z. Zhai, and H. B. Shu. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727-740. [DOI] [PubMed] [Google Scholar]
- 78.Yamamoto, M., S. Sato, K. Mori, K. Hoshino, O. Takeuchi, K. Takeda, and S. Akira. 2002. Cutting edge: a novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the Toll-like receptor signaling. J. Immunol. 169:6668-6672. [DOI] [PubMed] [Google Scholar]
- 79.Yoneyama, M., M. Kikuchi, K. Matsumoto, T. Imaizumi, M. Miyagishi, K. Taira, E. Foy, Y. M. Loo, M. Gale, Jr., S. Akira, S. Yonehara, A. Kato, and T. Fujita. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851-2858. [DOI] [PubMed] [Google Scholar]
- 80.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]
- 81.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang, P., and C. E. Samuel. 2008. Induction of protein kinase PKR-dependent activation of interferon regulatory factor 3 by vaccinia virus occurs through adapter IPS-1 signaling. J. Biol. Chem. 283:34580-34587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhong, B., Y. Yang, S. Li, Y. Y. Wang, Y. Li, F. Diao, C. Lei, X. He, L. Zhang, P. Tien, and H. B. Shu. 2008. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29:538-550. [DOI] [PubMed] [Google Scholar]
- 84.Zhu, H., J. P. Cong, G. Mamtora, T. Gingeras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. U. S. A. 95:14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhu, H., J. P. Cong, and T. Shenk. 1997. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc. Natl. Acad. Sci. U. S. A. 94:13985-13990. [DOI] [PMC free article] [PubMed] [Google Scholar]