Abstract
In this report the basis for the structural architecture of the envelope of hantaviruses, family Bunyaviridae, is systematically studied by the interactions of two glycoproteins N and C (Gn and Gc, respectively) and their respective disulfide bridge-mediated homo- and heteromeric oligomerizations. In virion extracts Gn and Gc associated in both homo- and hetero-oligomers which were, at least partially, thiol bridge mediated. Due to strong homo-oligomerization, the hetero-oligomers of Gn and Gc are likely to be mediated by homo-oligomeric subunits. A reversible pH-induced disappearance of a neutralizing epitope in Gc and dissociation of the Gn-Gc complex at pH values below 6.2 provide proteochemical evidence for the fusogenicity of Gc. Incomplete inactivation of virions at acidic pH indicates that additional factors are required for hantavirus fusion, as in the case of pestiviruses of the Flaviviridae. Based on similarities to class II fusion proteins, a structure model was created of hantavirus Gc using the Semliki Forest virus E1 protein as a template. In total, 10 binding regions for Gn were found by peptide scanning, of which five represent homotypic (GnI to GnV) and five represent heterotypic (GcI to GcV) interaction sites that we assign as intra- and interspike connections, respectively. In conclusion, the glycoprotein associations were compiled to a model wherein the surface of hantaviruses is formed of homotetrameric Gn complexes interconnected with Gc homodimers. This organization would create the grid-like surface pattern described earlier for hantaviruses in negatively stained electron microscopy specimens.
Hantaviruses, a genus in the family Bunyaviridae, are rodent- and insectivore-borne zoonotic viruses that are seemingly apathogenic to the carrier rodents (39, 57). A number of hantaviruses are human pathogens that in areas of endemicity are responsible for two diseases: hemorrhagic fever with renal syndrome in Eurasia and hantavirus cardiopulmonary syndrome in the Americas (49, 57, 61). Hantaviruses are enveloped viruses and have a trisegmented, single-stranded, negative-sense RNA genome that encodes an RNA-dependent RNA polymerase, two glycoproteins, and a nucleocapsid protein (22, 34, 49, 60). During assembly, the four proteins and the RNA genome are packed into a round or a pleomorphic particle enveloped with a lipid bilayer. The interactions among the structural components of hantavirus have not been described in sufficient detail to construct the basic architecture of the virus particle or to understand the mechanisms of its assembly and entry.
The envelope glycoproteins are expressed as a precursor polypeptide, which is cotranslationally cleaved after a conserved pentapeptide WAASA into an N- and C-terminal portion prior to maturation of the envelope glycoproteins proteins N and C (Gn and Gc, respectively) (27). In the family Bunyaviridae the transport of newly synthesized glycoproteins from endoplasmic reticulum to the Golgi apparatus requires the presence of both Gn and Gc (36, 37, 50, 53). Recombinant coexpression of the hantavirus glycoproteins is sufficient to achieve proper folding and the expected cellular localization at the Golgi even when the glycoproteins are not expressed from a common precursor (6, 36, 50). This suggests that the expression of the precursor is not a prerequisite for interactions between Gn and Gc during maturation in which the formation of a Gn-Gc complex results in exposure of a conformational Golgi apparatus-targeting signal, present only in the heterodimeric Gn-Gc complex (6, 50).
Entry of enveloped viruses via recognition of the cell surface receptors and subsequent fusion of the virus and cell membranes are accomplished by viral glycoproteins which often appear in homo- and/or heteromeric complexes. For example, the E1 and E2 of Semliki Forest virus (SFV) form a trimer of heterodimers (45), and the E protein of tick-borne encephalitis virus (TBEV) forms a homodimer (41) while the hemagglutinin of influenza A virus (67) and the S protein of severe acute respiratory syndrome coronavirus associate in homotrimers (4, 5). The mature glycoproteins extracted from virions of Uukuniemi phlebovirus exist as homodimers (44), whereas glycoprotein complex formations of many other members of the Bunyaviridae have not been defined. The viral fusion proteins can be classified into class I, class II, and class III (25). Between classes I and II, a distinguishing property is the orientation of a fusion protein in the metastable state. The class I proteins are oriented perpendicular to the viral membrane, and the class II protein is parallel to the viral membrane (7). The class II viral fusion proteins assemble in virions as metastable homo- or heterodimeric complexes which, upon exposure to low pH, fuse the viral and target cellular membranes (7). This process begins with a conformational change in the fusion protein, leading to the revelation of its fusion loop, which binds to the cellular target membrane (7). Additionally, the formation of a homotrimeric fusion protein complex and structural changes that drive the fusion into completion occur (7).
Understanding the multimeric status, protein-protein interactions, and pH-dependent conformational changes of glycoproteins is paramount to our understanding of selectivity in cell receptor binding and mechanisms of virus entry. It is unknown whether higher-order oligomeric complexes are found in hantavirus particles. Many neutralizing monoclonal antibodies (MAbs) have been isolated and by MAb escape mutants shown to recognize epitopes in both Gn and Gc, typically localized at discontinuous sites (15). Different neutralization mechanisms for hantavirus MAbs have been elucidated. These range from inhibiting receptor binding to inhibition of virus fusion (2, 23, 28, 30, 65). It is known that hantaviral glycoproteins possess fusogenic activity. Glycoproteins of hantaviruses that cause hemorrhagic fever with renal syndrome can induce syncytia when subjected to low pH (32, 35), and infection by Hantaan virus was shown to use low-pH-dependent clathrin-mediated endocytosis (19). Hantavirus Gc is suggested to be a class II fusion protein (13, 55), and the N-linked glycosylation of Gc is essential for cell fusion activity (70); but no clear understanding exists of the fusion mechanism or conformational changes that mediate uncoating of virions after entry.
Our study supports the hypothesis that the Gc of hantaviruses is a class II fusion protein. We show the interaction between Gn and Gc to be pH sensitive and dissociation to start at a pH below 6.4. The low-pH-induced Gc dissociation from Gn was reversible, suggesting that the conformational changes in Gc are also reversible. Both glycoproteins were found to form homodimeric and hetero-oligomeric complexes in virion extracts through thiol bridging. Interaction studies further suggested that the protruding part of the spike complex in the hantavirus virion consists of four Gn subunits and that the spike complexes interconnect with homodimeric Gc subunits. Finally, we mapped and compiled the interaction sites of Gn and Gc proteins in a class II fusion protein three-dimensional (3D) model of Gc. The identified Gn-Gn, Gn-Gc, and Gc-Gc interaction sites may play an important role in glycoprotein folding and maturation, spike assembly, virus fusion, and neutralization of infection.
MATERIALS AND METHODS
Cell cultures and viruses.
The Puumala virus (PUUV) Sotkamo strain and Tula virus (TULV) Moravia strain 5302 were cultivated in Vero E6 green monkey kidney epithelial cells (ATCC 94 CRL-1586). Cells were grown in minimal essential medium (MEM) supplemented with 5 to 10% heat-inactivated fetal calf serum, 2 mM l-glutamine, 100 IU/ml of penicillin, and 100 μg/ml of streptomycin at 37°C (supplemented MEM) in a humidified atmosphere containing 5% CO2. For virus production, subconfluent (∼80 to 90%) cell monolayers in 75-cm2 flasks were inoculated for 1 h at 37°C to absorb the virus, followed by addition of growth medium (15 to 20 ml/flask). The supernatant virus was collected 7 to 10 days postinfection (dpi) with TULV and 12 to 21 dpi with PUUV. The virus titers in the culture medium were determined as described previously (21) as the number of focus-forming units (FFU). The TULV titers were 106 to 107 FFU/ml, and PUUV titers were 104 to 105 FFU/ml.
The effect of acidic pH on the infectivity of TULV was studied with virus collected from infected Vero E6 monolayers and concentrated approximately 10-fold by centrifugation in a 30-kDa-cutoff Microcon filter device (Millipore). The concentrated medium was brought to serial pH values of 5.5, 6.0, 6.5, 7.0, and 7.5 through 1:10 dilutions with 100 mM Bis-Tris-150 mM NaCl. Virions were kept in a low-pH buffer for 30 min at room temperature (RT) and readjusted to pH 8.0 with 2 M Tris-150 mM NaCl or left unadjusted prior to the serial dilutions in supplemented MEM and virus titration. The average titer and standard deviation for each experiment were derived from two parallel wells of two 10-fold dilutions.
Antibodies and immunoblotting.
Neutralizing bank vole MAb 5A2 raised against PUUV Gn, MAb 4G2 to PUUV Gc, and the neutralizing human MAb 1C9 to PUUV Gc (28, 29) were kindly provided by Åke Lundkvist (Swedish Institute for Infectious Disease Control). Polyclonal antisera used were raised against glutathione-S-transferase and PUUV Gn and Gc ectodomain fusion proteins (59) and used at a 1:400 dilution. Goat polyclonal anti-rabbit IRDye 800CW (Li-Cor Biosciences) diluted 1:10,000 in 50 mM Tris-buffered saline (pH 8.0)-0.05% Triton X-100 ([TX-100] T-TBS) and 3% skim milk was used in conventional immunoblotting recorded with an Odyssey infrared imaging system (Li-Cor Biosciences).
Radioactive labeling of PUUV proteins.
PUUV-infected Vero E6 monolayers in 75-cm2 culture flasks were starved in cysteine- and methionine-depleted growth medium at 10 dpi for 1 h at 37°C and labeled with 1 mCi of [35S]cysteine (Wallac, PerkinElmer) in 15 ml of supplemented MEM. Conditioned medium with released virus was collected 5 days postlabeling and concentrated approximately 10-fold using a 100-kDa-cutoff Amicon ultrafiltration device (Millipore).
Purification of viruses.
Growth medium collected between 3 and 14 dpi was cleared through a 0.22-μm-pore-size filter (Millipore), and virus was concentrated by pelleting through a 3-ml 30% (wt/vol) sucrose cushion (Beckman SW28 rotor; 27,000 rpm for 2 h at 4°C) in 10 mM HEPES-100 mM NaCl, pH 7.4 (HN-buffer). The metabolically [35S]Cys-labeled PUUV was purified by pelleting through a 1-ml 30% (wt/vol) sucrose cushion (Beckman SW51 rotor; 40,000 rpm for 2 h at 4°C) and solubilized in 50 mM TBS with 1% TX-100. For cryo-electron microscopy (cryo-EM) TULV was pelleted between 30 and 70% (wt/vol) sucrose cushions (Beckman SW28 rotor; 27,000 rpm for 2 h at 4°C) and additionally purified in a 10 to 70% (wt/vol) sucrose gradient (Beckman SW41 rotor; 40,000 rpm for 2 h at 4°C). The gradient, fractionated by a dropwise collection of 500- to 700-μl fractions, was immunoblotted for Gc, Gn, and N proteins as virus markers. Pooled virus fractions were exchanged to HN buffer in a 100-kDa-cutoff Microcon centrifugal filter device (Millipore).
Analysis of complexes formed by Gn and Gc proteins.
Covalent cross-linking of glycoprotein complexes in intact virions was mediated with the membrane-impermeable cross-linker 3,3′-dithiobis[sulfosuccinimidylpropionate] (DTSSP), a cleavable analog of the commonly used bis-(sulfosuccinimidyl)suberate. TULV virions pelleted through sucrose cushion in HEPES-buffered saline (HBS) buffer were cross-linked at 0.1, 0.4 and 0.8 mM DTSSP for 3 min at RT, and the reactions were terminated by addition of 2 M Tris-HCl to 200 mM, pH 8.0. The complexes formed were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under reducing or nonreducing conditions and analyzed by immunoblotting with anti-Gc and anti-Gn sera and by silver staining (PageSilver Silver Staining Kit, Fermentas).
The appearance of thiol-bridged covalent complexes between Gn and Gc was studied by SDS-PAGE. Artificial cross-linking was prevented with N-ethylmaleimide (NEM) during sample preparation and SDS-PAGE by blocking the free thiols of virions by incubation with 2 mM NEM in HBS medium for 15 to 20 min at RT with or without addition of 1% TX-100.
To induce thiol bridging between glycoproteins in virions, pelleted TULV in HBS buffer was treated with 10 mM CuCl2 for 15 to 30 min at RT, and the reaction was terminated with 25 mM EDTA. Virus proteins were separated by SDS-PAGE under reducing and nonreducing conditions and immunoblotted with anti-Gn and anti-Gc sera.
pH dependence of glycoprotein interactions by coprecipitation.
The pH effects were studied by immunoprecipitations with the PUUV Gn-specific MAb 5A2 and the Gc-specific MAbs 1C9 and 4G2 preloaded on protein G-Sepharose (GE Healthcare, Amersham Biosciences) with 10 μg of MAb per 30 μl of beads. The 35S-labeled PUUV lysate was preabsorbed with unloaded Sepharose before incubation with the MAb-loaded Sepharose by an overnight incubation at 4°C. The default buffer in preloading and immunoprecipitations was T-TBS at pH 8.0. The buffers used were T-bTBS (25 mM bis-Tris, 150 mM NaCl, 0.1% TX-100) for pH values of 6.0 to 7.2 and T-TBS (25 mM Tris, 150 mM NaCl, 0.1% TX-100) for pH values of 7.3 to 8.0.
The effect of pH on dissociations of Gn and Gc from MAb 4G2 and MAb 5A2 complexes was determined by washing the resin containing prebound proteins at pH values of 6.0 to 8.0. The material that remained bound to beads was eluted in Laemmli sample buffer (1% SDS, 5% 2-mercaptoethanol, 0.1 M Tris, pH 6.8). Proteins and mobility markers (Precision Plus Protein Standards; Bio-Rad) were separated by SDS-PAGE, and gels were treated with Amersham Amplify reagent (GE Healthcare) and dried on Whatman No. 1 paper for autoradiography. Metabolically 35S-labeled proteins were detected by exposing films (SuperRX; Fuji Medical) for 1 to 3 days at −70°C. The band intensities of scanned films were quantified with ImageJ software (http://rsbweb.nih.gov/ij/) by selecting equally sized rectangles for the ratio of Gn (monomer) to Gc (monomer), which was calculated at each pH.
The effect of pH on the ability of neutralizing antibody to prevent a putative change in Gc protein conformation was studied with the Gc-specific MAb 4G2. The preloaded resins after low-pH washes were compared to proteins that were immunoprecipitated at pH values of 6.0 to 7.0. Proteins separated by SDS-PAGE were immunoblotted with anti-Gc serum. To assess whether the observed Gn-Gc complex dissociation and changes in Gc conformation are reversible, the 35S-labeled PUUV lysate was exposed to pH 5.8 and 8.0 for 1 to 2 h at RT. After exposure, the pH was adjusted to 8.0, and immunoprecipitations were performed with the Gc-specific MAbs 4G2 and 1C9. The bound material was separated by SDS-PAGE and detected by autoradiography.
Molecular mass determination of glycoprotein complexes by sedimentation in sucrose density gradients.
TULV was concentrated from medium collected from infected cells by pelleting through a 30% (wt/vol) sucrose cushion. Concentrated virus was lysed in 25 mM HBS-1% (vol/vol) TX-100 for 15 min at RT and diluted with HBS to 0.1% TX-100. Proteins from extract were separated by ultracentrifugation (SW41 rotor; 40,000 rpm for 20.5 h at 5°C) in 0 to 21% (wt/vol) sucrose gradients (25 mM HEPES, 100 mM NaCl, 0.1% TX-100), and the gradient was fractionated from the bottom to ∼600-μl fractions, for which sucrose concentration was determined using a refractometer. An aliquot (100 μl) of each fraction was concentrated ∼5 to 10-fold using a 30-kDa-cutoff Microcon centrifuge concentrator (Millipore), separated on SDS-PAGE, and immunoblotted with anti-Gn and anti-Gc sera. Molecular mass markers thyroglobulin (669 kDa; Pharmacia Biotech, GE Healthcare), ferritin (440 kDa; Pharmacia Biotech, GE Healthcare), rabbit anti-mouse immunoglobulin G1 (IgG1) fraction (∼155 kDa; Zymed Laboratories), and bovine serum albumin ([BSA] dimer, ∼132 kDa; monomer, ∼66.4 kDa; Sigma-Aldrich product number A7906) were detected by Coomassie staining, and their molecular masses were compared to those calculated from density sedimentations in sucrose.
To determine the molecular weights of protein sedimentations from the density of sucrose, precalculated concentration tables for linear sucrose gradients were used (33). We chose tables calculated for a particle density of 1.4 g/cm3, which is the estimation of protein density (9). The extrapolated sucrose concentration at the center of rotation (z0) of the gradient conditions used was determined to be −15 for the virus preparation and −17 for the marker centrifugation. The standard sedimentation coefficient (s20,w) value for each fraction was calculated from the following equation: s20,w × ω2 (t2 − t1) = I(z2) − I(z1), where ω is angular velocity (1/s), I is the sedimentation integral, and z1 and z2 are sucrose concentrations. Values for I(z1) and I(z2) were taken from the time integral table at 5°C, with the determined z0 value and particle density of 1.4 (33). The s20,w values were transformed to molecular weight using the following empirical relationship: s = 0.00242 M0.67 (42).
Analysis of Gc homodimers by gel filtration.
Proteins from TULV, concentrated and lysed as described for density gradient sedimentation analysis, were separated in a Sephacryl S-200HR (GE Healthcare Bio-Sciences) chromatography column in a BioCAD Vision Workstation (Perceptive BioSystems, Applied Biosystems). A column of 16/60 (bed volume, 120 ml) was loaded with a sample of 300 μl, and gel permeation was performed in 25 mM HEPES, 100 mM NaCl, and 0.1% TX-100 at a flow rate of 0.5 ml/min under absorbance monitoring at wavelengths of 260 and 280 nm. The gel filtration molecular weight of the Gc complex was estimated from its elution volume relative to a BSA dimer and monomer. The chromatograms of sample and marker were scaled to the highest peaks, creating an arbitrary absorbance scale at 280 nm for comparison of the elution volumes of proteins of interest as overlay. The viral proteins separated by SDS-PAGE were detected by Coomassie blue staining at the peak region using 100 μl of each fraction.
Gn-Gc interaction site mapping by peptide scanning.
The glycoprotein precursor encoded by the M-segment of PUUV (Sotkamo strain) was synthesized in 18-amino-acid-long, 15-residue overlapping peptides on an amino-functionalized cellulose membrane with 8- to 12-unit polyethylene glycol spacer arms (AIMS Scientific) (10). Peptide synthesis was carried out with an Abimed Autospot Robot ASP222 according to the manufacturer's protocol for fluorenylmethoxycarbonyl chemistry (Intavis Bioanalytical Instruments AG), and a spot protein overlay binding assay was performed according to protocols, with modifications, that were originally provided by Genosys (Cambridge, United Kingdom), as recently detailed (17). Prior to the interaction site mapping, the spot assay was calibrated for the background binding of MAbs used in detection. Briefly, the membrane was incubated for 1 h at RT with a MAb (Gn-specific MAb 5A2 and Gc-specific MAb 4G2) at 1 μg/ml in T-TBS containing 3% skim milk (blocking and dilution buffer for MAbs), washed with T-TBS (washing buffer), incubated for 1 h at RT with horseradish peroxidase-labeled rabbit anti-mouse immunoglobulins (DakoCytomation) used at a 1:1,000 dilution, and washed. Binding to peptides was recorded on X-ray film (SuperRX, Fuji Medical) by enhanced chemiluminescence (ECL). After the background binding of the MAb was recorded, membrane regeneration was performed. The spot binding assay to map the interaction sites was then performed with a 50-fold concentration of PUUV lysate as the source of Gn and Gc. The membrane was overlaid overnight with a 1:50 dilution of the concentrated lysate in T-TBS containing 5 mg/ml BSA and washed, and the spot assay was performed as described for detection MAbs. The result of the interaction site mapping was also recorded on X-ray film using ECL, and the membrane was regenerated for reuse in the next MAb background interaction site mapping cycle. Spot intensities of X-ray films were analyzed visually.
Modeling of Gc 3D structure.
To visualize the Gn-binding sites in Gc, we constructed a 3D model structure of PUUV Gc using the Swiss-Model program (http://swissmodel.expasy.org//SWISS-MODEL.html) for comparative modeling. We attempted to model the structures of two known class II viral fusion proteins: SFV E1 protein (Protein Data Bank [PDB] accession code 2ALA) (45) and TBEV E protein (PDB accession code 1SVB) (41) along with a class III fusion protein of vesicular stomatitis virus G (PDB accession code 2J6J) (43). The selected template sequences were aligned with PUUV Gc sequence lacking the last 42 residues using regular T-Coffee multiple alignment (http://tcoffee.vital-it.ch). The query sequences as T-Coffee alignments in FASTA format were submitted to the Swiss-Model program (http://swissmodel.expasy.org) utilizing the alignment interface option (3). The use of vesicular stomatitis virus G protein as a template was unsuccessful due to low sequence similarity, but the PUUV Gc structure models based on class II fusion proteins were obtained from Swiss-Model. The obtained PBD files were analyzed in VADAR (66) using default settings of version 1.5 (http://redpoll.pharmacy.ualberta.ca/vadar). The Ramachandran plots and dihedral angles showed the PUUV Gc model based on SFV E1 protein to be more acceptable than the model based on TBEV E. The models and the respective templates were also compared in TM-align (69) in pairwise structure and sequence similarity (http://zhang.bioinformatics.ku.edu). The TM-score lies between 0 and 1; a score of less than 0.2 indicates no similarity between structures, and a score above 0.5 indicates that the structures share the same fold. Visualization of the 3D model and the interaction data was done with YASARA View (http://www.yasara.org).
Cryo-EM of TULV particles.
Aliquots of freshly prepared TULV were pipetted onto 400-mesh copper grids covered with a holey carbon film (Quantifoil R 2/2) and vitrified by rapid plunging into liquid ethane (1). Samples were maintained at −180°C in a Gatan 626 cryoholder while images were recorded using an FEI Tecnai F20 field emission gun transmission EM (Electron Microscopy Unit, Institute of Biotechnology, University of Helsinki, Finland) operated at 200 kV under low-dose conditions. Micrographs were recorded at a nominal magnification of ×50,000 on Kodak SO163 film and developed in full-strength Kodak D19 for 12 min.
RESULTS
Interaction between Gn and Gc is pH dependent.
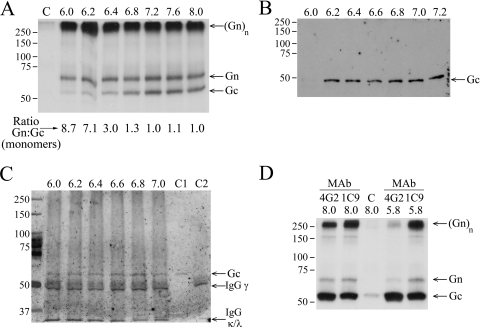
The hantavirus glycoproteins possess fusogenic activity with a capacity to induce syncytia at low pH (32, 35). Since the membrane fusion activity of enveloped viruses is mediated by conformational changes in envelope glycoproteins to form fusion-active complexes, we first investigated changes in the formation of glycoprotein complexes at low pH. To begin with, we monitored interactions of PUUV Gn and Gc in a series of coimmunoprecipitations with neutralizing MAbs. Hantaviruses, typical of Bunyaviridae, have an abundance of conserved cysteine residues: PUUV has 33 in Gn and 28 in Gc (40, 60). Therefore, [35S]cysteine was chosen to label the glycoproteins effectively in order to assess immunoprecipitation in virus lysates. The glycoprotein complexes coimmunoprecipitated with the Gn-specific, neutralizing MAb 5A2 are shown in Fig. 1A. MAb 5A2-Sepharose-bound material was washed with buffers covering a pH range of 6.0 to 8.0. The profiles of 35S-labeled Gn and Gc that remained bound after the washes along with the ratio of monomers in SDS-PAGE gels indicate their equal abundance in complexes at neutral to basic pH (ratio, ∼1.0), while the majority of Gn migrated at the high-molecular-weight region in 10% SDS-PAGE gels. The amount of high-molecular-weight Gn was not affected by washes at acidic pH, indicating tight, pH-independent binding of Gn to MAb 5A2-Sepharose. Strikingly, Gc dissociated from the Gn-Gc complex in a pH-dependent manner. The dissociation started at pH 6.2 to 6.4 (Gn-to-Gc ratio increasing from 1.0 to 3.0) and was complete at pH 6.0 (Gn-to-Gc ratio of 8.7).
FIG. 1.
Effect of low-pH treatment on PUUV Gn-Gc complex in immunoprecipitation of virion lysates. (A) Gn-specific immunoprecipitation with MAb 5A2. Autoradiography of MAb 5A2-Sepharose-bound, [35S]cysteine-labeled Gn and Gc after washing at indicated pH values. The control lane C represents background binding to protein G Sepharose at pH 8.0 without MAb. SDS-PAGE was run under reducing conditions with 2-mercaptoethanol as the reducing agent. The band labeled (Gn)n may also contain Gc. (B) Loss of PUUV Gc recognition by Gc-specific MAb 4G2. Gc in nonlabeled PUUV lysate was bound to MAb 4G2-Sepharose in buffers at the indicated pH. Proteins were separated by reducing SDS-PAGE and immunoblotted with anti-Gc serum. (C) MAb 4G2 binding to Gc prevents dissociation at low pH. Nonlabeled PUUV lysate was bound to MAb 4G2-Sepharose at pH 8.0 and washed in buffers at the indicated pH. The material remaining bound was separated by reducing SDS-PAGE and immunoblotted with anti-Gc serum. C1 and C2 represent controls; C1 is a virus preparation used in immunoprecipitation, and C2 is MAb 4G2-Sepharose without addition of virus. IgG(γ) (∼50 kDa) and IgG(κ/λ) (25 kDa; migrating at front) chains are indicated. (D) The low-pH-induced dissociation of Gn-Gc complex is reversible. Lysates of metabolically ([35S]cysteine) labeled PUUV treated at pH 5.8 or kept at pH 8.0, as indicated, were immunoprecipitated with Gc-specific MAbs 4G2 and 1C9 at pH 8.0. Autoradiography of SDS-PAGE-separated proteins and the control (lane C) was performed as described for panel A. The protein mobility estimation in panels A and D was done using Precision Plus protein standards (Bio-Rad). The band labeled (Gn)n may also contain Gc.
Next, we carried out coimmunoprecipitation with the Gc-specific MAb 4G2 at pH 6.0 to 7.2 (Fig. 1B). The MAb 4G2 epitope in Gc disappeared at pH values below 6.2. Both the loss of coprecipitation of Gc with Gn-specific MAb 5A2 with the low-pH wash and the loss of Gc recognition by MAb 4G2 at pH below 6.2 support the hypothesis of a conformational change in Gc which leads to dissociation of the Gn-Gc complex. Interestingly, the low-pH exposure had no disruptive effect on Gc bound to MAb 4G2 prior to acid washes, not even at a pH below 6.2 (Fig. 1C). This suggests that MAb 4G2 protects its epitope in Gc from low-pH-induced structural changes and as a result prevents fusion activity of Gc.
Dissociation of Gn-Gc complex is reversible.
The low-pH-induced changes in conformation during the fusion event of some class II fusion proteins result in irreversible changes in the structure of the protein (7). To assess whether this holds true for hantaviruses, PUUV lysate was exposed to pH 5.8 for 2 h at RT. After the pH treatment, immunoprecipitations with Gc-specific MAbs 1C9 and 4G2 were carried out at pH 8.0 (Fig. 1D). The nearly identical MAb 1C9 binding profiles of treated and untreated samples at pH 5.8 indicated that Gn and Gc are able to reassociate even though the time of low-pH exposure was considerably long. In contrast, the MAb 4G2 binding profile of proteins treated at pH 5.8 was altered, showing little or no bound Gn but full recovery of Gc (and MAb 4G2 epitope). The MAbs 4G2 and 1C9 have been reported to partially compete with each other (16, 29), indicating an overlap in their epitopes. It is possible that MAb 4G2 interferes with the reformation of the Gc-Gn complex by binding to a region in Gc that is close to the Gn-Gc interaction surface. Furthermore, our results propose that both Gn and Gc contribute to the MAb 1C9 epitope since their association in coimmunoprecipitation recruited nearly equimolar amounts of both proteins (Fig. 1D). The MAb 1C9 epitope is therefore suggested to be a viral neotope (48, 58) consisting of amino acids from both Gn and Gc.
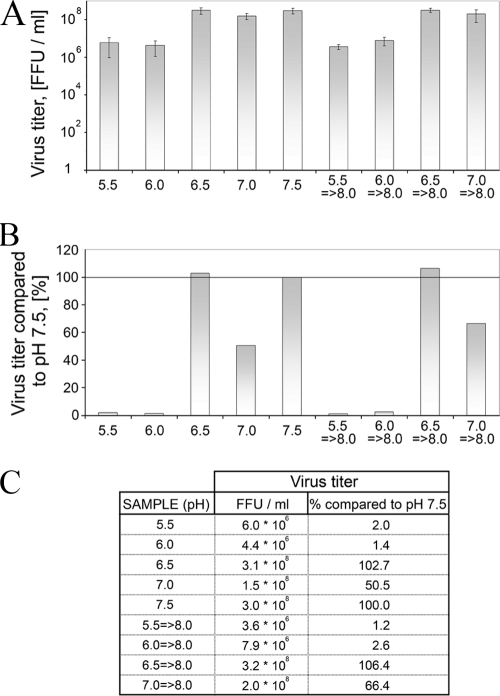
To further confirm the reversibility of virus envelope protein interactions after low-pH treatment when the viral membrane is also present, we followed the infectivity of TULV virions subjected to a serial acidic pH treatment (Fig. 2). The virus titers were determined as FFU/ml (Fig. 2A) and related to the treatment conducted at pH 7.5 (Fig. 2B) as summarized in Fig. 2C. Exposure to pH 5.5 to 7.5 caused a 100-fold decrease in virus titer at pH 5.5 to 6.0 (from ∼108 FFU/ml to ∼106 FFU/ml). This 99% inactivation of virus at acidic pH is in line with the Gn-Gc dissociation at the same low-pH range. However, when virions were subjected to acidic pH, the inactivation was incomplete since titers were not lowered further between pH 6.0 and 5.5. The observed, partial decrease in infectivity could be due to the high initial virus titer (∼108 FFU/ml), or, alternatively, a certain population of virions may tolerate the acidic pH.
FIG. 2.
Effect of low-pH treatment on the infectivity of TULV virions. (A) Titers of TULV virions subjected to low pH followed by pH adjustment to pH 8.0 (right bars) or left unadjusted (left bars) in FFU/ml determined by 10-fold serial dilutions. The error bars represent ± standard deviations of two parallel samples in two 10-fold dilutions. (B) The virus titers in panel A as relative titers to titers determined at pH 7.5. (C) Table of the titers (FFU/ml) and relative titers (%) in graphs in panels A and B.
Cross-linking of glycoproteins on the surface of intact TULV virions.
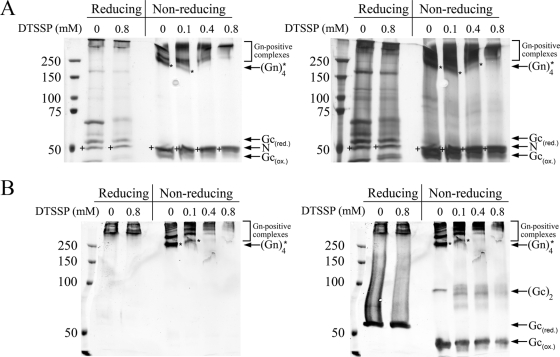
Quaternary glycoprotein complexes in virions can in some cases be visualized by chemical cross-linking of purified virus. We subjected TULV virions to cross-linking in increasing concentrations of cleavable, membrane-impermeable DTSSP and separated the Gn-Gc complexes by SDS-PAGE under both reducing and nonreducing conditions without boiling the samples. Detection of proteins was done by silver staining (Fig. 3A) and immunoblotting (Fig. 3B). Immunoblots revealed the complexes at 250 kDa and above to contain Gn protein; however, we were unable to identify a monomeric (or dimeric) Gn. Gc appeared as a monomer and a dimer in nonreducing SDS-PAGE gels and as a monomer only in reducing SDS-PAGE separation. The DTSSP-untreated sample, in a nonreduced state, had most of Gn at ∼250 kDa, probably as a tetrameric Gn complex. This appeared to be the simplest subunit composition of Gn extracted from the virion, present in Laemmli sample buffer, if the sample was not boiled. Addition of DTSSP resulted quickly in the disappearance of the Gn complex, which had practically vanished at a 0.4 mM concentration of the cross-linker, as judged by both silver staining and immunoblotting. At the same time, treatment of virions with DTSSP resulted in an altered mobility effect of dimeric Gc, as judged by anti-Gc immunoblotting. The amount of monomeric Gc decreased clearly only with the highest DTSSP concentration in immunoblotting and silver staining experiments. The Gn protein appeared more prone to cross-linking than Gc, but both could be cross-linked via lysines to higher-molecular-weight complexes.
FIG. 3.
Cross-linking of the surface of TULV virions with membrane-impermeable cross-linker. Native TULV virions, treated with three concentrations of membrane-impermeable cleavable DTSSP linker, were solubilized in Laemmli sample buffer, and proteins were separated by 6% SDS-PAGE under both reducing and nonreducing conditions (samples not boiled). (A) Silver staining (PageSilver Silver Staining Kit; Fermentas) shows bands resolved upon exposure to short (left panel) and long (right panel) periods of staining. (B) Detection by immunoblotting shows Gn (left panel) overlaid with Gc (right panel) as recorded in an Odyssey infrared imager with goat anti-rabbit IR800CW-conjugated secondary antibody. In both panels A and B the nucleocapsid protein (N) is indicated by a plus sign, and tetrameric Gn complex is indicated by an asterisk. ox., oxidized; red., reduced.
Mobility of glycoprotein complexes in SDS-PAGE.
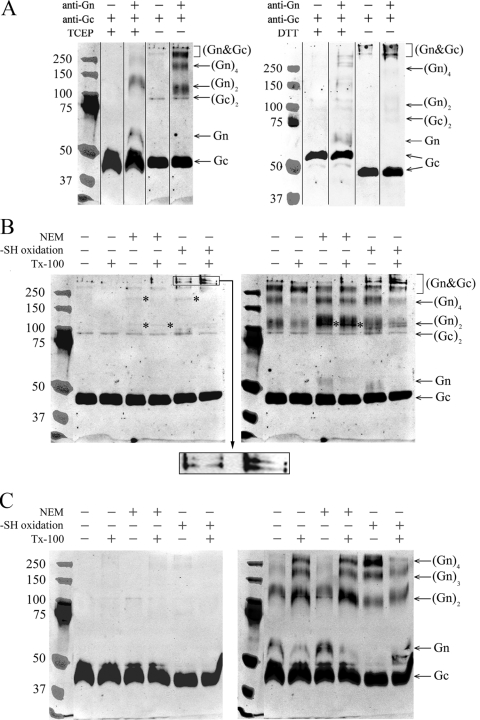
The tendency of hantavirus glycoproteins to make disulfide-bridges in SDS-PAGE was systematically studied using purified TULV (Fig. 4). Intact virions were reduced with either a membrane-impermeable disulfide-breaking agent, Tris(2-carboxyethyl) phosphine (TCEP), or a membrane-permeable reductant dithiothreitol (DTT) and subjected to SDS-PAGE. The proteins were immunoblotted first with antiserum to Gc, followed by antiserum to Gn, and the digital images of immunoblots detected with an Odyssey infrared imaging system were overlaid in Photoshop CS3 (Adobe), with the marker bands visible in antibody detections (Fig. 4A). The Gc in nonreducing SDS-PAGE gels appeared mainly as a monomer with a mobility of ∼45 kDa. In the TCEP-treated sample Gc migrated at the same rate as in the untreated sample (Fig. 4A, left panel), while a clear shift of Gc monomer from ∼45 kDa to ∼55 kDa was caused by DTT (Fig. 4A, right panel). We concluded that in contrast to DTT, TCEP did not break the intramolecular thiol bridges at the concentrations used, thus making Gc migrate faster due to preserved globularity. Under nonreducing SDS-PAGE separation, Gc-positive bands of ∼45 kDa, ∼90 kDa, and >250 kDa were observed. Both of the reducing agents, TCEP and DTT, caused a disappearance of the higher-molecular-mass complexes. Overall, it was evident then that the multimeric Gc complexes seen in SDS-PAGE gels are mediated by intermolecular thiol bridges. The molecular masses of ∼45 kDa and ∼90 kDa clearly represent monomeric and dimeric Gc. The band appearing above the 250-kDa marker detected by both Gc and Gn antisera represents a hetero-oligomeric glycoprotein complex. Monomeric Gn was not observed under nonreducing conditions, whereas the complexes positive with Gn antiserum, but not with Gc antiserum, were 110 to 120 kDa and 200 to 220 kDa, most likely representing a homodimeric and homotetrameric Gn. Monomeric Gn was seen only when proteins from the virion were separated by reducing SDS-PAGE, indicating that Gn exists in thiol bridge-mediated homo- and hetero-oligomeric complexes. For a summary of the observed complexes, see Fig. 9A.
FIG. 4.
Glycoprotein complexes of TULV formed under reducing and nonreducing conditions and under separation by SDS-PAGE (samples boiled in panels A, B, and C). (A) Glycoprotein mobilities of TULV pelleted through sucrose cushion, treated with the reductant (TCEP or DTT) or left untreated, and separated by SDS-PAGE. The composite immunoblots of Gc and Gn shown were overlaid as indicated. (B) TULV concentrated by ultracentrifugation was oxidized with 10 mM CuCl2 or thiol acetylated with NEM either with or without detergent TX-100 as indicated, and proteins were separated by SDS-PAGE without reductant as shown. Gc- and Gn-positive bands of ∼110 kDa and ∼200 kDa are indicated by asterisks. An enlargement of the region of >250 kDa from -SH (where SH is sulfhydryl) oxidized samples is shown as insert from the Gc immunoblot, highlighting novel bands positive for both Gn and Gc that appeared after CuCl2 treatment. (C) The virions after treatments, as in panel B, were reduced by the addition of 2 mM TCEP prior to SDS-PAGE. The composite immunoblots in panels B and C show Gc (left) overlaid with Gn (right). The images were recorded in an Odyssey infrared imaging system with goat anti-rabbit IR800CW-conjugated secondary antibody. The anti-Gc and anti-Gn serum reactivities were overlaid in PhotoShop CS using protein markers (Precision Plus Protein Standards; Bio-Rad) visible in both detections.
FIG. 9.
Cryo-EM image of TULV particles. (A) Summary of glycoprotein complexes detected using different methods. The first row shows the estimated molecular mass of each species in kDa; the lower value represents the molecular mass observed by nonreducing SDS-PAGE, and the higher value is calculated from primary sequence. Estimations for oligomeric complexes were calculated by multiplying the molecular mass of monomeric units. The other rows show the molecular masses of observed complexes. (B) Electron cryo-micrograph of TULV virions taken at approximately 3 μm under focus. The box indicates an area where the spikes are clearly visible projecting from the surface of the virion. An arrow in the magnified inset of the same area highlights an apparently symmetrical, dimeric spike. Bar, 300 nm. (C) Hypothetical surface arrangement of the glycoproteins in hantavirus particles. The illustration shows the three possible glycoprotein compositions seen in the plane of the viral membrane, where they produce the spike structures highlighted in panel A. The surface architecture of the virion was generated based on the proposed arrangement of Gn and Gc (oriented perpendicular to virus surface). The described interactions favor hypothesis 3, and the spike is proposed to be formed of two or four Gn protein units, forming a grid-like pattern as in negatively stained specimens (31).
Induced oxidation and intermolecular disulfide bridges between Gn and Gc.
Viral glycoproteins form stable complexes with each other to ensure proper organization of the viral surface and envelope. The Gn of Uukuniemi virus, if cross-linked by the introduction of thiol bridges, forms homodimers (44). We induced thiol bond formation by CuCl2 oxidation on TULV virions to follow formation of complexes between glycoproteins of the hantavirus envelope. To evaluate whether thiol-bridged associations could form during SDS-PAGE, we used NEM to block free thiol groups in virus preparations. The Gn and Gc complexes resulting from CuCl2 treatment of intact and detergent-solubilized purified virus were compared with untreated and NEM-treated samples. Proteins were separated by SDS-PAGE under both nonreducing (Fig. 4B) and reducing (Fig. 4C) conditions to further monitor thiol-bridged complexes that were formed. TCEP was previously seen to break intermolecular disulfide bridges in Gc, possibly leaving most intramolecular bridges intact; therefore “back-reduction” for SDS-PAGE of induced oxidations was done by the addition of TCEP into the samples prior to Laemmli buffer (Fig. 4C). Immunoblots generated as previously (Fig. 4A) were of Gc (Fig. 4B and C, left) overlaid with Gn (Fig. 4B and C, right). New, distinct bands resulted from CuCl2 treatment, which shifted the protein complexes to higher-molecular-mass regions. The induced oxidation produced two high-molecular-mass glycoprotein oligomers (470 kDa and 550 kDa, as estimated by extrapolation from marker mobility), which clearly contained Gn and Gc, as highlighted in Fig. 4B (magnified section of the left panel). Both in oxidized and NEM-treated samples, the sharp bands recognized by both antisera correspond in mobility to a Gn and Gc heterodimer (∼105 to 110 kDa) and heterotetramer (∼200 kDa), possibly in the form of (Gn-Gc)2 or Gn2-Gc2 glycoprotein complexes (Fig. 4B). Independent of the treatment, covalent Gc homodimer was present in nonreducing but not in reducing SDS-PAGE gels (Fig. 4B versus C). The heteromeric complexes largely disappeared after the back-reduction of samples for SDS-PAGE, demonstrating the induced hetero-oligomerization of Gn and Gc to be thiol bridge mediated. Reduction was required to obtain monomeric Gn (the amount of monomeric Gn was insignificant under nonreducing SDS-PAGE separation). Under reducing conditions with TCEP, proteins that were separated by SDS-PAGE from intact virions, either NEM treated or untreated, dissociated to a Gc monomer, Gn monomer, and Gn-dimer (Fig. 4C, right panel). The homodimeric Gn was not seen (Fig. 3A and B) unless the samples in Laemmli buffer were subjected to high temperatures (boiling or 56°C) prior to separation by SDS-PAGE, as in Fig. 4B and C. Oxidation with CuCl2 resulted in the disappearance of the Gn monomer accompanied by appearance of a Gn trimer and an abundant Gn tetramer, demonstrating Gn to have the capacity to associate as multiples of monomers and homodimers (Fig. 4C, right panel).
Treatment of the virus with nonionic detergent prior to SDS-PAGE affected the separation of Gn. This was obvious under reducing conditions of SDS-PAGE (Fig. 4C), but slight changes were also seen in nonreducing separation (Fig. 4B). Detergent treatment clearly induced formation of higher-order Gn oligomers (trimers and tetramers) in both NEM-treated and untreated samples but caused a decrease in the intensity of these bands in CuCl2-treated samples. This implies lipid solubilization to enforce thiol bridging of Gn into high-molecular-weight complexes that do not migrate in SDS-PAGE. Treatment of virions with nonionic detergent under reducing conditions seemed to increase the total amount of Gn migrating in SDS-PAGE (Fig. 4C, right panel).
In light of these results, SDS-PAGE separation especially under nonreducing conditions was observed to induce thiol-bridged associations between glycoproteins. Both Gn and Gc were observed to form mainly thiol-bridged homo-oligomers, but indications of possible hetero-oligomers were also seen.
Composition of complexes formed by glycoproteins in solution.
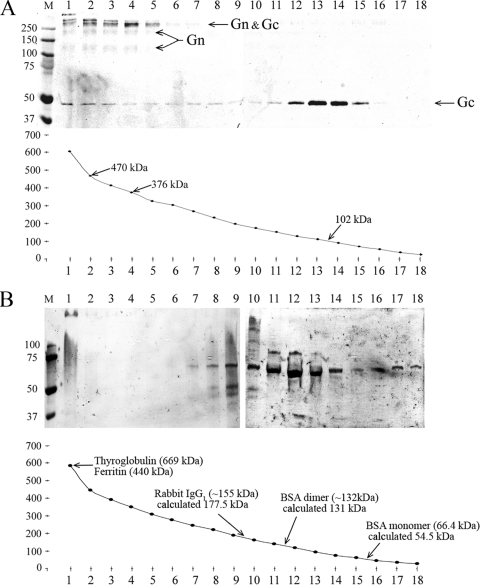
The complexes between macromolecules can be studied in density gradient ultracentrifugation to relate the mobility of complexes to density and volume. To verify that the glycoprotein complexes observed by cross-linking and by their migratory behavior in SDS-PAGE exist when glycoproteins are extracted from intact virions using nonionic detergent, the molecular masses of glycoprotein complexes were assessed from density gradient sedimentations. TULV proteins in lysates containing detergent were sedimented by ultracentrifugation in a 0 to 21% sucrose gradient. Fractions collected from the bottom of gradient were analyzed by nonreducing SDS-PAGE by immunoblotting with anti-Gn and anti-Gc sera (Fig. 5A). To obtain the curve of sedimentation using molecular mass versus fraction, the mean molecular mass was calculated from the measured sucrose concentration of each fraction. Most Gc migrated at 92 to 112 kDa (fractions 13 and 14), giving an average molecular mass of ∼102 kDa for solubilized Gc, which would correspond to a Gc dimer. The Gn was expected in detergent-solubilized lysates to form homo-oligomers (dimers and tetramers) and to associate with Gc in high-molecular-mass complexes of >250 kDa. The Gn-Gc complexes migrated to the bottom of the gradient with Gn tetramers (fractions 1 to 5), and Gc sedimented across the gradient from Gc dimer to the bottom of the gradient, indicating active adherence to larger complexes. SDS-resistant high-molecular-weight complexes of Gn and Gc (fractions 1 to 5) appeared as separated in fractions 2 and 4 since the uppermost band in fraction 2 was missing from fraction 4, and the Gc monomer band was more abundant in fraction 2 than in fraction 4. The difference between the calculated molecular masses of these fractions was ∼94 kDa, corresponding to the mobility of homodimeric Gc (Gc2). The estimated molecular mass of 376 kDa in fraction 4 could represent complexes of Gn4Gc2 (385 kDa by sequence), and that of 470 kDa in fraction 2 could indicate complexes of Gn4Gc4 (493 kDa by sequence). To verify that the accuracy of the molecular masses derived from the sucrose density was adequate, the gradient conditions were calibrated by sedimentation of a cocktail of molecular mass markers including thyroglobulin (669 kDa), ferritin (440 kDa), rabbit IgG1 (∼155 kDa), and BSA dimer (∼132 kDa) and monomer (66.4 kDa). The molecular masses of marker proteins, as estimated by calculation from the sucrose concentration after sedimentation in a density gradient, are presented in Fig. 5B. The molecular mass estimations from sucrose concentration differ from the published molecular masses by ∼10 to 20%, likely due to the preparative conditions used. Based on the mobility and molecular mass estimations of marker proteins, Gc exists in solution as a homodimer. The BSA migrated mainly as a dimer at the ∼40 μM concentration applied, which is above the reported dissociation constant (Kd) of BSA dimer of 10 ± 2 μM (26).
FIG. 5.
Complexes of TULV Gn and Gc after sedimentation in a sucrose density gradient by ultracentrifugation. (A) Glycoproteins extracted from concentrated TULV with nonionic detergent (TX-100) were separated by sedimentation in a 0 to 21% sucrose gradient (in 25 mM HEPES, 100 mM NaCl, 0.1% TX-100). The fractions from ultracentrifugation (40,000 rpm at +5°C for 20.5 h in an SW41 rotor) were collected dropwise from the tube bottom and are shown on the x axis (1 to 18). The nonreducing SDS-PAGE gel, immunoblotted with anti-Gn and anti-Gc sera, represents 100 μl of each fraction, and the molecular mass approximations corresponding to the peak fractions of Gc and Gn-Gc complexes calculated from refractivity of sucrose concentration in each fraction are as shown. (B) Molecular mass markers as indicated were sedimented and fractionated; fractions were analyzed by Coomassie-stained SDS-PAGE, and molecular sizes were calculated from sucrose concentration as in panel A. The experimental molecular masses in the peak fraction are shown with the values reported elsewhere.
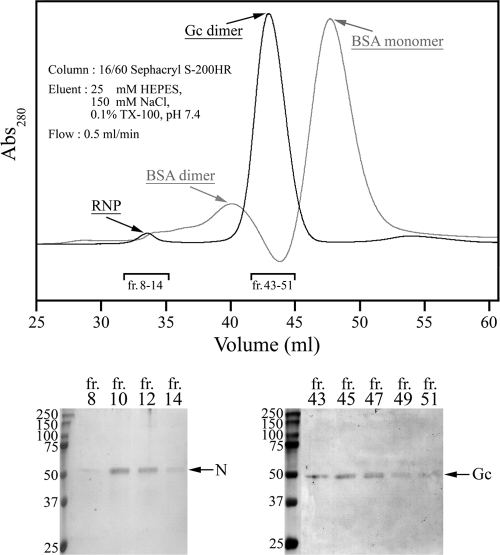
To further confirm that Gc exists as a homodimer, the TULV virion proteins were separated by gel permeation under identical extraction and buffer conditions as in the density sedimentation. Eluted TULV proteins monitored at A260 and A260 were collected, and fractions were analyzed in Coomassie blue-stained SDS-PAGE gels as shown (Fig. 6). The N protein eluted as a ribonucleoprotein complex next to the void volume (fractions 8 to 14) based on an observed high A260-to-A280 ratio (1.95), and Gc (fraction 43 to 45) was found in between BSA dimer and monomer, confirming that Gc exists in virion extracts as a homodimer with an approximate molecular mass of 100 kDa.
FIG. 6.
Complexes of TULV Gn and Gc analyzed by gel filtration. Proteins extracted from TULV virions with detergent (TX-100), as in sedimentation analysis, were separated in Sephacryl S-200HR chromatography. Elution of viral proteins detected by nonreducing SDS-PAGE and Coomassie blue staining of peak fractions is shown below, as indicated in the chromatogram. Detection of ribonucleoprotein is based on the presence of nucleocapsid protein (N) in fractions 8 to 14 (bottom left). An overlay of the virion sample (black) and BSA marker (gray) in an arbitrary 280-nm absorbance scale is presented to allow comparison of the elution volumes. fr, fraction.
Mapping of the Gn-Gc interaction sites.
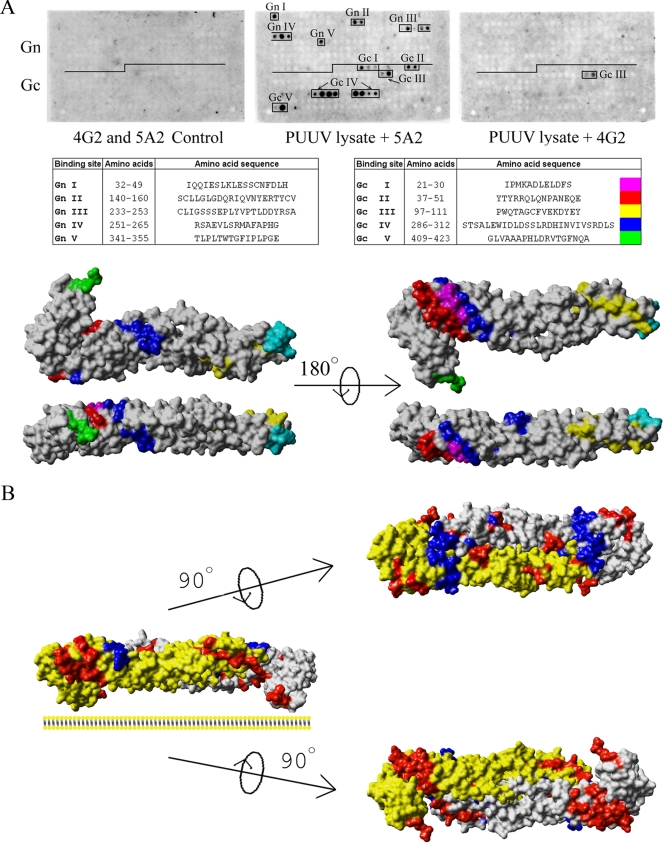
The glycoproteins of hantaviruses are known to interact with one another; however, these interaction sites have not been mapped. The location of interacting regions in Gn and Gc will further understanding of the overall architecture of hantaviruses. Since recombinant expression of the glycoproteins is complex and since making constructs with adequate deletions is challenging, we decided to use peptide scanning as the means of interaction mapping (10, 17). In order to locate the binding sites of Gn and Gc proteins in the glycoprotein precursor, a set of overlapping 18-mer peptides covering the PUUV glycoprotein precursor with a shift of 3 amino acids on the spot membrane were synthesized, and a spot assay was performed principally as previously described in epitope mapping of the PUUV glycoprotein neutralizing MAbs (15). The bindings were monitored with the Gn-specific MAb 5A2 and the Gc-specific MAb 4G2, used at dilutions which were shown not to react with the peptides (see Fig. 8A). The mappings were repeated with two different virus extracts and spot membranes with different starting residues in the overlapping peptides. Five binding sites in both Gn (GnI to GnV) and Gc (GcI to GcV) were identified for the Gn protein, implying that Gn forms both hetero- and homo-oligomeric complexes. The interaction sites (shown in Fig. 8A as peptide sequences in PUUV Gn or Gc) were located in regions that, by prediction, are in the ectodomains of both glycoproteins.
FIG. 8.
PUUV Gn-Gc interaction sites by peptide scanning. (A) Fine-mapping of the Gn-Gc interaction sites by peptide scanning of the glycoprotein precursor of PUUV with 18-mer peptides in a shift of 3 amino acids (spot method). PUUV lysate in the protein overlay assay was monitored for binding of Gn (middle) and Gc (right) to peptides on the membrane. In spot assay detection, MAbs 5A2 and 4G2 were used at concentrations at which they did not give unspecific binding in ECL signals on X-ray film (left). The interaction sites are indicated in the membranes, and the corresponding peptide sequences are listed. An illustration of the Gn binding sites in Gc (model) on a solvent-accessible surface structure is shown on the right. The coloring of interaction sites is the same as in the table of sequences, and the putative fusion peptide of PUUV Gc protein is shown in cyan (13, 55). (B) Hypothetical dimer of Gc (model), aligned such that the known epitopes of Gc-specific MAb 4G2 form a uniform surface. The docked dimer is shown as a solvent-accessible surface, the Gc molecules in gray highlight binding sites of MAb 4G2 (16) in cyan, and the mapped Gn interaction sites are shown as a monomer structure as in panel A. Docking of two Gc molecules in this fashion interestingly resembles the reported homodimer of TBEV E protein (41).
To illustrate the binding sites, we used comparative modeling to build the PUUV Gc structure in Swiss-Model (http://swissmodel.expasy.org/) (3). Structure models were obtained with class II viral fusion proteins (SFV E1 and TBEV E) as templates and were tried on a class III fusion protein template without success. The models created were analyzed by comparing Ramachandran plots and dihedral angles of the model to the template in VADAR, version 1.5 (66). The models were compared to the template also in the TM-align algorithm (server at http://zhang.bioinformatics.ku.edu) (69). The TM-score of TM-align between the PUUV Gc model and SFV E1 template was as high as 0.9266, and the amino acid identity between the sequences was 18.2%. The corresponding values for a model based on TBEV E were 0.8251 and 19.4%. The lower TM-score obtained for the TBEV E model implied that the model based on SFV E1 template was more acceptable for illustrating the interaction mapping results. The created 3D structure model of PUUV Gc and the template structure of SFV E1, PDB accession code 2ALA (45), are presented with the respective Ramachandran plots in Fig. 7. The values for the dihedral angles of the Gc model and E1 template shown in Table 1 are comparable to each other. The domains in modeled Gc that had the strongest homology to the SFV E1 were, in adoption of nomenclature by F. Rey et al. (41), domain I (DI) and DIII. DII, containing the internal fusion loop, differed the most from the template, showing partially disordered regions.
FIG. 7.
3D structure model of PUUV Gc with SFV E1 as a template. The Swiss-Model-generated PDB coordinate file of PUUV Gc and the SFV E1 used as a template (PBD code, 2ALA) (45) were visualized with YASARA View. The model and template were superimposed, and the images shown were captured in the same respective angles. DI, DII, and DIII are shown in red, yellow, and blue, respectively, as suggested by Rey and collaborators (41). The PDB coordinates of the created Gc model were analyzed using VADAR, version 1.5, and the Ramachandran plots of the PUUV Gc model and the SFV E1 template obtained from the VADAR server (http://redpoll.pharmacy.ualberta.ca/vadar) are presented below for the respective structures.
TABLE 1.
Dihedral angles of the SFV E1 (template) and PUUV Gc (model)
| Protein and statistic | Value for the statistic |
|
|---|---|---|
| Observed | Expected | |
| SFV E1 (template)a | ||
| Mean helix phi (° [SD]) | −72.3 (12.1) | −65.3 (11.9) |
| Mean helix psi (° [SD]) | −26.9 (18) | −39.4 (25.5) |
| No. of residues (%) with gauche-plus chi | 115 (37) | 167 (55) |
| No. of residues (%) with gauche- minus chi | 66 (21) | 60 (20) |
| No of residues (%) with trans chi | 123 (40) | 76 (25) |
| Mean chi gauche plus (° [SD]) | −62.2 (13.3) | −66.7 (15.0) |
| Mean chi gauche minus (° [SD]) | 58.2 (13.1) | 64.1 (15.7) |
| Mean chi trans (° [SD]) | 167.8 (11.3) | 168.6 (16.8) |
| SD of chi pooled | 12.45 | 15.70 |
| Mean omega (|omega| > 90) (° [SD]) | 179.5 (6.9) | 180.0 (5.8) |
| No. of residues with |omega| < 90 (%) | 0 (0) | |
| PUUV Gc (model) | ||
| Mean helix phi (° [SD]) | −70.0 (14.3) | −65.3 (11.9) |
| Mean helix psi (° [SD]) | −46.7 (40.1) | −39.4 (25.5) |
| No. of residues (%) with gauche-plus chi | 118 (36) | 176 (55) |
| No. of residues (%) with gauche- minus chi | 82 (25) | 64 (20) |
| No. of residues (%) with trans chi | 120 (37) | 80 (25) |
| Mean chi gauche plus (° [SD]) | −63.4 (10.0) | −66.7 (15.0) |
| Mean chi gauche minus (° [SD]) | 61.5 (6.4) | 64.1 (15.7) |
| Mean chi trans (° [SD]) | 176.2 (3.1) | 168.6 (16.8) |
| SD of chi pooled | 6.49 | 15.70 |
| Mean omega (|omega| > 90) (° [SD]) | 179.2 (7.8) | 180 (5.8) |
| No. of residues with |omega| < 90 (%) | 5 (1) | |
PDB 2ALA.
The five binding sites of Gn, GcI to GcV, are highlighted in the 3D structure model of Gc and were found in a region that, according to the SFV E1 nomenclature, is the central DI over which the DI-DII-DIII linker region spans. Short interaction sites were also found in the DII fusion region and in the DIII membrane stem linker. The interaction sites in DI were GcI, GcII, and GcIV (Fig. 8A), corresponding to the beta strands Bo-Co-Io in the 3D structure of SFV E1. The GcIV-binding site (Fig. 8A) also included sites that correspond to the flexible linker regions k, l, (Io), and η3 seen in the SFV E1 structure. The fusion domain interaction site GcIII overlapped with SFV E1 region c, and the GcV binding site was next to the DIII stem linker nearby the membrane anchor site. The Gc-specific MAb 4G2 gave only two positive peptides when the membrane was probed with PUUV lysate. These peptides represented the same GcIII site which was detected in the PUUV lysate with the MAb.
Hypothetical structure of the PUUV Gc homodimer.
According to the density sedimentation rate in ultracentrifugation (Fig. 5), gel permeation (Fig. 6), and protein complex formation chemistries (Fig. 4), the Gc of hantaviruses forms homodimers. For the SFV E1 protein, a homodimer was seen when the protein was crystallized in the absence of glycoprotein E2 (25). The E protein of TBEV exists as a homodimer in intact virions, and the fusion peptide is buried due to homodimeric interaction (41). To test the hypothesis that Gc exists as a homodimer, two 3D models of Gc were docked head to tail in a manner similar to that described for the SFV E1 and TBEV E (25). For docking, we highlighted the mapped Gn and Gc interaction sites, which by peptide scanning were located at both ends of the Gc 3D model (Fig. 8A). The previously described peptide sequences forming the discontinuous epitope of neutralizing MAb 4G2 (15) were also highlighted in the monomer structures. Docking to form a homodimer was done manually in order to meet the requirement of a uniform MAb 4G2 epitope surface (Fig. 8B). This is achieved when monomers are facing one another (as in TBEV) rather than being back to back (as in SFV E1), thereby more closely resembling in quaternary structure the TBEV E protein than SFV E1. This arrangement of proteins would allow a high degree of symmetry to the assembly and formation of both Gc-Gc and Gn-Gc complexes. Thus, it would also enable the proteins to undergo dynamic changes and changes in organization upon membrane fusion.
Cryo-EM was used to study the glycoprotein organization in the virion. A typical micrograph is shown in Fig. 9B. The TULV virions were mainly round or pleomorphic with a diameter varying from 100 to 120 nm. In cryo-EM preparations virus particles appeared in random orientations. In favorable views, the spikes were clearly visible and found to create a regular surface structure seen on the edge of the particle. A spike formed of two seemingly identical subunits is highlighted in Fig. 9B. The appearance of this protruding spike suggests that it could consist of either a homo-oligomer of one glycoprotein or a hetero-oligomer of two. The previously described grid-like surface pattern of Hantaan virus (31) was not observed in cryo-EM preparations, obviously because no surface stain is used in cryo-EM. To study the surface organization of glycoproteins using cryo-EM, tomography needs to be applied, but the virus density in our preparations was insufficient for that.
DISCUSSION
Here, we present the biochemical characterization of interactions between hantavirus glycoproteins. The approaches used included coimmunoprecipitation, chemical cross-linking, velocity sedimentation, gel permeation, and peptide interaction mapping of Gn and Gc. Glycoprotein interactions and known discontinuous neutralizing antibody epitopes were compiled into a model structure of PUUV Gc generated using comparative modeling with a known class II fusion protein as a template. We aimed to provide a description of the surface architecture of hantaviruses and of the spikes formed by glycoproteins in virions.
First, we showed that the complex between PUUV Gn and Gc dissociates in a pH-dependent manner. The Gc of hantaviruses has been suggested to be a class II viral fusion protein (13, 55). Viral fusion proteins of class II generally exist as homo- or heterodimers, while upon activation in acidic pH rearrangement to homotrimers occurs (7). By means of immunoprecipitation with a monoclonal antibody to PUUV Gn (MAb 5A2), we observed that the hetero-oligomeric Gn-Gc complex dissociates at a pH below 6.2 and reassociates at neutral to slightly basic pH. Concomitant to Gn-Gc complex dissociation, the epitope of PUUV Gc-specific MAb 4G2 disappeared at pH 6.0. Both the observed dissociation of the Gn-Gc complex at pH 6.2 to 6.4 and the loss of recognition of the Gc-specific MAb epitope imply a change in the conformation of Gc at pH 6.0 to 6.4. This is in line with evidence showing that the fusion of hantavirus with cellular membranes takes place at low pH and would occur in early (pH 6.1 to 6.2) or late endosomes (pH ∼5.5) of susceptible cells (47), as proposed for endocytosis of hantavirus (19). By analogy to either the SFV or the TBEV fusion event (14, 52, 68), the pH-inducible changes in Gc conformation and the simultaneous dissociation of Gc from the hetero- or homo-oligomeric complex would reveal a fusion loop. Interestingly, low-pH exposure of the MAb 4G2-bound Gc did not result in dissociation of the Gc from MAb resin. This suggests that the neutralization mechanism of MAb 4G2 is through prevention of the low-pH-induced structural changes in Gc, thereby hindering the fusion activity of Gc.
The immunoprecipitations with PUUV Gn- and Gc-specific neutralizing MAbs 5A2, 4G2, and 1C9 imply that the low-pH-induced changes to Gc and dissociation of the Gn-Gc complex are reversible since Gc precipitation and coprecipitation of Gn with Gc were lost at acidic pH and were both restored at neutral to slightly basic pH. In contrast to this, there was a 99% decrease in the infectivity of TULV treated at pH 6.0 or at pH 5.5. This suggests that the Gn and Gc interactions in virions are affected at a pH between 6.0 to 6.5. However, the low-pH treatment of TULV virions did not result in complete inactivation of virus, as determined by virus titration. The observed decrease in infectivity was 100-fold, which compared to the initial virus titer (∼108 FFU/ml), still suggests a considerable resistance to acidic pH. Further investigations are needed to determine whether hantaviruses in fact tolerate acidic pH. A previous report shows that hantaviruses are prone to inactivation in cell culture supernatants (21), but the reason for this inactivation has not been shown. Based on both the observed dissociation of Gn-Gc complexes in solution and incomplete inactivation of virions in an acidic environment, we suggest that low pH alone is not sufficient to render hantaviruses inactive. An acid resistance is described for bovine viral diarrhea virus, a member of the Flaviviridae, suggesting that additional factors could be required for the fusion of hantaviruses as for fusion entry of pestiviruses, which, like hantaviruses, form complex intra- and intermolecular disulfide bonds between glycoproteins (24, 56).
The composition of complexes formed by Gn and Gc has not been characterized previously in detail. Reports characterizing the epitopes of MAbs against hantavirus glycoproteins (2, 18, 28, 29, 65) have shed light on the coimmunoprecipitation properties of Gn and Gc. Additionally, coprecipitation of the glycoproteins from two different hantavirus species has been demonstrated, which in theory enables generation of a reassortant virus (6). However, no previous reports on the quaternary organization between Gn and Gc in hantaviruses have been published. Our report describes the composition of the SDS-resistant Gn mobilities, regularly seen when hantavirus proteins are separated by gel electrophoresis (46, 54). The high-molecular-weight complexes of Gn and Gc were generated by thiol bridging which, at least to some extent, occurs during SDS-PAGE separation and persists upon protein extraction. It seems that, once formed, the thiol bridges resist reduction, possibly because oxidation leads to formation of tight Gn-Gc clusters. We also conclude that the homo- and hetero-oligomeric complexes can be maintained by intramolecular thiol bridges, which affect the tertiary structure of individual proteins, thereby strengthening intermolecular interactions. When separation of glycoproteins was done under nonreducing SDS-PAGE conditions, dimeric and tetrameric Gn proteins along with homodimeric Gc were observed. Complexes containing both Gn and Gc were over 250 kDa, indicating that the complex consists of more than four subunits. Both the dimeric Gc and its high-molecular-weight clusters were abolished under reducing conditions, but the subunit associations of Gn remained. In ultracentrifugation and gel filtration, the Gc protein sedimented primarily as a homodimer of ∼100 kDa, whereas Gn was found in virion extracts primarily in a size range above 250 kDa together with Gc. Based on the molecular mass calculations in velocity sedimentation, these clusters were 376 kDa and 470 kDa, thus consisting of six and eight glycoprotein subunits, which most probably consisted of four units of Gn binding either two or four Gc units, respectively.
By peptide scanning, we were able to map interactions between glycoproteins in several regions. Five binding sites for homotypic Gn-Gn interaction were identified (GnI to GnV) and five for heterotypic Gn-Gc (GcI to GcV). One of the latter (GcIII) was found also to mediate homodimeric Gc interaction and was a potentially interesting site for the formation of Gn-Gc complexes. To visualize the detected Gn-binding sites to Gc peptides, we created a 3D model of Gc based on the SFV E1 crystal structure. The GcIII-interaction site at 755PWQTAGCFVEKDYEY769 of the PUUV precursor (97P to Y111 in Gc) contains the 761Cys residue (underlined; 103Cys in Gc) which is conserved among hantaviruses and located in the part of the 3D model (Fig. 8A) that corresponds to the region c in the SFV E1 structure (41). Additionally, GcIII partially overlaps with the putative fusion loop (Fig. 8A) at 762FVEKDYEYETGWGCNPPDCPGVGTGC787 (104F to C129 in Gc) (55), centered at the 773Trp residue conserved among hantaviruses (underlined; 115W in Gc). The N-terminal part of the fusion loop has been described as part of the MAb 4G2 epitope (15). The peptide epitopes of MAb 4G2 in PUUV Gc are 754YPWQTAGCFVEK765 (96Y to K107 in Gc), 772GWGCNPPD779 (114G to D121 in Gc), 910TPVCQF915 (252T to F257 in Gc), and 1057DTKCSSTGLVAA1068 (399D to A410 in Gc). Of these, the first two overlap with the putative fusion loop residues (55). Several Hantaan virus neutralizing recombinant antibodies have been mapped to Gc, residues 916KVMATIDSF924 and 954LVTKDIDFD963 (23), of which the first overlaps with the MAb 4G2 epitope. The MAb 4G2 binding site 1057DTKCSSTGLVAA1068 (399D to A410 in Gc) locates adjacent to the interaction site GcV 1067GLVAAAPIILDRVTGFNQA1081 (409G to A423 in Gc), suggesting that the MAb 4G2 binding sites are near the Gn-Gc interaction interface and that its discontinuous epitope may cover the proposed fusion loop. The cross-competition of MAbs 1C9, 4G2, and 5A2 (16, 28-30) suggests that they all bind close to the Gn-Gc interaction surface in the spike. It appears likely that the MAb 1C9 epitope is a neotope formed by the Gn-Gc complex. Mutation 944S → F (286S in Gc protein), a reported MAb escape mutant of 1C9 (18), is the first residue of GcIV site 944STSALEWIDLDSSLRDHINVIVSRDLS970 (286S to S312 in Gc) and may be important for Gn-Gc interactions. The N-glycosylation site of Gc located seven residues away from the GcIV at 937N (279N in Gc) is essential for cell fusion activity (70). The MAbs 4G2 and 1C9 share at least partially the peptide epitope located inside the fusion loop (DII) (15). The MAb escape mutant (249D → V) of the Gn-specific MAb 5A2 (15, 18) was located to the GnIII site, and the GnI, GnII, and GnIV sites have been assigned as discontinuous epitope binding sites of this MAb (15). Altogether, the epitope and interaction data suggest that prevention of Gn-Gc or Gc-Gc complex dissociations mediates hantavirus neutralization and demonstrate mechanisms for inter- and intramolecular relationships of glycoproteins which could prevent fusion.
Based on the interaction results and location of known antibody epitopes in the 3D model of PUUV Gc created using the SFV E1 crystal structure as a template, we hypothesize that there is structural homology between the SFV E1 and hantavirus Gc protein. The spike complex of SFV is a trimer of heterodimers (E1-E2)3 mediated by glycoprotein E2 transmembrane domain interactions, which are considered responsible for the three-fold symmetry of the spike (25, 38). The oligomerization between SFV glycoproteins E1 and E2 was first described in covalent cross-linking of glycoproteins on the surface of intact virions (11, 12), and the components forming the spike were later confirmed in the virion structure solved by cryo-EM tomography (62, 63). Detergent treatment of SFV showed an association of glycoproteins, called rosettes (20, 51). Our data suggest that the detergent-soluble, large hantavirus glycoprotein complexes formed under reducing conditions (Fig. 4C) most likely represent the spikes seen on the surface of virions in negatively stained specimens. The surface architecture in negatively stained EM is grid-like, and glycoproteins seem to form a diamond- or square-like structure consisting of perhaps four subunits (31). Membrane-impermeable cross-linking of intact virions made the Gn susceptible to the higher oligomerizations found in complexes, a property that was not as apparent for Gc. Our hypothesis is that reduction of existing thiol bonds in the region of transmembrane helices would form a long hydrophobic region, resulting in formation of SDS-stable Gn complexes. The known aggregation tendency of hantavirus Gn is reported to reside between residues 500 to 520 of Sin Nombre virus (54). By prediction, this region in Gn (amino acids 450 to 520) contains one or three transmembrane helices. The domain that is responsible for SFV spike trimerization in the E2 glycoprotein has two transmembrane segments. Thus, we hypothesize that the Gn homo-oligomerization is responsible for the spike formation and that Gn could be more solvent exposed in virions than Gc. Based on this, we suggest that the spike seen on the virus surface (Fig. 9C) is Gn as it is oriented perpendicular to the membrane while Gc lies parallel to the membrane. Accordingly, Gn was observed to exist in oligomeric complexes of at least two and four subunits. Strong evidence favoring Gc homodimers led to the interpretation of how glycoproteins organize on the surface of a virion. The complexes observed upon virion extraction (Fig. 9A) could be derived from only one type of Gn and Gc organization to provide the surface model of hantavirus (Fig. 9C). The results suggest that the protein spike consists of an equimolar ratio of Gn and Gc, implying that the smallest interacting unit can be a heterodimer. Strong homo-oligomeric (dimeric and tetrameric) interactions between Gn proteins and the presence of Gc dimers indicated that homotetrameric Gn and homodimeric Gc are the basic assembly units of hantaviruses. In contrast to the homodimers observed in phleboviruses (44), the Gn homotetrameric subunits, through interactions with homodimeric Gc subunits, would interconnect the spike structures on the surface of hantavirus. The contacts between Gn subunits most probably resemble the contacts between E2 subunits in SFV virions that are maintained by interactions between domains at or close to transmembrane helices (25, 45), with the exception that four instead of three Gn subunits appear to form the foundation of the spike in hantaviruses. In the case of hantaviruses the contacts between homo-oligomeric (dimeric and tetrameric) Gn subunits would create the spike, and contacts between Gc subunits would contribute to the overall surface architecture (Fig. 9C, hypothesis 3). We also hypothesize that hantavirus Gc shares qualities of both SFV E1 and TBEV E, i.e., that it is structurally analogous to SFV E1 but with an orientation in the virion similar to that of TBEV E.
For a virus spread via excreta, survival outside the rodent host is extremely critical. Virus particles need to overcome drought and other mechanical stresses caused by the environment outside of the host in order to maintain the integrity of viral RNA. It is possible that the observed cross-linking of glycoproteins via free thiol groups would provide mechanical stability to the envelope of hantaviruses. The appearance of thiol bridges between complexes involving Gc are of special interest. Covalent bonding of Gc could interfere with the structural changes required during fusion, such as homotrimerization. We did not observe homotrimers of Gc when intact viruses were treated at low pH; membranes were disrupted and analyzed by sedimentation in a density gradient experiment (results not shown). Accordingly, similar low-pH treatment of viruses at pH 5.5 or 6.0 did not inactivate the virus. If thiol bridging provides mechanical stability, there has to be a mechanism for the virus to regulate the formation and breakage of cysteines to inactivate and reactivate the fusion protein. Gc of hantaviruses contains a well-conserved disulfide bond isomerase motif adjacent to the putative fusion loop, residues 787Cys-X-X-Cys790 (129 to 132 in Gc) (8). This type of motif is essential for prevention of the premature fusion of retroviruses (64). This motif might also be active in hantaviruses and prevent virus inactivation under conditions of low-pH treatment; curiously, we have observed TULV to be sensitive to DTT and TCEP, an observation that requires further investigation.
Acknowledgments
This work was supported by Academy of Finland grant 102371, project number 200921, to H.L.
Juha Huiskonen (Department of Molecular Structural Biology, Max-Planck-Institute of Biochemistry, Martinsried, Germany), Sarah Butcher (Institute of Biotechnology, Helsinki University), and Esa Kuismanen (Department of Biosciences, Division of Biochemistry, University of Helsinki) are thanked for fruitful discussions along with suggestions and comments during preparation of the manuscript. We thank the Electron Microscopy Unit (Institute of Biotechnology, Helsinki University) and Pasi Laurinmäki, Engineering Institute of Biotechnology, for expertise in cryo-EM sample preparation and imaging. Leena Kostamovaara and Tytti Manni are thanked for skilled technical assistance at the Haartman Institute. Scientific English proofreading was by Robert M. Badeau (Scienceproof Proofreading Services).
Footnotes
Published ahead of print on 14 October 2009.
REFERENCES
- 1.Adrian, M., J. Dubochet, J. Lepault, and A. W. McDowall. 1984. Cryo-electron microscopy of viruses. Nature 308:32-36. [DOI] [PubMed] [Google Scholar]
- 2.Arikawa, J., A. L. Schmaljohn, J. M. Dalrymple, and C. S. Schmaljohn. 1989. Characterization of Hantaan virus envelope glycoprotein antigenic determinants defined by monoclonal antibodies. J. Gen. Virol. 70:615-624. [DOI] [PubMed] [Google Scholar]
- 3.Arnold, K., L. Bordoli, J. Kopp, and T. Schwede. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195-201. [DOI] [PubMed] [Google Scholar]
- 4.Beniac, D. R., A. Andonov, E. Grudeski, and T. F. Booth. 2006. Architecture of the SARS coronavirus prefusion spike. Nat. Struct. Mol. Biol. 13:751-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delmas, B., and H. Laude. 1990. Assembly of coronavirus spike protein into trimers and its role in epitope expression. J. Virol. 64:5367-5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deyde, V. M., A. A. Rizvanov, J. Chase, E. W. Otteson, and S. C. St Jeor. 2005. Interactions and trafficking of Andes and Sin Nombre Hantavirus glycoproteins G1 and G2. Virology 331:307-315. [DOI] [PubMed] [Google Scholar]
- 7.Earp, L. J., S. E. Delos, H. E. Park, and J. M. White. 2005. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 285:25-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrari, D. M., and H. D. Soling. 1999. The protein disulphide-isomerase family: unravelling a string of folds. Biochem. J. 339:1-10. [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer, H., I. Polikarpov, and A. F. Craievich. 2004. Average protein density is a molecular-weight-dependent function. Protein Sci. 13:2825-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank, R. 2002. The SPOT-synthesis technique. Synthetic peptide arrays on membrane supports-principles and applications. J. Immunol. Methods 267:13-26. [DOI] [PubMed] [Google Scholar]
- 11.Garoff, H. 1974. Cross-linking of the spike glycoproteins in Semliki Forest virus with dimethylsuberimidate. Virology 62:385-392. [DOI] [PubMed] [Google Scholar]
- 12.Garoff, H., and K. Simons. 1974. Location of the spike glycoproteins in the Semliki Forest virus membrane. Proc. Natl. Acad. Sci. U. S. A. 71:3988-3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garry, C. E., and R. F. Garry. 2004. Proteomics computational analyses suggest that the carboxyl terminal glycoproteins of Bunyaviruses are class II viral fusion protein (beta-penetrenes). Theor. Biol. Med. Model 1:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammar, L., S. Markarian, L. Haag, H. Lankinen, A. Salmi, and R. H. Cheng. 2003. Prefusion rearrangements resulting in fusion peptide exposure in Semliki forest virus. J. Biol. Chem. 278:7189-7198. [DOI] [PubMed] [Google Scholar]
- 15.Heiskanen, T., A. Lundkvist, R. Soliymani, E. Koivunen, A. Vaheri, and H. Lankinen. 1999. Phage-displayed peptides mimicking the discontinuous neutralization sites of Puumala hantavirus envelope glycoproteins. Virology 262:321-332. [DOI] [PubMed] [Google Scholar]
- 16.Heiskanen, T., A. Lundkvist, A. Vaheri, and H. Lankinen. 1997. Phage-displayed peptide targeting on the Puumala hantavirus neutralization site. J. Virol. 71:3879-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hilpert, K., D. F. Winkler, and R. E. Hancock. 2007. Peptide arrays on cellulose support: SPOT synthesis, a time and cost efficient method for synthesis of large numbers of peptides in a parallel and addressable fashion. Nat. Protoc. 2:1333-1349. [DOI] [PubMed] [Google Scholar]
- 18.Horling, J., and A. Lundkvist. 1997. Single amino acid substitutions in Puumala virus envelope glycoproteins G1 and G2 eliminate important neutralization epitopes. Virus Res. 48:89-100. [DOI] [PubMed] [Google Scholar]
- 19.Jin, M., J. Park, S. Lee, B. Park, J. Shin, K. J. Song, T. I. Ahn, S. Y. Hwang, B. Y. Ahn, and K. Ahn. 2002. Hantaan virus enters cells by clathrin-dependent receptor-mediated endocytosis. Virology 294:60-69. [DOI] [PubMed] [Google Scholar]
- 20.Kaariainen, L., K. Simons, and C. H. von Bonsdorff. 1969. Studies in subviral components of Semliki Forest virus. Ann. Med. Exp. Biol. Fenn. 47:235-248. [PubMed] [Google Scholar]
- 21.Kallio, E. R., J. Klingstrom, E. Gustafsson, T. Manni, A. Vaheri, H. Henttonen, O. Vapalahti, and A. Lundkvist. 2006. Prolonged survival of Puumala hantavirus outside the host: evidence for indirect transmission via the environment. J. Gen. Virol. 87:2127-2134. [DOI] [PubMed] [Google Scholar]
- 22.Khaiboullina, S. F., S. P. Morzunov, and S. C. St Jeor. 2005. Hantaviruses: molecular biology, evolution and pathogenesis. Curr. Mol. Med. 5:773-790. [DOI] [PubMed] [Google Scholar]
- 23.Koch, J., M. Liang, I. Queitsch, A. A. Kraus, and E. K. Bautz. 2003. Human recombinant neutralizing antibodies against Hantaan virus G2 protein. Virology 308:64-73. [DOI] [PubMed] [Google Scholar]
- 24.Krey, T., H. J. Thiel, and T. Rumenapf. 2005. Acid-resistant bovine pestivirus requires activation for pH-triggered fusion during entry. J. Virol. 79:4191-4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lescar, J., A. Roussel, M. W. Wien, J. Navaza, S. D. Fuller, G. Wengler, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137-148. [DOI] [PubMed] [Google Scholar]
- 26.Levi, V., and F. L. Gonzalez Flecha. 2002. Reversible fast-dimerization of bovine serum albumin detected by fluorescence resonance energy transfer. Biochim. Biophys. Acta 1599:141-148. [DOI] [PubMed] [Google Scholar]
- 27.Lober, C., B. Anheier, S. Lindow, H. D. Klenk, and H. Feldmann. 2001. The Hantaan virus glycoprotein precursor is cleaved at the conserved pentapeptide WAASA. Virology 289:224-229. [DOI] [PubMed] [Google Scholar]
- 28.Lundkvist, A., J. Horling, L. Athlin, A. Rosen, and B. Niklasson. 1993. Neutralizing human monoclonal antibodies against Puumala virus, causative agent of nephropathia epidemica: a novel method using antigen-coated magnetic beads for specific B cell isolation. J. Gen. Virol. 74:1303-1310. [DOI] [PubMed] [Google Scholar]
- 29.Lundkvist, A., and B. Niklasson. 1992. Bank vole monoclonal antibodies against Puumala virus envelope glycoproteins: identification of epitopes involved in neutralization. Arch. Virol. 126:93-105. [DOI] [PubMed] [Google Scholar]
- 30.Lundkvist, A., C. Scholander, and B. Niklasson. 1993. Anti-idiotypic antibodies against Puumala virus glycoprotein-specific monoclonal antibodies inhibit virus infection in cell cultures. Arch. Virol. 132:255-265. [DOI] [PubMed] [Google Scholar]
- 31.Martin, M. L., H. Lindsey-Regnery, D. R. Sasso, J. B. McCormick, and E. Palmer. 1985. Distinction between Bunyaviridae genera by surface structure and comparison with Hantaan virus using negative stain electron microscopy. Arch. Virol. 86:17-28. [DOI] [PubMed] [Google Scholar]
- 32.McCaughey, C., X. Shi, R. M. Elliot, D. E. Wyatt, H. J. O'Neill, and P. V. Coyle. 1999. Low pH-induced cytopathic effect—a survey of seven hantavirus strains. J. Virol. Methods 81:193-197. [DOI] [PubMed] [Google Scholar]
- 33.McEwen, C. R. 1967. Tables for estimating sedimentation through linear concentration gradients of sucrose solution. Anal. Biochem. 20:114-149. [DOI] [PubMed] [Google Scholar]
- 34.Nichol, S. T., B. J. Beaty, R. M. Elliott, et al. 2005. Bunyaviridae, p. 695-716. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy: classification and nomenclature of viruses. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier, Amsterdam, The Netherlands.
- 35.Ogino, M., K. Yoshimatsu, H. Ebihara, K. Araki, B. H. Lee, M. Okumura, and J. Arikawa. 2004. Cell fusion activities of Hantaan virus envelope glycoproteins. J. Virol. 78:10776-10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pensiero, M. N., and J. Hay. 1992. The Hantaan virus M-segment glycoproteins G1 and G2 can be expressed independently. J. Virol. 66:1907-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Persson, R., and R. F. Pettersson. 1991. Formation and intracellular transport of a heterodimeric viral spike protein complex. J. Cell Biol. 112:257-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pletnev, S. V., W. Zhang, S. Mukhopadhyay, B. R. Fisher, R. Hernandez, D. T. Brown, T. S. Baker, M. G. Rossmann, and R. J. Kuhn. 2001. Locations of carbohydrate sites on alphavirus glycoproteins show that E1 forms an icosahedral scaffold. Cell 105:127-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plyusnin, A., and S. P. Morzunov. 2001. Virus evolution and genetic diversity of hantaviruses and their rodent hosts. Curr. Top. Microbiol. Immunol. 256:47-75. [DOI] [PubMed] [Google Scholar]
- 40.Plyusnin, A., O. Vapalahti, and A. Vaheri. 1996. Hantaviruses: genome structure, expression and evolution. J. Gen. Virol. 77:2677-2687. [DOI] [PubMed] [Google Scholar]
- 41.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 42.Rickwood, D. (ed.) 1984. Centrifugation: a practical approach. IRL Press Limited, Oxford, United Kingdom.
- 43.Roche, S., F. A. Rey, Y. Gaudin, and S. Bressanelli. 2007. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science 315:843-848. [DOI] [PubMed] [Google Scholar]
- 44.Ronka, H., P. Hilden, C. H. Von Bonsdorff, and E. Kuismanen. 1995. Homodimeric association of the spike glycoproteins G1 and G2 of Uukuniemi virus. Virology 211:241-250. [DOI] [PubMed] [Google Scholar]
- 45.Roussel, A., J. Lescar, M. C. Vaney, G. Wengler, G. Wengler, and F. A. Rey. 2006. Structure and interactions at the viral surface of the envelope protein E1 of Semliki Forest virus. Structure 14:75-86. [DOI] [PubMed] [Google Scholar]
- 46.Ruusala, A., R. Persson, C. S. Schmaljohn, and R. F. Pettersson. 1992. Coexpression of the membrane glycoproteins G1 and G2 of Hantaan virus is required for targeting to the Golgi complex. Virology 186:53-64. [DOI] [PubMed] [Google Scholar]
- 47.Rybak, S. L., and R. F. Murphy. 1998. Primary cell cultures from murine kidney and heart differ in endosomal pH. J. Cell. Physiol. 176:216-222. [DOI] [PubMed] [Google Scholar]
- 48.Saunal, H., and M. H. Van Regenmortel. 1995. Mapping of viral conformational epitopes using biosensor measurements. J. Immunol. Methods 183:33-41. [DOI] [PubMed] [Google Scholar]
- 49.Schmaljohn, C., and B. Hjelle. 1997. Hantaviruses: a global disease problem. Emerg. Infect. Dis. 3:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi, X., and R. M. Elliott. 2002. Golgi localization of Hantaan virus glycoproteins requires coexpression of G1 and G2. Virology 300:31-38. [DOI] [PubMed] [Google Scholar]
- 51.Simons, K., A. Helenius, and H. Garoff. 1973. Solubilization of the membrane proteins from Semliki Forest virus with Triton X100. J. Mol. Biol. 80:119-133. [DOI] [PubMed] [Google Scholar]
- 52.Sollner, T. H. 2004. Intracellular and viral membrane fusion: a uniting mechanism. Curr. Opin. Cell Biol. 16:429-435. [DOI] [PubMed] [Google Scholar]
- 53.Spiropoulou, C. F. 2001. Hantavirus maturation. Curr. Top. Microbiol. Immunol. 256:33-46. [DOI] [PubMed] [Google Scholar]
- 54.Spiropoulou, C. F., C. S. Goldsmith, T. R. Shoemaker, C. J. Peters, and R. W. Compans. 2003. Sin Nombre virus glycoprotein trafficking. Virology 308:48-63. [DOI] [PubMed] [Google Scholar]
- 55.Tischler, N. D., A. Gonzalez, T. Perez-Acle, M. Rosemblatt, and P. D. Valenzuela. 2005. Hantavirus Gc glycoprotein: evidence for a class II fusion protein. J. Gen. Virol. 86:2937-2947. [DOI] [PubMed] [Google Scholar]
- 56.Tscherne, D. M., M. J. Evans, M. R. Macdonald, and C. M. Rice. 2008. Transdominant inhibition of bovine viral diarrhea virus entry. J. Virol. 82:2427-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vaheri, A., O. Vapalahti, and A. Plyusnin. 2008. How to diagnose hantavirus infections and detect them in rodents and insectivores. Rev. Med. Virol. 18:277-288. [DOI] [PubMed] [Google Scholar]
- 58.Van Regenmortel, M. H., D. Altschuh, and G. Zeder-Lutz. 1993. Tobacco mosaic virus: a model antigen to study virus-antibody interactions. Biochimie 75:731-739. [DOI] [PubMed] [Google Scholar]
- 59.Vapalahti, O., H. Kallio-Kokko, A. Narvanen, I. Julkunen, A. Lundkvist, A. Plyusnin, H. Lehvaslaiho, M. Brummer-Korvenkontio, A. Vaheri, and H. Lankinen. 1995. Human B-cell epitopes of Puumala virus nucleocapsid protein, the major antigen in early serological response. J. Med. Virol. 46:293-303. [DOI] [PubMed] [Google Scholar]
- 60.Vapalahti, O., H. Kallio-Kokko, E. M. Salonen, M. Brummer-Korvenkontio, and A. Vaheri. 1992. Cloning and sequencing of Puumala virus Sotkamo strain S and M RNA segments: evidence for strain variation in hantaviruses and expression of the nucleocapsid protein. J. Gen. Virol. 73:829-838. [DOI] [PubMed] [Google Scholar]
- 61.Vapalahti, O., J. Mustonen, A. Lundkvist, H. Henttonen, A. Plyusnin, and A. Vaheri. 2003. Hantavirus infections in Europe. Lancet Infect. Dis. 3:653-661. [DOI] [PubMed] [Google Scholar]
- 62.Venien-Bryan, C., and S. D. Fuller. 1994. The organization of the spike complex of Semliki Forest virus. J. Mol. Biol. 236:572-583. [DOI] [PubMed] [Google Scholar]
- 63.Vogel, R. H., S. W. Provencher, C. H. von Bonsdorff, M. Adrian, and J. Dubochet. 1986. Envelope structure of Semliki Forest virus reconstructed from cryo-electron micrographs. Nature 320:533-535. [DOI] [PubMed] [Google Scholar]
- 64.Wallin, M., M. Ekstrom, and H. Garoff. 2004. Isomerization of the intersubunit disulphide-bond in Env controls retrovirus fusion. EMBO J. 23:54-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang, M., D. G. Pennock, K. W. Spik, and C. S. Schmaljohn. 1993. Epitope mapping studies with neutralizing and non-neutralizing monoclonal antibodies to the G1 and G2 envelope glycoproteins of Hantaan virus. Virology 197:757-766. [DOI] [PubMed] [Google Scholar]
- 66.Willard, L., A. Ranjan, H. Zhang, H. Monzavi, R. F. Boyko, B. D. Sykes, and D. S. Wishart. 2003. VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Res. 31:3316-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature 289:366-373. [DOI] [PubMed] [Google Scholar]
- 68.Wu, S. R., L. Haag, M. Sjoberg, H. Garoff, and L. Hammar. 2008. The dynamic envelope of a fusion class II virus. E3 domain of glycoprotein E2 precursor in Semliki Forest virus provides a unique contact with the fusion protein E1. J. Biol. Chem. 283:26452-26460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang, Y., and J. Skolnick. 2005. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33:2302-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng, F., L. Ma, L. Shao, G. Wang, F. Chen, Y. Zhang, and S. Yang. 2007. Defining the N-linked glycosylation site of Hantaan virus envelope glycoproteins essential for cell fusion. J. Microbiol. 45:41-47. [PubMed] [Google Scholar]