Abstract
Removal of genome-bound viral DNA polymerase ought to be an essential step in the formation of hepadnavirus covalently closed circular DNA (cccDNA). We previously demonstrated that deproteinized (DP) relaxed circular DNA (rcDNA) of hepatitis B virus (HBV) existed in both the cytoplasm and nuclei of infected cells and the vast majority of cytoplasmic DP rcDNA was associated with DNase I-permeable nucleocapsids. In our efforts to investigate the role of the cytoplasmic DP rcDNA in cccDNA formation, we demonstrated that rcDNA deproteinization could occur in an endogenous DNA polymerase reaction with either virion-derived or intracellular nucleocapsids. As observed in the cytoplasm of virally infected cells, in vitro deproteinization requires the maturation of plus-strand DNA and results in changes in nucleocapsid structure that render the DP rcDNA susceptible to DNase I digestion. Remarkably, we found that the cytoplasmic DP rcDNA-containing nucleocapsids could be selectively immunoprecipitated with an antibody against the carboxyl-terminal peptide of HBV core protein and are associated with cellular nuclear transport receptors karyopherin-α and -β. Moreover, transfection of small interfering RNA targeting karyopherin-β1 mRNA or expression of a dominant-negative karyopherin-β1 in a stable cell line supporting HBV replication resulted in the accumulation of DP rcDNA in cytoplasm and reduction of nuclear DP rcDNA and cccDNA. Our results thus favor a hypothesis that completion of plus-strand DNA synthesis triggers the genomic DNA deproteinization and structural changes of nucleocapsids, which leads to the exposure of nuclear localization signals in the C terminus of core protein and mediates the nuclear transportation of DP rcDNA via interaction with karyopherin-α and -β.
Hepatitis B virus (HBV) is the prototype member of the Hepadnaviridae family and contains a relaxed circular (rc) partially double-stranded DNA (3.2 kb in length) genome with its DNA polymerase protein covalently attached to the 5′ terminus of minus-strand DNA (10, 26, 38). One of the most intriguing biological features of hepadnaviruses is that the viral genomic DNA is replicated via protein-primed reverse transcription of an RNA intermediate called pregenomic RNA (pgRNA) in the cytoplasmic nucleocapsids (37). However, unlike classical retroviruses, the integration of hepadnavirus genomic DNA into host cellular chromosomes is not an obligatory step in its life cycle. Instead, a nuclear episomal covalently closed circular DNA (cccDNA) is formed from the rcDNA genome in nucleocapsids, either from incoming virions during initial infection or from the pool of progeny nucleocapsids formed in the cytoplasm during replication (40, 42). Those two pathways culminate in the formation of a regulated steady-state population of 10 to 50 cccDNA molecules per infected cell (3, 29, 34). The cccDNA exists as a minichromosome in the nucleus and serves as the template for the transcription of viral RNAs (47). The stability of this key replication intermediate is still in debate, but a continued productive hepadnavirus infection clearly requires a persistent population of cccDNA as the source of viral RNAs for viral replication and production of virions (27, 40, 42, 44). Thus far, therapeutic elimination of cccDNA with highly active viral DNA polymerase inhibitors has not been achieved in chronically HBV-infected patients and remains a major challenge for a cure of chronic hepatitis B (18, 20, 23, 45).
Concerning the molecular mechanism of cccDNA formation from its precursor, the cytoplasmic nucleocapsid-associated rcDNA, one of the most obvious biochemical reactions that ought to occur is the removal of genome-bound viral DNA polymerase. In principle, the resulting protein-free or deproteinized (DP) rcDNA could be an essential intermediate of cccDNA formation. Recently, we and others rigorously demonstrated that such predicted DP rcDNA species indeed exist in the hepadnavirus-infected cells (9, 12). Detailed analysis of the structural features revealed that DP rcDNA contained exclusively complete plus-strand DNA, suggesting that the removal of covalently genome-bound polymerase may require the completion of plus-strand DNA synthesis (9, 12).
In an effort to determine where rcDNA deproteinization may occur and the role of DP rcDNA in cccDNA formation, we found previously that (i) the DP rcDNA existed in both the cytoplasm and the nucleus; (ii) while the majority of the cytoplasmic DP rcDNA presented in DNase I-permeable nucleocapsids, a small portion (∼10%) of cytoplasmic DP rcDNA was located in DNase I-resistant, presumably intact nucleocapsids; (iii) the nuclear DP rcDNA was DNase I sensitive and did not associate with nucleocapsids. Moreover, we showed that the DP rcDNA appeared earlier than cccDNA during hepadnavirus DNA replication and that transfection of purified duck HBV (DHBV) DP rcDNA into chicken hepatoma cells initiated cccDNA formation and viral DNA replication (12).
Based on the experimental evidence summarized above, we proposed that the removal of genome-bound polymerase protein initiates inside the nucleocapsid and may even trigger nucleocapsid disassembly, which, in turn, leads to the exposure of a nuclear localization signal (NLS) at the carboxyl terminus of capsid protein to mediate the import of the DP rcDNA into the nucleus through the nuclear pore complex (31, 46). Subsequently, the DP rcDNA is converted into cccDNA by cellular DNA repair machinery (15).
In order to test this hypothesis, we focused our research efforts on elucidating the molecular mechanism of the production, uncoating, and nuclear transportation of cytoplasmic DP rcDNA. Our results for the first time demonstrate that hepadnavirus nucleocapsid contains sufficient information and factors to allow for deproteinization of the associated viral genome and provide evidence suggesting that the deproteinization reaction requires activities of both a viral DNA polymerase and a putative serine protease. Consistent with the notion that the cytoplasmic DP rcDNA is the functional precursor of cccDNA formation (12), we obtained evidence showing that rcDNA deproteinization was tightly linked with nucleocapsid disassembly and exposure of an NLS located at the carboxyl-terminal portion of the core protein. Furthermore, we showed that the transfection of small interfering RNA (siRNA) targeting karyopherin-β1 mRNA or expression of a dominant-negative karyopherin-β1 in a stable cell line supporting HBV replication resulted in the accumulation of cytoplasmic DP rcDNA and reduction of nuclear DP rcDNA and cccDNA.
Our findings presented herein provide insight on the molecular mechanism of cccDNA formation and clues on the development of novel intervention strategies to control chronic HBV infection.
MATERIALS AND METHODS
Preparation of DHBV virions and intracellular DHBV nucleocapsids.
DHBV virions were prepared from sera of congenitally infected ducks (kindly provided by William S. Mason, Fox Chase Cancer Center, Philadelphia, PA) by sucrose gradient centrifugation (24). Briefly, 1 ml of duck sera was layered onto a 5-ml 10-to-20% (wt/vol) sucrose gradient in 0.15 M NaCl-0.02 M Tris-HCl (pH 7.4) and centrifuged for 3 h at 45,000 rpm in SW55 rotor at 4°C. The supernatant fluid was removed, and the pellet was resuspended in TNE buffer (0.15 M NaCl, 0.01 M Tris-HCl [pH 7.4], and 0.1 mM EDTA).
To prepare intracellular immature DHBV nucleocapsids, Dstet5 cells were cultured in medium without tetracycline, but in the presence of 1 mM foscarnet (PFA), for 3 days with daily medium change and followed by removal of PFA for 4 h to resume viral DNA synthesis (13). The cells were then lysed with chilled lysis buffer containing 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 0.1% Nonidet P-40, and 8% sucrose on ice for 10 min, the lysate was centrifuged at 10,000 × g for 5 min to remove the nuclei and cell debris. The clarified supernatant was overlaid onto a 10 to 55% (wt/wt) sucrose gradient and centrifuged at 24,000 rpm for 16 h at 4°C using a Beckman SW28 rotor. Nineteen 2-ml fractions were collected from the bottom of the cushion. Ten microliters of each fraction was dot-blotted onto the nitrocellulose membrane, and DHBV core protein was detected by sequential incubation with a rabbit antibody against DHBV core protein and horseradish peroxidase-labeled antibody against rabbit immunoglobulin G (IgG). The bound antibody is revealed by enhanced chemiluminescence (12). DHBV core protein-positive fractions were pooled together, and the sucrose concentration was adjusted to 10% by addition of TNE buffer. The diluted sample was overlaid on a 20% sucrose cushion and centrifuged at 45,000 rpm for 3 h at 4°C using the Beckman SW55 rotor. The pellet was resuspended in TNE buffer.
Endogenous DNA polymerase reaction.
A typical endogenous DNA polymerase reaction (EPR) mixture was assembled with 40 μl of DHBV virion preparation, 50 μl of 2× EPR buffer which consisted of 0.3 M NaCl, 0.1 M Tris-HCl (pH 8.0), 20 mM MgCl2, 2 mM dithiothreitol, 0.2% (vol/vol) Nonidet P-40, and 0.2 mM of deoxynucleoside triphosphate (dNTP). DNA polymerase and/or protease inhibitors (Pierce and Calbiochem) were added as indicated below, and water was provided to bring the reaction volume to 100 μl. After incubation at 37°C for the indicated period of time, the reaction volume was subjected to the extraction of viral DNA with or without prior DNase I digestion. Occasionally, DHBV DNA polymerase activity is measured by a [α-32P]dCTP incorporation assay. Briefly, 0.2 mM of dCTP in the 2× EPR buffer was replaced with 10 μM [α-32P]dCTP and followed by incubation of the endogenous DNA polymerase reaction volume at 37°C for 1 h. The acid insoluble 32P was counted with a liquid scintillation counter (PerkinElmer).
Analysis of viral DNA.
Total viral DNA from the EPR sample was extracted by adding equal volumes of DNA extraction buffer that contained 20 mM EDTA, 20 mM Tris-HCl (pH 8.0), 0.2% sodium dodecyl sulfate (SDS), and 1 mg/ml pronase, followed by incubation for 1 h at 37°C. The digestion mixture was extracted twice with phenol, and DNA was precipitated with ethanol and dissolved in TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). Protein-free or DP DHBV DNA from the EPR samples were prepared by direct phenol extraction without prior pronase digestion (Hirt procedure) (12). Briefly, EPR mixture was brought up to 1.5 ml by addition of TE buffer (10 mM Tris-HCl [pH 7.5], 10 mM EDTA) and mixed with 100 μl of 10% SDS. After 30 min incubation at room temperature, 5 M NaCl was added to bring the final concentration of NaCl to 1 M and continued to be incubated at 4°C overnight. The sample was then clarified by centrifugation at 12,000 × g for 30 min at 4°C and extracted twice with phenol and once with phenol-chloroform. DNA was precipitated with 2 volumes of ethanol overnight at room temperature and dissolved in TE buffer. One-half of the DNA sample from each EPR was resolved by electrophoresis into a 1.5% agarose gel. The gel was then subjected to denaturation in a solution containing 0.5 M NaOH and 1.5 M NaCl, followed by neutralization in a buffer containing 1 M Tris-HCl (pH 7.4) and 1.5 M NaCl. DNA was then blotted onto a Hybond-XL membrane (GE Healthcare) in 20× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). For the detection of HBV DNA, membranes were probed with an [α-32P]UTP (800 Ci/mmol; Perkin Elmer)-labeled minus-strand specific full-length HBV riboprobe. Hybridization was carried out in 5 ml Ekono hybridization buffer (Genotech) with 1-h prehybridization at 65°C and overnight hybridization at 65°C, followed by a 1-h wash with 0.1× SSC and 0.1% SDS at 65°C. The membrane was exposed to a phosphorimager screen, and hybridization signals were quantified with QuantityOne software (Bio-Rad).
Immunoprecipitation assay.
The cytoplasmic fractions of HepDES19 cells that were cultured in the absence of tetracycline for 12 days were prepared with a Qproteome cell compartment kit (Qiagen) by following the manufacturer's directions (12). For the immunoprecipitation assay, 1 ml of cytoplasmic lysate prepared from approximately 5 × 106 cells was mixed with 35 μl of protein A/G plus beads (Santa Cruz) that were preabsorbed with antibodies against karyopherin-α1 (Zymed), karyopherin-α2 (Santa Cruz), karyopherin-β1 (Abcam), or HBsAg (Dako), respectively. The mixtures were incubated at 4°C overnight. Beads were washed four times with TNE buffer. Core and DP DNA were extracted with or without prior digestion of DNase I, respectively. Viral DNA was analyzed by Southern blot hybridization.
Plasmid DNA and siRNA transfection.
HepDES19 cells cultured in a collagen-coated 35-mm dish were transfected by Lipofectamine 2000 (Invitrogen) with a 4 μg control vector plasmid or a plasmid expressing dominant negative karyopherin-β1, dominant negative karyopherin-α1, or 90 nM of control siRNA (Santa Cruz), or Smartpool siRNA targeting karyopherin-β1 (Dharmacon). The full-length karyopherin-β1 was amplified from the cDNA synthesized from HepG2 cell total RNA and cloned into pcDNA3.1 (Invitrogen). The C-terminal fragment from aa 256 to aa 876 of karyopherin-β1, which can bind to karyopherin α but fails to bind Ran GTPase (17), was PCR amplified from the karyopherin-β1 plasmid and cloned into pcDNA3.1-TOPO-V5 (Invitrogen) to express the dominant negative karyopherin-β1 with a C-terminal V5 tag. All the cDNA clones were confirmed by DNA sequencing. N-FLAG-tagged dominant negative karyopherin-α1 was kindly provided by Christopher Basler (Mount Sinai School of Medicine, New York, NY) (32). The transfected cells were cultured in tetracycline-free medium for 5 days, followed by a second round of transfection and continued culture with tetracycline-free media for another 5 days. Total core DNA, DP DNA, and cccDNA were extracted and analyzed as described previously (12).
Western blot assay.
The transfected cells were lysed in 300 μl of 1 × Lamini buffer, a total of 30 μl of the cell lysate was resolved on an SDS-12% polyacrylamide gel and transferred onto Immobilon-FL polyvinylidene difluoride membrane (Millipore). The membrane was blocked with Western Breeze blocking buffer (Invitrogen) and probed with specific antibody against karyopherin-β1 (SC-1919; Santa Cruz), V5 epitope (Invitrogen), FLAG peptide (Sigma), and HBc170 (produced in Genscript Facility, NJ), and bound antibody was revealed by IRDye secondary antibodies and visualized by the Li-COR Odyssey system.
RESULTS
DHBV virion-associated rcDNA deproteinization and nucleocapsid disassembly occur following endogenous DNA polymerase reaction in vitro.
As stated above, hepadnavirus contains a partially double-stranded rcDNA genome that the viral DNA polymerase covalently attaches to the 5′ terminus of its minus strand, and the plus strand is incompletely synthesized and heterogeneous in length (38). It has been demonstrated several decades ago that incubation of purified virion particles in solution containing nonionic detergent and dithiothreitol (DTT) to disrupt viral envelope and dNTP as substrates and that viral DNA polymerase can elongate the incomplete plus strand DNA chain in vitro. This is the so-called endogenous DNA polymerase reaction (33). Based on our observation that DP rcDNA contains full-length plus-strand DNA, we hypothesized that the completion of the plus-strand DNA synthesis might serve as a trigger for rcDNA deproteinization and/or nucleocapsid disassembly (12).
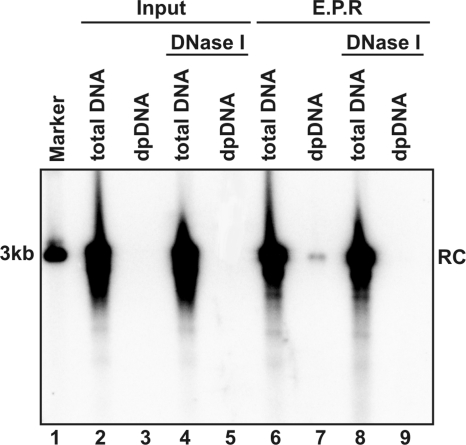
To test this hypothesis, endogenous DNA polymerase reactions were performed with purified DHBV virions. The reaction mixtures were either without incubation (Fig. 1, lanes 2 to 5) or incubated at 37°C for 16 h (Fig. 1, lanes 6 to 9). Total viral DNA was extracted by protease digestion and phenol extraction. DP viral DNA was extracted via the Hirt procedure (14). The Hirt procedure involves lysis of cells and/or viral particles with SDS and high concentrations of salts to disassociate physically associated proteins, such as capsid proteins, from DNA, and was followed by direct phenol extraction of DNA from the lysates without protease digestion (14). Therefore, DNA covalently linked to protein is partitioned into phenol or interphase, and only the DNA molecules that are not covalently attached to protein are selectively extracted. Hence, the viral DNA species in the Hirt preparations should be free of covalently genome-bound viral DNA polymerase. To monitor the disassembly of nucleocapsids, the indicated samples were subjected to DNase I digestion for 30 min at 37°C prior to total and DP DNA extraction.
FIG. 1.
Removal of genome-bounded polymerase and capsid disassembly occur following endogenous polymerase reaction in vitro. Approximately 108 DHBV virion particles prepared from DHBV-positive duck serum were contained in each 100-μl EPR mixture including 150 mM NaCl, 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 1 mM DTT, 0.1% NP-40, and 0.1 mM dNTP. The reaction mixtures were either without incubation (lanes 2 to 5) or incubated at 37°C for 16 h (lanes 6 to 9), followed by total DNA and DP DNA extraction without (lanes 2, 3, 6, and 7) or with (lanes 4, 5, 8 and 9) prior DNase I digestion, respectively. The viral DNA were resolved in agarose gel and detected by Southern blot hybridization with full-length riboprobe recognizing minus-strand DNA. One hundred picograms of 3.0-kb unit-length DHBV DNA served as the quantification standard and molecular weight marker (lane 1). The position of rcDNA (RC) is indicated.
Consistent with previous observations, there is no detectable DP DNA in nonincubated virions, suggesting that all DHBV rcDNA in virion particles is covalently attached to DNA polymerase (11, 26). Interestingly, after a prolonged endogenous polymerase reaction, a small amount of DP rcDNA could be detected. As observed in DHBV-infected duck livers and Dstet5 cells, the DP rcDNA species is exclusively DNase I sensitive (12). These results hence indicate that the rcDNA deproteinization and capsid disassembly occur following the in vitro endogenous DNA polymerase reaction, albeit at a very low efficiency.
DHBV rcDNA deproteinization requires viral DNA polymerase activity.
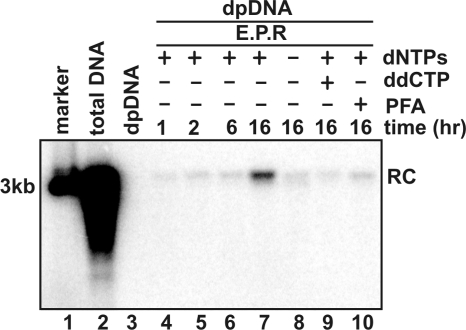
To further characterize the requirements for this in vitro genomic DNA deproteinization reaction, the endogenous polymerase assay was performed under the condition that dNTPs were omitted or DNA polymerase inhibitor ddCTP or PFA were added into the reaction and incubated at various periods of time. DP DNA were then extracted by the Hirt procedure. Total virion DNA and DP virion DNA were extracted and served as positive and negative controls. The results revealed the following observations. First, as expected, untreated DHBV virion particles do not contain any detectable level of DP DHBV DNA (Fig. 2, lane 3). Second, DP DHBV DNA is detectable at 1 h and accumulated to a higher level after 16 h of incubation (Fig. 2, lanes 4 to 7). Third, both omission of dNTPs and addition of DNA polymerase inhibitors reduced the levels of DP DHBV DNA (Fig. 2, lanes 8 to 10), indicating that the deproteinization reaction requires DNA polymerase activity.
FIG. 2.
DHBV virion-associated rcDNA deproteinization requires DNA polymerase activity. Approximately 108 DHBV virion particles prepared from DHBV-positive duck serum were contained in each 100-μl EPR mixture as described in the legend of Fig. 1. The reaction mixtures were either without incubation (lanes 2 and 3) or incubated (lanes 4 to 10) at 37°C for the indicated periods of time. For the samples analyzed in lanes 8 to 10, dNTP was omitted (lane 8), or dCTP was replaced by ddCTP (lane 9), or 1 mM PFA was added into the EPR mixture (lane 10). Core and DP DNA were extracted from the reaction mixtures and analyzed by Southern blot hybridization assay. One hundred picograms of unit-length DHBV DNA served as the molecular weight marker (lane 1). The position of rcDNA (RC) is indicated.
DHBV DNA deproteinization and nucleocapsid disassembly require the completion of plus-strand DNA synthesis.
One simple explanation for the requirement of viral DNA polymerase activity in the deproteinization reaction is that the completion or extension of plus-strand DNA synthesis beyond a certain length is an essential signal to trigger the biochemical reaction. However, because DHBV virion-associated rcDNA contains an almost full-length plus-strand DNA (21) and the observed deproteinization reaction is at such a low efficiency, it is difficult to rule out the possibility that the observed deproteinization reaction might occur randomly in capsids after a long period of incubation. To firmly establish the role of plus-strand DNA synthesis in triggering DHBV genome deproteinization, we took advantage of our previous observation that the heterogeneous lengths of DHBV DNA, from less-than-full-length minus-strand DNA to full-length double-stranded DNA, could be synthesized in purified intracellular pgRNA-containing DHBV nucleocapsids via an in vitro endogenous DNA polymerase reaction (13).
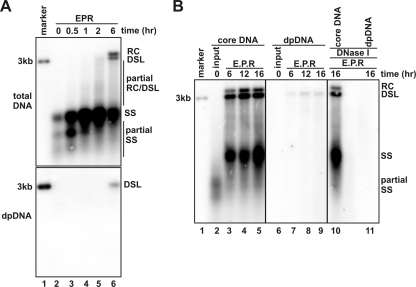
Accordingly, DHBV intracellular nucleocapsids were purified from Dstet5 cells that were cultured in the absence of tetracycline and presence of PFA for 3 days to allow the accumulation of pgRNA-containing capsids and were then cultured in the absence of PFA for an additional 4 h to resume minus-strand DNA synthesis (13). Such nucleocapsids contained only pgRNA and heterogeneous lengths of minus-strand DHBV DNA, based upon their characteristic mobility in the agarose gel (Fig. 3A, lane 2). However, incubation of the nucleocapsids in the endogenous DNA polymerase reaction resulted in the sequential appearance of more full-length minus-strand DNA (Fig. 3A, upper panel, lanes 3 to 5), partially double-stranded DNA (the smear between single stranded [ss] and double-stranded linear [dsl] DNA bands [Fig. 3A, upper panel, lanes 4 to 6] and full-length rc- and dslDNA [Fig. 3A, upper panel, lane 6]). Compared with that observed in DHBV-replicating cells where rcDNA is the predominant form (approximately 90%) of mature double-stranded viral DNA (36), the in vitro endogenous DNA polymerase reaction with purified intracellular nucleocapsids produced more dslDNA, but less rcDNA (Fig. 3A and B), suggesting a less efficient primer translocation during the initiation of plus-strand DNA synthesis under the in vitro conditions (36). Interestingly, as shown in the lower panel of Fig. 3A, the DP DNA appeared only after 6 h of incubation and was presented exclusively as full-length dslDNA. While the result clearly indicates that completion or at least near completion of the plus-strand DNA synthesis is an essential signal to trigger deproteinization, it is not yet clear why DP rcDNA could not be produced in this in vitro reaction, although mature rcDNA was made.
FIG. 3.
Mature double-stranded DNA synthesis and deproteinization occur in purified intracellular DHBV nucleocapsids following endogenous DNA polymerase reaction. (A) EPRs were performed with immature intracellular nucleocapsids prepared from Dstet5 cells (see Materials and Methods for details) in a 100-μl EPR mixture as described in the legend of Fig. 1. The reaction mixtures were either without incubation (lane 2) or incubated at 37°C for the indicated periods of time (lanes 3 to 6). Core DNA and DP DNA were analyzed by Southern blot hybridization. The amount of core DNA (upper panel) and DP DNA (lower panel) loaded onto each lane was derived from 4 × 106 and 4 × 107 cells. (B) EPRs were performed with immature intracellular DHBV nucleocapsids, and the reaction mixtures were either without incubation (lanes 2 and 6) or incubated at 37°C for the indicated periods of time (lanes 3 to 5 and 7 to 11). Core DNA and DP DNA were extracted without (lanes 2 to 9) or with (lanes 10 and 11) prior DNase I digestion and detected by Southern blot hybridization with a DHBV minus-strand specific α-32P-riboprobe. Fifty picograms of 3.0-kb unit-length DHBV DNA was loaded as the hybridization size marker (lane 1). The positions of rcDNA (RC), dslDNA (DSL), single-stranded DNA (SS), and partial single-strand DNA are indicated.
To test the possibility that rcDNA deproteinization may require a longer time of incubation and determine whether dslDNA deproteinization could also result in nucleocapsid disassembly, the purified intracellular DHBV nucleocapsids, as described above, were incubated under standard EPR conditions for 6, 12, and 16 h, respectively. Total and DP viral DNA were extracted and analyzed by Southern blot hybridization. In agreement with the results presented in Fig. 3A, after 6 h of incubation, a large amount of full-length minus-strand DNA, heterogeneous lengths of double-stranded DNA and mature forms of dslDNA and rcDNA were synthesized. Further extension of the incubation period did not produce a significant additional amount of matured forms of double-stranded viral DNA (Fig. 3B, compare lanes 3 to 5). The DP dslDNA could be detected after 6 h, and the amount slightly increased with additional incubation time (Fig. 3B, compare lanes 7 to 9). Although the mature rcDNA were made under these experimental conditions, the DP rcDNA was not detected even after 16 h of incubation. Furthermore, while the vast majority of viral DNA synthesized under the EPR conditions were resistant to DNase I digestion (Fig. 3B, lane 10), suggesting their existence in intact nucleocapsids, the DP dslDNA is completely susceptible to DNase I treatment (Fig. 3B, lane 11). Hence, as observed for rcDNA, the dslDNA deproteinization was also associated with nucleocapsid disassembly.
Taken together, our results presented in the above sections clearly demonstrate that DHBV nucleocapsids contain sufficient information and factors to allow for deproteinization of the associated viral genomes. Although the sequence of the events remains to be determined, the DNA deproteinization and nucleocapsid disassembly are tightly linked events. Moreover, while our results strongly support the notion that the maturation of plus-strand DNA synthesis is an essential signal to trigger the DNA deproteinization and nucleocapsid disassembly, an additional signal(s) may be required for the deproteinization reaction to occur. This conclusion is based upon the following two experimental evidence: (i) less than 1% of mature rcDNA and/or dslDNA underwent deproteinization under the in vitro EPR conditions (Fig. 1 to 3), and (ii) only the deproteinization of mature dslDNA, but not rcDNA, occurred under the EPR conditions with purified intracellular nucleocapsids.
DHBV DNA deproteinization appears to require a serine protease activity.
As extensively discussed in our previous report (12) and the report by Gao and Hu (9), removal of the genome-bound DNA polymerase could potentially occur via the following three biochemical reactions: (i) endonucleolytic cleavage of DNA sequence near the 5′ terminus of the minus-strand DNA; (ii) hydrolysis of the phosphodiester bond between tyrosine residue in the terminal protein (TP) domain of polymerase and the 5′ end of the minus-strand DNA; (iii) proteolytic cleavage of DNA polymerase. The three reactions will yield DP DNA bearing distinct structural features at the 5′ end of the minus-strand DNA. For instance, the hydrolysis of phosphodiester bond and proteolytic cleavage of polymerase will yield DP rcDNA with its 5′ end at its authentic position, and an endonuclease reaction will trim the 5′ end of the minus-strand DNA. We had previously demonstrated by a primer extension assay that the minus strand of DP rcDNA contains an authentic 5′ end (12). Thus, the possibility of endonuclease reaction for the removal of genome-bound DNA polymerase can be ruled out. To further distinguish the two other possibilities, we took the advantage of our in vitro deproteinization assay and determined whether the deproteinization reaction could be inhibited by protease inhibitors.
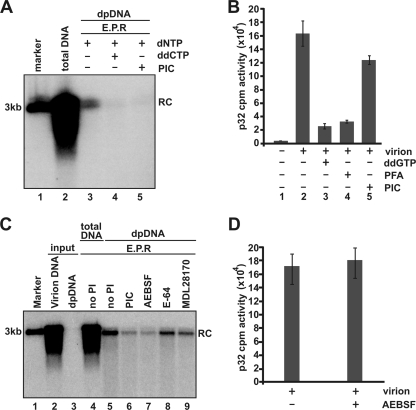
To this end, the in vitro deproteinization assay was performed in the presence of the Halt protease inhibitor cocktail (PIC; without EDTA). DHBV DNA polymerase inhibitor ddCTP was used as a positive control. As shown in Fig. 4A, ddCTP efficiently inhibited DHBV DNA deproteinization, and similarly, the PIC also prevented viral DNA deproteinization. The result appeared to suggest that the deproteinization was mediated by proteolytic reaction. However, one argument is that the PIC may actually inhibit the polymerase activity. To address this issue, effects of the PIC on DHBV DNA polymerase activity was measured by [α-32P]dCTP incorporation. As shown in Fig. 4B, while the polymerase inhibitors ddGTP and PFA potently inhibited [α-32P]dCTP incorporation, the polymerase activity was only modestly reduced in the presence of PIC.
FIG. 4.
A serine protease inhibitor inhibits DHBV DNA deproteinization and nucleocapsid disassembly in vitro (A) EPRs were performed with DHBV virions in a 100-μl EPR mixture as described in the legend of Fig. 1. The reaction mixtures were either without incubation (lane 2) or incubated at 37°C for 16 h (lanes 3 to 5). For the samples analyzed in lanes 4 and 5, dCTP was replaced by ddCTP (lane 4) or 1× Halt PIC (Pierce) was added in the EPR mixture (lane 5). Core DNA and DP DNA were analyzed by Southern blot hybridization. (B) EPRs were performed with 2 × 107 DHBV virions in a 10-μl EPR mixture including 150 mM NaCl, 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 1 mM DTT, 0.1% NP-40, 0.1 mM of dATP, dGTP, and dTTP, and 5 μM [α-32P]dCTP (800 Ci/mmole; Perkin Elmer). When indicated, dGTP was replaced by ddGTP or 1 mM PFA, or 1× EDTA-free Halt PIC was added. The reaction mixture was incubated at 37°C for 1 h; the mixture was blotted onto the 3MM Whatman filter, rinsed by 10% acetic acid for 15 min three times and briefly washed by 95% ethanol three times; the filter was then air dried; and the incorporated [32P]dCTP was counted with a liquid scintillation counter (Perkin Elmer). (C) EPRs were performed in the absence or presence of 1× Halt PIC (lane 6) or its three individual active components—1 mM of wide-spectrum serine protease inhibitor AEBSF (lane 7), 15 μM of cysteine protease inhibitor E-64 (lane 8), and 10 μM of calpain protease inhibitor MDL28170 (lane 9)—at 37°C for 16 h. Core DNA (lanes 2, 4, and 5) and DP DNA (lanes 3 and 6 to 9) were extracted and detected by Southern blot hybridization. One hundred picograms of 3.0-kb unit-length DHBV DNA served as the quantification standard and molecular weight marker (lane 1). The position of rcDNA (RC) is indicated. (D) EPRs were performed as described in legend for panel B of this figure in the absence or presence of 1 mM AEBSF. The incorporated [32P]dCTP was counted with a liquid scintillation counter as described above.
To determine the active components in the Halt PIC, each individual component was tested, and we demonstrated that the in vitro DHBV rcDNA deproteinization reaction can be specifically inhibited by a serine protease inhibitor AEBSF, but not the cysteine (E-64) or calpain (MDL28170) protease inhibitor (Fig. 4C). Moreover, AEBSF did not apparently inhibit DHBV DNA polymerase activity, as revealed by [α-32P]dCTP incorporation assay (Fig. 4D). Our results hence suggest that the removal of genome-bound DNA polymerase might be catalyzed by a cellular serine protease. However, due to the cell toxicity of the compound, its effect on deproteinization and cccDNA formation of DHBV in cultured cells could not be determined. Moreover, it should be pointed out that removal of the polymerase by such a proteolytic reaction will leave a short peptide or at least one amino acid residue (tyrosine) that still attaches onto the 5′ terminus of the minus-strand DNA. Therefore, to create a 5′ end of minus-strand DNA that is able to be ligated, the DNA end must be further processed by either an endonucleolytic cleavage of DNA sequence near the 5′ terminus of the minus-strand DNA or a reaction that cleave the phosphodiester bond between the tyrosine residue and first nucleotide of minus strand DNA. The later reaction could potentially been catalyzed by a cellular DNA repair enzyme called tyrosyl-DNA phosphodiesterase 1 (TDP1), which is able to remove topoisomerase (Topo) II from Topo II inhibitors induced DNA strand break by cleavage of the tyrosine and 5′-phosphotyrosyl linkage between Topo II and chromosomal DNA (2, 30). Involvement of TDP1 in DP-DNA production and cccDNA synthesis is currently under investigation in our laboratory.
The carboxyl terminus of the core protein is exposed on HBV DP rcDNA-associated nucleocapsids.
The results obtained from our previous cell fractionation studies (12) and the in vitro deproteinization assays presented above clearly demonstrate that the deproteinization of the mature forms of hepadnavirus DNA were initiated inside the nucleocapsids in the cytoplasm and associated with nucleocapsid disassembly. However, considering the fact that although the cytoplasmic DP rcDNA was largely DNase I sensitive, the DNA species was still associated with the core protein and cosedimented with nucleocapsids by sucrose gradient centrifugation, it was reasonable to believe that the deproteinization reaction most likely resulted in a drastic structural change in the nucleocapsids, but not a complete capsid disassembly (12).
Biologically, to serve as a functional precursor for cccDNA formation, the cytoplasmic DP rcDNA must be transported into the nucleus. The deproteinization-associated nucleocapsid structural change may thus be essential for delivery of the DP rcDNA into the nuclei for cccDNA formation. For instance, it was demonstrated previously that the HBV core protein (HBcAg) contained a classical bipartite NLS at its C-terminal portion (7, 22, 43). This portion was not required for empty capsid assembly but was essential for pgRNA assembly into nucleocapsid and viral DNA replication (22). However, cryo-electron microscopy study of nucleocapsids assembled with full-length HBcAg in vitro revealed that the C-terminal portion of HBcAg was localized inside of the capsid shell (46). Hence, we hypothesized that the C-terminal tail of HBcAg ought to flip out upon deproteinization of the matured capsid DNA, which will lead to a selective recognition and nuclear pore complex binding of DP rcDNA-containing nucleocapsids by cellular nuclear transport receptor karyopherins (4).
To determine the localization of the C-terminal portion of the HBV core protein in the various forms of nucleocapsids, we raised a rabbit polyclonal antibody against a peptide derived from the 14 C-terminal amino acid residues, which was designated HBc170 (Fig. 5A). Western blot analysis demonstrated that the antibody specifically recognizes the HBV core protein in HepDES19 cell lysates (Fig. 5B). As input material for an immunoprecipitation assay, nucleocapsids in the cytoplasmic lysate of HepDES19 cells contain heterogenous lengths of HBV DNA replicative intermediates, and the cytoplasmic DP HBV DNA exists only as matured rcDNA. Consistent with our previous report (12), approximately 10% cytoplasmic DP rcDNA is DNase I resistant (Fig. 5C, lanes 1 to 4), suggesting that a deproteinization reaction might occur prior to nucleocapsid partial disassembly. Interestingly, the immunoprecipitation assay demonstrated that while a rabbit polyclonal antibody against the native HBV core protein (Dako) efficiently precipitated nucleocapsids containing all forms of HBV DNA intermediates and DP rcDNA (Fig. 5C, lanes 5 to 8), the antibody HBc170 precipitated only nucleocapsids containing mature rcDNA and DP rcDNA, but not the immature DNA replicative intermediates (Fig. 5C, lanes 9 to 12). Because HepDES19 cells are deficient in producing viral envelope proteins (12), as expected, antibody against HBsAg failed to precipitate any form of HBV DNA (Fig. 5C, lanes 13 and 14). Taken together, these results are in agreement with the notion that the C-terminal portion of HBcAg is exposed upon the completion of plus-strand DNA synthesis and/or viral DNA deproteinization-induced capsid partial disassembly.
FIG. 5.
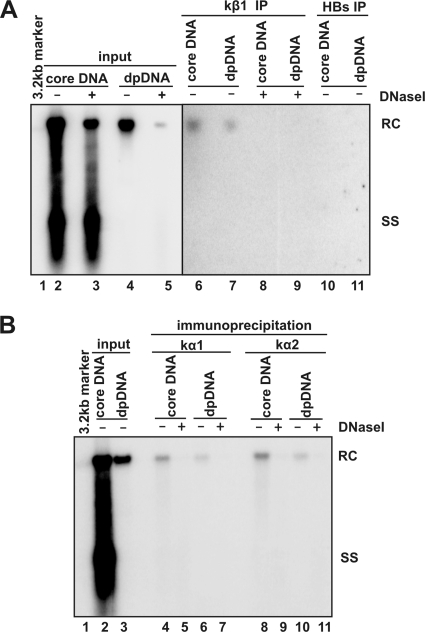
The carboxyl terminus of HBcAg is exposed on the surface of DP rcDNA-containing nucleocapsid. (A) A rabbit polyclonal antibody (HBc170) was raised against the synthetic peptide corresponding to the carboxy-terminal 14 amino acid residues of the HBV core protein. The underlined 12-amino-acid peptide within the C-terminal 14 amino acid residues is one of the bipartite NLSs of HBcAg. (B) Proteins in cell lysates of HepDES19 cells cultured in the presence or absence of tetracycline for 8 days were resolved by SDS-polyacrylamide gel electrophoresis, transferred onto the membrane, probed with the antibody HBc170, and visualized by Li-COR. A cross-reactive cellular protein band is indicated with an asterisk, and β-actin served as the loading controls. (C) Cytoplasmic lysate of HepDES19 cells were subjected to immunoprecipitation with antibodies against the HBV core protein (Dako), C-terminal 14-amino-acid peptide (HBc170), and HBsAg. Core DNA and DP DNA were extracted with or without prior DNase I digestion from the original lysate (input) and immunocomplexes on beads (IP) and analyzed by Southern blot hybridization. Lanes 1 and 2 were loaded with 1/10 of core DNA from the cytoplasmic fraction of one 60-mm dish; lanes 3 to 14 were loaded with half of the indicated DNA samples from the cytoplasmic fraction of one 60-mm dish. The positions of rcDNA (RC) and single-stranded DNA (SS) are indicated. MW, molecular size.
Cytoplasmic HBV DP rcDNA associates with karyopherin-α and kβ.
To investigate whether the C-terminal exposure of HBcAg on DP rcDNA-containing nucleocapsid resulted in the accessibility of the capsids to cellular nuclear transport receptors, an immunoprecipitation assay was performed with antibodies against karyopherin-β1 and HBsAg in the lysates of HepDES19 cells. The results shown in Fig. 6A revealed the following. (i) Comparing lanes 2 and 3 shows that prior DNase I treatment of the input lysate reduces only the level of mature rcDNA, but not that of immature DNA replicative intermediates. Moreover, consistent with our previous observation, the majority of DP rcDNA in the cytoplasm is DNase I sensitive (compare lanes 4 and 5) (12). These results indicate that a significant amount of intracellular mature rcDNA is actually DP rcDNA and exists in DNase I-permeable nucleocapsids. (ii) Only mature rcDNA can be immunoprecipitated and present in both total core DNA and DP DNA preparations (lanes 6 and 7). The difference in amount of rcDNA in the total cytoplasmic core DNA and Hirt DNA preparations could be due to a higher recovery rate of core DNA extraction procedure. Finally, the immunoprecipitated rcDNA in both the total core DNA and DP DNA preparations are DNase I sensitive (lanes 8 and 9). As a negative control, antibody against HBsAg failed to precipitate any form of HBV DNA. In addition, similar immunoprecipitation assays were performed with antibodies against two types of karyopherin-α and demonstrated that like karyopherin-β1, both karyopherin-α1 and karyopherin-α2 selectively associated with only the cytoplasmic mature rcDNA and/or DP rcDNA, but not nucleocapsids containing immature viral DNA (Fig. 6B).
FIG. 6.
Cytoplasmic DP rcDNA were associated with karyopherins. One milliliter of cytoplasmic lysate prepared from 5 × 105 HepDES19 cells that were cultured in the absence of tetracycline for 12 days was mixed with 35 μl of protein A/G plus beads (Santa Cruz) preabsorbed with antibodies against karyopherin-β1 (kβ1) (Abcam) or HBsAg (Dako) (panel A) and against karyopherin-α1 (kα1; clone 114-E12; Zymed) or karyopherin-α2 (kα2; sc-55537; Santa Cruz) (panel B). The mixtures were incubated at 4°C overnight. Beads were washed four times with TNE buffer, and core DNA and DP DNA were extracted from the beads with or without prior DNase I digestion as indicated. Viral DNA were analyzed by Southern blot hybridization. Lane 1 was loaded with 50 pg of 3.2-kb HBV DNA. In panel A, lanes 2 and 3 were loaded with 1/20 of core DNA from the cytoplasmic fraction of one 60-mm dish, and lanes 4 and 5 were loaded with half of DP DNA from the cytoplasmic fraction of one 60-mm dish. In panel B, lane 2 was loaded with 1/30 of core DNA from the cytoplasmic fraction of one 60-mm dish, and lane 3 was loaded with 1/10 of DP DNA from the cytoplasmic fraction of one 60-mm dish. Half the volume of immunoprecipitated core DNA and DP DNA recovered from one 60-mm dish was loaded onto the gel (panel A, lanes 6 to 11; panel B, lanes 4 to 11) The positions of rcDNA (RC) and single-stranded DNA (SS) are indicated.
In summary, the results presented in the above two sections demonstrate that the deproteinization of viral genomic DNA resulted in drastic structural changes in nucleocapsids, which lead to the exposure of the C-terminal portion of HBcAg and interaction with cellular nuclear transport receptors, such as karyopherin-α and -β.
Role of cytoplasmic DP rcDNA in cccDNA formation.
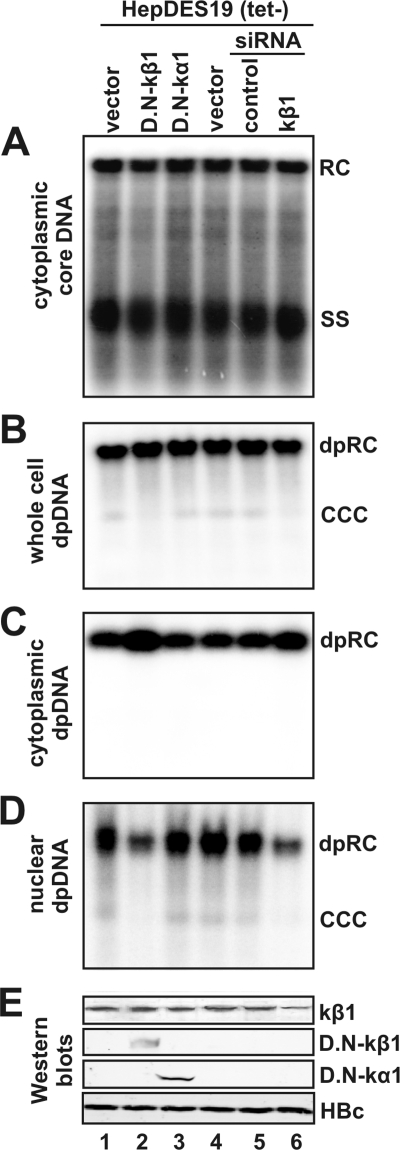
To investigate the functional role of the cytoplasmic DP rcDNA in cccDNA formation, the interaction of cytoplasmic DP rcDNA-containing nucleocapsid with karyopherin-β and DP rcDNA nuclear transportation in HepDES19 cells were disrupted either by expression of a dominant-negative form of karyopherin-β1 or by reducing karyopherin-β1 expression by siRNA transfection. The accumulation of DP rcDNA in the cytoplasm and nuclei and cccDNA formation were determined by a Southern blot hybridization assay. As a confirmation, we have demonstrated that the truncated karyopherin-β1 was properly expressed in the transfected cells and expression of the protein was reduced by approximately 50% in siRNA-transfected cells by Western blot analyses (Fig. 7E). As expected, neither expression of truncated karyopherin-β1 nor reducing its expression by siRNA transfection affected HBV DNA replication (Fig. 7A), core protein synthesis (Fig. 7E, lower panel), or total cellular DP rcDNA production (Fig. 7B). However, both inhibition of karyopherin-β1 and reducing its expression led to the accumulation of cytoplasmic DP rcDNA (Fig. 7C, lanes 2 and 6) and reduction in the amounts of nuclear DP rcDNA and cccDNA (Fig. 7E, lanes 2 and 6). However, we noticed that unlike dominant negative karyopherin-β, expression of dominant negative forms of karyopherin-α1 did not affect DP rcDNA nuclear transportation or cccDNA formation. Considering that the karyopherin-α family contains six isoform members (32) and both karyopherin-α1 and -α2 efficiently bound DP rcDNA (Fig. 6B), it is possible that inhibition of function of a single species of karyopherin-α can be compensated by other subtypes of karyopherin-α. Nevertheless, our results demonstrated that karyopherins are essential for DP rcDNA nuclear import and cccDNA formation. This observation is consistent with the model that the cytoplasmic DP rcDNA is a precursor of nuclear DP rcDNA and cccDNA.
FIG. 7.
Nuclear transportation of HBV DP rcDNA and cccDNA formation was inhibited by expression of dominant negative karyopherin-β1 or knockdown of endogenous karyopherin-β1 expression. HepDES19 cells in a collagen-coated 35-mm dish were transfected by Lipofectamine 2000 (Invitrogen) with 4 μg plasmid of control vector (lanes 1 and 4), dominant negative karyopherin-β1 (D.N-kβ1; lane 2), dominant negative karyopherin-α1 (D.N-kα1; lane 3), 90 nM of control siRNA (lane 5; Santa Cruz), or Smartpool siRNA for karyopherin-β1 (lane 6; Dharmacon). The transfected cells were cultured in tetracycline-free medium for 5 days, followed by a second round of transfection and continued culture under tetracycline-free conditions for another 5 days. The intracellular core DNA (A), whole-cell Hirt DNA (B), cytoplasmic DP DNA (C), and nuclear DP DNA (D) were extracted as previously described and subjected to Southern blot and DNA hybridization. The positions of rcDNA (RC), single-stranded DNA (SS), and cccDNA are indicated. Expression of wild-type or recombinant proteins and HBcAg was assessed by a Western blot assay (E).
DISCUSSION
cccDNA is the first viral product made from the incoming nucleocapsid-associated rcDNA during initial infection of hepadnaviruses (39). In addition, cccDNA can also be produced from the rcDNA in the progeny nucleocapsids in the cytoplasm of infected cells via an intracellular amplification pathway (40, 42). Considering the molecular pathway of cccDNA formation, the essential events should include genomic rcDNA uncoating (nucleocapsid disassembly), nuclear transportation, and conversion of rcDNA into cccDNA (19, 28). Biochemically, removal of genome-bound viral DNA polymerase is among the most obvious reactions required for the conversion of rcDNA into cccDNA (9, 12). Detailed understanding of the underlying molecular mechanisms of these molecular events should not only advance our knowledge in HBV biology, but also provide a basis for the development of therapeutic strategies to inhibit cccDNA formation, which is essential to cure chronic HBV infection (16).
Based on our previous discovery and characterization of DP rcDNA, a product derived from the removal of genome-linked viral DNA polymerase, we proposed that cytoplasmic DP rcDNA is the functional precursor of cccDNA. In the studies presented in this report, we further elucidated the molecular mechanism of cytoplasmic DP rcDNA production, uncoating, and nuclear transportation and investigated its role in cccDNA formation. Our results demonstrated that the deproteinization of DHBV rcDNA could occur in a prolonged endogenous DNA polymerase reaction with either virion-derived or intracellular nucleocapsids (Fig. 1 and 3). We also showed that the in vitro deproteinization reaction could be inhibited by viral reverse transcriptase inhibitors (Fig. 2) and a broad-spectrum serine protease inhibitor (Fig. 4). Moreover, as observed in virally infected cells, hepadnavirus genome deproteinization was tightly linked with nucleocapsid disassembly which leads to the exposure of the C-terminal polypeptide of core protein and NLS on DP rcDNA-associated nucleocapsids. This was demonstrated by the following two independent experiments. First, the cytoplasmic HBV DP rcDNA-containing nucleocapsids could be selectively immunoprecipitated with an antibody against carboxyl-terminal peptide of capsid protein (Fig. 5). Second, DP rcDNA could be selectively immunoprecipitated with antibodies against cellular nuclear transport receptors karyopherin-α and -β (Fig. 6). Furthermore, consistent with the proposed role that the cytoplasmic DP rcDNA is a functional precursor of cccDNA formation, we provided evidence showing that inhibition of karyopherin-mediated nuclear transportation by either transfection of siRNA targeting karyopherin-β1 mRNA or expression of a dominant negative karyopherin-β1 in a stable cell line supporting HBV replication resulted in the accumulation of cytoplasmic DP rcDNA and reduction of nuclear DP rcDNA and cccDNA (Fig. 7).
One of the most remarkable phenomena in cccDNA biosynthesis is that cccDNA can be formed from the rcDNA in the progeny nucleocapsids in the cytoplasm of infected cells. This phenomenon suggests that the disassembly of progeny rcDNA-containing nucleocapsids occurs in the host cells without an extracellular virion phase (40, 42). This is in marked contrast to many other animal viruses whose genome uncoating strictly relies on extracellular virion phase, due to the fact that the nucleocapsids of these viruses either become matured only during budding and secretion process (8, 25) or require interaction with viral receptors or an acid bath in endosomes during entry of host cells to trigger disassembly (1, 6, 35). Hence, the ability of intracellular progeny nucleocapsid to uncoat in host cells suggests that the disassembly of hepadnaviral capsids may be triggered directly either by maturation of DNA synthesis from inside of capsids or by attacking cellular factors that recognize structural features presented only on the surface of mature capsids. Our results obtained from the in vitro deproteinization assay strongly support the notion that virion-associated or intracellular nucleocapsids contain all the information and factors required for the viral genomic DNA deproteinization and uncoating. Moreover, we provided compelling evidence suggesting that the completion of plus-strand DNA synthesis is an essential, but not sufficient, signal to trigger genome deproteinization and capsid disassembly.
Considering the biochemical mechanism of deproteinization, our results indicated that a serine protease activity was required for the deproteinization reaction. A simple explanation for this observation is that a host cellular serine protease may be packaged in nucleocapsid and cleave the polymerase upon the completion of plus-strand DNA synthesis. Alternatively, the completion of plus-strand DNA synthesis may trigger nucleocapsid partial disassembly that makes rcDNA accessible to a copurified cellular serine protease. Nevertheless, the proteolytic removal of DNA polymerase is consistent with our previous observation that DP rcDNA purified from DHBV replicating cells has an authentic 5′ end (12).
The results presented herein (Fig. 6 and 7) and in our previous report demonstrated that approximately 15% of the total intracellular mature rcDNA were DP rcDNA (9, 12). However, the deproteinization in our in vitro endogenous DNA polymerase reaction occurred at a very low efficiency. We estimated that after 16 h of incubation, only <1% of mature rc- or dslDNA were deproteinized. While this is most likely due to the in vitro reaction that does not recapitulate the intracellular environment, it is also entirely possible that efficient deproteinization requires recruitment of additional cellular factors which did not exist in virion particle or were lost during purification of intracellular nucleocapsids.
Macromolecular traffic between the cytoplasm and nucleoplasm occurs through nuclear pore complexes (5). As for many other viruses, the genomic DNA nuclear import is an obligatory step in the life cycle of hepadnaviruses (15, 41). Consistent with a previous report (31), our results obtained from this study help to reveal a very important feature of hepadnavirus nucleocapsids—that the carboxyl-terminal portion of core protein is exposed only in mature double-stranded DNA-containing nucleocapsids. In agreement with this observation, we showed in this report that only DP rcDNA-containing nucleocapsids are selectively associated with nuclear import receptor karyopherins and karyopherin-β1 is essential for DP rcDNA nuclear transportation and cccDNA formation.
Our work reported herein further elucidated the molecular mechanism of cytoplasmic DP rcDNA production and provided additional evidence supporting the hypothesis that cytoplasmic DP rcDNA is the functional precursor of cccDNA formation. The molecular pathway of hepadnavirus cccDNA formation proposed in this report will guide our future investigation toward understanding the mechanism of cccDNA formation and development of antiviral strategies to cure chronic HBV infections.
Acknowledgments
We thank Jinhong Chang and Pamela Norton for critical reading of the manuscript and helpful suggestions.
This work was supported by grants from the Commonwealth of Pennsylvania through the Hepatitis B Foundation. H.G. and J.-T.G. are Bruce Witte fellow and scholar of Hepatitis B Foundation, respectively.
Footnotes
Published ahead of print on 28 October 2009.
REFERENCES
- 1.Alain, T., T. S. Kim, X. Lun, A. Liacini, L. A. Schiff, D. L. Senger, and P. A. Forsyth. 2007. Proteolytic disassembly is a critical determinant for reovirus oncolysis. Mol. Ther. 15:1512-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barthelmes, H. U., M. Habermeyer, M. O. Christensen, C. Mielke, H. Interthal, J. J. Pouliot, F. Boege, and D. Marko. 2004. TDP1 overexpression in human cells counteracts DNA damage mediated by topoisomerases I and II. J. Biol. Chem. 279:55618-55625. [DOI] [PubMed] [Google Scholar]
- 3.Beck, J., and M. Nassal. 2007. Hepatitis B virus replication. World J. Gastroenterol. 13:48-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chook, Y. M., and G. Blobel. 2001. Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 11:703-715. [DOI] [PubMed] [Google Scholar]
- 5.D'Angelo, M. A., and M. W. Hetzer. 2008. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 18:456-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Day, P. M., D. R. Lowy, and J. T. Schiller. 2003. Papillomaviruses infect cells via a clathrin-dependent pathway. Virology 307:1-11. [DOI] [PubMed] [Google Scholar]
- 7.Eckhardt, S. G., D. R. Milich, and A. McLachlan. 1991. Hepatitis B virus core antigen has two nuclear localization sequences in the arginine-rich carboxyl terminus. J. Virol. 65:575-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganser-Pornillos, B. K., M. Yeager, and W. I. Sundquist. 2008. The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 18:203-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao, W., and J. Hu. 2007. Formation of hepatitis B virus covalently closed circular DNA: removal of genome-linked protein. J. Virol. 81:6164-6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerlich, W. H., M. A. Feitelson, P. L. Marion, and W. S. Robinson. 1980. Structural relationships between the surface antigens of ground squirrel hepatitis virus and human hepatitis B virus. J. Virol. 36:787-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerlich, W. H., and W. S. Robinson. 1980. Hepatitis B virus contains protein attached to the 5′ terminus of its complete DNA strand. Cell 21:801-809. [DOI] [PubMed] [Google Scholar]
- 12.Guo, H., D. Jiang, T. Zhou, A. Cuconati, T. M. Block, and J. T. Guo. 2007. Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: an intermediate of covalently closed circular DNA formation. J. Virol. 81:12472-12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo, J. T., M. Pryce, X. Wang, M. I. Barrasa, J. Hu, and C. Seeger. 2003. Conditional replication of duck hepatitis B virus in hepatoma cells. J. Virol. 77:1885-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 15.Kann, M., A. Schmitz, and B. Rabe. 2007. Intracellular transport of hepatitis B virus. World J. Gastroenterol. 13:39-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krastev, Z. A. 2006. The “return” of hepatitis B. World J. Gastroenterol. 12:7081-7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kutay, U., E. Izaurralde, F. R. Bischoff, I. W. Mattaj, and D. Gorlich. 1997. Dominant-negative mutants of importin-beta block multiple pathways of import and export through the nuclear pore complex. EMBO J. 16:1153-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai, C. L., M. Rosmawati, J. Lao, H. Van Vlierberghe, F. H. Anderson, N. Thomas, and D. Dehertogh. 2002. Entecavir is superior to lamivudine in reducing hepatitis B virus DNA in patients with chronic hepatitis B infection. Gastroenterology 123:1831-1838. [DOI] [PubMed] [Google Scholar]
- 19.Levrero, M., T. Pollicino, J. Petersen, L. Belloni, G. Raimondo, and M. Dandri. 2009. Control of cccDNA function in hepatitis B virus infection. J. Hepatol. 51:581-592. [DOI] [PubMed] [Google Scholar]
- 20.Liaw, Y. F., N. W. Leung, T. T. Chang, R. Guan, D. I. Tai, K. Y. Ng, R. N. Chien, J. Dent, L. Roman, S. Edmundson, C. L. Lai, et al. 2000. Effects of extended lamivudine therapy in Asian patients with chronic hepatitis B. Gastroenterology 119:172-180. [DOI] [PubMed] [Google Scholar]
- 21.Lien, J., D. J. Petcu, C. E. Aldrich, and W. S. Mason. 1987. Initiation and termination of duck hepatitis B virus DNA synthesis during virus maturation. J. Virol. 61:3832-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mabit, H., and H. Schaller. 2000. Intracellular hepadnavirus nucleocapsids are selected for secretion by envelope protein-independent membrane binding. J. Virol. 74:11472-11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marcellin, P., T. T. Chang, S. G. Lim, M. J. Tong, W. Sievert, M. L. Shiffman, L. Jeffers, Z. Goodman, M. S. Wulfsohn, S. Xiong, J. Fry, and C. L. Brosgart. 2003. Adefovir dipivoxil for the treatment of hepatitis B e antigen-positive chronic hepatitis B. N. Engl. J. Med. 348:808-816. [DOI] [PubMed] [Google Scholar]
- 24.Mason, W. S., G. Seal, and J. Summers. 1980. Virus of Pekin ducks with structural and biological relatedness to human hepatitis B virus. J. Virol. 36:829-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mettenleiter, T. C., B. G. Klupp, and H. Granzow. 2009. Herpesvirus assembly: an update. Virus Res. 143:222-234. [DOI] [PubMed] [Google Scholar]
- 26.Molnar-Kimber, K. L., J. Summers, J. M. Taylor, and W. S. Mason. 1983. Protein covalently bound to minus-strand DNA intermediates of duck hepatitis B virus. J. Virol. 45:165-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moraleda, G., J. Saputelli, C. E. Aldrich, D. Averett, L. Condreay, and W. S. Mason. 1997. Lack of effect of antiviral therapy in nondividing hepatocyte cultures on the closed circular DNA of woodchuck hepatitis virus. J. Virol. 71:9392-9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nassal, M. 2008. Hepatitis B viruses: reverse transcription a different way. Virus Res. 134:235-249. [DOI] [PubMed] [Google Scholar]
- 29.Newbold, J. E., H. Xin, M. Tencza, G. Sherman, J. Dean, S. Bowden, and S. Locarnini. 1995. The covalently closed duplex form of the hepadnavirus genome exists in situ as a heterogeneous population of viral minichromosomes. J. Virol. 69:3350-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nitiss, K. C., M. Malik, X. He, S. W. White, and J. L. Nitiss. 2006. Tyrosyl-DNA phosphodiesterase (Tdp1) participates in the repair of Top2-mediated DNA damage. Proc. Natl. Acad. Sci. USA 103:8953-8958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rabe, B., A. Vlachou, N. Pante, A. Helenius, and M. Kann. 2003. Nuclear import of hepatitis B virus capsids and release of the viral genome. Proc. Natl. Acad. Sci. USA 100:9849-9854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reid, S. P., C. Valmas, O. Martinez, F. M. Sanchez, and C. F. Basler. 2007. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin α proteins with activated STAT1. J. Virol. 81:13469-13477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson, W. S., and R. L. Greenman. 1974. DNA polymerase in the core of the human hepatitis B virus candidate. J. Virol. 13:1231-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seeger, C., and W. S. Mason. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, J. L., S. K. Campos, A. Wandinger-Ness, and M. A. Ozbun. 2008. Caveolin-1-dependent infectious entry of human papillomavirus type 31 in human keratinocytes proceeds to the endosomal pathway for pH-dependent uncoating. J. Virol. 82:9505-9512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Straprans, S., D. D. Loeb, and D. Ganem. 1991. Mutations affecting hepadnavirus plus-strand DNA synthesis dissociate primer cleavage from translocation and reveal the origin of linear viral DNA. J. Virol. 65:1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Summers, J., and W. S. Mason. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403-415. [DOI] [PubMed] [Google Scholar]
- 38.Summers, J., A. O'Connell, and I. Millman. 1975. Genome of hepatitis B virus: restriction enzyme cleavage and structure of DNA extracted from Dane particles. Proc. Natl. Acad. Sci. USA 72:4597-4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tagawa, M., M. Omata, and K. Okuda. 1986. Appearance of viral RNA transcripts in the early stage of duck hepatitis B virus infection. Virology 152:477-482. [DOI] [PubMed] [Google Scholar]
- 40.Tuttleman, J. S., C. Pourcel, and J. Summers. 1986. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell 47:451-460. [DOI] [PubMed] [Google Scholar]
- 41.Whittaker, G. R., M. Kann, and A. Helenius. 2000. Viral entry into the nucleus. Annu. Rev. Cell Dev. Biol. 16:627-651. [DOI] [PubMed] [Google Scholar]
- 42.Wu, T. T., L. Coates, C. E. Aldrich, J. Summers, and W. S. Mason. 1990. In hepatocytes infected with duck hepatitis B virus, the template for viral RNA synthesis is amplified by an intracellular pathway. Virology 175:255-261. [DOI] [PubMed] [Google Scholar]
- 43.Yeh, C. T., Y. F. Liaw, and J. H. Ou. 1990. The arginine-rich domain of hepatitis B virus precore and core proteins contains a signal for nuclear transport. J. Virol. 64:6141-6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, Y. Y., B. H. Zhang, D. Theele, S. Litwin, E. Toll, and J. Summers. 2003. Single-cell analysis of covalently closed circular DNA copy numbers in a hepadnavirus-infected liver. Proc. Natl. Acad. Sci. USA 100:12372-12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu, Y., T. Yamamoto, J. Cullen, J. Saputelli, C. E. Aldrich, D. S. Miller, S. Litwin, P. A. Furman, A. R. Jilbert, and W. S. Mason. 2001. Kinetics of hepadnavirus loss from the liver during inhibition of viral DNA synthesis. J. Virol. 75:311-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zlotnick, A., N. Cheng, S. J. Stahl, J. F. Conway, A. C. Steven, and P. T. Wingfield. 1997. Localization of the C terminus of the assembly domain of hepatitis B virus capsid protein: implications for morphogenesis and organization of encapsidated RNA. Proc. Natl. Acad. Sci. USA 94:9556-9561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zoulim, F. 2005. New insight on hepatitis B virus persistence from the study of intrahepatic viral cccDNA. J. Hepatol. 42:302-308. [DOI] [PubMed] [Google Scholar]