Abstract
The envelope glycoprotein of human immunodeficiency virus type 1 (HIV-1) has several adaptations that allow the virus to evade antibody neutralization. Nevertheless, a few broadly cross-reactive neutralizing antibodies as well as reagents containing portions of CD4, the HIV receptor, have demonstrated partial efficacy in suppressing viral replication. One type of reagent designed for improved HIV neutralization fuses the CD4 D1-D2 domains to the variable regions of an antibody recognizing the CD4-induced (CD4i) coreceptor binding site on the gp120 portion of the HIV envelope spike. We designed, expressed, purified, and tested the neutralization potencies of CD4-CD4i antibody reagents with different architectures, antibody combining sites, and linkers. We found that fusing CD4 to the heavy chain of the CD4i antibody E51 yields a bivalent reagent including an antibody Fc region that expresses well, is expected to have a long serum half-life, and has comparable or greater neutralization activity than well-known broadly neutralizing anti-HIV antibodies. A CD4 fusion with the anti-HIV carbohydrate antibody 2G12 also results in a potent neutralizing reagent with more broadly neutralizing activity than 2G12 alone.
The envelope spike of human immunodeficiency virus type 1 (HIV-1), a trimer of gp120/gp41 heterodimers, utilizes a number of strategies to avoid antibodies (Abs) elicited by the humoral immune response. These include variable loops, heavy glycosylation (36), conformational masking of key functional sites (19), and an architecture and surface density that reduce bivalent Ab engagement (18). Nevertheless, a small number of broadly cross-reactive neutralizing Abs have been found and extensively characterized (5, 32, 41). The targets of these Abs include the membrane proximal region of gp41 (24, 42), a cluster of high-mannose carbohydrates on gp120 (29), and the HIV receptor (CD4)-binding site (3, 28). A combination of several of these Abs has been evaluated in clinical trials as a passive immunotherapy to reduce viral rebound during an interruption of antiretroviral therapy (34).
Several CD4-containing proteins have also been explored clinically as possible therapeutics for treating HIV-1: soluble CD4 (13, 16), a CD4-Fc fusion protein (7), and the tetravalent CD4-immunoglobulin G2 (CD4-IgG2; PRO 542) reagent (1, 17). In patients with advanced disease, CD4-IgG2 treatment led to a ∼0.5 log10 mean reduction in viral load (17). In addition, D1D2-Igαtp, an approximately dodecameric CD4 reagent created as a chimeric IgG1/IgA fusion protein (2), exhibited very potent HIV neutralization activity and targeted HIV-infected cells for lysis by natural killer cells (14).
Another approach to targeting gp120 is a fusion protein composed of CD4 linked to the variable regions of a CD4-induced (CD4i) Ab (11). CD4i Abs represent a potentially promising class of Abs because they bind to the conserved HIV-1 coreceptor binding site on gp120, which is exposed after a conformational change resulting from binding to CD4 (25, 27, 38). Examples of CD4i Abs include 17b (33), E51 (39), m9 (40), 412d (8), and 21c (38). These Abs are often broadly cross-reactive but generally show little neutralization potency in vivo due to limited steric accessibility when gp120 on the viral membrane is bound to CD4 on the surface of the target cell (20). Fusing CD4 to the combining site of a CD4i Ab solves the accessibility problem since the Ab epitope would be exposed by CD4 binding when the virion is not bound to the target cell. This class of reagent has two other favorable features: bivalent binding and targeting of functionally critical epitopes on gp120, the CD4 and coreceptor binding sites. One such reagent, sCD4-17b (referred to here as CD4-scFv17b), contains the first two domains of CD4 linked to the single-chain fragment variable (scFv) form of the CD4i Ab 17b (Fig. 1) (11). This reagent was shown to potently neutralize multiple primary strains of HIV-1 (11), suggesting that CD4-CD4i Ab fusion proteins are promising candidates for passive immunization or gene therapy trials.
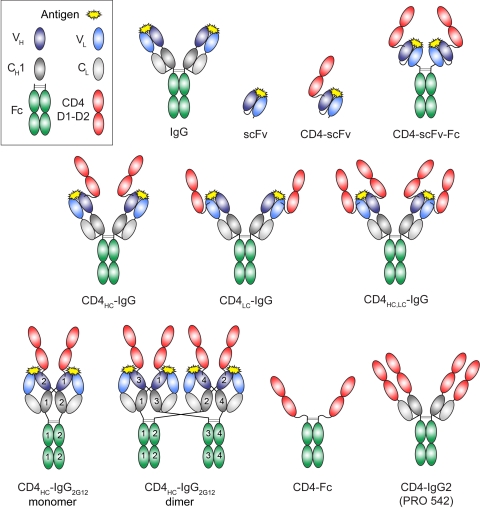
FIG. 1.
Schematic depiction of CD4-CD4i reagents and related molecules. VH, variable domain of the IgG heavy chain (HC); VL, variable domain of the IgG light chain (LC), CH1, constant region 1 of the HC; CL, constant region of the LC; Fc, CH2 and CH3 domains of dimerized HCs; CD4 D1-D2, N-terminal two domains of CD4; scFv, single-chain fragment variable (VH and VL domains of an IgG); CD4HC, CD4 linked to the VH domain of an IgG; CD4LC, CD4 linked to the VL domain of an IgG; CD4HC,LC, CD4 linked to the VH and VL domains of an IgG.
Critical properties for CD4-containing reagents include their breadth of neutralization activity, half-life, and, for reagents used in a gene therapy context, their expression level. We have undertaken a systematic effort to develop the optimal architecture for a CD4-CD4i Ab reagent by designing, constructing, and testing reagents with different CD4i Ab combining sites and including an Ab Fc region to increase valency and serum half-life (7). We varied the arrangements of the Ab combining sites; the lengths, attachments, and forms of the linking regions; and the ways in which CD4 was fused to the CD4i Ab (Fig. 1). CD4-CD4i Ab reagents were evaluated using in vitro neutralization assays across a broad range of clade A, B, and C HIV-1 strains. One promising reagent, a fusion of CD4 domains 1 and 2 (D1-D2) to the heavy chain of the E51 CD4i Ab, was expressed at high levels in mammalian cells and exhibited neutralization potencies that compared favorably with or exceeded those of known broadly neutralizing Abs such as 4E10, b12, 2G12, and 2F5.
Since much of the activity of our CD4-CD4i reagents resulted from the CD4 component, we also explored the effects of attaching CD4 to an Ab with a different quaternary structure. The anticarbohydrate Ab 2G12 is unusual in that its heavy chains are involved in a domain swap creating a rigid (Fab)2 unit in which the combining sites are separated by ∼35Å (6). This domain swapping tendency also leads to the formation of 2G12 dimers containing two (Fab)2 units and two Fc regions, which form when the domain swapping occurs intermolecularly between two IgGs rather than intramolecularly between the two Fab arms of a single IgG (37). The 2G12 dimer is 50- to 80-fold more potent than monomeric 2G12 in neutralizing clade B 2G12-sensitive strains; however, neither form of 2G12 neutralizes clade C strains of HIV-1 (37). In order to assess the effects of adding CD4 to the 2G12 monomer and dimer architectures and to explore whether addition of CD4 would broaden the range of HIV-1 strains that are sensitive to 2G12, we constructed CD4-2G12 fusion proteins and tested their neutralization potencies (Fig. 1). We found that these hybrid reagents had potent neutralizing activities with both the CD4 and Ab combining site components apparently contributing to this behavior.
MATERIALS AND METHODS
Isolation of genes encoding the 21c Ab.
21c IgG was purified from an Epstein-Barr virus-transformed human B-cell line obtained from James Robinson (Tulane University). N-terminal amino acid sequencing was performed after treatment with Pfu pyroglutamate aminopeptidase to remove blocking pyroglutamate, and 5′ primers were designed based on these sequences. Heavy and light chain genes were obtained by cDNA amplification using a SMART RACE kit (Clontech). The 3′ primers were the human CH1 (constant region 1 of the heavy chain) domain primer CAGCTCCACCCTCTTGTCCACCTTGGTGTTGCTGGG and the human λ constant domain primer CTAAGAACATTCTGCATGGGCCATTGTCTTCTCC. Bridge PCR was used to generate the full-length heavy chain.
Materials.
Genes encoding the variable regions (variable heavy and variable light, VH and VL, or the intact light chain VL-CL, where CL refers to the constant light domain) of the E51, m9, and 412d Abs were synthesized (BlueHeron Biotechnologies or Integrated DNA Technologies). Intact IgG genes were constructed by subcloning the relevant variable sequences onto a human IgG1 sequence. The 2G12 and anti-glycoprotein D (gD) IgG genes were obtained from Dennis Burton (The Scripps Research Institute). The CD4-scFv17b gene (11) was obtained from Ed Berger (Laboratory of Viral Diseases, NIAID, NIH). The CD4-IgG2 (PRO 542) protein (1) was obtained from Progenics.
Sequences for all of the constructs are in Fig. S1 in the supplemental material; schematic structures are shown in Fig. S1 and Table S1 in the supplemental material. Genes encoding scFv versions of Abs E51, m9, and 21c were synthesized as the VH domain followed by a (Gly4Ser)3 linker, the VL domain, and a C-terminal six-His tag. CD4-scFv genes were constructed to be similar to the CD4-scFv17b gene (11) by fusing the DNA encoding the CD4 hydrophobic leader sequence and first two domains (D1-D2; residues 1 to 182 of the mature CD4 protein) to a (Gly4Ser)7 linker sequence followed by the His-tagged scFv construct. The CD4-scFv17b gene was fused to the human IgG1 Fc domain sequence by bridge PCR to create CD4-scFv17b-Fc.
CD4-IgG heavy chain constructs (denoted by the prefix CD4HC-) were made by fusing the CD4-VH region of the 21c and E51 CD4-scFv constructs to the remaining domains (CH1, CH2, and CH3) of a human IgG1 heavy chain gene using bridge PCR. Analogous constructs for 412d and m9 were synthesized. Bridge PCR was used to construct CD4-Fc [CD4 D1-D2 domains linked with (Gly4Ser)7, denoted GS7 in the construct names, to the Fc region of IgG1]. CD4HC-(GS7)-IgG2G12 and CD4HC-(GS7)-IgGanti-gD were constructed by inserting sequences encoding the relevant VH and CH1 domains into the CD4-IgG scaffold. The light chain (denoted LC in the constructs) of CD4LC-(GS7)-IgGE51 was constructed by bridge PCR.
A gene encoding a flexible linker [(Gly4Ser)9, denoted GS9] was synthesized with NheI and BamHI restriction enzyme sites. The linker region sequence was inserted between the CD4 and VH sequences of the CD4HC-(GS7)-IgGE51 construct by subcloning, replacing the (Gly4Ser)7 sequence with the new linker sequence.
Protein expression and purification.
The CD4-scFv17b gene was subcloned into baculovirus expression vector pBacPAK8 (BD Biosciences), and CD4-scFv17b protein was purified from insect cell supernatants by Ni-nitrilotriacetic acid (NTA) chromatography. All other constructs were subcloned into the mammalian expression vector pTT5 (NRC Biotechnology Research Institute), and the corresponding proteins were expressed transiently in suspension HEK 293-6E cells (NRC Biotechnology Research Institute) using 25-kDa linear polyethylenimine (PEI) (Polysciences) as described previously (12). When the heterodimeric constructs were expressed, the heavy chain and light chain vectors were mixed at a 1:1 ratio by mass. Cell culture supernatants were collected 6 days posttransfection. For Fc-containing constructs, supernatants were passed over protein A resin (Thermo Fisher Scientific), eluted using pH 3.0 citrate buffer, and then immediately neutralized. The scFv and CD4-scFv reagents were purified using Ni-NTA chromatography and eluted using 300 mM imidazole. All reagents were then subjected to size exclusion chromatography in 20 mM Tris (pH 8.0)-150 mM NaCl using a Superdex 200 16/60 or 10/30 column (GE Healthcare). Final yields of purified reagents are given in Table S1 in the supplemental material.
In vitro neutralization assays.
A previously described pseudovirus neutralization assay, which measures the reduction in luciferase reporter gene expression in the presence of a potential inhibitor following a single round of pseudovirus infection in TZM-bl cells (21, 23), was used to evaluate the neutralization potencies of the reagents. Pseudoviruses were generated by cotransfection of HEK 293T cells with an Env expression plasmid and a replication-defective backbone plasmid. Neutralization assays were performed either by our laboratory (see Tables 1, 2, 4, and 5) or by the Collaboration for AIDS Vaccine Discovery (CAVD) core neutralization facility (see Table 3) using the same protocol (21, 23). Briefly, each sample was tested in triplicate (our assays) or duplicate (CAVD assays), with 200 infectious viral units per well incubated with a threefold dilution series and with 75 μg/ml DEAE-dextran. For experiments testing inhibition of 2G12 binding by fructose, the 1-h incubation at 37°C of reagent plus pseudovirus was done in the presence of 3% (wt/vol) fructose. For all assays, after a 1-h incubation at 37°C, 10,000 TZM-bl cells were added to each well and incubated for 2 days. Cells were then lysed and assayed for luciferase expression using BriteLite plus (PerkinElmer) and a Victor3 luminometer (PerkinElmer) (our assays) or equivalent equipment (CAVD assays). Percentage neutralization was determined by calculating the difference in luminescence between test wells (cells plus virus plus reagent) and cell control wells (cells only), dividing this value by the difference between the virus control wells (cells plus virus) and cell control wells, subtracting from 1, and multiplying by 100.
TABLE 1.
IC50s for CD4-scFv reagents
| Env | Clade | IC50 (nM)a |
|||||
|---|---|---|---|---|---|---|---|
| CD4-scFv17b | CD4-scFvm9 | CD4-scFv21c | CD4-scFvE51 | CD4 | scFvE51 | ||
| SF162.LS | B | 3.5 | 1.7 | 1.40 | 0.75 | 16 | 15 |
| SC422661.8 | B | 91 | ND | 1200 | 24 | 480 | 61 |
| TRJO4551.58 | B | 1000 | 720 | 1500 | 500 | 940* | 320 |
| QH0692.42 | B | 59 | ND | ND | 31 | 110 | 450 |
| RHPA4259.7 | B | 110 | ND | 370 | 48 | 170 | 76 |
| ZM53M.PB12 | C | 200 | 150 | 880 | 60 | 390* | 66 |
| ZM214M.PL15 | C | 91 | 110 | 330 | 23 | 480 | 120 |
| Du172.17 | C | 24 | 66 | 89 | 26 | 150 | 190 |
| Geometric mean | 70 | 66 | 220 | 29 | 210 | 110 | |
| Arithmetic mean | 200 | 210 | 620 | 89 | 340 | 160 | |
Results are from in-house neutralization assays except for values marked with an asterisk, which are from CAVD core neutralization facility assays. ND, not determined.
TABLE 2.
IC50s for CD4-IgGE51 constructs compared to CD4-scFvE51
| Env | Clade | IC50 (nM)a |
|||
|---|---|---|---|---|---|
| CD4-scFvE51 | CD4HC-(GS7)-IgGE51 | CD4LC-(GS7)-IgGE51 | CD4HC,LC-(GS7)-IgGE51 | ||
| SF162.LS | B | 0.75 | 0.47 | 1.3 | 1.7 |
| SC422661.8 | B | 24 | 16 | 30 | 47 |
| QH0692.42 | B | 31 | 2.1 | ND | ND |
| RHPA4259.7 | B | 48 | 3.5 | ND | ND |
| Du172.17 | C | 26 | 4.9 | 5.5 | 32 |
| Geometric mean | 15 | 3.1 | 6.0 | 14 | |
| Arithmetic mean | 26 | 5.4 | 12 | 27 | |
ND, not determined.
TABLE 4.
IC50s for CD4-2G12 constructs compared with 2G12-only and CD4-only reagents
| Env | Clade | IC50 (nM)a |
||||
|---|---|---|---|---|---|---|
| Monomer |
Dimer |
CD4- IgG2 | ||||
| 2G12 | CD4HC-IgG2G12 | 2G12 | CD4HC-IgG2G12 | |||
| PVO.4 | B | 33* | 4.2* | <0.17* | 0.30* | 27* |
| SC422661.8 | B | 140 | 4.4 | 0.38 | 0.46 | 7.3 |
| QH0692.42 | B | 72 | 1.8 | 0.71 | 0.60 | 0.83 |
| TRO.11 | B | 13 | 6.8 | 0.11 | 0.63 | 92 |
| Du156.12 | C | >670* | >530* | >330* | >260* | 40* |
| Du172.17 | C | >670 | 6.3 | >330 | 1.1 | 2.5 |
| Geometric mean | 110 | 9.5 | 2.9 | 1.6 | 11 | |
| Arithmetic mean | 270 | 92 | 110 | 44 | 28 | |
Results are from in-house neutralization assays except for values marked with an asterisk, which are from CAVD core neutralization facility assays. In calculating average IC50s, measurements outside of the range of the assay (indicated by a > symbol) were assigned to that limiting value.
TABLE 5.
IC50s for CD4HC-IgG2G12, 2G12, and CD4-CD4i reagents in the presence and absence of 3% fructose
| Env | Effect on IC50 of fructosea |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| CD4HC-IgG2G12 monomer |
2G12 monomer |
CD4HC-(GS7)-IgGE51 |
|||||||
| IC50 (nM) |
Fold change | IC50 (nM) |
Fold change | IC50 (nM) |
Fold change | ||||
| + | − | + | − | + | − | ||||
| WITO4160.33 | 200 | 18 | 11 | 650 | 65 | 10 | 110 | 91 | 1.2 |
| SC422661.8 | 430 | 46 | 9.3 | 260 | 64 | 4.1 | 310 | 300 | 1.0 |
| QH0692.42 | 16 | 6 | 2.7 | 130 | 44 | 3.0 | 18 | 27 | 0.7 |
| TRO.11 | 28 | 5.7 | 4.9 | 33 | 5.3 | 6.2 | ND | ND | ND |
+, treatment with 3% fructose; −, no fructose; ND, not determined.
TABLE 3.
Comparison of IC50s for CD4 reagents and commonly studied broadly neutralizing anti-HIV-1 Abs
| Clade and Env | IC50 (nM)a |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD4HC-(GS7)-IgGE51 | CD4HC-(GS9)-IgGE51 | CD4HC,LC-(GS7)-IgGE51 | CD4HC-(GS7)-IgG17b | CD4HC-(GS7)-IgGm9 | CD4HC-(GS7)-IgG412d | CD4HC-(GS7)- IgGanti-gD | CD4-IgG2 | CD4-Fc | CD4 | 4E10 | b12 | 2G12 | 2F5 | PG9 | PG16 | |
| Clade B | ||||||||||||||||
| SF162.LS | 0.37 | <0.26 | ND | <0.26 | ND | ND | ND | ND | ND | 2.3 | 0.07 | 0.06 | 4.0 | 0.68 | >330 | >330 |
| PVO.4 | 12 | 7.1 | 9.8 | 7.0 | 27 | 14 | 5.2 | 27 | 300 | 310 | 45 | >330 | 8.0 | >330 | 27 | 36 |
| CAAN5342.A2 | 14 | 6.8 | ND | 7.5 | 110 | 90 | 25 | 41 | 870 | 750 | 19 | >330 | >330 | 24 | 38 | 59 |
| WITO4160.33 | 3.2 | 1.6 | 5.5 | 1.4 | 17 | 8.2 | 4.6 | 8.0 | 120 | 250 | 2.1 | 20 | 7.3 | 4.1 | ||
| AC10.2.29 | 7.3 | 2.8 | >420 | 3.5 | 51 | ND | 14 | 11 | 230 | 400 | 2.1 | 12 | >330 | 8.8 | ||
| SC422661.8 | 7.0 | 5.7 | 10 | 7.4 | 28 | 14 | 6.7 | 14 | 240 | 220 | 6.2 | 1.3 | 14 | 4.8 | 5.3 | 7.5 |
| 6535.30 | 1.7 | 1.1 | 9.8 | 0.8 | 6.8 | ND | 1.5 | 13 | 29 | 37 | 1.4 | 9.1 | 13 | 13 | 1.5 | 250 |
| THRO4156.18 | 2.8 | 1.8 | ND | ND | 4.7 | 5.1 | 2.6 | 2.3 | 12 | 14 | 2.1 | 3.2 | >330 | >330 | 83 | 8.9 |
| REJO4541.67 | 2.3 | 1.4 | 30 | ND | 8.4 | ND | 3.1 | 2.9 | 17 | 23 | 4.9 | 4.5 | >330 | 4.1 | ||
| TRJO4551.58 | 20 | 11 | ND | ND | 66 | ND | 35 | 19 | 500 | 940 | 31 | >330 | >330 | >330 | 2.9 | 7.7 |
| QH0692.42 | 3.1 | 1.8 | 16 | ND | 10 | 9.7 | 1.0 | 2.3 | 14 | 23 | 9.7 | 1.9 | 19 | 6.8 | >330 | >330 |
| TRO.11 | 86 | 52 | 4.7 | ND | 31 | ND | 9.3 | 45 | 770 | 540 | 2.1 | >330 | 2.7 | >330 | 36 | 1.5 |
| RHPA4259.7 | 3.2 | 1.5 | 370 | ND | 34 | 20 | 5.2 | 5.2 | 82 | 84 | 48 | 0.6 | >330 | 82 | ||
| Clade C | ||||||||||||||||
| MW965.26 | <0.26 | <0.26 | ND | <0.26 | ND | ND | ND | ND | ND | |||||||
| ZM197M.PB7 | 22 | 12 | >420 | 7.2 | 61 | 63 | 35 | 91 | >1,000 | 180 | 3.5 | 130 | >330 | 84 | ||
| ZM249.PL1 | 13 | 4.6 | ND | 4.1 | 83 | 42 | 19 | 30 | 1000 | 450 | 15 | 21 | >330 | >330 | ||
| ZM53M.PB12 | 2.8 | 1.1 | ND | 1.5 | 28 | 31 | 4.6 | 12 | 350 | 390 | 49 | 170 | >330 | >330 | ||
| ZM214M.PL15 | 7.0 | 6.0 | >420 | 5.2 | 22 | 16 | 13 | 16 | 470 | 370 | 28 | 19 | >330 | >330 | ||
| Du156.12 | 51 | 35 | >420 | 35 | 120 | 53 | 87 | 40 | >1,000 | 630 | 1.4 | 5.2 | >330 | >330 | ||
| Du422.1 | 1.6 | 0.8 | >420 | 1.3 | 150 | 85 | 68 | 40 | 630 | 420 | 4.9 | 1.3 | >330 | >330 | ||
| Du172.17 | 5.2 | 2.9 | 160 | ND | 21 | 28 | 8.8 | 8.6 | 56 | 79 | 2.1 | 6.5 | >330 | >330 | ||
| CAP45.2.00.G3 | 3.6 | 2.0 | ND | ND | 46 | 11 | 20 | 6.3 | >1,000 | 1200 | 18 | 4.5 | >330 | >330 | ||
| CAP210.2.00.E8 | 6.3 | 2.9 | ND | ND | 18 | 20 | 7.7 | 3.4 | 50 | 160 | 8.3 | 130 | >330 | >330 | ||
| ZM233M.PB6 | 6.6 | 2.3 | ND | ND | 42 | 40 | 11 | 33 | 460 | 140 | 8.3 | >330 | >330 | >330 | ||
| ZM109F.PB4 | 0.52 | <0.26 | ND | ND | 2.1 | 2.0 | 0.52 | 0.29 | 3.1 | 9 | 4.2 | >330 | >330 | >330 | ||
| ZM135M.PL10a | 13 | 9.3 | ND | ND | 71 | 43 | 22 | 8.0 | 420 | 280 | 4.2 | >330 | >330 | >330 | ||
| Clade A | ||||||||||||||||
| DJ263.8 | <0.26 | <0.26 | ND | <0.26 | ND | ND | ND | ND | ND | |||||||
| Q23.17 | 3.6 | 2.1 | >420 | 1.8 | 69 | 53 | 15 | 18 | 530 | 120 | >330 | >330 | 50 | |||
| Q842.d12 | 45 | 43 | >420 | 33 | 200 | 86 | 110 | 90 | >1,000 | 97 | >330 | >330 | 59 | |||
| Q259.d2.17 | 2.7 | 1.8 | >420 | 1.8 | 43 | 38 | 12 | 13 | 93 | 99 | >330 | >330 | 72 | |||
| 3718.v3.c11 | 19 | 9.4 | >420 | 19 | ND | 94 | 55 | 41 | >1,000 | 80 | >330 | >330 | 23 | |||
| 0330.v4.c3 | 2.4 | 1.3 | 9.3 | 1.0 | ND | 12 | ND | 13 | 130 | 40 | >330 | 4.7 | 65 | |||
| 3415.00 | 16 | 8.1 | 12 | 10 | ND | 18 | ND | 90 | 640 | 160 | 160 | 14 | 250 | |||
| Geometric mean | 5.0 | 2.9 | 75 | 2.9 | 32 | 25 | 11 | 14 | 210 | 150 | 10 | 33 | 110 | 78 | 27 | 33 |
| Arithmetic mean | 12 | 7.2 | 220 | 7.1 | 51 | 36 | 22 | 25 | 440 | 320 | 30 | 150 | 240 | 190 | 96 | 110 |
IC50s for the 4E10, b12, 2F5 Abs and CD4 were taken from previously reported results (21, 22) or were provided by the CAVD neutralization core. IC50s for 2G12 were taken from references 21 and 22 and converted to molar concentrations, assuming the monomeric form. PG9 and PG16 were tested on a larger panel of viruses (35), but only those that overlapped with our panel are reported in this table. Over a 162-pseudovirus panel, PG9 and PG16 had geometric mean IC50s of 5.7 and 5.5 nM, respectively. In calculating average IC50s, measurements outside of the range of the assay (indicated by > or < symbol) were assigned to that limiting value. ND, not determined.
Nonlinear regression analysis was used to calculate concentrations at which half-maximal inhibition was observed (IC50s). Average IC50s across multiple HIV-1 strains are reported as both arithmetic and geometric means. Calculation of geometric means is suitable for data sets covering multiple orders of magnitude (31), as is the case for neutralization data across multiple viral strains. IC50s are reported in Tables 1 to 5 as molar concentrations to compensate for different molecular weights of the reagents. IC50s in μg/ml are presented for the Table 3 data in Table S2 in the supplemental material.
Calculation of the IC50eff of a bivalent reagent.
We calculated the effective IC50 (IC50eff) for a reagent containing independently acting components with individual IC50s of IC50a and IC50b. The fraction of Env binding sites occupied by component a is [c]/(IC50a + [c]), where [c] is the concentration of the reagent. Likewise, the fraction of sites occupied by component b is [c]/(IC50b + [c]). The fraction occupied by components a and/or b is calculated as follows: [c]/(IC50a + [c]) + [c]/(IC50b + [c]) − [c]2/((IC50a + [c]) × (IC50b + [c])). Solving for [c] when the fraction occupied by a and/or b equals 0.5 gives the following IC50eff: IC50eff = 0.5 × (−IC50a − IC50b + √(IC50a2 + IC50b2 + 6 × IC50a × IC50b)). From Table 1, the geometric mean IC50s for CD4-scFvE51, CD4, and scFvE51 are 29, 210, and 110 nM, respectively. For IC50a of 210 nM (CD4) and IC50b of 110 nM (scFvE51), we calculate that the IC50eff is 61 nM. Hence, the average IC50 for CD4-scFvE51, 29 nM, is about twofold better than the independent model would predict. Using the IC50 of scFvE51 for the activity of the CD4i component of CD4-scFvE51 might underestimate the synergy of the components since larger CD4i reagents are thought to be significantly less potent than smaller ones (20).
Nucleotide sequence accession numbers.
The 21c sequences have been deposited in GenBank under accession numbers GU179344 and GU179345.
RESULTS
To determine which CD4i Ab combining sites formed effective CD4-scFv CD4i reagents, we expressed fusions of 17b, E51, and m9 and evaluated these using an in vitro neutralization assay (Table 1). In addition, we made a CD4-scFv reagent using the CD4i Ab 21c, which was reported to be cross-reactive and highly potent against HIV-2 (10), after isolating the heavy and light chain genes encoding the Ab. All CD4-scFv reagents consisted of the first two domains of CD4 (D1 and D2) fused to an scFv with a (Gly4Ser)7 linker sequence (Fig. 1). Of these, 17b, E51, and m9 gave reagents with comparable potencies. The geometric mean molar IC50 for CD4-scFvE51 (29 nM) is significantly lower than that of CD4 alone (210 nM) or scFvE51 alone (110 nM) (Table 1), implying that both components are contributing to the neutralization potency of this reagent. The more weakly neutralizing 21c reagent was less effective (IC50 of 220 nM) than the other CD4-scFv reagents (IC50s of 29 to 70 nM). The lower efficacy of this reagent might result if the linker in the CD4-scFv21c construct did not permit simultaneous binding by the CD4 and 21c components, perhaps due to a difference in the relative orientation of 21c on gp120 compared to other CD4i antibodies.
Next, we evaluated the effects of including an Ab Fc domain, which would convert a CD4-CD4i reagent into a dimer and increase the serum half-life through binding of the Fc to the protection receptor FcRn (7). Our initial Fc construct consisted of the CD4-scFv17b sequence (11) fused to the N terminus of the human IgG1 Fc domain to make CD4-scFv17b-Fc (Fig. 1). In addition, we tried to express a two-chain reagent in which CD4 was fused to the N terminus of the constant domain of the light chain (CL), and the 17b scFv was fused to the N terminus of the Fc domain. This two-chain form gave only trace quantities of the desired protein product (data not shown), whereas the single chain form, CD4-scFv17b-Fc, yielded a small but testable quantity for neutralization assays (see Table S1 in the supplemental material). These assays demonstrated that CD4-scFv17b-Fc had a molar neutralization potency about twofold better than CD4-scFv17b (for strain QH-0692, the IC50s were 30 nM versus 59 nM, and for strain SF162.LS, the IC50s were 2.5 nM versus 3.5 nM), as would be expected if each half of the dimeric reagent functioned independently. Hence, no enhancement due to avidity was observed for this reagent.
Although the CD4-scFv17b-Fc reagent maintained its antiviral activity and likely would have an increased serum half-life due to the Fc domain, it had a relatively poor expression yield (see Table S1 in the supplemental material), and its neutralization properties suggested no evidence for an avidity effect. Since we had observed that two-chain IgG reagents were often expressed well, we designed a CD4-CD4i reagent with a more IgG-like architecture by attaching CD4 to the N terminus of a complete Ab heavy chain, which would be expressed together with a conventional Ab light chain. The CD4HC-(GS7)-IgGE51 construct was expressed well (∼10 mg/liter), and we compared the neutralization potencies of this reagent with CD4-scFvE51 (Table 2). CD4HC-(GS7)-IgGE51 had a molar neutralization potency (mean IC50 of 3.1 nM) higher than what would be expected for monovalent binding by only one arm of the reagent at a time (expected IC50 of ∼7.5 nM), suggesting that this geometry allowed an avidity enhancement resulting from simultaneous binding of both arms. Simultaneous binding might have been facilitated by the greater distance between the CD4 arms in this reagent compared to the CD4-scFv17b-Fc construct.
To fully explore the potential of the IgG-like architecture, we made additional reagents in which CD4 was fused to the N terminus of the Ab light chain (CD4LC-(GS7)-IgGE51) or to both the heavy chain and the light chain [CD4HC,LC-(GS7)-IgGE51]. These reagents were expressed at yields of ∼30% and ∼10% of the heavy chain CD4HC-(GS7)-IgGE51 form, and the average neutralization potencies of these reagents were modestly reduced compared to the original reagent (Table 2).
Using the CD4HC-(GS7)-IgGE51 reagent as a benchmark, we next varied the linker between CD4 and the variable domain of the heavy chain (VH). We constructed a longer flexible glycine-serine linker (Gly4Ser)9 to make CD4HC-(GS9)-IgGE51. To more thoroughly evaluate these reagents, as well as reagents containing 17b, m9, and 412d Ab combining sites in place of E51, we compared neutralization potencies over a panel of clade A, B, and C HIV-1 strains (Table 3). The neutralization potency of the longer CD4HC-(GS9)-IgGE51 construct was slightly, but consistently, ∼30% better than CD4HC-(GS7)-IgGE51 (P < 0.001 [two-tailed paired t test on log10-transformed values]).
To determine the degree to which the combining site of the CD4i Ab contributed to the activity of CD4HC-(GS7)-IgGE51, we constructed a similar reagent using an Ab against an irrelevant antigen (the herpes simplex virus gD protein) (4) to create CD4HC-(GS7)-IgGanti-gD. Control experiments demonstrated that the parental anti-gD Ab was inactive in HIV-1 neutralization assays (data not shown); thus, any potential neutralization activity of the CD4HC-(GS7)-IgGanti-gD reagent would be derived solely from its CD4 component. We also evaluated the neutralization potencies of other reagents whose neutralization activities would be solely due to CD4: the tetravalent CD4-IgG2 reagent (also known as PRO 542) (1) and a bivalent CD4-Fc construct similar to a previously described CD4-immunoadhesin reagent (7) (Fig. 1). CD4HC-(GS7)-IgGE51 had a geometric mean IC50 of 5 nM and was about twofold more potent than the corresponding reagent lacking CD4i Ab activity, CD4HC-(GS7)-IgGanti-gD, which had a geometric mean IC50 of 11 nM (P < 0.02, by a two-tailed paired t test on log10-transformed values). We also found that the CD4HC-(GS9)-IgGE51 and CD4HC-(GS7)-IgG17b reagents were about threefold more potent than the CD4HC-(GS7)-IgGanti-gD and CD4-IgG2 reagents lacking CD4i Ab combining sites (all pairwise comparisons have P values of <0.0001) (Table 3). However, the m9 and 412d reagents (CD4HC-(GS7)-IgGm9 and CD4HC-(GS7)-IgG412d) were two- to threefold less potent than either CD4-IgG2 or CD4HC-(GS7)-IgGanti-gD, suggesting that the m9 and 412d combining sites were not paired with CD4 to achieve optimal binding of the CD4 or Ab components to gp120. All of these reagents were considerably more potent than CD4-Fc, which had a geometric mean IC50 of 210 nM, compared with mean values of <10 nM for the E51 and 17b CD4HC reagents (Table 3). A direct comparison between the two bivalent CD4-containing proteins with no CD4i components [CD4HC-(GS7)-IgGanti-gD and CD4-Fc] demonstrates that increasing the potential distance between the CD4 components yields a far more effective reagent (mean IC50 of 11 nM versus 210 nM). The fact that the bivalent CD4HC-(GS7)-IgGanti-gD has a potentially longer CD4 separation distance than the tetravalent CD4-IgG2 reagent (Fig. 1) may explain their similar neutralization potencies (mean IC50s of 11 nM and 14 nM) despite the greater number of CD4 moieties in CD4-IgG2. Similarly, a longer CD4 separation distance combined with the inclusion of CD4i components is likely to account for the increased neutralization potencies of CD4HC-(GS7)-IgG17b (mean IC50 of 2.9 nM) and CD4HC-(GS9)-IgGE51 (mean IC50 of 2.9 nM) compared with CD4-IgG2 (mean IC50 of 14 nM) (Table 3).
In order to test a multivalent CD4 architecture with a different geometry, we constructed a fusion of CD4 D1-D2 with the non-CD4i Ab 2G12, which recognizes a cluster of carbohydrates on gp120 via a rigid (Fab)2 created by three-dimensional domain swapping (6). Expression and purification of a CD4 fusion to the 2G12 heavy chain yielded monomeric and dimeric fractions similar to those observed with unmodified 2G12 IgG (Fig. 1) (37). The neutralization potencies of these reagents (Table 4) suggested that both the CD4 components and the antibody combining sites of these reagents contribute to neutralization since (i) CD4HC-(GS7)-IgG2G12 is active on a clade C stain (Du172.17) that is not neutralized by unmodified 2G12 monomers or dimers and since (ii) CD4HC-(GS7)-IgG2G12 neutralizes the clade B TRO.11 strain 13-fold (2G12 monomer) and 150-fold (2G12 dimer) better than the pure CD4-reagent CD4-IgG2 (PRO 542). The second observation implies that either the 2G12 anticarbohydrate binding activity is functional in CD4HC-(GS7)-IgG2G12 and/or that the geometry of the CD4 components is more favorable than in the CD4-IgG2 reagent. The more similar neutralization potencies observed between CD4HC-(GS7)-IgG2G12 and CD4-IgG2 on other strains suggest that differences in the CD4 geometries are not likely to explain the TRO.11 results, suggesting that carbohydrate recognition contributes to the neutralization potencies of the CD4HC-(GS7)-IgG2G12 reagents against 2G12-sensitive viral strains.
To confirm that antigen binding by 2G12 contributes to the activity of CD4HC-(GS7)-IgG2G12, we conducted additional neutralization assays in the presence of fructose, which has been reported to inhibit 2G12 binding to its carbohydrate epitope on gp120 (6). In order to observe a potential inhibitory effect of fructose addition, we chose clade B strains that were sensitive to 2G12. In the presence of 3% fructose, we observed 3- to 11-fold weaker neutralization of 2G12 IgG and CD4HC-(GS7)-IgG2G12, whereas neutralization by CD4HC-(GS7)-IgGE51, a CD4-CD4i reagent, was unaffected by including fructose (0.7- to 1.2-fold differences) (Table 5).
DISCUSSION
CD4-CD4i reagents are designed to allow both the CD4 and CD4i components to bind gp120 simultaneously, in which case neutralization of HIV-1 should occur at a lower concentration because the effective affinity of this type of bivalent reagent would be increased compared to its components. In other words, if binding of the CD4 portion of the reagent to gp120 is in equilibrium, then binding of the CD4i portion to gp120 would reduce the rate at which the reagent dissociates from the virus by either stabilizing the CD4-bound conformation or by permitting rebinding of the CD4 component.
Our comparative study of the neutralization potencies of various CD4-CD4i reagents allows an estimate of the enhanced potencies of these reagents resulting from potential bivalent binding. Analysis of the neutralization activity of CD4-scFvE51 compared to its CD4 and scFvE51 components suggests about a twofold enhanced potency for the combined reagent (see Materials and Methods), similar to the increased potency of CD4HC-(GS7)-IgGE51 compared to CD4HC-(GS7)-IgGanti-gD (Table 3). The relatively modest enhancement we see for CD4-CD4i reagents compared to other bivalent reagents (neutralization potencies of IgG versus Fab can be up to 1,000-fold different for some viruses) (30) may be due to gp120 conformational changes associated with CD4 binding. First, the CD4i Ab component is unlikely to bind unless CD4 is already bound. Second, dissociation of CD4 may be quickly followed by a conformational change in gp120 that eliminates the CD4i binding site and thereby causes dissociation of the whole CD4-CD4i reagent. In theory, the CD4-CD4i Ab fusion protein can reduce the rate at which the reagent dissociates from the virus by either stabilizing the CD4-bound conformation or by permitting rebinding of the CD4 component. These mechanisms may be limited by the kinetics of gp120 conformational change.
Although it is not yet clear if it is possible to enhance the synergy between the components of a CD4-CD4i fusion protein, we were able to construct reagents with increased neutralization potencies compared with CD4-scFv reagents. By comparing the expression levels and efficacies of different architectures of CD4-CD4i Ab fusion proteins (Fig. 1; see also Table S1 in the supplemental material) in neutralization of a broad range of primary HIV-1 strains, we found that including an IgG Fc region improved the neutralization potency of a CD4-CD4i reagent. In addition to dimerizing a CD4-CD4i reagent, addition of Fc would also be expected to improve a reagent's serum half-life due to rescue from degradation via binding to the neonatal Fc receptor, which serves as a protection receptor for IgGs and other Fc-containing proteins (26). After testing several architectures for CD4-IgG constructs, we found that reagents in which CD4 was linked to the heavy chain of a CD4i Ab [i.e., CD4HC-(linker)-IgG] showed favorable expression characteristics and a low nanomolar geometric mean IC50 across a broad range of primary HIV strains (Table 3). The polymeric reagent D1D2-Igαtp has also been reported to neutralize HIV strains very efficiently at low-nanomolar concentrations (2), and even without binding to gp120 or virus, this reagent exhibited a functionally irreversible association with CD16 on natural killer cells (14). Although D1D2-Igαtp should be a powerful reagent for elimination of virally infected cells, our monomeric reagents may have advantages for providing passive immunity (e.g., as smaller molecules there should be more complete distribution to potential sites of exposure).
We also compared our reagents to an existing set of broadly neutralizing Abs (Table 3). We were unable to fully compare our reagents with recently described highly potent broadly neutralizing Abs (35) because the new Abs were tested on a different panel of primary HIV-1 strains than used in our study; however, IC50s are presented for the common strains that were tested (Table 3). Three reagents, CD4HC-(GS7)-IgGE51, CD4HC-(GS9)-IgGE51, and CD4HC-(GS7)-IgG17b, neutralized a range of clade A, B, and C HIV-1 strains with comparable if not higher potencies than the broadly neutralizing Abs and CD4-IgG2. Of the three new CD4-CD4i reagents, we achieved the highest expression levels for CD4HC-(GS7)-IgGE51, suggesting that this reagent would be the best suited for applications involving expression by gene therapy.
In addition to the CD4-CD4i fusion proteins, we tried fusing CD4 to 2G12, an Ab that binds to a constellation of carbohydrates on gp120 using a domain-swapped (Fab)2 unit (6). We previously showed that 2G12 forms stable dimers that exhibit a 50- to 80-fold increase in neutralization potency compared with monomeric 2G12 against a collection of clade B HIV-1 strains, but that 2G12 dimers, like their monomeric counterparts, were unable to neutralize clade C HIV-1 strains (37). We found that the CD4-2G12 fusion protein, similar to unmodified 2G12, was expressed as a mixture of monomers and dimers, with two and four antibody combining sites, respectively (Fig. 1). The CD4HC-(GS7)-IgG2G12 proteins showed nearly comparable (in the case of the dimer) or enhanced (in the case of the monomer) neutralization potencies against clade B HIV-1 strains, but most promisingly, fusion of CD4 to 2G12 conferred the ability to neutralize some clade C HIV-1 strains (Table 4).
The IgG-based CD4-CD4i and CD4-2G12 reagents reported here represent a new class of anti-HIV-1 protein therapeutics of potential use for passive immunization or gene therapy efforts. The addition of CD4 conferred a broader range of neutralization activity and/or facilitated binding of a CD4i Ab. Comparison of CD4-CD4i reagents with CD4-only reagents suggested that the addition of a CD4i Ab enhanced the potency of CD4 reagents two- to threefold. The moderate degree of synergy may be related to a nonoptimal geometry between the CD4 and CD4i Ab components; thus, alternative linkers may be more efficient for promoting simultaneous binding. Also, since we sometimes observed partial degradation of the CD4-CD4i reagents on storage (A. P. West, Jr., R. P. Galimidi, and P. J. Bjorkman, unpublished observations), improvement of the linker design may yield more stable and potent reagents.
The maximal potential potency of CD4-CD4i reagents may be inherently limited by kinetic factors or other properties; for example, in the trimeric envelope spike, engagement of multiple CD4 molecules might be required to elicit the conformational change that exposes the coreceptor binding sites. Furthermore, binding studies involving trimeric Env proteins and CD4 and/or a CD4i Ab have revealed unexpected stoichiometries. For example, only a single CD4i Fab X5 bound to an HIV-1 Env trimer complexed with three CD4 molecules (9), apparently reflecting steric constraints on CD4i Ab binding to Env trimers. Recently, a specific mechanism of action for soluble CD4 and CD4-mimetic compounds was discovered; engagement with these reagents causes a short-lived activated state of Env trimers that is followed by irreversible decay into a nonfunctional form (15). A greater understanding of steric and other factors that limit the activity of CD4-CD4i reagents would facilitate design of bispecific reagents with increased potency relative to the individual components.
Supplementary Material
Acknowledgments
We thank the CAVD Neutralizing Antibody Core Laboratories for performing in vitro neutralization assays, the Caltech Protein and Peptide Microanalysis Facility for performing N-terminal sequencing, Noreen Tiangco for cloning the 21c genes, Maria Politzer for subcloning and DNA preparation, Marta Murphy for figure preparation, Edward Berger (National Institutes of Health) for the CD4-scFv17b gene, William Olson at Progenics Pharmaceuticals for providing CD4-IgG2, Dennis Burton (Scripps Research Institute) for the anti-gD and 2G12 genes, and James Robinson (Tulane University) for the 21c cell line.
This work was supported by a grant from the Bill and Melinda Gates Foundation through the Grand Challenges in Global Health Initiative.
Footnotes
Published ahead of print on 28 October 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Allaway, G. P., K. L. Davis-Bruno, G. A. Beaudry, E. B. Garcia, E. L. Wong, A. M. Ryder, K. W. Hasel, M. C. Gauduin, R. A. Koup, J. S. McDougal, et al. 1995. Expression and characterization of CD4-IgG2, a novel heterotetramer that neutralizes primary HIV type 1 isolates. AIDS Res. Hum. Retroviruses 11:533-539. [DOI] [PubMed] [Google Scholar]
- 2.Arthos, J., C. Cicala, T. D. Steenbeke, T. W. Chun, C. Dela Cruz, D. B. Hanback, P. Khazanie, D. Nam, P. Schuck, S. M. Selig, D. Van Ryk, M. A. Chaikin, and A. S. Fauci. 2002. Biochemical and biological characterization of a dodecameric CD4-Ig fusion protein: implications for therapeutic and vaccine strategies. J. Biol. Chem. 277:11456-11464. [DOI] [PubMed] [Google Scholar]
- 3.Barbas, C. F., III, T. A. Collet, W. Amberg, P. Roben, J. M. Binley, D. Hoekstra, D. Cababa, T. M. Jones, R. A. Williamson, G. R. Pilkington, and et al. 1993. Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J. Mol. Biol. 230:812-823. [DOI] [PubMed] [Google Scholar]
- 4.Burioni, R., R. A. Williamson, P. P. Sanna, F. E. Bloom, and D. R. Burton. 1994. Recombinant human Fab to glycoprotein D neutralizes infectivity and prevents cell-to-cell transmission of herpes simplex viruses 1 and 2 in vitro. Proc. Natl. Acad. Sci. U. S. A. 91:355-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burton, D. R., R. L. Stanfield, and I. A. Wilson. 2005. Antibody vs. HIV in a clash of evolutionary titans. Proc. Natl. Acad. Sci. U. S. A. 102:14943-14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calarese, D. A., C. N. Scanlan, M. B. Zwick, S. Deechongkit, Y. Mimura, R. Kunert, P. Zhu, M. R. Wormald, R. L. Stanfield, K. H. Roux, J. W. Kelly, P. M. Rudd, R. A. Dwek, H. Katinger, D. R. Burton, and I. A. Wilson. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300:2065-2071. [DOI] [PubMed] [Google Scholar]
- 7.Capon, D. J., S. M. Chamow, J. Mordenti, S. A. Marsters, T. Gregory, H. Mitsuya, R. A. Byrn, C. Lucas, F. M. Wurm, J. E. Groopman, et al. 1989. Designing CD4 immunoadhesins for AIDS therapy. Nature 337:525-531. [DOI] [PubMed] [Google Scholar]
- 8.Choe, H., W. Li, P. L. Wright, N. Vasilieva, M. Venturi, C. C. Huang, C. Grundner, T. Dorfman, M. B. Zwick, L. Wang, E. S. Rosenberg, P. D. Kwong, D. R. Burton, J. E. Robinson, J. G. Sodroski, and M. Farzan. 2003. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell 114:161-170. [DOI] [PubMed] [Google Scholar]
- 9.Crooks, E. T., P. L. Moore, D. Richman, J. Robinson, J. A. Crooks, M. Franti, N. Schulke, and J. M. Binley. 2005. Characterizing anti-HIV monoclonal antibodies and immune sera by defining the mechanism of neutralization. Hum. Antibodies 14:101-113. [PMC free article] [PubMed] [Google Scholar]
- 10.Decker, J. M., F. Bibollet-Ruche, X. Wei, S. Wang, D. N. Levy, W. Wang, E. Delaporte, M. Peeters, C. A. Derdeyn, S. Allen, E. Hunter, M. S. Saag, J. A. Hoxie, B. H. Hahn, P. D. Kwong, J. E. Robinson, and G. M. Shaw. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 201:1407-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dey, B., C. S. Del Castillo, and E. A. Berger. 2003. Neutralization of human immunodeficiency virus type 1 by sCD4-17b, a single-chain chimeric protein, based on sequential interaction of gp120 with CD4 and coreceptor. J. Virol. 77:2859-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durocher, Y., S. Perret, and A. Kamen. 2002. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 30:E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher, R. A., J. M. Bertonis, W. Meier, V. A. Johnson, D. S. Costopoulos, T. Liu, R. Tizard, B. D. Walker, M. S. Hirsch, R. T. Schooley, et al. 1988. HIV infection is blocked in vitro by recombinant soluble CD4. Nature 331:76-78. [DOI] [PubMed] [Google Scholar]
- 14.Gupta, N., J. Arthos, P. Khazanie, T. D. Steenbeke, N. M. Censoplano, E. A. Chung, C. C. Cruz, M. A. Chaikin, M. Daucher, S. Kottilil, D. Mavilio, P. Schuck, P. D. Sun, R. L. Rabin, S. Radaev, D. Van Ryk, C. Cicala, and A. S. Fauci. 2005. Targeted lysis of HIV-infected cells by natural killer cells armed and triggered by a recombinant immunoglobulin fusion protein: implications for immunotherapy. Virology 332:491-497. [DOI] [PubMed] [Google Scholar]
- 15.Haim, H., Z. Si, N. Madani, L. Wang, J. R. Courter, A. Princiotto, A. Kassa, M. DeGrace, K. McGee-Estrada, M. Mefford, D. Gabuzda, A. B. Smith III, and J. Sodroski. 2009. Soluble CD4 and CD4-mimetic compounds inhibit HIV-1 infection by induction of a short-lived activated state. PLoS Pathog. 5:e1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hussey, R. E., N. E. Richardson, M. Kowalski, N. R. Brown, H. C. Chang, R. F. Siliciano, T. Dorfman, B. Walker, J. Sodroski, and E. L. Reinherz. 1988. A soluble CD4 protein selectively inhibits HIV replication and syncytium formation. Nature 331:78-81. [DOI] [PubMed] [Google Scholar]
- 17.Jacobson, J. M., R. J. Israel, I. Lowy, N. A. Ostrow, L. S. Vassilatos, M. Barish, D. N. Tran, B. M. Sullivan, T. J. Ketas, T. J. O'Neill, K. A. Nagashima, W. Huang, C. J. Petropoulos, J. P. Moore, P. J. Maddon, and W. C. Olson. 2004. Treatment of advanced human immunodeficiency virus type 1 disease with the viral entry inhibitor PRO 542. Antimicrob. Agents Chemother. 48:423-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klein, J. S., P. N. Gnanapragasam, R. P. Galimidi, C. P. Foglesong, A. P. West, Jr., and P. J. Bjorkman. 2009. Examination of the contributions of size and avidity to the neutralization mechanisms of the anti-HIV antibodies b12 and 4E10. Proc. Natl. Acad. Sci. USA 106:7385-7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwong, P. D., M. L. Doyle, D. J. Casper, C. Cicala, S. A. Leavitt, S. Majeed, T. D. Steenbeke, M. Venturi, I. Chaiken, M. Fung, H. Katinger, P. W. Parren, J. Robinson, D. Van Ryk, L. Wang, D. R. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678-682. [DOI] [PubMed] [Google Scholar]
- 20.Labrijn, A. F., P. Poignard, A. Raja, M. B. Zwick, K. Delgado, M. Franti, J. Binley, V. Vivona, C. Grundner, C. C. Huang, M. Venturi, C. J. Petropoulos, T. Wrin, D. S. Dimitrov, J. Robinson, P. D. Kwong, R. T. Wyatt, J. Sodroski, and D. R. Burton. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 77:10557-10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, M., F. Gao, J. R. Mascola, L. Stamatatos, V. R. Polonis, M. Koutsoukos, G. Voss, P. Goepfert, P. Gilbert, K. M. Greene, M. Bilska, D. L. Kothe, J. F. Salazar-Gonzalez, X. Wei, J. M. Decker, B. H. Hahn, and D. C. Montefiori. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108-10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, M., J. F. Salazar-Gonzalez, C. A. Derdeyn, L. Morris, C. Williamson, J. E. Robinson, J. M. Decker, Y. Li, M. G. Salazar, V. R. Polonis, K. Mlisana, S. A. Karim, K. Hong, K. M. Greene, M. Bilska, J. Zhou, S. Allen, E. Chomba, J. Mulenga, C. Vwalika, F. Gao, M. Zhang, B. T. Korber, E. Hunter, B. H. Hahn, and D. C. Montefiori. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J. Virol. 80:11776-11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montefiori, D. C. 2005. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr. Protoc. Immunol. doi: 10.1002/0471142735.im1211s46. [DOI] [PubMed]
- 24.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Ruker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 26.Roopenian, D. C., and S. Akilesh. 2007. FcRn: the neonatal Fc receptor comes of age. Nat. Rev. Immunol. 7:715-725. [DOI] [PubMed] [Google Scholar]
- 27.Salzwedel, K., E. D. Smith, B. Dey, and E. A. Berger. 2000. Sequential CD4-coreceptor interactions in human immunodeficiency virus type 1 Env function: soluble CD4 activates Env for coreceptor-dependent fusion and reveals blocking activities of antibodies against cryptic conserved epitopes on gp120. J. Virol. 74:326-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saphire, E. O., P. W. Parren, R. Pantophlet, M. B. Zwick, G. M. Morris, P. M. Rudd, R. A. Dwek, R. L. Stanfield, D. R. Burton, and I. A. Wilson. 2001. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science 293:1155-1159. [DOI] [PubMed] [Google Scholar]
- 29.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. Ollmann Saphire, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schofield, D. J., J. R. Stephenson, and N. J. Dimmock. 1997. Variations in the neutralizing and haemagglutination-inhibiting activities of five influenza A virus-specific IgGs and their antibody fragments. J. Gen. Virol. 78:2431-2439. [DOI] [PubMed] [Google Scholar]
- 31.Sheskin, D. 2004. Handbook of parametric and nonparametric statistical procedures, 3rd ed. Chapman and Hall/CRC, Boca Raton, FL.
- 32.Srivastava, I. K., J. B. Ulmer, and S. W. Barnett. 2005. Role of neutralizing antibodies in protective immunity against HIV. Hum. Vaccin. 1:45-60. [DOI] [PubMed] [Google Scholar]
- 33.Sullivan, N., Y. Sun, Q. Sattentau, M. Thali, D. Wu, G. Denisova, J. Gershoni, J. Robinson, J. Moore, and J. Sodroski. 1998. CD4-Induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J. Virol. 72:4694-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trkola, A., H. Kuster, P. Rusert, B. Joos, M. Fischer, C. Leemann, A. Manrique, M. Huber, M. Rehr, A. Oxenius, R. Weber, G. Stiegler, B. Vcelar, H. Katinger, L. Aceto, and H. F. Gunthard. 2005. Delay of HIV-1 rebound after cessation of antiretroviral therapy through passive transfer of human neutralizing antibodies. Nat. Med. 11:615-622. [DOI] [PubMed] [Google Scholar]
- 35.Walker, L. M., S. K. Phogat, P. Y. Chan-Hui, D. Wagner, P. Phung, J. L. Goss, T. Wrin, M. D. Simek, S. Fling, J. L. Mitcham, J. K. Lehrman, F. H. Priddy, O. A. Olsen, S. M. Frey, P. W. Hammond, G. Miiro, J. Serwanga, A. Pozniak, D. McPhee, O. Manigart, L. Mwananyanda, E. Karita, A. Inwoley, W. Jaoko, J. Dehovitz, L. G. Bekker, P. Pitisuttithum, R. Paris, S. Allen, S. Kaminsky, T. Zamb, M. Moyle, W. C. Koff, P. Poignard, and D. R. Burton. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 37.West, A. P., Jr., R. P. Galimidi, C. P. Foglesong, P. N. Gnanapragasam, K. E. Huey-Tubman, J. S. Klein, M. D. Suzuki, N. E. Tiangco, J. Vielmetter, and P. J. Bjorkman. 2009. Design and expression of a dimeric form of human immunodeficiency virus type 1 antibody 2G12 with increased neutralization potency. J. Virol. 83:98-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiang, S. H., N. Doka, R. K. Choudhary, J. Sodroski, and J. E. Robinson. 2002. Characterization of CD4-induced epitopes on the HIV type 1 gp120 envelope glycoprotein recognized by neutralizing human monoclonal antibodies. AIDS Res. Hum. Retroviruses 18:1207-1217. [DOI] [PubMed] [Google Scholar]
- 39.Xiang, S. H., L. Wang, M. Abreu, C. C. Huang, P. D. Kwong, E. Rosenberg, J. E. Robinson, and J. Sodroski. 2003. Epitope mapping and characterization of a novel CD4-induced human monoclonal antibody capable of neutralizing primary HIV-1 strains. Virology 315:124-134. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, M. Y., Y. Shu, D. Rudolph, P. Prabakaran, A. F. Labrijn, M. B. Zwick, R. B. Lal, and D. S. Dimitrov. 2004. Improved breadth and potency of an HIV-1-neutralizing human single-chain antibody by random mutagenesis and sequential antigen panning. J. Mol. Biol. 335:209-219. [DOI] [PubMed] [Google Scholar]
- 41.Zolla-Pazner, S. 2004. Identifying epitopes of HIV-1 that induce protective antibodies. Nat. Rev. Immunol. 4:199-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.