Abstract
Cytosolic chaperones are a diverse group of ubiquitous proteins that play central roles in multiple processes within the cell, including protein translation, folding, intracellular trafficking, and quality control. These cellular proteins have also been implicated in the replication of numerous viruses, although the full extent of their involvement in viral replication is unknown. We have previously shown that the heat shock protein 40 (hsp40) chaperone encoded by the yeast YDJ1 gene facilitates RNA replication of flock house virus (FHV), a well-studied and versatile positive-sense RNA model virus. To further explore the roles of chaperones in FHV replication, we examined a panel of 30 yeast strains with single deletions of cytosolic proteins that have known or hypothesized chaperone activity. We found that the majority of cytosolic chaperone deletions had no impact on FHV RNA accumulation, with the notable exception of J-domain-containing hsp40 chaperones, where deletion of APJ1 reduced FHV RNA accumulation by 60%, while deletion of ZUO1, JJJ1, or JJJ2 markedly increased FHV RNA accumulation, by 4- to 40-fold. Further studies using cross complementation and double-deletion strains revealed that the contrasting effects of J domain proteins were reproduced by altering expression of the major cytosolic hsp70s encoded by the SSA and SSB families and were mediated in part by divergent effects on FHV RNA polymerase synthesis. These results identify hsp70 chaperones as critical regulators of FHV RNA replication and indicate that cellular chaperones can have both positive and negative regulatory effects on virus replication.
The compact genomes of viruses relative to those of other infectious agents restrict their ability to encode all proteins required to complete their replication cycles. To circumvent this limitation, viruses often utilize cellular factors or processes to complete essential steps in replication. One group of cellular proteins frequently targeted by viruses are cellular chaperones, which include a diverse set of heat shock proteins (hsps) that normally facilitate cellular protein translation, folding, trafficking, and degradation (18, 64). The connection between viruses and cellular chaperones was originally identified in bacteria, where the Escherichia coli hsp40 and hsp70 homologues, encoded by dnaJ and dnaK, respectively, were identified as bacterial genes essential for bacteriophage λ DNA replication (62). Research over the past 30 years has further revealed the importance of cellular chaperones in viral replication, such that the list of virus-hsp connections is now quite extensive and includes viruses from numerous families with diverse genome structures (4, 6, 7, 16, 19, 20, 23, 25, 40, 41, 44, 51, 54, 60). These studies have demonstrated the importance of cellular chaperones in multiple steps of the viral life cycle, including entry, viral protein translation, genome replication, encapsidation, and virion release. However, the list of virus-hsp connections is likely incomplete. Further studies to explore this particular host-pathogen interaction will shed light on virus replication mechanisms and pathogenesis, and potentially highlight targets for novel antiviral agents.
To study the role of cellular chaperones in the genome replication of positive-sense RNA viruses, we use flock house virus (FHV), a natural insect pathogen and well-studied member of the Nodaviridae family. The FHV life cycle shares many common features with other positive-sense RNA viruses, including the membrane-specific targeting and assembly of functional RNA replication complexes (37, 38), the exploitation of various cellular processes and host factors for viral replication (5, 23, 60), and the induction of large-scale membrane rearrangements (24, 28, 38, 39). FHV virions contain a copackaged bipartite genome consisting of RNA1 (3.1 kb) and RNA2 (1.4 kb), which encode protein A, the viral RNA-dependent RNA polymerase, and the structural capsid protein precursor, respectively (1). During active genome replication, FHV produces a subgenomic RNA3 (0.4 kb), which encodes the RNA interference inhibitor protein B2 (12, 29, 32). These viral characteristics make FHV an excellent model system to study many aspects of positive-sense RNA virus biology.
In addition to the benefits of a simple genome, FHV is able to establish robust RNA replication in a wide variety of genetically tractable eukaryotic hosts, including Drosophila melanogaster (38), Caenorhabditis elegans (32), and Saccharomyces cerevisiae (46). The budding yeast S. cerevisiae has been an exceptionally useful model host to study the mechanisms of viral RNA replication complex assembly and function with FHV (31, 37, 39, 45, 53, 55, 56, 60) as well as other positive-sense RNA viruses (11). The facile genetics of S. cerevisiae, along with the vast array of well-defined cellular and molecular tools and techniques, make it an ideal eukaryotic host for the identification of cellular factors required for positive-sense RNA virus replication. Furthermore, readily available yeast libraries with deletions and regulated expression of individual proteins have led to the completion of several high-throughput screens to provide a global survey of host factors that impact virus replication (26, 42, 52). An alternative approach with these yeast libraries that reduces the inherently high false-negative rates associated with high-throughput screens is to focus on a select set of host genes associated with a particular cellular pathway, process, or location previously implicated in virus replication.
We have utilized such a targeted approach and focused on examining the impact of cytosolic chaperones on FHV RNA replication. Previously, we have shown that the cellular chaperone hsp90 facilitates protein A synthesis in Drosophila cells (5, 23), and the hsp40 encoded by the yeast YDJ1 gene facilitates FHV RNA replication in yeast, in part through effects on both protein A accumulation and function (60). In this report, we further extend these observations by examining FHV RNA accumulation in a panel of yeast strains with deletions of known or hypothesized cytosolic chaperones. We demonstrate that cytosolic chaperones can have either suppressive or enhancing effects on FHV RNA accumulation. In particular, related hsp70 members encoded by the SSA and SSB yeast chaperone families have marked and dramatically divergent effects on both genomic and subgenomic RNA accumulation and viral polymerase synthesis. These results highlight the complexities of the host-pathogen interactions that influence positive-sense RNA virus replication and identify the hsp70 family of cytosolic chaperones as key regulators of FHV replication.
MATERIALS AND METHODS
Yeast strains, transformation, and culture conditions.
The parental wild-type diploid S. cerevisiae strain BY4743 (MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/+ met15Δ0/+ ura3Δ0/ura3Δ0) and corresponding mutant strains with homozygous deletions of individual cytosolic chaperones (Table 1) were purchased from the American Type Culture Collection (Manassas, VA). Genotypes of BY4743-based deletion strains were verified by PCR. The Δssa1 Δssa2 strain MW123 (MATα his3 leu2 lys2 Δtrp2 ura3 ssa1::HIS3 ssa2::LEU2) and corresponding wild-type strain DS10 have been described previously (61). The Δssz1 strain IDA2 (MATα ura3 leu2 his3 trp1 ade2 ssz1::LEU2), the Δssb1 Δssb2 strain IDA56A (MATα ura3 leu2 his3 trp1 ade2 ssb1::kanR ssb2::HIS3), and the corresponding wild-type strain MH272-3f have also been described previously (14). Yeast were transformed and cultured as previously described (60).
TABLE 1.
Analysis of cellular cytosolic chaperone impact on FHV RNA replication in S. cerevisiae
| Family | Chaperone genea | Cellular functionb | Growth (fold change in doubling time)c | Plus-strand RNA3 accumulation (fold change in level)d |
|---|---|---|---|---|
| hsp90 | HSC82 | Specialized folding | 1.1 ± 0.0 | 0.8 ± 0.3 |
| HSP82 | Specialized folding | 1.1 ± 0.0 | 0.8 ± 0.3 | |
| hsp70 | SSA1 | Protein folding/sorting | 1.1 ± 0.0 | 2.5 ± 0.8 |
| SSA2 | Protein folding/sorting | 1.1 ± 0.0 | 1.5 ± 0.4 | |
| SSA3 | Protein folding/sorting | 1.0 ± 0.0 | 1.6 ± 0.3 | |
| SSA4 | Protein folding/sorting | 1.0 ± 0.0 | 1.3 ± 0.1 | |
| SSB1 | Translation and folding | 1.1 ± 0.1 | 1.3 ± 0.2 | |
| SSB2 | Translation and folding | 1.3 ± 0.1 | 1.9 ± 0.3 | |
| SSE1 | Protein folding | 1.4 ± 0.3 | 4.1 ± 1.6 | |
| SSE2 | Protein folding | 1.0 ± 0.0 | 1.4 ± 0.5 | |
| SSZ1 | Translation | 1.8 ± 0.2 | 34.3 ± 10.9 | |
| hsp40 | APJ1 | Unknown | 1.2 ± 0.0 | 0.4 ± 0.1 |
| (JDP) | DJP1 | Peroxisome biogenesis | 1.1 ± 0.0 | 1.3 ± 0.3 |
| XDJ1 | Protein folding | 1.0 ± 0.0 | 0.9 ± 0.3 | |
| YDJ1e | Protein folding/sorting | |||
| JJJ1 | Ribosome biogenesis | 2.4 ± 0.3 | 7.9 ± 3.1 | |
| JJJ2 | Unknown | 1.0 ± 0.1 | 3.5 ± 0.7 | |
| JJJ3 | Unknown | 1.2 ± 0.1 | 1.2 ± 0.3 | |
| SIS1f | Protein folding | ND | ND | |
| SWA2 | Vesicle transport | 1.2 ± 0.1 | 2.0 ± 0.6 | |
| ZUO1 | Translation | 1.9 ± 0.2 | 44.1 ± 8.9 | |
| NAC | BTT1 | Protein folding | 1.1 ± 0.1 | 1.6 ± 0.3 |
| EGD1 | Protein folding | 1.3 ± 0.0 | 0.7 ± 0.2 | |
| EGD2 | Protein folding | 1.3 ± 0.0 | 1.1 ± 0.2 | |
| Others | CPR1 | Protein metabolism | 0.9 ± 0.0 | 2.9 ± 1.6 |
| CPR6 | Protein folding | 1.0 ± 0.0 | 1.7 ± 0.1 | |
| CPR7 | Protein folding | 1.2 ± 0.1 | 9.6 ± 3.5 | |
| GIM4 | Tubulin assembly | 1.1 ± 0.0 | 1.0 ± 0.3 | |
| GIM5 | Tubulin assembly | 1.9 ± 0.1 | 8.5 ± 4.0 | |
| HSP26 | Protein folding | 1.0 ± 0.0 | 1.1 ± 0.2 | |
| HSP42 | Cytoskeletal organization | 1.1 ± 0.0 | 0.9 ± 0.2 | |
| HSP104 | Protein folding | 1.0 ± 0.0 | 1.2 ± 0.4 | |
| SBA1e | Specialized folding | |||
| STI1e | Specialized folding |
Diploid yeast strains with homozygous deletions of the listed cellular chaperone genes were transformed with pF1 and assessed for growth and FHV RNA replication after galactose induction. We examined yeast mutants in groups of 8 to 10 individual deletion strains and included wild-type BY4743 yeast with each set. Results are the composite of two or three biological replicates for each strain.
Cellular functions indicate gene ontology annotations (biological processes) obtained from the Saccharomyces Genome Database (http://www.yeastgenome.org).
Compared to wild-type BY4743 yeast. Growth differences with P values of <0.05 are indicated in bold italics.
Compared to wild-type BY4743 yeast, determined by strand-specific Northern blotting of total RNA recovered from equivalent numbers of cells. We quantitated viral RNA accumulation by densitometry and normalized values to rRNA levels determined by parallel ethidium bromide staining. Accumulation differences with P values of <0.05 are indicated in bold italics.
Effects of chaperones encoded by YDJ1, SBA1, and STI1 on FHV RNA replication have been reported previously (60).
Nonviable homozygous deletion. ND, not determined.
Plasmids.
Yeast plasmids pF1, pF1fs, pFA-C/HA, and pGAL-LacZ/HA have been described previously (60). Complementation plasmids encoding yeast J domain proteins (JDPs) were generated by moving open reading frames from a previously described panel of JDP expression plasmids (50) into a centromeric pRS416-based yeast shuttle vector that contains a constitutive GPD promoter. Yeast overexpression plasmids pSSA1, pZUO1, and pSSZ1 have been described previously (13, 61).
Antibodies and reagents.
Rabbit polyclonal antibodies against yeast Ssb1p and Ssz1p have been described previously (13, 14). Rabbit polyclonal antibodies against the hemagglutinin (HA) epitope tag were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and mouse monoclonal antibodies against 3-phosphoglycerate kinase (Pgk1p) were purchased from Molecular Probes (Eugene, OR). All secondary antibodies for immunoblotting were purchased from Jackson Immunoresearch (West Grove, PA).
Immunoblot and Northern blot analysis.
Total protein and RNA were isolated from equivalent numbers of yeast cells by post-alkaline extraction (27) and hot acidic phenol (60), respectively, and samples were stored at −20°C or −80°C until analysis. Protein and RNA samples were analyzed as previously described, by immunoblotting and Northern blotting, respectively (60). Digital images were obtained with an Alpha Innotech Fluorchem 8900 instrument and then quantified with AlphaEase FC software or with a Typhoon phosphorimager and then quantified with Image Quant Total lab software. Final images were prepared with Adobe Photoshop software. All contrast adjustments to the final images were done prior to cropping.
Statistics.
A two-tailed t test assuming equal variances was used for all statistical analyses, and a P value of <0.05 was considered statistically significant. Log10 transformation prior to statistical analyses was used for data sets that included fold change comparisons. Unless otherwise indicated, all quantitative data represent results from at least three independent experiments and are presented as the means ± standard errors of the means.
RESULTS
Targeted analysis of cellular cytosolic chaperones and their impact on FHV RNA accumulation in S. cerevisiae.
We have previously shown that deletion of the hsp40 chaperone encoded by YDJ1 suppresses FHV RNA replication in yeast (60). To further explore the impact of cellular chaperones on FHV RNA replication, we conducted a targeted analysis of RNA accumulation in yeast strains with homozygous deletions of genes encoding cytosolic proteins with a known or hypothesized chaperone function. We identified 34 yeast genes in the Saccharomyces Genome Database (www.yeastgenome.org) that encoded cytosolic chaperones (Table 1). We have previously described results with yeast strains containing a deletion of YDJ1, SBA1, or STI1 (60); in addition, the chaperone encoded by SIS1 is essential, and therefore a homozygous deletion strain is not available. We transformed the remaining 30 yeast strains with pF1, which encodes an authentic FHV RNA1 replicon under the control of the inducible GAL1 promoter (37, 60), induced the strains with galactose for 24 h, and measured cell growth by spectrophotometry and viral RNA accumulation by quantitative strand-specific Northern blotting. Due to the large number of strains, we have presented the results as quantitative increases or decreases in subgenomic RNA3 accumulation compared to that in wild-type BY4743 yeast and describe results centered on specific chaperone families (Table 1).
(i) hsp90 family.
The yeast hsp90 family consists of two functionally redundant genes, HSC82 and HSP82 (3). Deletion of either gene did not significantly alter FHV RNA3 accumulation. These results were consistent with the observation that deletion of STI1 or SBA1, two hsp90 chaperone complex component genes required for full activity (58), did not significantly alter FHV RNA replication in yeast (60).
(ii) hsp70 family.
The hsp70 family is a large family of chaperones in yeast, comprised of nine members, encoded by four SSA genes, two SSB genes, two SSE genes, and SSZ1. There is considerable functional redundancy in the hsp70 family, particularly in the SSA subfamily (61), and single deletions of SSA1, SSA2, SSA3, SSA4, SSB1, SSB2, SSE1, and SSE2 did not significantly alter FHV RNA3 accumulation from wild-type levels. However, deletion of SSZ1, which encodes an atypical hsp70 involved in translational fidelity as part of the ribosome-associated complex (RAC) (14), resulted in a 30- to 40-fold increase in FHV RNA3 accumulation.
(iii) hsp40 family (JDPs).
The hsp40 family members examined in this report were JDPs, so named due to the presence of a motif present in the E. coli hsp40 protein encoded by dnaJ (50, 57). Often referred to as cochaperones, hsp40 JDPs associate with partner hsp70s and influence chaperone function, in part by stimulating hsp70 ATPase activity. We have previously shown that deletion of YDJ1, the most abundant JDP gene in the cell (50), suppresses FHV RNA replication (60). Additionally, deletion of the JDP encoded by APJ1 also resulted in decreased FHV RNA3 accumulation, whereas deletions of XDJ1, DJP1, SWA2, and JJJ3 had no effect. In contrast, deletion of JJJ1, JJJ2, or ZUO1 markedly increased FHV RNA3 accumulation, although cell doubling times were also slightly increased in Δjjj1 and Δzuo1 yeast strains. The dramatic increase in FHV RNA3 accumulation with Δzuo1 yeast was very similar to that observed in Δssz1 yeast and was interesting in light of the observation that both Zuo1p and Ssz1p are RAC members involved in protein translation (14).
(iv) Nascent polypeptide-associated complex family.
In contrast to results with deletion of the translation-associated RAC members encoded by ZUO1 and SSZ1, deletions of EGD1, EGD2, and BTT1, which encode members of another chaperone complex associated with ribosomes called the nascent polypeptide-associated complex (48), had minimal impact on FHV RNA3 accumulation.
(v) Miscellaneous.
Deletion of the stress response chaperones encoded by HSP26, HSP42, and HSP104, which are involved in preventing aggregation or refolding proteins under stress conditions (17, 43, 49), had no significant effect on FHV RNA3 accumulation. Similarly, deletion of the immunophilin gene CPR1 or CPR6 had no significant effect. In contrast, deletion of CPR7, which encodes an hsp90 complex-associated immunophilin related to the CPR6 gene product, but with distinct functional characteristics (35), significantly increased FHV RNA3 accumulation. Finally, deletion of the chaperonin complex member encoded by GIM5, which is associated with microtubule biogenesis (15), also caused a significant increase in FHV RNA3 accumulation, whereas deletion of the closely related GIM4 gene had a minimal effect.
Hsp40 JDPs have contrasting effects on FHV RNA accumulation.
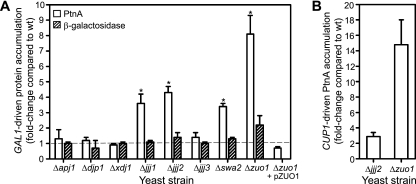
The majority of yeast strains with single deletions of known or hypothesized cytosolic chaperones had similar levels of FHV RNA3 accumulation compared to wild-type BY4743 yeast (Table 1). However, results with hsp40 JDP deletion strains were particularly interesting because they revealed dramatic decreases (YDJ1 and APJ1) and increases (JJJ1, JJJ2, and ZUO1) in FHV RNA3 accumulation, and therefore we further explored this subset of cytosolic chaperones. To exclude potential confounding effects of JDP function on GAL1 promoter activity (10), we transformed select deletion strains with a GAL1 promoter-driven lacZ construct (pGAL-LacZ/HA) and measured β-galactosidase expression after galactose induction (Fig. 1A). To also determine the effects of JDP deletions on FHV RNA polymerase expression independent of RNA replication, we examined parallel yeast strains transformed with pFA-C/HA, a GAL1 promoter-driven expression vector that produces a translation-optimized capped and polyadenylated mRNA encoding protein A that cannot function as a replication template (37, 60). Yeast strains with deletions of JJJ1, JJJ2, SWA2, and ZUO1 had four- to eightfold increases in FHV protein A accumulation after galactose induction compared to wild-type BY4743 yeast, and the increase in protein A accumulation in Δzuo1 yeast was reversed by complementation with a Zuo1p expression plasmid (Fig. 1A, open bars). The increases in FHV protein A accumulation in JDP deletion strains correlated with increases in RNA3 accumulation (Table 1), with the exception of Δswa2 yeast. However, RNA3 accumulation was increased twofold in Δswa2 yeast, although the increase was not statistically significant (Table 1). In contrast to protein A accumulation, we saw an increase in β-galactosidase accumulation of only twofold or less in yeast strains with a deletion of JJJ2 or ZUO1 (Fig. 1A, hatched bars). We also saw significant increases in FHV protein A accumulation in Δjjj2 and Δzuo1 yeast strains when we substituted the copper-inducible CUP1 promoter for the GAL1 promoter (Fig. 1B). These results suggested that differences in GAL1 promoter activity in JDP deletion strains were not primarily responsible for the differences in FHV RNA3 accumulation observed with these strains (Table 1). In addition, they suggested that the increases in FHV RNA3 accumulation in Δjjj1, Δjjj2, and Δzuo1 strains were due in part to enhanced viral polymerase accumulation.
FIG. 1.
Promoter activity in yeast strains with JDP deletions. (A) Yeast strains with individual deletions of the indicated JDPs were transformed with either pFA-C/HA or pGAL-LacZ/HA and induced for 24 h with galactose, and HA-tagged FHV protein A (PtnA) or β-galactosidase accumulation levels were determined by quantitative immunoblotting and normalized to Pgk1p loading controls. Results are expressed as changes relative to the BY4743 wild-type (wt) yeast level and are from two or three independent experiments. *, P < 0.05 compared to GAL1 promoter-driven β-galactosidase accumulation level. (B) Yeast strains were transformed with pFA-CUP-C/HA and induced for 24 h with 0.5 mM copper sulfate, and protein A levels were determined as described above.
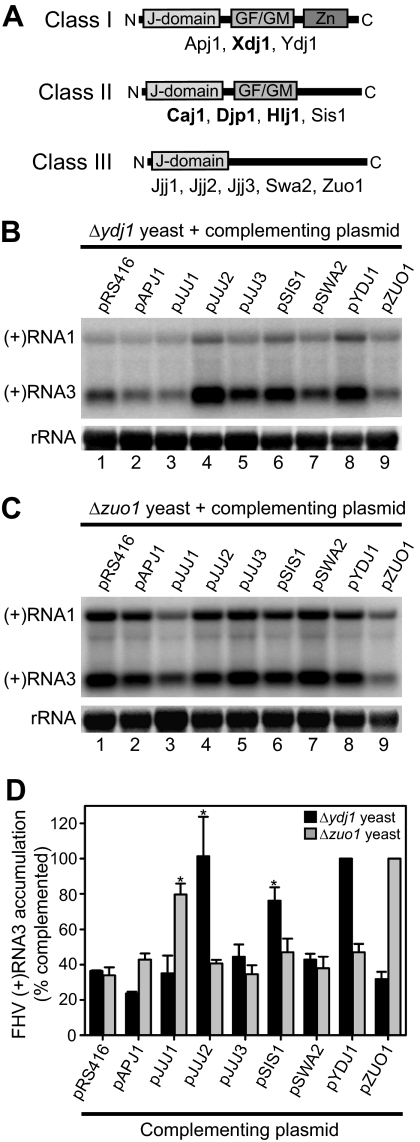
Yeast JDPs can be grouped into three classes based on the presence of common sequence and structural motifs, including the J domain motif, a glycine/phenylalanine- or glycine/methionine-rich motif, and a zinc finger motif (Fig. 2A). The shared overlapping activities of JDPs allow for cross-complementation analyses to potentially reveal informative functional characteristics (50, 57), an approach we used to further examine the impact of JDPs on FHV RNA accumulation. We generated a series of JDP overexpression plasmids by using a stable low-copy-number centromeric backbone and a constitutive GPD promoter, cotransformed either Δydj1 or Δzuo1 yeast with pF1 and individual JDP overexpression plasmids, induced the strains with galactose for 24 h, and examined FHV RNA accumulation by quantitative Northern blotting (Fig. 2B to D). For studies with Δydj1 yeast, induction was achieved with a galactose-raffinose combination, as this deletion strain has a severe growth defect when galactose is used as the sole carbon source (60). There were no differences in growth rate under induction conditions for either deletion strain cross complemented with the various JDPs (data not shown). We assayed for cross complementation that would either increase or decrease FHV RNA accumulation in Δydj1 (Fig. 2B) or Δzuo1 (Fig. 2C) yeast, respectively. For Δydj1 yeast, we observed significant increases in viral RNA3 accumulation with plasmids encoding Ydj1p, Jjj2p, and Sis1p (Fig. 2B and D). SIS1 encodes an essential protein that has been shown to regulate the SSA family of hsp70 chaperones and shares overlapping functions with Ydj1p (9, 22, 34), whereas the function of Jjj2p is unknown. For Δzuo1 yeast, we observed significant decreases in viral RNA3 accumulation with plasmids encoding Zuo1p and Jjj1p (Fig. 2C and D). JJJ1 encodes a protein that shares overlapping functions with Zuo1p, is involved in ribosomal biogenesis, and partially complements defects in cells lacking SSB family hsp70 chaperones (36). Since basal expression levels vary greatly between individual JDPs (50) and since antibodies against the majority of them were not available, we could not definitively exclude differences in overexpression levels as a potential explanation for the ability of specific JDPs to functionally complement defects in Δydj1 or Δzuo1 yeast. Nevertheless, the cross-complementation results suggested that the phenotypes of yeast strains lacking JDPs with respect to their ability to support FHV RNA accumulation were due, at least in part, to the activities of their partner hsp70s.
FIG. 2.
Cross complementation of Δydj1 and Δzuo1 yeast strains with JDP expression plasmids. (A) Schematics of JDP classes. CAJ1 and HLJ1 encode nonessential JDPs that localize to the nucleus (57) and endoplasmic reticulum (63), respectively, and therefore Δcaj1 and Δhlj1 yeast strains were not included in the initial cytosolic chaperone analysis (Table 1) but are included in the figure for completeness. JDPs whose overexpression was toxic to cells (50) are indicated in bold. GF/GM, glycine/phenylalanine- or glycine/methionine-rich regions; Zn, zinc finger. (B) FHV RNA replication in Δydj1 yeast cross complemented with JDP expression plasmids. Yeast cells were transformed with pF1 and either empty vector (lane 1) or the indicated JDP expression plasmid (lanes 2 to 9) and induced with galactose-raffinose for 24 h, and FHV RNA accumulation was analyzed by strand-specific Northern blotting. The positions of (+)RNA1 and (+)RNA3 are indicated on the left, and rRNA is shown as a loading control. FHV (+)RNA1 accumulation is the sum of both viral RNA replication and host RNA polymerase-directed transcription, whereas (+)RNA3 accumulation is dependent solely on viral polymerase activity. Thus, Δydj1 yeast has a more pronounced defect in (+)RNA3 accumulation (60). (C) FHV RNA replication in Δzuo1 yeast cross complemented with JDP cochaperones. Yeast cells were transformed and induced with galactose, and RNA replication was analyzed and presented as described above. Since both (+)RNA1 and (+)RNA3 are products of viral RNA replication, Δzuo1 yeast showed increases in both viral RNAs, in contrast to the preferential decrease in (+)RNA3 seen with Δydj1 yeast (see above). (D) Quantitative analysis of (+)RNA3 accumulation normalized to rRNA in cross-complemented Δydj1 or Δzuo1 yeast, where results are presented as the degree of complementation compared to pYDJ1- or pZUO1-transformed yeast, respectively. Inverse values were used for Δzuo1 yeast to facilitate direct comparisons with results from Δydj1 yeast, and in both cases successful cross complementation is indicated by increased values. *, P < 0.05 compared to control yeast transformed with pRS416.
Deletion of the yeast hsp70 chaperones encoded by SSA1 and SSA2 suppresses FHV RNA accumulation.
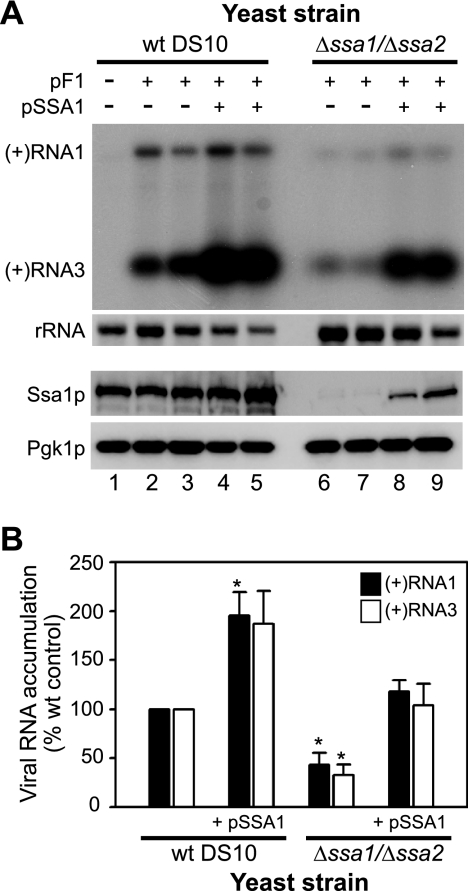
To directly examine the impact of hsp70 chaperones on FHV RNA accumulation in yeast and to eliminate the confounding effects of SSA family member redundancy (2, 61), we used the yeast strain MW123, which contains a codeletion of both the SSA1 and SSA2 genes (61). The SSA family has four members, all of which are >70% homologous to each other (2), but the SSA1 and SSA2 genes encode the most abundantly expressed hsp70s in yeast (61). We transformed the Δssa1 Δssa2 strain and corresponding wild-type DS10 yeast with pF1, induced the strains with galactose for 24 h, and measured FHV RNA accumulation by Northern blotting (Fig. 3). Deletion of both SSA1 and SSA2 resulted in a dramatic growth defect when galactose was used as the sole carbon source (data not shown), similar to the case with Δydj1 yeast (60). We therefore included raffinose during the induction period, as described above for cross-complementation studies with Δydj1 yeast. Under galactose-raffinose induction conditions, there was no difference in doubling time between wild-type DS10 and Δssa1 Δssa2 yeast (17.5 ± 1.9 and 17.3 ± 1.5 h, respectively). However, deletion of both SSA1 and SSA2 reduced FHV RNA1 and RNA3 accumulation by 60 to 70% (Fig. 3A, lanes 6 and 7, and Fig. 3B), which could be rescued by exogenous expression of Ssa1p (Fig. 3A, lanes 8 and 9, and Fig. 3B). Furthermore, overexpression of Ssa1p in wild-type DS10 yeast enhanced FHV RNA accumulation approximately twofold (Fig. 3A, lanes 4 and 5, and Fig. 3B). These results indicate that the hsp70 chaperones encoded by SSA1 and SSA2 are required for efficient FHV RNA accumulation in yeast.
FIG. 3.
Codeletion of SSA1 and SSA2 suppresses FHV RNA replication in S. cerevisiae. (A) Wild-type DS10 (lanes 1 to 5) and Δssa1 Δssa2 (lanes 6 to 9) yeast strains were transformed with pF1 (lanes 2, 3, 6, and 7) or pF1 plus pSSA1 (lanes 4, 5, 8, and 9) and induced with galactose-raffinose for 24 h, and FHV RNA accumulation was analyzed by Northern blotting as described in the legend to Fig. 2. Total protein was also isolated from an equivalent number of cells and immunoblotted for Ssa1p and Pgk1p. (B) Quantitative analysis of FHV (+)RNA1 and (+)RNA3 accumulation normalized to rRNA levels, where results are presented as percentages of wild-type DS10 yeast levels in the absence of Ssa1p overexpression. *, P < 0.05.
Deletion of the yeast hsp70 chaperones encoded by SSZ1 or SSB1/SSB2 enhances FHV RNA accumulation.
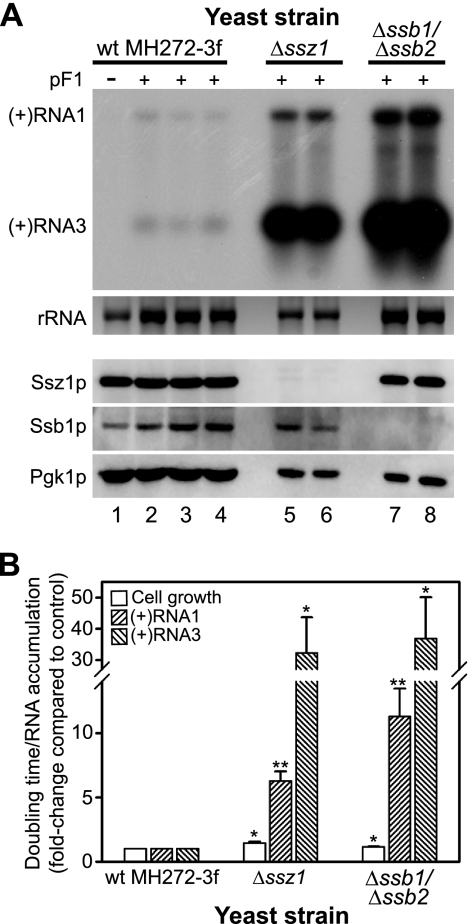
We initially showed that deletion of SSZ1, which encodes an atypical hsp70 chaperone (14), resulted in a 30- to 40-fold increase in FHV RNA3 accumulation in the BY4743 strain background (Table 1). Furthermore, similar to results with Δzuo1 yeast (Fig. 1A), Δssz1 yeast had a 14.5-fold ± 8.0-fold increase in protein A accumulation in cells transformed with pFA-C/HA 24 h after galactose induction. Both of these phenotypes were reversed to wild-type levels by expression of exogenous Ssz1p (data not shown). To further explore these observations and to determine whether the increased FHV RNA accumulation in Δzuo1 and Δssz1 yeast strains was due in part to their roles as RAC partners for SSB family members, we used yeast strain IDA56A, which contains a codeletion of both the SSB1 and SSB2 genes on the MH272-3f background (14). The hsp70s encoded by SSB1 and SSB2 are 99% identical (2). Since the single deletion library was on the BY4743 background (Table 1), we also examined yeast strain IDA2, which is a companion MH272-3f-based strain with a deletion of SSZ1 (14). We transformed yeast strains with pF1, induced them with galactose and raffinose for 24 h, as Δssb1 Δssb2 yeast had a moderate to severe growth defect when galactose was used as the sole carbon source (data not shown), and measured FHV RNA accumulation by Northern blotting (Fig. 4). The baseline FHV RNA accumulation in wild-type MH272-3f yeast (Fig. 4A, lanes 2 to 4) was lower than the levels seen previously in other yeast strains, which accentuated the dramatic increase in both RNA1 and RNA3 accumulation in Δssz1 (Fig. 4A, lanes 5 and 6) and Δssb1 Δssb2 (Fig. 4A, lanes 7 and 8) yeast strains. Both Δssz1 and Δssb1 Δssb2 yeast strains had a 20 to 40% increase in doubling time compared to wild-type MH272-3f yeast. The lower growth rates of these mutant yeast strains may have accounted for a portion, but likely not the majority, of the observed 5- to 10-fold increases in RNA1 and 30- to 40-fold increases in RNA3 accumulation (Fig. 4B). These results, in conjunction with the initial targeted chaperone analysis (Table 1) and cross-complementation studies (Fig. 2), indicated that deletion of the RAC component Zuo1p or Ssz1p or the partner hsp70 chaperones Ssb1p and Ssb2p markedly enhanced FHV RNA accumulation.
FIG. 4.
Deletion of SSZ1 or codeletion of SSB1 and SSB2 enhances FHV RNA replication in S. cerevisiae. (A) Wild-type MH272-3f (lanes 1 to 4) and Δssz1 (lanes 5 and 6) and Δssb1 Δssb2 (lanes 7 and 8) yeast strains were transformed with pF1 and induced with galactose-raffinose for 24 h, and FHV RNA accumulation was analyzed by Northern blotting as described in the legend to Fig. 2. Total protein was also isolated from an equivalent number of cells and immunoblotted for Ssz1p, Ssb1p, and Pgk1p. (B) Quantitative analysis of cell growth (doubling time) and rRNA-normalized FHV (+)RNA1 and (+)RNA3 accumulation levels, where results are presented as changes compared to levels in wild-type MH272-3f yeast. The y axis is broken to highlight the small but statistically significant increase in doubling times in both Δssz1 and Δssb1 Δssb2 yeast strains. *, P < 0.005; **, P < 0.0005.
Deletions of the yeast hsp70 chaperones encoded by SSA1/SSA2 and SSB1/SSB2 have divergent effects on FHV RNA polymerase synthesis.
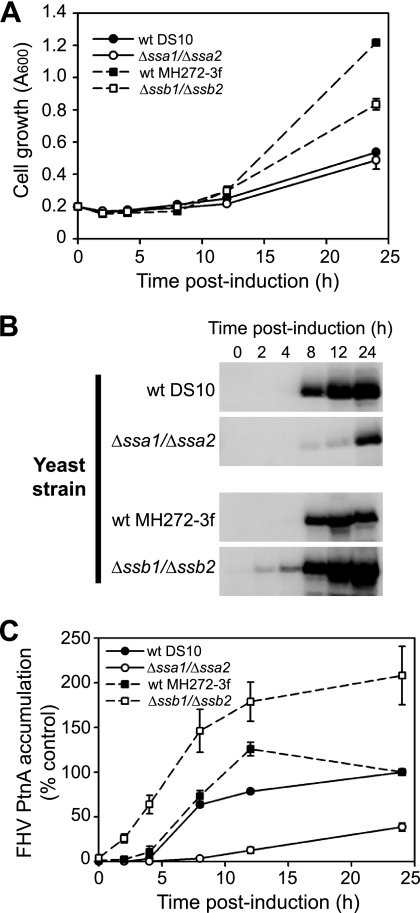
We found that deletion of JDPs encoded by JJJ1, JJJ2, SWA2, and ZUO1 increased FHV RNA polymerase accumulation even in the absence of RNA replication (Fig. 1) and that deletion of YDJ1 also resulted in reduced protein A accumulation (60). To determine whether the contrasting phenotypes of Δssa1 Δssa2 (Fig. 3) and Δssb1 Δssb2 (Fig. 4) yeast strains with respect to FHV RNA accumulation were due in part to reduced viral polymerase production, we transformed these mutant strains and the corresponding wild-type DS10 and MH272-3f yeast strains with pFA-C/HA, induced them with galactose-raffinose, and measured cell growth and the temporal appearance of full-length FHV RNA polymerase (Fig. 5). There was no difference in the growth rates of Δssa1 Δssa2 and wild-type DS10 yeast strains (Fig. 5A, circles), whereas Δssb1 Δssb2 yeast showed a slight decrease in cell growth between 12 and 24 h after induction compared to wild-type MH272-3f yeast (Fig. 5A, squares). In both wild-type strains, full-length FHV protein A was first readily visible 8 h after induction (Fig. 5B, first and third blots). Although protein A was also visible in Δssa1 Δssa2 yeast at this early time point, the level of accumulation was markedly reduced and remained at less than 40% of wild-type DS10 levels after 24 h (Fig. 5B, second blot, and Fig. 5C). In contrast, protein A accumulation was readily visible in Δssb1 Δssb2 yeast 2 h after induction and peaked at levels that were >2-fold above the wild-type MH272-3f yeast level (Fig. 5B, fourth blot, and Fig. 5C). The decrease in FHV protein A accumulation in Δssa1 Δssa2 yeast was not due to globally suppressed GAL1 promoter activity, as published studies have demonstrated equivalent GAL1 promoter-driven expression of some viral proteins in MW123 (Δssa1 Δssa2) and control wild-type yeast strains (59). Furthermore, the increase in protein A accumulation in Δssb1 Δssb2 yeast was not due to enhanced GAL1 promoter activity, as β-galactosidase expression in Δssb1 Δssb2 yeast transformed with pGAL-LacZ/HA (Fig. 1A) was similar to that in wild-type MH272-3f yeast at both 12 and 24 h after induction (86.7% ± 8.3% and 111.0% ± 13.4%, respectively; P > 0.25 for both). In addition, protein A accumulation in Δssb1 Δssb2 yeast was increased three- to fivefold compared to that in wild-type MH272-3f yeast when we used a CUP1 promoter-driven expression plasmid (Fig. 1B) (data not shown).
FIG. 5.
Codeletions of SSA1/SSA2 and SSB1/SSB2 have contrasting effects on FHV RNA polymerase accumulation in S. cerevisiae. (A) Cell growth after galactose-raffinose induction. (B) Temporal appearance of full-length protein A accumulation in wild-type DS10, Δssa1 Δssa2, wild-type MH272-3f, and Δssb1 Δssb2 yeast strains. Total protein was harvested from an equivalent number of cells and immunoblotted with HA-specific antibodies. (B) Quantitative analysis of protein A (PtnA) accumulation normalized to Pgk1p levels, where results are presented as percentages of the control wild-type DS10 or MH272-3f yeast level 24 h after induction.
Finally, we examined whether altered FHV protein A degradation in Δssa1 Δssa2 or Δssb1 Δssb2 yeast was responsible for the dramatic differences in polymerase accumulation. We transformed yeast strains with pFA-C/HA, induced them with galactose-raffinose for 24 h, subsequently incubated cells with 2% glucose to repress GAL1 promoter-driven transcription and with 100 μg/ml cycloheximide to inhibit translation, and harvested cells 6 h later for analysis by immunoblotting. There were no significant differences in the fraction of protein A recovered after 6 h in Δssa1 Δssa2 (94.4% ± 9.7%) or wild-type DS10 (88.6% ± 3.8%) yeast (P = 0.61). Similarly, protein A recovery in Δssb1 Δssb2 (97.0% ± 1.0%) and wild-type MH272-3f (87.0% ± 4.3%) yeast strains was not significantly different (P = 0.09). These results indicated that the contrasting effects of the yeast hsp70 chaperones encoded by SSA1/SSA2 and SSB1/SSB2 with respect to FHV RNA accumulation were due in part to divergent effects on viral polymerase synthesis.
DISCUSSION
In this report, we used a targeted genetic approach to examine the roles of cellular cytosolic chaperones in viral RNA replication. We identified several genes, including APJ1, CPR7, GIM5, JJJ1, JJJ2, SSZ1, and ZUO1, whose individual deletion significantly altered FHV RNA accumulation in S. cerevisiae. Interestingly, none of these genes were identified as encoding host factors that influenced viral RNA replication in the two published screens that examined the entire S. cerevisiae homozygous deletion library, which were conducted with replicon systems derived from the plant viruses brome mosaic virus (BMV) (26) and tomato bushy stunt virus (TBSV) (42). This suggests that either dissimilar sets of genes may be involved in FHV, BMV, and TSBV RNA replication or the rigorous selection criteria that are employed in high-throughput genomic functional screens, which are necessary to reduce the number of false-positive results, also inadvertently eliminate potentially interesting genes. Both explanations are probably correct to some degree, which emphasizes that parallel and complementary approaches with different viruses are likely to provide the maximal amount of information to identify and characterize those cellular processes that are crucial for efficient positive-sense RNA virus replication.
Despite the identification of different specific genes, the targeted analysis described in this report and published genome-wide analyses (21, 26, 42) had one similar and interesting characteristic: both approaches identified genes whose deletion either decreased or increased viral RNA replication. In the case of FHV and cytosolic chaperones, these opposite responses were particularly evident in the analysis of the hsp40 JDPs, where deletion of APJ1 or YDJ1 severely reduced, while deletion of JJJ1, JJJ2, or ZUO1 dramatically increased, FHV RNA accumulation. The JDP family of hsp40s are ubiquitous cellular proteins that function as coupling factors to stimulate the ATPase activity of a partner hsp70 during protein translation, folding, assembly, or transport (57). Although they share certain sequence and structural motifs, the family is quite diverse, such that there is some selectivity in the interactions of particular JDPs with unique hsp70s (57). However, enough functional overlap exists that hsp70-JDP interactions can be analyzed and characterized as networks rather than specific individual protein-protein interactions (50). The functional overlap of hsp70-JDP interactions facilitated interpretation of the FHV replication cross-complementation studies, where Sis1p overexpression rescued the viral RNA accumulation defect in Δydj1 yeast, while Jjj1p overexpression repressed the enhanced viral RNA accumulation in Δzuo1 yeast. Functional overlaps between Sis1p/Ydj1p and Jjj1p/Zuo1p have been demonstrated previously (9, 34, 36). However, there was not always a direct correlation between previously identified JDP functions and FHV RNA accumulation cross-complementation results. For example, although deletion of JJJ2 resulted in enhanced FHV RNA accumulation, overexpression of Jjj2p rescued the RNA accumulation defect in Δydj1 yeast but did not suppress the enhanced RNA accumulation in Δzuo1 yeast. In addition, APJ1 encodes a class I JDP similar to the YDJ1 gene product, Δapj1 yeast has a marked defect in FHV RNA accumulation similar to that in Δydj1 yeast (60), and Apj1p complements the growth defect of Δydj1 yeast when expressed at high enough levels (50), but Apj1p overexpression did not rescue FHV RNA accumulation in Δydj1 yeast. However, we cannot exclude the possibility that these contrasting cross-complementation results were due in part to differences in Apj1p expression levels, as we used low-copy-number centromeric plasmids for JDP expression studies, whereas published growth phenotype studies (50) employed high-copy-number 2μ plasmids. Nevertheless, these observations highlight the complexities of virus-chaperone interactions and suggest that further studies with FHV may help to define additional functional characteristics of cellular chaperone networks.
The observation that a Sis1p overexpression plasmid complemented the FHV RNA replication defect in Δydj1 yeast was particularly enlightening. Sis1p shares overlapping cellular functions with Ydj1p (9, 22, 34), and both JDPs have been shown to physically interact with SSA hsp70 family members in yeast (33), which suggested that SSA family hsp70s may be involved in FHV RNA replication. Indeed, codeletion of both SSA1 and SSA2 resulted in a significant reduction in FHV RNA accumulation. These results were consistent with a series of studies showing the essential nature of yeast SSA family chaperones in the replication of two closely related tombusviruses, TSBV (44, 59) and cucumber necrosis virus (51), which suggests possible parallels in cellular chaperone requirements between unrelated viruses. In support of this hypothesis, the cochaperone encoded by JJJ1 has also been shown to influence TBSV replication (30). However, the mechanisms whereby cellular chaperones facilitate viral RNA replication potentially differ between FHV and the tombusviruses. Overexpression of Jjj1p increases TBSV RNA accumulation (30), whereas deletion of JJJ1 increased FHV RNA accumulation. In addition, deletion of SSA1 and SSA2 primarily alters tombusvirus replicase protein localization and membrane insertion rather than synthesis (59), whereas codeletion of these two hsp70 chaperones suppressed FHV protein A accumulation without altering its posttranslational stability. This suggests that Ssa1p and Ssa2p facilitate translation of the FHV RNA polymerase, similar to the impact of hsp90 in Drosophila cells (5). Furthermore, Ssa1p and Ssa2p physically interact with the tombusvirus replication complex (51) and are required for its in vitro assembly (44), suggesting a direct mechanism of action. In contrast, we have not been able to demonstrate a physical interaction between yeast hsp70s and FHV protein A, despite repeated attempts under diverse conditions (S. Weeks and D. Miller, unpublished results), suggesting a potential indirect mechanism of action via an as yet unidentified intermediate cellular protein or proteins.
The JDP and hsp70 analyses also revealed previously unreported suppressive effects of cellular chaperones on FHV RNA replication. The demonstration of enhanced viral RNA accumulation in yeast strains with single deletions of SSZ1 and ZUO1, the ability of Jjj1p overexpression to restore baseline levels of viral RNA accumulation in Δzuo1 yeast, and the enhanced accumulation in Δssb1 Δssb2 yeast all point toward an important role of the RAC/SSB yeast chaperone complex in FHV RNA replication. This heterotrimeric complex consists of the JDP Zuo1p, the atypical hsp70 Ssz1p, and the more typical hsp70s Ssb1p and Ssb2p (14) and is responsible for maintaining translational fidelity in yeast (47). The mechanism whereby the RAC/SSB chaperone complex functions during FHV replication is unknown, but the marked increase in protein A accumulation associated with deletion of its components, in the absence of altered posttranslational stability, suggests an unidentified suppressor activity operative during viral polymerase synthesis. However, overexpression of Zuo1p or Ssz1p in wild-type BY4743 yeast did not reduce FHV RNA or protein A accumulation (W. Shield and D. Miller, unpublished results), which would suggest the absence of direct cellular chaperone-mediated suppression. It is tempting to speculate that the increase in FHV protein A accumulation in RAC/SSB-deficient yeast is related to the role of this cellular chaperone complex in maintaining translation fidelity, where FHV RNA polymerase synthesis may be less sensitive to the normal cellular translational quality control mechanisms. Experimental systems to examine translation fidelity in both intact yeast cells and cell extracts in vitro are available (47) and can be used to directly test this intriguing hypothesis.
In summary, we have demonstrated that hsp70 chaperones can have contrasting effects on FHV RNA accumulation, in part due to their differential impact on viral RNA polymerase synthesis. Interestingly, many of the yeast genes identified in genome-wide screens with BMV (26) and TBSV (21, 42) replicons that altered viral replication encode ribosomal proteins or cellular components directly involved in protein translation, which reinforces the concept that hijacking or actively manipulating the cellular translation apparatus is a common and likely crucial attribute of positive-sense RNA viruses (8).
Acknowledgments
We thank Kathryn Castorena, Allison Simms, Nina Nwachukwa, and Donna Gschwend for assistance and all members of the Miller lab for their helpful comments on the research and manuscript.
This work was funded by National Institutes of Health grant R01-AI062749 to D.J.M. and by grants SFB 746 and EXC 294 to S.R. S.A.W. was supported by training grant T32-AI007528 and by a departmental Willison fellowship.
Footnotes
Published ahead of print on 14 October 2009.
REFERENCES
- 1.Ball, L. A., and K. L. Johnson. 1998. Nodaviruses of insects, p. 225-267. In L. K. Miller and L. A. Ball (ed.), The insect viruses. Plenum Publishing Corporation, New York, NY.
- 2.Boorstein, W. R., T. Ziegelhoffer, and E. A. Craig. 1994. Molecular evolution of the HSP70 multigene family. J. Mol. Evol. 38:1-17. [DOI] [PubMed] [Google Scholar]
- 3.Borkovich, K. A., F. W. Farrelly, D. B. Finkelstein, J. Taulien, and S. Lindquist. 1989. Hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol. 9:3919-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burch, A. D., and S. K. Weller. 2005. Herpes simplex virus type 1 DNA polymerase requires the mammalian chaperone hsp90 for proper localization to the nucleus. J. Virol. 79:10740-10749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castorena, K. M., S. A. Weeks, K. A. Stapleford, A. M. Cadwallader, and D. J. Miller. 2007. A functional heat shock protein 90 chaperone is essential for efficient flock house virus RNA polymerase synthesis in Drosophila cells. J. Virol. 81:8412-8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chromy, L. R., J. M. Pipas, and R. L. Garcea. 2003. Chaperone-mediated in vitro assembly of polyomavirus capsids. Proc. Natl. Acad. Sci. USA 100:10477-10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Connor, J. H., M. O. McKenzie, G. D. Parks, and D. S. Lyles. 2007. Antiviral activity and RNA polymerase degradation following Hsp90 inhibition in a range of negative strand viruses. Virology 362:109-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dreher, T. W., and W. A. Miller. 2006. Translational control in positive strand RNA plant viruses. Virology 344:185-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan, C. Y., S. Lee, H. Y. Ren, and D. M. Cyr. 2004. Exchangeable chaperone modules contribute to specification of type I and type II Hsp40 cellular function. Mol. Biol. Cell 15:761-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Floer, M., G. O. Bryant, and M. Ptashne. 2008. Hsp90/70 chaperones are required for rapid nucleosome removal upon induction of the GAL genes of yeast. Proc. Natl. Acad. Sci. USA 105:2975-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galao, R. P., N. Scheller, I. Alves-Rodrigues, T. Breinig, A. Meyerhans, and J. Diez. 2007. Saccharomyces cerevisiae: a versatile eukaryotic system in virology. Microb. Cell Fact. 6:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galiana-Arnoux, D., C. Dostert, A. Schneemann, J. A. Hoffmann, and J. L. Imler. 2006. Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila. Nat. Immunol. 7:590-597. [DOI] [PubMed] [Google Scholar]
- 13.Gautschi, M., H. Lilie, U. Funfschilling, A. Mun, S. Ross, T. Lithgow, P. Rucknagel, and S. Rospert. 2001. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc. Natl. Acad. Sci. USA 98:3762-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gautschi, M., A. Mun, S. Ross, and S. Rospert. 2002. A functional chaperone triad on the yeast ribosome. Proc. Natl. Acad. Sci. USA 99:4209-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geissler, S., K. Siegers, and E. Schiebel. 1998. A novel protein complex promoting formation of functional α- and γ-tubulin. EMBO J. 17:952-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilmore, R., M. C. Coffey, and P. W. Lee. 1998. Active participation of Hsp90 in the biogenesis of the trimeric reovirus cell attachment protein σ1. J. Biol. Chem. 273:15227-15233. [DOI] [PubMed] [Google Scholar]
- 17.Haslbeck, M., N. Braun, T. Stromer, B. Richter, N. Model, S. Weinkauf, and J. Buchner. 2004. Hsp42 is the general small heat shock protein in the cytosol of Saccharomyces cerevisiae. EMBO J. 23:638-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hohfeld, J., D. M. Cyr, and C. Patterson. 2001. From the cradle to the grave: molecular chaperones that may choose between folding and degradation. EMBO Rep. 2:885-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu, J., and C. Seeger. 1996. Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc. Natl. Acad. Sci. USA 93:1060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung, J. J., C. S. Chung, and W. Chang. 2002. Molecular chaperone Hsp90 is important for vaccinia virus growth in cells. J. Virol. 76:1379-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang, Y., E. Serviene, J. Gal, T. Panavas, and P. D. Nagy. 2006. Identification of essential host factors affecting tombusvirus RNA replication based on the yeast Tet promoters Hughes Collection. J. Virol. 80:7394-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, J. L., and E. A. Craig. 2001. An essential role for the substrate-binding region of Hsp40s in Saccharomyces cerevisiae. J. Cell Biol. 152:851-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kampmueller, K. M., and D. J. Miller. 2005. The cellular chaperone heat shock protein 90 facilitates flock house virus RNA replication in Drosophila cells. J. Virol. 79:6827-6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kopek, B. G., G. Perkins, D. J. Miller, M. H. Ellisman, and P. Ahlquist. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 5:e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar, M., and D. Mitra. 2005. Heat shock protein 40 is necessary for human immunodeficiency virus-1 Nef-mediated enhancement of viral gene expression and replication. J. Biol. Chem. 280:40041-40050. [DOI] [PubMed] [Google Scholar]
- 26.Kushner, D. B., B. D. Lindenbach, V. Z. Grdzelishvili, A. O. Noueiry, S. M. Paul, and P. Ahlquist. 2003. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc. Natl. Acad. Sci. USA 100:15764-15769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kushnirov, V. V. 2000. Rapid and reliable protein extraction from yeast. Yeast 16:857-860. [DOI] [PubMed] [Google Scholar]
- 28.Lanman, J., J. Crum, T. J. Deerinck, G. M. Gaietta, A. Schneemann, G. E. Sosinsky, M. H. Ellisman, and J. E. Johnson. 2008. Visualizing flock house virus infection in Drosophila cells with correlated fluorescence and electron microscopy. J. Struct. Biol. 161:439-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, H., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 30.Li, Z., D. Barajas, T. Panavas, D. A. Herbst, and P. D. Nagy. 2008. Cdc34p ubiquitin-conjugating enzyme is a component of the tombusvirus replicase complex and ubiquitinates p33 replication protein. J. Virol. 82:6911-6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindenbach, B. D., J. Y. Sgro, and P. Ahlquist. 2002. Long-distance base pairing in flock house virus RNA1 regulates subgenomic RNA3 synthesis and RNA2 replication. J. Virol. 76:3905-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu, R., M. Maduro, F. Li, H. W. Li, G. Broitman-Maduro, W. X. Li, and S. W. Ding. 2005. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature 436:1040-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu, Z., and D. M. Cyr. 1998. Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J. Biol. Chem. 273:27824-27830. [DOI] [PubMed] [Google Scholar]
- 34.Luke, M. M., A. Sutton, and K. T. Arndt. 1991. Characterization of SIS1, a Saccharomyces cerevisiae homologue of bacterial DnaJ proteins. J. Cell Biol. 114:623-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayr, C., K. Richter, H. Lilie, and J. Buchner. 2000. Cpr6 and Cpr7, two closely related Hsp90-associated immunophilins from Saccharomyces cerevisiae, differ in their functional properties. J. Biol. Chem. 275:34140-34146. [DOI] [PubMed] [Google Scholar]
- 36.Meyer, A. E., N. J. Hung, P. Yang, A. W. Johnson, and E. A. Craig. 2007. The specialized cytosolic J-protein, Jjj1, functions in 60S ribosomal subunit biogenesis. Proc. Natl. Acad. Sci. USA 104:1558-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, D. J., and P. Ahlquist. 2002. Flock house virus RNA polymerase is a transmembrane protein with amino-terminal sequences sufficient for mitochondrial localization and membrane insertion. J. Virol. 76:9856-9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, D. J., M. D. Schwartz, and P. Ahlquist. 2001. Flock house virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J. Virol. 75:11664-11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, D. J., M. D. Schwartz, B. T. Dye, and P. Ahlquist. 2003. Engineered retargeting of viral RNA replication complexes to an alternative intracellular membrane. J. Virol. 77:12193-12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Momose, F., T. Naito, K. Yano, S. Sugimoto, Y. Morikawa, and K. Nagata. 2002. Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J. Biol. Chem. 277:45306-45314. [DOI] [PubMed] [Google Scholar]
- 41.Okamoto, T., Y. Nishimura, T. Ichimura, K. Suzuki, T. Miyamura, T. Suzuki, K. Moriishi, and Y. Matsuura. 2006. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J. 25:5015-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Panavas, T., E. Serviene, J. Brasher, and P. D. Nagy. 2005. Yeast genome-wide screen reveals dissimilar sets of host genes affecting replication of RNA viruses. Proc. Natl. Acad. Sci. USA 102:7326-7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parsell, D. A., A. S. Kowal, M. A. Singer, and S. Lindquist. 1994. Protein disaggregation mediated by heat-shock protein Hsp104. Nature 372:475-478. [DOI] [PubMed] [Google Scholar]
- 44.Pogany, J., J. Stork, Z. Li, and P. D. Nagy. 2008. In vitro assembly of the tomato bushy stunt virus replicase requires the host heat shock protein 70. Proc. Natl. Acad. Sci. USA 105:19956-19961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price, B. D., M. Roeder, and P. Ahlquist. 2000. DNA-directed expression of functional flock house virus RNA1 derivatives in Saccharomyces cerevisiae, heterologous gene expression, and selective effects on subgenomic mRNA synthesis. J. Virol. 74:11724-11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Price, B. D., R. R. Rueckert, and P. Ahlquist. 1996. Complete replication of an animal virus and maintenance of expression vectors derived from it in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:9465-9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rakwalska, M., and S. Rospert. 2004. The ribosome-bound chaperones RAC and Ssb1/2p are required for accurate translation in Saccharomyces cerevisiae. Mol. Cell. Biol. 24:9186-9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rospert, S., Y. Dubaquie, and M. Gautschi. 2002. Nascent-polypeptide-associated complex. Cell. Mol. Life Sci. 59:1632-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rossi, J. M., and S. Lindquist. 1989. The intracellular location of yeast heat-shock protein 26 varies with metabolism. J. Cell Biol. 108:425-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sahi, C., and E. A. Craig. 2007. Network of general and specialty J protein chaperones of the yeast cytosol. Proc. Natl. Acad. Sci. USA 104:7163-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Serva, S., and P. D. Nagy. 2006. Proteomics analysis of the tombusvirus replicase: Hsp70 molecular chaperone is associated with the replicase and enhances viral RNA replication. J. Virol. 80:2162-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Serviene, E., Y. Jiang, C. P. Cheng, J. Baker, and P. D. Nagy. 2006. Screening of the yeast yTHC collection identifies essential host factors affecting tombusvirus RNA recombination. J. Virol. 80:1231-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stapleford, K. A., D. Rapaport, and D. J. Miller. 2009. Mitochondrion-enriched anionic phospholipids facilitate flock house virus RNA polymerase membrane association. J. Virol. 83:4498-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tomita, Y., T. Mizuno, J. Diez, S. Naito, P. Ahlquist, and M. Ishikawa. 2003. Mutation of host dnaJ homolog inhibits brome mosaic virus negative-strand RNA synthesis. J. Virol. 77:2990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Wynsberghe, P. M., and P. Ahlquist. 2009. 5′ cis elements direct nodavirus RNA1 recruitment to mitochondrial sites of replication complex formation. J. Virol. 83:2976-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Wynsberghe, P. M., H. R. Chen, and P. Ahlquist. 2007. Nodavirus RNA replication protein A induces membrane association of genomic RNA. J. Virol. 81:4633-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walsh, P., D. Bursac, Y. C. Law, D. Cyr, and T. Lithgow. 2004. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 5:567-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wandinger, S. K., K. Richter, and J. Buchner. 2008. The Hsp90 chaperone machinery. J. Biol. Chem. 283:18473-18477. [DOI] [PubMed] [Google Scholar]
- 59.Wang, R. Y., J. Stork, and P. D. Nagy. 2009. A key role for heat shock protein 70 in the localization and insertion of tombusvirus replication proteins to intracellular membranes. J. Virol. 83:3276-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weeks, S. A., and D. J. Miller. 2008. The heat shock protein 70 cochaperone YDJ1 is required for efficient membrane-specific flock house virus RNA replication complex assembly and function in Saccharomyces cerevisiae. J. Virol. 82:2004-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Werner-Washburne, M., D. E. Stone, and E. A. Craig. 1987. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 7:2568-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yochem, J., H. Uchida, M. Sunshine, H. Saito, C. P. Georgopoulos, and M. Feiss. 1978. Genetic analysis of two genes, DnaJ and DnaK, necessary for Escherichia coli and bacteriophage λ DNA replication. Mol. Gen. Genet. 164:9-14. [DOI] [PubMed] [Google Scholar]
- 63.Youker, R. T., P. Walsh, T. Beilharz, T. Lithgow, and J. L. Brodsky. 2004. Distinct roles for the Hsp40 and Hsp90 molecular chaperones during cystic fibrosis transmembrane conductance regulator degradation in yeast. Mol. Biol. Cell 15:4787-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Young, J. C., V. R. Agashe, K. Siegers, and F. U. Hartl. 2004. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell. Biol. 5:781-791. [DOI] [PubMed] [Google Scholar]