Abstract
Killer immunoglobulin-like receptors (KIRs) are related to the activation and inhibition of NK cells and may play an important role in the innate response against infection with viruses such as hepatitis C virus (HCV). We examined whether the different combinations of KIRs with their HLA class I ligands influenced the response to combined treatment (pegylated alpha interferon and ribavirin) of patients infected by HCV. A total of 186 consecutive patients diagnosed with chronic HCV infection were analyzed. Seventy-seven patients exhibited HCV RNA levels at 6 months posttreatment and were called nonresponders (NR), while 109 cleared viral RNA and were named sustained viral responders (SVR). Patients were typed for HLA-B, HLA-Cw, KIR genes, and HCV genotype. In our study, the frequency of the KIR2DL2 allele was significantly increased in NR (P < 0.001; odds ratio [OR] = 1.95), as was the frequency of the KIR2DL2/KIR2DL2 genotype (P < 0.005; OR = 2.52). In contrast, the frequencies of the KIR2DL3 genotype (P < 0.001) and KIR2DL3/KIR2DL3 genotype (P < 0.05; OR = 0.54) were significantly increased in the SVR. Different combinations of KIR2DL2 and KIR2DL3 alleles with their ligands were analyzed. The frequency of the KIR2DL2/KIR2DL2-HLA-C1C2 genotype was significantly increased in the NR (P < 0.01; OR = 3.15). Additionally, we found a higher frequency of the KIR2DL3/KIR2DL3-HLA-C1C1 genotype in the SVR group (P < 0.05; OR = 0.33). These results were not affected by the HCV genotype. In conclusion, patients who carried the KIR2DL2/KIR2DL2-HLA-C1C2 genotype were less prone to respond to treatment. However, the KIR2DL3/KIR2DL3-HLA-C1C1 genotype clearly correlated with a satisfactory response to treatment, defined by the clearance of HCV RNA.
Hepatitis C virus (HCV) infection is a common chronic disease affecting over 170 million people worldwide (48). Around 80% of these individuals evolve to chronic infection, and 10 to 20% of patients develop cirrhosis over a 20-year period. A minority (2%) progresses to hepatocellular carcinoma annually (18). Several host factors including age, body mass index (BMI), gender, fibrosis, cirrhosis, or the absence of cirrhosis and several viral factors including viral genotype and viral load can influence the response to treatment (6, 32, 42). Pegylated alpha interferon (Peg-IFN-α) plus ribavirin (combined therapy) constitute the most effective therapy for the treatment of chronic HCV infection (13). Since this treatment carries serious side effects, it is necessary to identify those patients who can clear HCV infection in order to reduce the period of this aggressive therapy.
Natural killer (NK) cells are lymphocytes that play an important role in the host defense against HCV infection (14). NK cell activity is determined by the balance of different signals received and the equilibrium between inhibitory and activating receptors (3). Some receptors are specific for human leukocyte antigen (HLA) class I molecules (4). NK cells check the surface of the surrounding cells, detect the presence of their HLA class I molecules, and then discriminate between healthy, infected, or transformed cells (10). When NK cells contact target cells, the resulting interactions of their receptors produce either activating or inhibitory signals. If the expression of HLA class I molecules on the target cell is absent or reduced, the inhibitory signal is not generated (25).
Killer cell immunoglobulin-like receptors (KIRs) are members of a group of regulatory molecules expressed on NK cells and a subset of T cells (30). This family of polymorphic genes is located on chromosome 19 (19q13.4), within the leukocyte receptor complex. The leukocyte receptor complex also encodes a number of genetically and functionally related genes. KIRs with long cytoplasmic tails are inhibitors, based on the presence of immunoreceptor tyrosine-based inhibition motifs in their cytoplasmic domains. KIRs with short tails interact with adaptor molecules such as DAP-12 (DNAX activation protein), which contain immunoreceptor tyrosine-based activation motifs and transmit activating signals (24). Several inhibitory KIRs have been well defined. KIR2DL1 binds the subset of HLA-Cw molecules with lysine at position 80 of the heavy chain (HLA-C2 group). KIR2DL2 and KIR2DL3 bind the subset of HLA-Cw molecules with asparagine at position 80 (HLA-C1 group) (34).
Studies that have associated KIR genotypes with diseases have identified mainly viral infections and autoimmune diseases (22, 45). The importance of NK cells in the resolution of viral infections has prompted studies that correlate KIRs and their ligands with outcomes (12). Some studies identified a relationship between KIR genotypes and outcomes with several infectious agents such as human immunodeficiency virus (27, 29), cytomegalovirus (7), hepatitis B virus (28), and HCV (21, 26).
Recently, a protective association of the inhibitory receptor KIR2DL3 with HLA-CAsn80 (HLA-C1) and its effect on the course of HCV infection were described (21). The prevalence of KIR2DL3 and its ligand HLA-C1 is increased in individuals who eliminate HCV spontaneously, in contrast to those who remain chronically infected. The protective effect of KIR2DL3/HLA-CAsn80 was observed only among individuals who carried both homozygous genes and had received a low HCV exposure dose. Recently, we found that the frequency of HLA-Bw4I80 ligand and the activating receptor KIR3DS1 was increased healthy in HCV carriers compared to patients who had developed hepatocellular carcinoma (26).
The aim of this study was to investigate the influence of KIR genes and KIR-HLA combinations on the response to combined therapy with Peg-IFN-α-2b and ribavirin in a group of patients with HCV infection.
MATERIALS AND METHODS
Patients.
A group of 186 consecutive, unrelated Caucasian patients diagnosed with chronic HCV infection were enrolled in the study between January 2004 and December 2005. They were diagnosed by the Gastroenterology Service of the Hospital Universitario Central de Asturias in Oviedo, Spain, and by the Clínica Universitaria de Navarra in Pamplona, Spain. All patients were positive for anti-HCV antibodies and HCV RNA in serum.
The patients received the same standard treatment of Peg-IFN-α-2b (1.5 μg/kg of body weight/week) and ribavirin (<65 kg, 800 mg/day; 65 to 85 kg, 1,000 mg/day; >85 kg, 1,200 mg/day). The duration of the treatment was 24 weeks for HCV genotype 2 or 3 and 48 weeks for HCV genotype 1 or 4. All patients were monitored for at least 6 months posttreatment in order to establish the response, in accordance with previously reported virological criteria (11). Patients were classified into two treatment outcomes. Patients who did not mount a sufficient anti-HCV response (nonresponders [NR]) were defined by a consistent positive viral load during treatment, at its end, or at 6 months posttreatment. Another group of patients was classified as sustained virological responders (SVR). The SVR were defined by consistent undetectable HCV RNA levels in serum during 6 months posttreatment.
The protocol was approved by the ethics committees of both hospitals, and all patients gave written informed consent before enrollment.
Immunological tests.
Genomic DNA was extracted from peripheral blood with the Magtration-MagaZorb DNA Common Kit-200 N by using the Magtration 12GC system (Precision System Science Co., Ltd., Woerrstadt, Germany). The HLA-B, HLA-Cw, and KIR genes were typed by using Lifecodes HLA-SSO and KIR-SSO typing kits (Tepnel Lifecodes Corporation, Stamford, United Kingdom) based on Luminex xMAP technology (Luminex Corp., Austin, TX) according to the manufacturer's instructions. Ambiguities in KIR typing were resolved by PCR-single specific primer (SSP) in accordance with a method previously described (16, 19).
Microbiological tests.
Serum samples were obtained at least every month before, during, and after treatment. These samples were frozen at −80°C within 4 h of collection. The RNA viral levels in serum and HCV genotypes were determined. The HCV genotype was identified by the Versant HCV Genotype 2.0 assay (Bayer HealthCare, Tarrytown, NY). HCV RNA was quantified during the treatment and follow-up periods by real-time PCR (Cobas TaqMan 48; Roche Diagnostics, Mannheim, Germany) according to the manufacturer's instructions.
Statistical analysis.
We used the chi-square (χ2) test to examine whether there were observable differences between qualitative factors and the Student t test for quantitative factors in independent samples. The different factors that may contribute to the nonresponse to the treatment were compared by using a backward stepwise logistic regression analysis. The effect of the KIR-HLA genotypes on the clearance of HCV RNA in serum during treatment was estimated by the Kaplan-Meier method and compared using the log-rank test. In order to perform this statistical analysis, we have defined the HCV RNA death event as the mean between the last positive and the first negative HCV RNA results, under the prerequisite that the patient remained negative through the end of the period of treatment (31). Sera were collected at intervals of 4 weeks, and samples with undetectable levels of HCV RNA during treatment from patients with a “breakthrough effect” (39) were not considered for statistical analysis. Data were censored when patients finished their treatment. A P value of <0.05 was considered to be significant. Statistical data were calculated by using the SPSS 15.0 program (SPSS Inc., Chicago, IL).
RESULTS
Patient characteristics.
The clinical and demographic characteristics of the 186 patients are shown in Table 1. Seventy-seven patients (41.4%) were listed as NR to the treatment, and 109 patients (58.6%) comprised the SVR group because they exhibited no detectable HCV RNA levels throughout the 6 months posttreatment. The distribution of viral genotypes in our cohort showed that HCV genotype 1 was the most prevalent in our study population and was significantly more resistant to this treatment regimen (P < 0.001; odds ratio [OR] = 4.22; 95% confidence interval [CI] = 2.07 to 8.78). In contrast, HCV genotype 3 was significantly more susceptible to this treatment regimen (P < 0.001; OR = 0.12; 95% CI = 0.05 to 0.33). Moreover, the body mass index (BMI) was significantly higher for the NR group than for the SVR group (P < 0.05).
TABLE 1.
Clinical and HCV characteristics of infected patients included in the present study
| Characteristica | Value for group |
P valueb | ||
|---|---|---|---|---|
| All patients (n = 186) | SVR (n = 109) | NR (n = 77) | ||
| Mean age (yr) ± SD | 47.2 ± 12.9 | 43.6 ± 7.5 | 49.5 ± 10.4 | |
| Gender distribution [no. of patients (%)] | ||||
| Male | 135 (72.6) | 83 (76.1) | 52 (67.5) | |
| Female | 51 (27.4) | 26 (23.9) | 25 (32.5) | |
| Mean wt (kg) ± SD | 70.5 ± 13 | 68.3 ± 11 | 72.1 ± 13 | |
| Mean BMI (kg/m2) ± SD | 23.2 ± 3.6 | 22.7 ± 3.4 | 24.1 ± 4.1 | <0.05 |
| Median viral load before treatment (106 IU/ml) (IR) | 5.3 (4.67) | 5.38 (4.67) | 5.17 (3.9) | |
| Mean analytical levels before treatment (IU/liter) ± SD | ||||
| AST | 62.6 ± 16.3 | 59.8 ± 14.5 | 68.7 ± 17.5 | |
| ALT | 108.5 ± 19.4 | 98.5 ± 18.9 | 114.3 ± 20.5 | |
| γGT | 76.5 ± 24.3 | 64.7 ± 23.6 | 84.8 ± 26.7 | |
| Mean ferritin level (ng/ml) ± SD | 262.6 ± 46.3 | 247.7 ± 24.1 | 281.1 ± 35.2 | |
| Mean platelet level (103 U/ml) ± SD | 221.5 ± 57.6 | 212.7 ± 46.3 | 229.1 ± 60.4 | |
| Mean cholesterol level (mg/dl) ± SD | 183.6 ± 39.2 | 173.4 ± 32.8 | 180.3 ± 22.5 | |
| No. of patients (%) with: | ||||
| Cirrhosis | 5 (2.7) | 2 (1.8) | 3 (3.9) | |
| Source of HCV infection of: | ||||
| Blood transfusion | 61 (32.8) | 33 (30.3) | 28 (36.4) | |
| Drug use | 55 (29.6) | 36 (33) | 19 (24.7) | |
| Unknown | 70 (37.6) | 40 (36.7) | 30 (38.9) | |
| HCV genotype | ||||
| 1 | 126 (67.7) | 61 (55.9) | 65 (84.4) | <0.001 |
| 2 | 5 (2.8) | 4 (3.7) | 1 (1.3) | |
| 3 | 44 (23.6) | 39 (35.8) | 5 (6.5) | <0.001 |
| 4 | 11 (5.9) | 5 (4.6) | 6 (7.8) | |
IR, interquartile range; AST, aspartate aminotransferase; ALT, alanine aminotransferase; γGT, gamma-glutamyltranspeptidase.
All P values were not significant except as indicated.
HLA and KIR distribution in the cohort study.
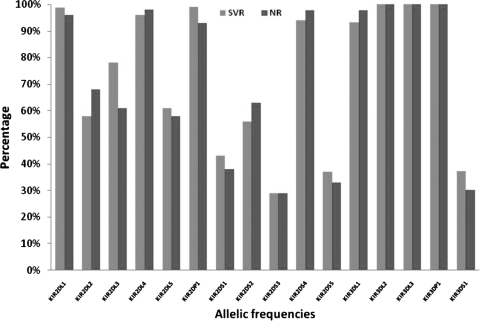
The frequencies of HLA-B and HLA-Cw alleles in our patients were not significantly different between the treatment outcomes. We then examined the frequencies of the different KIR genes (Fig. 1). No statistically significant differences were found for most allele combinations except for the KIR2DL2 and KIR2DL3 genes. Considering both genes as alleles of the same locus (20, 47), the frequency of KIR2DL2 was significantly increased in NR (P < 0.001; OR = 1.95; 95% CI = 1.26 to 3.03), whereas the frequency of KIR2DL3 was significantly increased in the SVR group (P < 0.001) (Table 2).
FIG. 1.
KIR allele frequencies for patients with chronic HCV compared to treatment outcome (for KIR2DL2, the P value was <0.001, the OR was 1.95, and the 95% CI was 1.26 to 3.03; for KIR2DL3, the P value was <0.001).
TABLE 2.
Comparison of frequencies of KIR2DL2 and KIR2DL3 and their respective ligands to viral responsesa
| Genotype | No. of patients with genotype (%) |
OR | 95% CI | P value | |
|---|---|---|---|---|---|
| SVR | NR | ||||
| 2n = 218 | 2n = 154 | ||||
| KIR2DL2 | 83 (38.1) | 84 (54.5) | 1.95 | 1.26-3.03 | <0.001 |
| KIR2DL3 | 135 (61.9) | 70 (45.5) | <0.001 | ||
| KIR alleles | n = 109 | n = 77 | |||
| 2DL2/2DL2 | 22 (20.2) | 30 (39) | 2.52 | 1.31-4.86 | <0.005 |
| 2DL2/2DL3 | 39 (35.8) | 24 (31.2) | NS | ||
| 2DL3/2DL3 | 48 (44) | 23 (29.8) | 0.54 | 0.28-0.99 | <0.05 |
| HLA-C alleles | n = 109 | n = 77 | |||
| HLA-C1C1 | 42 (38.5) | 27 (35) | NS | ||
| HLA-C1C2 | 46 (42.2) | 42 (54.6) | NS | ||
| HLA-C2C2 | 21 (19.3) | 8 (10.4) | NS | ||
| KIR-HLA combinations | n = 109 | n = 77 | |||
| 2DL2/2DL2 + HLA-C1C1 | 10 (9.2) | 10 (13) | NS | ||
| 2DL2/2DL2 + HLA-C1C2 | 9 (8.3) | 17 (22.1) | 3.15 | 1.32-7.51 | <0.01 |
| 2DL2/2DL2 + HLA-C2C2 | 3 (2.7) | 3 (3.9) | NS | ||
| 2DL2/2DL3 + HLA-C1C1 | 13 (11.9) | 12 (15.6) | NS | ||
| 2DL2/2DL3 + HLA-C1C2 | 17 (15.6) | 11 (14.2) | NS | ||
| 2DL2/2DL3 + HLA-C2C2 | 9 (8.3) | 1 (1.3) | NS | ||
| 2DL3/2DL3 + HLA-C1C1 | 19 (17.4) | 5 (6.5) | 0.33 | 0.1-0.99 | <0.05 |
| 2DL3/2DL3 + HLA-C1C2 | 20 (18.3) | 14 (18.2) | NS | ||
| 2DL3/2DL3 + HLA-C2C2 | 9 (8.3) | 4 (5.2) | NS | ||
Statistical significance was calculated by using the χ2 test. For KIR2DL2, an OR of >1 indicates a poor response to treatment. NS, not significant.
We also analyzed the distribution of the homozygous and heterozygous genotypes of both alleles. When the KIR2DL2 allele was homozygous, we found significant differences between the treatment outcomes. The KIR2DL2/KIR2DL2 genotype significantly correlated with NR (P < 0.005; OR = 2.52; 95% CI = 1.31 to 4.86). In contrast, the frequency of the KIR2DL3/KIR2DL3 genotype was significantly associated with viral clearance (P < 0.05; OR = 0.54; 95% CI = 0.28 to 0.99). No significant differences were found between treatment outcomes when both alleles were heterozygous.
Next, we analyzed the different interactions of KIR2DL2 and KIR2DL3 alleles with their HLA-Cw group 1 (HLA-C1) ligands. When KIR2DL2 was homozygous and its ligand HLA-C1 was heterozygous (KIR2DL2/KIR2DL2-HLA-C1C2), we observed a significant increase in the frequency of a poor response to treatment (P < 0.01; OR = 3.15; 95% CI = 1.32 to 7.51). In contrast, the KIR2DL3/KIR2DL3-HLA-C1C1 genotype was associated with at least a 6-month clearance of HCV (P < 0.05; OR = 0.33; 95% CI = 0.1 to 0.99). We analyzed the remaining combinations of heterozygous KIR2DL2 and KIR2DL3 alleles with their ligands, but no significant differences were observed.
We then decided to analyze the progressive effect of KIR2DL3/KIR2DL3-C1C1 on the response to treatment. Patients were sorted according to KIR2DL3 and HLA-C genotypes. The first group contained patients who carried the KIR2DL3/KIR2DL3-HLAC1C1 genotype; the second one contained patients who carried the KIR2DL3/KIR2DL3-HLA-C1C2, KIR2DL2/KIR2DL3-HLA-C1C1, or KIR2DL2/KIR2DL3-HLA-C1C2 genotype; and the last group consisted of the remaining patients who were homozygous for KIR2DL2, were homozygous for HLA-C2, or carried the KIR2DL2/KIR2DL2-HLA-C2C2 genotype. This analysis revealed a linear trend between the number of KIR2DL3-HLA-C1 interactions and the odds of an SVR (χ2 for trend of 4.736; P < 0.05).
The distribution of the frequencies of KIR3DL1 and KIR3DS1 alleles and the interaction with their ligands (HLA-B Bw4 allotypes) was compared with outcome. No associations were found between these genes and the response to the treatment (data not shown).
Influence of KIR2DL genotype in combination with other risk factors in the nonresponse to treatment.
As previously mentioned, the response to treatment is particularly ineffective for the most prevalent HCV genotype, genotype 1 (23), but patients with HCV genotype 2 or 3 are expected to have a higher probability of treatment success. Other factors that could contribute to the achievement of positive outcome are gender, age, BMI, extent of fibrosis, HCV RNA levels, and the presence or absence of cirrhosis. Using a backward stepwise logistic regression model, we initially analyzed the influence of these host and viral variables with KIR and HLA-Cw genetic factors on the nonresponse to treatment. The resulting comparative model is represented in Table 3. The KIR2DL2/KIR2DL2-HLA-C1C2 genotype had a correlation with poor outcome (P < 0.01; OR = 4.12; 95% CI = 1.68 to 10.1). Moreover, consistent with data from previous studies, HCV genotype 1 (P < 0.005; OR = 3.32; 95% CI = 1.49 to 7.42) contributed clearly to the nonresponse of this treatment. Significant interactions between the different variables analyzed were not detected, including BMI, which was statistically significant in the univariate analysis. In this multivariate analysis, the KIR2DL3/KIR2DL3-HLA-C1C1 genotype was not significantly associated with nonresponse to treatment.
TABLE 3.
Final step of a backward logistic regression analysis of risk factors associated with nonresponse to anti-HCV treatmenta
| Variable | OR | 95% CI | P value |
|---|---|---|---|
| KIR2DL2/KIR2DL2-HLA-C1C2 genotype | 4.12 | 1.68-10.1 | <0.01 |
| HCV genotype 1 | 3.32 | 1.49-7.42 | <0.005 |
In the initial analysis, factors such as gender, age (>50 years), BMI (>27 kg/m2), aspartate aminotransferase levels (more than two times the normal value), alanine amino transferase levels (more than two times the normal value), gamma-glutamyltranspeptidase levels (more than two times the normal value), HCV RNA levels (>400,000 IU/ml), and cirrhosis or absence of cirrhosis were also included. In this analysis, the KIR2DL3/KIR2DL3-HLA-C1C1 genotype was not significant.
Effect of the KIR-HLA genotype on the clearance of HCV RNA at the end of the treatment.
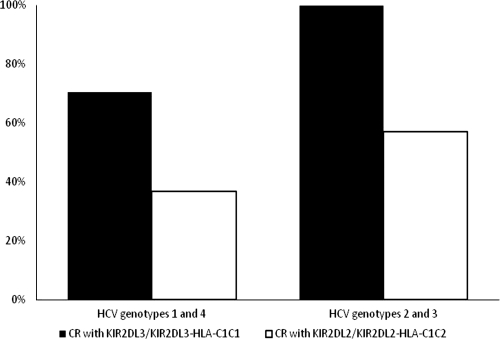
We determined the HCV RNA levels in sera of our patient cohort during the treatment period. For the Kaplan-Meier analysis, patients were divided into two groups: the first group was composed of patients with HCV genotype 1 or 4 (n = 137), and the second group was composed of patients with HCV genotype 2 or 3 (n = 49) (Fig. 2). In group 1, 70.6% of the patients who carried the KIR2DL3/KIR2DL3-HLA-C1C1 genotype had a complete response and cleared HCV RNA (P = 0.062 by log-rank test), which showed a trend for a protective effect of this KIR-HLA genotype. In contrast, most patients who carried the KIR2DL2/KIR2DL2-HLA-C1C2 genotype did not clear HCV RNA (P < 0.01 by log-rank test) by the end of the treatment. Group 2 patients infected with HCV genotype 2 or 3 exhibited similar associations between genotypes and outcomes. All patients who carried the KIR2DL3/KIR2DL3-HLA-C1C1 genotype cleared HCV RNA (P < 0.05 by log-rank test) after 24 weeks of treatment, whereas patients who carried the KIR2DL2/KIR2DL2-HLA-C1C2 genotype did not (P < 0.005 by log-rank test). Patients with the KIR2DL3/KIR2DL3-HLA-C1C1 genotype cleared HCV RNA at a higher frequency than did patients who carried the KIR2DL2/KIR2DL2-HLA-C1C2 genotype.
FIG. 2.
Percentage of complete response (CR) to combined treatment before the follow-up period. Note that for patients with HCV genotypes 2 and 3, there were 24 weeks of treatment. For patients with HCV genotypes 1 and 4, there were 48 weeks of treatment.
Moreover, at the end of the follow-up period, 65.4% of patients who carried the KIR2DL2/KIR2DL2-HLA-C1C2 genotype were poor responders, whereas 79.2% of all patients who carried the KIR2DL3/KIR2DL3-HLA-C1C1 genotype were able to clear HCV RNA in serum by the end of treatment. In a previous analysis, we did not find statistical differences when patients were divided into groups of early viral responders, SVR, and NR (data not shown).
In conclusion, the KIR-HLA genotype can influence the clearance of HCV RNA at the end of treatment independently of the HCV genotype.
DISCUSSION
Several studies have identified specific genetic factors that play an important role in the persistence of HCV infection (41). It was also observed previously that some HLA alleles clearly influence the resolution of other viral infections (1). Thus, HLA molecules have been the focus of numerous studies for establishing possible associations with the persistence or the elimination of HCV (9, 40). Some associations between certain KIRs with their HLA ligands and the progression of HCV infection were also described previously (26).
In the present study, we analyzed the influence of KIRs on the response to combined treatment. While the number of patients was relatively small, we considered the homogeneity of the group important. In spite of these limitations, we observed that KIR2DL3 was associated with a sustained viral immune response, while KIR2DL2 was clearly correlated with a nonresponse. Furthermore, we found that KIR2DL3/KIR2DL3-HLA-C1C1 was associated with a sustained virological response, whereas KIR2DL2/KIR2DL2-HLA-C1C2 had a significantly increased frequency in the NR population.
Previous studies suggested a model in which KIR-HLA combinations inhibit NK cells with different intensities (44, 46). According to this model, KIR2DL1-HLA-C2 has the strongest capacity of inhibition, followed by KIR2DL2-HLA-C1 and, finally, by KIR2DL3-HLA-C1. The weaker inhibition of KIR2DL3 may result in a greater activation of NK cells and, consequently, a more efficient resolution of viral infection. The effects of additional NK-activating receptors, such as NKG2D, may lead to a higher efficacy of control against HCV infection. The two KIR2DL1 (present in 98% of the individuals) and KIR2DL2 receptors may, however, induce a more intense inhibition of NK cells and therefore reduce the efficacy of resolving HCV infection. Moreover, the different KIR2DL-HLA-C interactions may further transmit inhibitory signals with different strengths (37).
In agreement with those studies, we suggest that the KIR2DL3/KIR2DL3-HLA-C1C1 genotype combination influences the generation of a sustained virological response, since the inhibitory signals produced by KIR2DL1 and KIR2DL2 are absent. Individuals who carry the KIR2DL3/KIR2DL3-HLA-C1C1 genotype more effectively activated NK cells in response to the administration of Peg-IFN-α during treatment, although NK cell functions have been impaired during chronic HCV infection (15). Furthermore, several studies described HCV mechanisms that inhibited the responses of NK cells and were correlated with the establishment of a chronic infection (17, 43). Nevertheless, recent findings demonstrate that a sustained response to combined treatment for patients with chronic HCV infection is closely associated with increased numbers of NK cells in the liver (49). Other KIR2DL-HLA genotypes have at least one stronger inhibitory signal, and their response to treatment was less effective. Within these genotypes, we suggest that the KIR2DL2/KIR2DL2-HLA-C1C2 genotype has the most inefficient response to treatment, because cumulative inhibitory signals are produced by KIR2DL1 and KIR2DL2. This stronger inhibition could be enhanced by treatment because Peg-IFN-α induces the expression of HLA class I molecules (38) and favors the interaction between the infected cells and NK cells.
We further observed that the homozygous KIR2DL3 allele in combination with homozygous HLA-C1 was found significantly more frequently in patients who responded to antiviral treatment. The nonresponding patients had a higher frequency of the homozygous KIR2DL2 allele with its heterozygous ligand, supporting our hypothesis. As previously mentioned, Khakoo et al. (21) described that the presence of the homozygous KIR2DL3 allele and its ligand is correlated to the spontaneous clearance of HCV infection. Our study shows a similar association, but in this case, KIR2DL3/KIR2DL3-HLA-C1C1 is significantly associated with a complete response to antiviral therapy, while the presence of the KIR2DL2/KIR2DL2-HLA-C1C2 genotype correlates with an inadequate response to the treatment and with persistent viral RNA. Despite the observation that the KIR2DL2/KIR2DL2-HLA-C2C2 genotype carries the strongest inhibitory signal mediated by KIR2DL1-HLA-C2, we did not detect statistical differences related to a nonresponse to treatment. Possible explanations include its low frequency in our population and/or the absence of an additional KIR2DL2-HLA-C1 inhibitory signal. Moreover, the bound peptides in MHC class I play an important role in the balanced recognition of NK cells, and the level of KIR2DL1 recognition of this complex may be reduced by some modified peptides during tumor transformation or viral infections (2).
The monitoring of HCV RNA levels in serum throughout the treatment period revealed a high percentage of patients carrying KIR2DL3/KIR2DL3-HLA-C1C1 who exhibited a complete response. In contrast, the majority of individuals carrying the KIR2DL2/KIR2DL2-HLA-C1C2 genotype were unable to clear HCV RNA. These data also suggested that treatment in the presence of the KIR2DL2/KIR2DL2-HLA-C1C2 genotype will be less effective in resolving HCV infection.
Analysis of the risk factors in the NR to the treatment showed significant associations only with BMI, the HCV genotype, the KIR genotype, and the KIR-HLA genotype. Previous studies have shown that HCV genotypes influence the response to treatment (50). For example, HCV genotype 1 is associated with a poor response to antiviral treatment (36), similar to our results. Several studies suggested that HCV genotype 1 is more resistant to treatment due to the interactions of certain viral proteins, such as NS5A, with the IFN-α signaling pathway (35). In fact, five HCV-infected patients carrying the KIR2DL3/KIR2DL3-HLA-C1C1 allele did not resolve their infections after therapy in our study, which suggests that additional modalities for resolving HCV infections are warranted. On the other hand, antiviral treatment is usually efficacious against HCV genotypes 2 and 3 and justifies a shorter treatment schedule (8).
CD8+ T lymphocytes, as NK cells, are crucial in the defense against viral infections. With regard to CD8+ T lymphocytes and HLA class I molecules, several studies indicated a central association with the resolution of viral diseases (33). On the other hand, CD8+ T lymphocytes can express KIRs, and the signals from these receptors can contribute to the control of viral infections (5).
In conclusion, the balance of activating and inhibitory signals on NK cells and CD8+ T lymphocytes, which is modulated by treatment and conditioned by genetics, helps to define the antiviral response. In addition to the viral genotype, KIRs play an important role in the immune response against HCV and may be key factors that modulate the progression of the infection and the response to treatment.
Despite the small number of subjects included in this study, we were able to demonstrate significant effects of KIRs and their ligands on the response to treatment of chronic HCV infection. Nevertheless, additional genetic and functional studies will be necessary in order to clarify the involvement of KIRs in HCV infection.
Acknowledgments
This study was supported by Spanish grants PI08-0566 from Institute Carlos III and grant PC-07/06 and was undertaken with the collaboration of CAJASTUR.
We declare that we have no financial interest.
Footnotes
Published ahead of print on 21 October 2009.
REFERENCES
- 1.Bailey, J. R., H. Zhang, B. W. Wegweiser, H. C. Yang, L. Herrera, A. Ahonkhai, T. M. Williams, R. F. Siliciano, and J. N. Blankson. 2007. Evolution of HIV-1 in an HLA-B*57-positive patient during virologic escape. J. Infect. Dis. 196:50-55. [DOI] [PubMed] [Google Scholar]
- 2.Betser-Cohen, G., G. Katz, T. Gonen-Gross, N. Stern, T. I. Arnon, H. Achdout R. Gazit, and O. Mandelboim. 2006. Reduced KIR2DL1 recognition of MHC class I molecules presenting phosphorylated peptides. J. Immunol. 176:6762-6769. [DOI] [PubMed] [Google Scholar]
- 3.Biassoni, R. 2008. Natural killer cell receptors. Adv. Exp. Med. Biol. 640:35-52. [DOI] [PubMed] [Google Scholar]
- 4.Bottino, C., M. Vitale, D. Pende, R. Biassoni, and A. Moretta. 1995. Receptors for HLA class I molecules in human NK cells. Semin. Immunol. 7:67-73. [DOI] [PubMed] [Google Scholar]
- 5.Byers, A. M., C. C. Kemball, N. P. Andrews, and A. E. Lukacher. 2003. Regulation of antiviral CD8+ T cells by inhibitory natural killer cell receptors. Microbes Infect. 5:169-177. [DOI] [PubMed] [Google Scholar]
- 6.Conjeevaram, H. S., M. W. Fried, L. J. Jeffers, N. A. Terrault, T. E. Wiley-Lucas, N. Afdhal, R. S. Brown, S. H. Belle, J. H. Hoofnagle, D. E. Kleiner, and C. D. Howell. 2006. Peginterferon and ribavirin treatment in African American and Caucasian American patients with hepatitis C genotype 1. Gastroenterology 131:470-477. [DOI] [PubMed] [Google Scholar]
- 7.Cook, M., D. Briggs, C. Craddock, P. Mahendra, D. Milligan, C. Fegan, P. Darbyshire, S. Lawson, E. Boxall, and P. Moss. 2006. Donor KIR genotype has a major influence on the rate of cytomegalovirus reactivation following T-cell replete stem cell transplantation. Blood 107:1230-1232. [DOI] [PubMed] [Google Scholar]
- 8.Dalgard, O., and A. Mangia. 2006. Short-term therapy for patients with hepatitis C virus genotype 2 or 3 infection. Drugs 66:1807-1815. [DOI] [PubMed] [Google Scholar]
- 9.de Arias, A. E., S. E. Haworth, L. S. Belli, P. Burra, G. Pinzello, M. Vangeli, E. Minola, M. Guido, P. Boccagni, T. M. De Feo, R. Torelli, M. Cardillo, M. Scalamogna, and F. Poli. 2009. Killer cell immunoglobulin-like receptor genotype and killer cell immunoglobulin-like receptor-human leukocyte antigen C ligand compatibility affect the severity of hepatitis C virus recurrence after liver transplantation. Liver Transpl. 15:390-399. [DOI] [PubMed] [Google Scholar]
- 10.Draghi, M., N. Yawata, M. Gleimer, M. Yawata, N. M. Valiante, and P. Parham. 2005. Single-cell analysis of the human NK cell response to missing self and its inhibition by HLA class I. Blood 105:2028-2035. [DOI] [PubMed] [Google Scholar]
- 11.Ferenci, P. 2004. Predicting the therapeutic response in patients with chronic hepatitis C: the role of viral kinetic studies. J. Antimicrob. Chemother. 53:15-18. [DOI] [PubMed] [Google Scholar]
- 12.French, A. R., and W. M. Yokoyama. 2003. Natural killer cells and viral infections. Curr. Opin. Immunol. 15:45-51. [DOI] [PubMed] [Google Scholar]
- 13.Fried, M. W., M. I. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Gonçales, Jr., D. Häussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon-α-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 14.Golden-Mason, L., and H. R. Rosen. 2006. Natural killer cells: primary target for hepatitis C virus immune evasion strategies? Liver Transpl. 12:363-372. [DOI] [PubMed] [Google Scholar]
- 15.Golden-Mason, L., L. Madrigal-Estebas, E. McGrath, M. J. Conroy, E. J. Ryan, J. E. Hegarty, C. O'Farrelly, and D. G. Doherty. 2008. Altered natural killer cell subset distributions in resolved and persistent hepatitis C virus infection following single source exposure. Gut 57:1121-1128. [DOI] [PubMed] [Google Scholar]
- 16.Gómez-Lozano, N., and C. Vilches. 2002. Genotyping of human killer-cell immunoglobulin-like receptor genes by polymerase chain reaction with sequence-specific primers: an update. Tissue Antigens 59:184-193. [DOI] [PubMed] [Google Scholar]
- 17.Herzer, K., C. S. Falk, J. Encke, S. T. Eichhorst, A. Ulsenheimer, B. Seliger, and P. H. Krammer. 2003. Upregulation of major histocompatibility complex class I on liver cells by hepatitis C virus core protein via p53 and TAP1 impairs natural killer cell cytotoxicity. J. Virol. 77:8299-8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoofnagle, J. H. 2002. Course and outcome of hepatitis C. Hepatology 36(Suppl. 5):S21-S29. [DOI] [PubMed] [Google Scholar]
- 19.Hsu, K. C., X. R. Liu, A. Selvakumar, E. Mickelson, R. J. O'Reilly, and B. Dupont. 2002. Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J. Immunol. 169:5118-5129. [DOI] [PubMed] [Google Scholar]
- 20.Hsu, K. C., S. Chida, D. E. Geraghty, and B. Dupont. 2002. The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism. Immunol. Rev. 190:40-52. [DOI] [PubMed] [Google Scholar]
- 21.Khakoo, S. I., C. L. Thio, M. P. Martin, C. R. Brooks, X. Gao, J. Astemborski, J. Cheng, J. J. Goedert, D. Vlahov, M. Hilgartner, S. Cox, A. M. Little, G. J. Alexander, M. E. Cramp, S. J. O'Brien, W. M. Rosenberg, D. L. Thomas, and M. Carrington. 2004. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science 305:872-874. [DOI] [PubMed] [Google Scholar]
- 22.Khakoo, S. I., and M. Carrington. 2006. KIR and disease: a model system or system of models? Immunol. Rev. 214:186-201. [DOI] [PubMed] [Google Scholar]
- 23.Lam, N. P., A. U. Neumann, D. R. Grech, T. E. Wiley, A. S. Perelson, and T. J. Layden. 1997. Dose-dependent acute clearance of hepatitis C genotype 1 virus with interferon alfa-2b. Hepatology 26:226-231. [DOI] [PubMed] [Google Scholar]
- 24.Lanier, L. L., B. C. Corliss, J. Wu, C. Leong, and J. H. Phillips. 1998. Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature 391:703-707. [DOI] [PubMed] [Google Scholar]
- 25.Lanier, L. L. 2005. NK cell recognition. Annu. Rev. Immunol. 23:225-274. [DOI] [PubMed] [Google Scholar]
- 26.López-Vázquez, A., L. Rodrigo, J. Martínez-Borra, R. Pérez, M. Rodríguez, J. L. Fdez-Morera, D. Fuentes, S. Rodríguez-Rodero, S. González, and C. López-Larrea. 2005. Protective effect of the HLA-Bw4I80 epitope and the killer cell immunoglobulin-like receptor 3DS1 gene against the development of hepatocellular carcinoma in patients with hepatitis C virus infection. J. Infect. Dis. 192:162-165. [DOI] [PubMed] [Google Scholar]
- 27.López-Vázquez, A., A. Miña-Blanco, J. Martínez-Borra, P. D. Njobvu, B. Suárez-Alvarez, M. A. Blanco-Gelaz, S. González, L. Rodrigo, and C. López-Larrea. 2005. Interaction between KIR3DL1 and HLA-B*57 supertype alleles influences the progression of HIV-1 infection in a Zambian population. Hum. Immunol. 66:285-289. [DOI] [PubMed] [Google Scholar]
- 28.Lu, Z., B. Zhang, S. Chen, Z. Gai, Z. Feng, X. Liu, Y. Liu, X. Wen, L. Li, Y. Jiao, C. Ma, S. Shao, X. Cui, G. Chen, J. Li, and Y. Zhao. 2008. Association of KIR genotypes and haplotypes with susceptibility to chronic hepatitis B virus infection in Chinese Han population. Cell. Mol. Immunol. 5:457-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin, M. P., X. Gao, J. H. Lee, G. W. Nelson, R. Detels, J. J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, J. Trowsdale, M. Wilson, S. J. O'Brien, and M. Carrington. 2002. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 31:429-434. [DOI] [PubMed] [Google Scholar]
- 30.Middleton, D., F. Williams, and I. A. Halfpenny. 2005. KIR genes. Transpl. Immunol. 14:135-142. [DOI] [PubMed] [Google Scholar]
- 31.Mimidis, K., V. P. Papadopoulos, I. Elefsiniotis, D. Koliouskas, I. Ketikoglou, E. Paraskevas, S. Kanatakis, D. Chrysagis, G. N. Dalekos, C. Tzathas, A. Protopapas, E. Gigi, E. Tsianos, and G. Kartalis. 2006. Hepatitis C virus survival curve analysis in naïve patients treated with peginterferon alpha-2b plus ribavirin. A randomized controlled trial for induction with high doses of peginterferon and predictability of sustained viral response from early virologic data. J. Gastrointestin. Liver Dis. 15:213-219. [PubMed] [Google Scholar]
- 32.National Institutes of Health. 2002. National Institutes of Health Consensus Development Conference Statement: management of hepatitis C. Gastroenterology 123:2082-2099. [DOI] [PubMed] [Google Scholar]
- 33.Neumann-Haefelin, C., S. McKiernan, S. Ward, S. Viazov, H. C. Spangenberg, T. Killinger, T. F. Baumert, N. Nazarova, I. Sheridan, O. Pybus, F. von Weizsäcker, M. Roggendorf, D. Kelleher, P. Klenerman, H. E. Blum, and R. Thimme. 2006. Dominant influence of an HLA-B27 restricted CD8+ T cell response in mediating HCV clearance and evolution. Hepatology 43:563-572. [DOI] [PubMed] [Google Scholar]
- 34.Parham, P. 2005. MHC class I molecules and KIRs in human history, health and survival. Nat. Rev. Immunol. 5:201-214. [DOI] [PubMed] [Google Scholar]
- 35.Pavio, N., and M. M. Lai. 2003. The hepatitis C virus persistence: how to evade the immune system? J. Biosci. 28:287-304. [DOI] [PubMed] [Google Scholar]
- 36.Pearlman, B. L., C. Ehleben, and S. Saifee. 2007. Treatment extension to 72 weeks of peginterferon and ribavirin in hepatitis C genotype 1-infected slow responders. Hepatology 46:1688-1694. [DOI] [PubMed] [Google Scholar]
- 37.Rajagopalan, S., and E. O. Long. 1998. Zinc bound to the killer cell-inhibitory receptor modulates the negative signal in human NK cells. J. Immunol. 161:1299-1305. [PubMed] [Google Scholar]
- 38.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sievert, W. 2002. Management issues in chronic viral hepatitis: hepatitis C. J. Gastroenterol. Hepatol. 17:415-422. [DOI] [PubMed] [Google Scholar]
- 40.Singh, R., R. Kaul, A. Kaul, and K. Khan. 2007. A comparative review of HLA associations with hepatitis B and C viral infections across global populations. World J. Gastroenterol. 13:1770-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thio, C. L., X. Gao, J. J. Goedert, D. Vlahov, K. E. Nelson, M. W. Hilgartner, S. J. O'Brien, P. Karacki, J. Astemborski, M. Carrington, and D. L. Thomas. 2002. HLA-Cw*04 and hepatitis C virus persistence. J. Virol. 76:4792-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tong, M. J., N. S. el-Farra, A. R. Reikes, and R. L. Co. 1995. Clinical outcomes after transfusion-associated hepatitis C. N. Engl. J. Med. 332:1463-1466. [DOI] [PubMed] [Google Scholar]
- 43.Tseng, C. T., and G. R. Klimpel. 2002. Binding of the hepatitis C virus envelope protein E2 to CD81 inhibits natural killer cell functions. J. Exp. Med. 195:43-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.VandenBussche, C. J., S. Dakshanamurthy, P. E. Posch, and C. K. Hurley. 2006. A single polymorphism disrupts the killer Ig-like receptor 2DL2/2DL3 D1 domain. J. Immunol. 177:5347-5357. [DOI] [PubMed] [Google Scholar]
- 45.Williams, A. P., A. R. Bateman, and S. I. Khakoo. 2005. Hanging in the balance. KIR and their role in disease. Mol. Interv. 5:226-240. [DOI] [PubMed] [Google Scholar]
- 46.Winter, C. C., J. E. Gumperz, P. Parham, E. O. Long, and N. Wagtmann. 1998. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J. Immunol. 161:571-577. [PubMed] [Google Scholar]
- 47.Witt, C. S., C. Dewing, D. C. Sayer, M. Uhrberg, P. Parham, and F. T. Christiansen. 1999. Population frequencies and putative haplotypes of the killer cell immunoglobulin-like receptor sequences and evidence for recombination. Transplantation 68:1784-1789. [DOI] [PubMed] [Google Scholar]
- 48.Wong, T., and S. S. Lee. 2006. Hepatitis C: a review for primary care physicians. CMAJ 174:649-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamagiwa, S., Y. Matsuda, T. Ichida, Y. Honda, M. Takamura, S. Sugahara, T. Ishikawa, S. Ohkoshi, Y. Sato, and Y. Aoyagi. 2008. Sustained response to interferon-alpha plus ribavirin therapy for chronic hepatitis C is closely associated with increased dynamism of intrahepatic natural killer and natural killer T cells. Hepatol. Res. 38:664-672. [DOI] [PubMed] [Google Scholar]
- 50.Yu, J. W., C. Q. Wang, L. J. Sun, X. G. Li, and S. C. Li. 2007. Predictive value of rapid virological response and early virological response on sustained virological response in HCV patients treated with pegylated interferon alpha-2a and ribavirin. J. Gastroenterol. Hepatol. 22:832-836. [DOI] [PubMed] [Google Scholar]