Abstract
West Nile virus capsid protein (WNVCp) displays pathogenic toxicity via the apoptotic pathway. However, a cellular mechanism protective against this toxic effect has not been observed so far. Here, we identified Makorin ring finger protein 1 (MKRN1) as a novel E3 ubiquitin ligase for WNVCp. The cytotoxic effects of WNVCp as well as its expression levels were inhibited in U2OS cells that stably expressed MKRN1. Immunoprecipitation analyses revealed an interaction between MKRN1 and WNVCp. Domain analysis indicated that the C terminus of MKRN1 and the N terminus of WNVCp were required for the interaction. MKRN1 could induce WNVCp ubiquitination and degradation in a proteasome-dependent manner. Interestingly, the WNVCp mutant with amino acids 1 to 105 deleted WNVCp was degraded by MKRN1, whereas the mutant with amino acids 1 to 90 deleted was not. When three lysine sites at positions 101, 103, and 104 of WNVCp were replaced with alanine, MKRN1-mediated ubiquitination and degradation of the mutant were significantly inhibited, suggesting that these sites are required for the ubiquitination. Finally, U2OS cell lines stably expressing MKRN1 were resistant to cytotoxic effects of WNV. In contrast, cells depleted of MKRN1 were more susceptible to WNVCp cytotoxicity. Confirming this, overexpression of MKRN1 significantly reduced, but depletion of MKRN1 increased, WNV proliferation in 293T cells. Taken together, our results suggest that MKRN1 can protect cells from WNV by inducing WNVCp degradation.
West Nile virus (WNV) is an arthropod-borne virus that is a member of the Flaviviridae family, which includes St. Louis encephalitis virus, Kunjin virus, yellow fever virus, dengue virus, and Murray Valley encephalitis virus (2). Since its first identification in the West Nile province of Uganda in 1937, WNV has spread quickly through Asia, Europe, and the United States and has caused a serious global health problem (34). The clinical manifestations of WNV usually entail neurological diseases such as meningitis and encephalitis. This might be caused by WNV genome replication after inoculation and its subsequent spread to lymph nodes and blood, followed by its entrance into the central nervous system through Toll-like receptor and tumor necrosis factor receptor (40).
WNV has the genome of a single positive-sense RNA containing one open reading frame. The encoded polypeptide is processed further by viral and cellular proteases into several nonstructural and structural proteins (2). Nonstructural (NS) proteins include NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5. NS1 is involved in synthesis of viral RNA, and NS3 mediates the cleavage of nonstructural proteins (22, 25, 30, 48). NS5 functions as an RNA polymerase and methyltransferase, which are required for viral replication (14, 17, 18). NS2A, NS2B, NS4A, and NS4B promote the organization of viral replication factors and membrane permeabilization (3, 5, 6, 13, 37). The capsid, envelope (E), and premembrane (prM) proteins are the structural proteins, which are involved in virus assembly (43). E protein is a virion surface protein that regulates binding and fusion to the cell membrane (1, 11, 32). The prM protein is a precursor of the M protein, which is translocated to the endoplasmic reticulum (ER) by capsid (2, 21). Viral assembly occurs mainly in the ER membrane following release of viral particles (23).
The capsid of WNV (WNVCp) localizes and is involved in nucleocapsid assembly on the ER membrane (15). However, extra roles of the flavivirus capsid in the nucleus has been reported. For example, capsid proteins of Japanese encephalitis virus (JEV) and hepatitis C virus (HCV), which are also members of the Flaviviridae family, participate in pathogenesis by localizing to the nucleus (33). Nucleolar and nuclear WNVCp is involved in pathogenesis via induction of the apoptotic process in cells through interaction with Hdm2, which results in the activation of the potent tumor suppressor p53 (47). It also induces apoptotic death of neuron cells via mitochondrial dysfunction and activation of caspase pathways when introduced into the brains of mice (46).
The Makorin ring finger protein 1 (MKRN1) gene was first reported as the source gene of introns for the intronless imprinted MKRN gene family (10). The protein is an ancient protein conserved from invertebrates to vertebrates, and it contains several zinc finger motifs, including C3H, C3HC4, and unique Cys-His motifs (10). Furthermore, this gene is constitutively expressed in most human tissues, including neurons (10). The role of MKRN1 as an E3 ligase was first identified by its ability to degrade hTERT (16). Interestingly, MKRN1 functions as a coregulator of androgen and retinoic acid receptor (27), suggesting possible diverse roles of MKRN1 in human cells.
In this study, we report on an ubiquitin (Ub) E3-ligase for WNVCp. MKRN1 was able to ubiquitinate and degrade WNVCp in a proteasome-dependent manner. Furthermore, degradation of WNVCp resulted in a reduction of WNV-induced cell death. Cells stably overexpressing MKRN1 were resistant to WNV-induced cell death. In contrast, ablation of MKRN1 by small interfering RNA (siRNA) renders cells more susceptible to the cytotoxicity of WNVCp. Furthermore, WNV proliferation was suppressed in 293T cells overexpressing MKRN1 but increased in MKRN1-depleted 293T cells. Based on these data, we suggest that MKRN1 might play a role in protection of cells against WNV infection.
MATERIALS AND METHODS
Cell culture.
H1299 (human lung carcinoma), U2OS (human osteoblast), 293T (human kidney carcinoma), HeLa (cervical carcinoma), and MKRN1 stable U2OS cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA) and 1% penicillin-streptomycin (Invitrogen). Lipofectamine Plus reagent (Invitrogen) and Wellfect (Welgene, Valencia, CA) were used for transfection according to the manufacturer's instructions. An MKRN1 stable U2OS cell line was established via transfection of U2OS with hemagglutinin (HA)-MKRN1, using 600 μg/ml of G418 as a selection marker. Viral amplification and infection were carried out as previously described (47).
Plasmids.
pcDNA3/HA-WNVCp, which contains the membrane anchor sequence, was constructed by subcloning a fragment digested from pcDNA3/his-WNVCp into the pcDNA3/HA vector. pcDNA3/HA-WNVCp deletion mutants (with amino acids 1 to 75, 1 to 90, and 1 to 105 deleted [mutants 1-75, 1-90, and 1-105, respectively) were constructed by PCR using pcDNA3/HA-WNVCp as a template. pcDNA3/HA-WNVCp3KA and pcDNA3/FLAG-MKRN1 H307E were constructed by use of the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). MKRN1 cDNA was obtained from FHGB (21C Frontier Human Gene Bank, Seoul, South Korea), provided by I. K. Chung, Yonsei University. pcDNA3/FLAG-MKRN1 was prepared by PCR of MKRN1 cDNA. pEGFP-MKRN1 deletion mutants (1-109, 110-263, and 264-482) were constructed by PCR using pcDNA3/FLAG-MKRN1 as a template.
Immunoprecipitation.
Cells were plated in a 100-mm-diameter dish. Plasmids were transfected using Wellfect, and after a defined time the cells were lysed with lysis buffer (150 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 0.5% Triton X-100, 0.1% sodium deoxycholate, 50 mM Tris-HCl [pH 7.5], 1 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, and a protease inhibitor mixture). After centrifugation for 10 min at 13,000 rpm, the supernatant was removed and the cell lysate was incubated with antibodies for 2 h at 4°C. The cells were incubated with protein G-Sepharose beads (GE Healthcare, Buckinghamshire, United Kingdom) for 2 h at 4°C. The samples were centrifuged and washed three times with lysis buffer. The samples were added to 2× sample buffer and then boiled.
Ubiquitination assay.
H1299 cells were first transfected with plasmids expressing FLAG-MKRN1, HA-WNVCp, and His-tagged Ub (His-Ub) and then treated with MG132. After 24 h of transfection, the cells were lysed with 6 M guanidine buffer containing 10 mM β-mercaptoethanol, 5 mM N-ethylmaleimide, and 5 mM imidazole prior to incubation with Ni2+-nitrilotriacetic acid (NTA) beads (Qiagen, Valencia, CA). The beads were washed with a buffer containing 8 M urea, 10 mM β-mercaptoethanol, and 0.2% Triton X-100. The beads were then eluted with a sample buffer containing 0.72 M β-mercaptoethanol and 200 mM imidazole.
Cell death analysis.
Cells were trypsinized and harvested with medium. After centrifugation, the supernatant was removed and the cells were suspended in phosphate-buffered saline. The suspended cells were fixed with 70% ethanol for 1 h at 4°C. The ethanol was removed by centrifugation, and the cells were washed. The washed cells were resuspended in phosphate-buffered saline, RNase (40 μg/ml) was added, and the cells were incubated for 30 min at 37°C. Propidium iodide (PI) was added to the cells prior to incubation for 1 h at 37°C. Stained cells were detected by flow cytometry.
Antibodies, chemicals, and siRNAs.
Monoclonal mouse anti-HA (F-7), polyclonal rabbit anti-HA (Y-11), and polyclonal rabbit anti-green fluorescent protein (anti-GFP) (FL) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal anti-FLAG (M2) mouse and anti-β-actin antibodies and PI were obtained from Sigma-Aldrich (St. Louis, MO). Polyclonal rabbit anti-MKRN1 antibody was purchased from Bethyl Laboratories (Montgomery, TX). MG132 was obtained from Calbiochem. MKRN1 siRNA (5′-CAGGCGAAGCTGAGTCAAGAA-3′) and control siRNA were synthesized by Qiagen-Xeragon (Valencia, CA).
Virus infection.
WNV strain NY385-99 (American Type Culture Collection, Manassas, VA) was isolated from a snowy owl in New York during the 1999 epizootic (38). The virus was grown in African green monkey kidney cells (Vero; ATCC CCL-81) in alpha minimum essential medium (Gibco BRL) supplemented with 5% fetal bovine serum (Gibco BRL) and an antibiotic-antimycotic mixture (Invitrogen, Carlsbad, CA). WNV manipulations were performed in a biosafety level 3 maximum-containment research laboratory at the National Veterinary Research and Quarantine Service in accordance with the regulations of the South Korean government. 293T cells transfected with plasmids were inoculated with WNV. Supernatants of infected cells were collected for virus titer determination at 24, 48, and 72 h postinfection.
Plaque assay.
The virus infectivity in sample supernatants was determined by titration in a plaque assay on Vero cells. Briefly, 0.1 ml/well of 10-fold serial dilutions of sample was inoculated onto Vero cell monolayers grown in 24-well tissue culture plates. After incubation for 1 h at 37°C with 5% CO2, unabsorbed residual virus was removed and the cells were overlaid with overlay medium (Earle's basic salts solution containing 1% Noble agar, 2% fetal bovine serum, and antibiotics) and further incubated at 37°C for 3 days while cytopathic effects were monitored. After 3 days, a second overlay medium containing 0.006% neutral red was added to each well. After an additional 18 to 24 h, the number of plaques in each well was counted.
RESULTS
U2OS cells stably expressing MKRN1 are resistant to the cytotoxic effects of WNVCp.
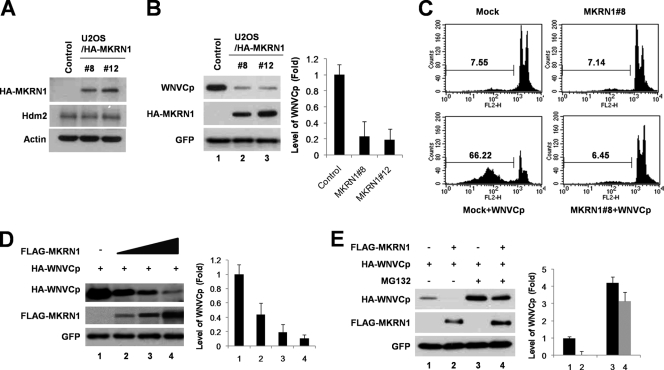
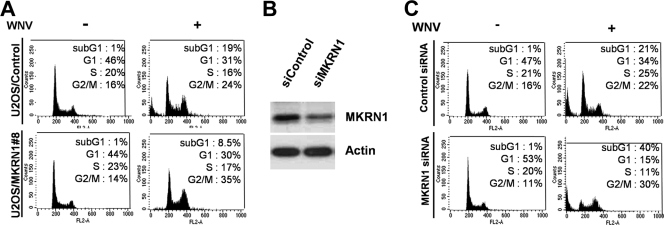
MKRN1 possesses a zinc finger motif involved in E3 ligase activities of p53 and hTERT (10, 16, 20). In previous studies, we reported that WNVCp is able to induce the accumulation of p53 and cellular apoptotic process (47). Since MKRN1 is also p53 E3 ligase, we tested whether WNVCp could be involved in the regulatory pathway of MKRN1. To identify the effect of MKRN1 on WNVCp, cell lines U2OS/HA-MKRN1-8 and -12 of U2OS human osteoblastoma cells, which stably express HA-MKRN1, were established (Fig. 1A) (20). The control cell line was established by introducing pcDNA3-HA into U2OS cells. Hdm2 was detected as an internal control (Fig. 1A). When WNVCp was transfected into the control or MKRN1 stable cell lines, a marked decrease in the levels of capsid (up to 80%) was observed in the MKRN1 stable cell lines compared to the control (Fig. 1B). The cytotoxic effects of WNVCp on the MKRN1 stable or control cell lines were further determined by transient transfection of WNVCp. WNVCp-induced cell death in the control cells after 72 h of transfection was around 66%, while cytotoxicity was prevented in MKRN1 stable cell line 8 (Fig. 1C). We obtained similar results with U2OS/MKRN1-12 (data not shown). Since MKRN1 was able to protect WNVCp-mediated cell death, we next ascertained whether it protected against cytotoxic effects of WNV. When the U2OS control cell line was first infected with WNV for 3 days, there was an approximately 20% increase in the sub-G1 phase, indicative of cell death. On the other hand, the U2OS/HA-MKRN1-8 cell line was quite resistant to the cytotoxic effect induced by WNV, resulting in <10% sub-G1 phase cells (Fig. 2A). Furthermore, we depleted the endogenous levels of MKRN1 using MKRN1 siRNA (Fig. 2B). Ablation of MKRN1 in U2OS cells increased the sub-G1 cell populations from 21% to 40%, suggesting that MKRN1 depletion renders cells more susceptible to the apoptotic effects of WNVCp (Fig. 2C). These observations led us to investigate whether the transient overexpression of MKRN1 was able to induce a reduction in the levels of WNVCp. The results showed that exogenous MKRN1 could induce a reduction in the levels of WNVCp in U2OS cells (Fig. 1D). The reduced level of WNVCp caused by MKRN1 was prevented by the addition of MG132, a proteasome inhibitor (Fig. 1E), suggesting that MKRN1 might help induce proteasome-dependent degradation of WNVCp. Overall, the data are consistent with MKRN1-mediated amelioration of the cytotoxic effects of WNVCp as well as WNV in U2OS cells by preventing the accumulation of WNVCp, possibly through induction of WNVCp degradation.
FIG. 1.
Constitutive expression of MKRN1 reduces levels of exogenous WNVCp and its cytotoxicity. (A) U2OS stable cell lines constitutively and stably expressing HA-MKRN1. U2OS/HA-MKRN1-8 and -12 stable cell lines and control cell lines were lysed, followed by the detection of MKRN1 using anti-HA antibodies. Actin and Hdm2 were detected using antiactin and anti-HDM2 antibodies as internal controls. (B) Low levels of WNVCp under constitutive expression of MKRN1. U2OS/HA-MKRN1-8 and -12 stable cell lines and control cell lines were transfected with a plasmid expressing HA-WNVCp. pEGFP-C2 expressing enhanced GFP was cotransfected as a transfection control. The cell lysates were exposed to anti-HA and anti-GFP antibodies. Error bars indicate standard deviations. (C) Protective effects of MKRN1 against WNVCp cytotoxic effects. U2OS/HA-MKRN1-8 and control cell lines were transfected with mock vector or plasmid expressing WNVCp. After 72 h, the cells were fixed and stained with PI. The stained cells were detected via flow cytometry and analyzed using Cell Quest Pro software. (D) Degradation of WNVCp by MKRN1. U2OS cells were cotransfected with plasmids expressing HA-WNVCp (0.5 μg) and FLAG-MKRN1 (1, 2, and 4 μg) and analyzed by Western blotting using anti-HA and anti-FLAG antibodies. pEGFP-C2 expressing enhanced GFP was cotransfected as a transfection control. Relative amounts of WNVCp were calculated after normalizing to GFP (right panel). The Image J program was used to measure the relative amounts of bands. (E) MKRN1-mediated degradation of WNVCp in a proteasome-dependent pathway. U2OS cells were transfected with the indicated plasmid and treated with 10 μM MG132 for 2 h (lanes 3 and 4). Western blotting was performed as described for panel D. The mean values in panels B, D, and E were quantified using densitometry analysis in three independent experiments.
FIG. 2.
MKRN1 reduces WNV-induced cell death. (A) Reduction of WNV-induced cell death in MKRN1 stable cell line. To assess the effects of MKRN1 overexpression against WNV cytotoxicity, U2OS cells constitutively expressing MKRN1 or a control were seeded at 8 × 105 in a 60-mm-diameter plate and infected with WNV at a multiplicity of infection of 1 PFU/cell. After 72 h, the cells were fixed, followed by PI staining. The stained cells were detected and analyzed via flow cytometry as described for Fig. 1C. (B) Ablation of MKRN1. U2OS cells were transfected using MKRN1 siRNA. After 72 h, the levels of the endogenous MKRN1 were measured using anti-MKRN1 antibodies. (C) Effects of MKRN1 depletion on WNV cytotoxicity. U2OS cells transfected with MKRN1 or control siRNA were seeded at 8 × 105 in a 60-mm-diameter plate and infected with WNV at a multiplicity of infection of 1 PFU/cell. After 72 h, the cells were fixed, followed by PI staining. The stained cells were detected and analyzed by flow cytometry.
MKRN1 and WNVCp bind to each other through the C terminus of MKRN1.
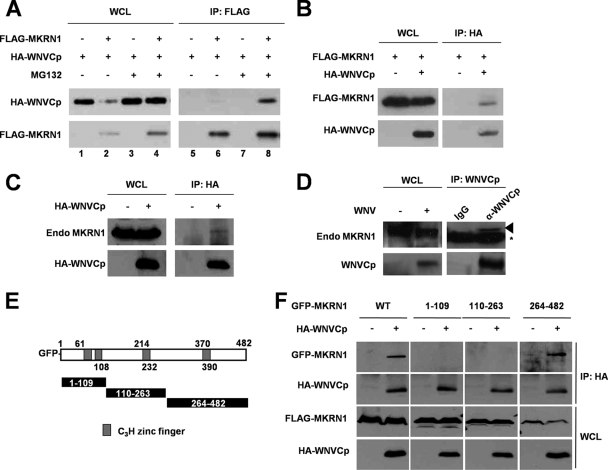
The inhibitory effect of MKRN1 on the expression levels and cytotoxicity of WNVCp suggests that these two proteins might interact with each other. To assess this, immunoprecipitation analysis was carried out. After transient transfection of FLAG-MKRN1 and HA-WNVCp in HeLa cells, FLAG antibodies were used to immunoprecipitate the complexes. MKRN1 and WNVCp were recovered in the same complexes in MG132-treated cells. However, only marginal interaction was detected in non-MG132-treated cells because of degradation of WNVCp by MKRN1 (Fig. 3A). Also, an interaction between WNVCp and MKRN1 was found when the complexes were precipitated using HA antibodies (Fig. 3B). To further confirm the interaction between the two proteins, transiently expressed HA-WNVCp was immunoprecipitated; endogenous MKRN1 bound to the WNVCp was detected, confirming that exogenous WNVCp was able to bind to endogenous MKRN1 (Fig. 3C). Finally, after infection of WNV into 293T cells, the cell extracts were immunoprecipitated using anti-WNVCp antibodies followed by immunoblotting. The data showed that the endogenous MKRN1 was able to interact with the capsid of WNV (Fig. 3D).
FIG. 3.
WNVCp interacts with MKRN1. (A) Interaction between MKRN1 and WNVCp. A plasmid expressing HA-WNVCp was cotransfected with a plasmid expressing FLAG-MKRN1 or the empty vector into HeLa cells in the absence or presence of 10 μM MG132. Cell lysates were immunoprecipitated with anti-FLAG mouse antibodies and immunoblotted with anti-HA rabbit antibodies. (B) Interaction between MKRN1 and WNVCp. Plasmids expressing FLAG-MKRN1 and HA-WNVCp were transfected into HeLa cells. Cells were treated with 10 μM MG132 for 2 h before harvest. Cell lysates were immunoprecipitated using monoclonal anti-HA mouse antibodies. Whole-cell lysates (WCL) and anti-FLAG immunoprecipitates (IP) were analyzed by Western blotting with anti-HA rabbit and anti-MKRN1 rabbit antibodies. (C) Interaction between HA-WNVCp and endogenous MKRN1. Lysates of 293T cells mock transfected or transfected with a plasmid expressing HA-WNVCp were immunoprecipitated using monoclonal anti-HA mouse antibodies. WCL and IP were immunoblotted and detected using polyclonal anti-HA and anti-MKRN1 rabbit antibodies. (D) Interaction between endogenous MKRN1 and WNVCp from infection. 293T cells were seeded at 1 × 106 in a 60-mm-diameter plate and infected with WNV at a multiplicity of infection of 1 PFU/cell. The cell lysates were immunoprecipitated using anti-WNVCp antibodies. WCL and IP were detected by anti-MKRN1 and anti-WNVCp antibodies. (E) Schematic diagrams of MKRN1 deletion mutants. (F) Interaction between MKRN1 fragments and WNVCp. The mock vector or plasmids expressing HA-WNVCp, GFP-MKRN1, or deletion mutants were transfected into 293T cells. The lysates were immunoprecipitated with monoclonal anti-HA mouse antibodies. WCL and IP samples were detected with polyclonal anti-HA and anti-GFP antibodies. WT, wild type.
To further elucidate the interaction between MKRN1 and WNVCp in detail, domain analysis was carried out. Three deletion mutants of MKRN1 labeled with GFP were constructed (Fig. 3E). Immunoprecipitation analysis indicated that the C-terminal fragment, 264-482, was able to interact with WNVCp (Fig. 3F).
MKRN1 induces degradation of WNVCp through ubiquitination.
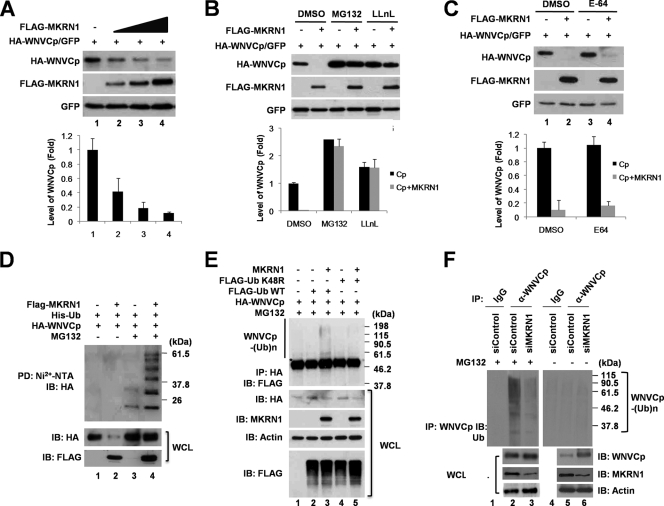
Since MKRN1 binds to WNVCp and induces its degradation in U2OS cells in a proteasome-dependent pathway (Fig. 1D and E), we next asked whether MKRN1-mediated polyubiquitination was involved. In HeLa cells, the degradation of WNVCp was accelerated as the concentration of MKRN1 was increased (Fig. 4A, graph). The same result was observed in H1299 cells (data not shown). Also, we confirmed that the degradation of WNVCp was prevented by MG132 or N-acetyl-l-leucinyl-l-leucinyl-norleucinal (LLnL) treatment (Fig. 4B, graph). On the other hand, the MKRN1-mediated degradation of WNVCp was not affected by E-64, a lysosome inhibitor, suggesting that WNVCp is degraded via a proteasome-dependent pathway (Fig. 4C, graph).
FIG. 4.
MKRN1 induces ubiquitination and degradation of WNVCp. (A) MKRN1-mediated degradation of WNVCp in HeLa cells. HeLa cells were transfected with a plasmid expressing HA-WNVCp (0.5 μg) with increasing amounts of a plasmid expressing FLAG-MKRN1 (1, 2, and 4 μg). The lysates were detected with anti-HA, anti-FLAG, and anti-GFP antibodies. Relative amounts of WNVCp were calculated as described for Fig. 1D. (B) MKRN1-mediated degradation of WNVCp via a proteasome-dependent pathway. HeLa cells transfected with a plasmid expressing HA-WNVCp alone (lanes 1, 3, and 5) or together with a plasmid expressing FLAG-MKRN1 (lanes 2, 4, and 6) were treated with 10 μM MG132 (lanes 3 and 4) or LLnL (lane 5 and 6) for 2 h. The lysates were immunoblotted and detected with anti-HA, anti-FLAG, and anti-GFP antibodies. DMSO, dimethyl sulfoxide. Error bars indicate standard deviations. (C) MKRN1-mediated degradation of capsid in the presence of E-64, a lysosome inhibitor. HeLa cells transfected with a plasmid expressing HA-WNVCp alone (lanes 1 and 3) or together with a plasmid expressing FLAG-MKRN1 (lanes 2 and 4) were treated with 10 μM E-64 (lanes 3 and 4) for 2 h. The lysates were immunoblotted and detected with anti-HA, anti-FLAG, and anti-GFP antibodies. (D) MKRN1-dependent ubiquitination of WNVCp in HeLa cells. Plasmids expressing HA-WNVCp, His-Ub, and FLAG-MKRN1 were transfected into HeLa cells. Cells were treated with MG132 2 h before harvest. The lysates were pulled down (PD) with Ni2+-NTA resin to purify His/Ub. WCL and PD samples were detected with anti-HA and anti-FLAG antibodies. The asterisks indicate nonspecific bands. IB, immunoblotting; WCL, whole-cell lysates. (E) MKRN1-dependent ubiquitination analysis of WNVCp using FLAG-Ub K48R. Plasmids expressing HA-WNVCp, FLAG-Ub, FLAG-Ub K48R, and MKRN1 were transfected into HeLa cells. Cells were treated with MG132 for 2 h before harvest. The lysates were immunoprecipitated using monoclonal anti-HA mouse antibodies. WCL and IP samples were detected with anti-HA, anti-FLAG, antiactin and anti-MKRN1 antibodies. (F) Ubiquitination of WNVCp during WNV infection by endogenous MKRN1. HeLa cells transfected with control siRNA and MKRN1 siRNA were seeded in a 60-mm-diameter plate and infected with WNV at a multiplicity of infection of 1 PFU/cell. At 48 hours after infection, cells were treated with 10 μM of MG132 (lanes 1 to 3) or were not treated (lanes 4 to 6). Cell lysates were immunoprecipitated with anti-WNVCp antibodies. WCL and IP samples were detected with anti-WNVCp, anti-MKRN1, antiactin, and anti-Ub antibodies. The protein quantification was performed as described for Fig. 1.
The ubiquitination analysis using His-Ub indicated that MKRN1 was able to induce polyubiquitinated bands of WNVCp only in the presence of MG132 (Fig. 4D, lanes 3 and 4). Without MG132, there were decreased levels of WNVCp, and the polyubiquitinated bands of WNVCp seemed to be absent due to MKRN1-mediated degradation of WNVCp (Fig. 4D, lanes 1 and 2). MKRN1-induced ubiquitination of WNVCp was observed in H1299 cells (data not shown). When FLAG-Ub K48R, in which residue 48 (lysine) is replaced by arginine and is unable to mediate K48-linked polyubiquitination, was employed for ubiquitination assays, no shifted band was observed (Fig. 4E). This observation suggested that MKRN1-mediated polyubiquitination indeed occurs with WNVCp. To identify the MKRN1-mediated polyubiquitination of WNVCp in a more physiological context, WNV was infected into 293T cells with depletion of MKRN1. Ablation of MKRN1 reduced endogenous polyubiquitinated WNVCp compared to the control (Fig. 4F, lanes 2 and 3).
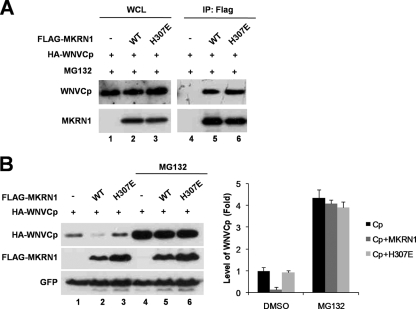
H307E is an MKRN1 point mutant that is defective for E3 ligase activities (20). This mutant was able to bind to WNVCp with an affinity similar to that of wild-type MKRN1 (Fig. 5A), while being unable to induce degradation of WNVCp (Fig. 5B, lanes 1 to 3). When cells were treated with MG132, the levels of WNVCp in the presence of MKRN1 recovered similarly to those of the control or the H307E mutant (Fig. 5B, lanes 4 to 6). These observations suggest that the E3 ligase activity of MKRN1 is required for WNVCp degradation. Overall, the data support the identity of MKRN1 as an E3 ligase of WNVCp which can induce polyubiquitination and degradation of WNVCp.
FIG. 5.
E3 ligase activity is required for degradation of WNVCp. (A) Interaction between MKRN1 H307E and WNVCp. A plasmid expressing HA-WNVCp was cotransfected with a plasmid expressing FLAG-MKRN1 WT or H307E into HeLa cells. Cells were treated with MG132 for 2 h prior to harvest. Immunoprecipitation (IP) assay was carried out as described in the legend to Fig. 2A. WCL, whole-cell lysates. WT, wild type. (B) Requirement of MKRN1 E3 ligase activity for the degradation of HA-WNVCp. Plasmids expressing HA-WNVCp, FLAG-MKRN1 WT, FLAG-MKRN1 H307E, and GFP were transfected into HeLa cells as indicated. Cells were treated with 10 micrograms of MG132 for 2 h (lanes 4 to 6) or not treated (lanes 1 to 3). The lysates were detected with anti-HA, anti-FLAG, and anti-GFP antibodies. The protein quantification was performed as described for FIG. 1. Error bars indicate standard deviations.
WNVCp lysines at sites 101, 103, and 104 are the targets for ubiquitination by MKRN1.
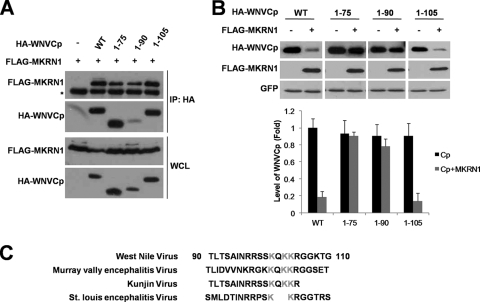
Since it was identified that MKRN1 was able to ubiquitinate WNVCp, we attempted to find a region of WNVCp required for ubiquitination. All three deletion mutants of WNVCp, 1-75, 1-90, and 1-105, were able to bind to MKRN1 (Fig. 6A). When these mutants were tested for MKRN1-dependent degradation in H1299 cells, the mutant 1-105 was significantly degraded compared to the wild type (Fig. 6B). On the other hand, the decrease in the levels of mutants 1-75 and 1-90 in the presence of MKRN1was insignificant compared to the case for the wild type or 1-105. These observations led us to assume that the region between positions 90 and 105 was the most critical for MKRN1-mediated polyubiquitination and degradation of WNVCp.
FIG. 6.
WNVCp contains the region from amino acid 90 to 105 responsible for MKRN1-dependent degradation. (A) Requirement of the N terminus of WNVCp for MKRN1 interaction. A plasmid expressing FLAG-MKRN1 was transfected with or without HA-WNVCp or its deletion mutants in 293T cells. Cell lysates were immunoprecipitated with anti-FLAG antibodies. Whole-cell lysates (WCL) and immunoprecipitates (IP) were detected with anti-HA and anti-FLAG antibodies. The asterisk indicates heavy chains of antibodies. WT, wild type. (B) Requirement of the 90-105 region of WNVCp for MKRN1-dependent degradation. Mock vector or plasmids expressing FLAG-MKRN1, HA-WNVCp, or HA-WNVCp deletion mutants were transfected into H1299 cells. pEGFP-C2 expressing enhanced GFP was cotransfected as a transfection control. The lysates were detected with anti-HA antibodies, anti-FLAG antibodies, and anti-GFP antibodies. The protein quantification was performed as described for Fig. 1. Error bars indicate standard deviations. (C) Alignment of WNVCp orthologs containing conserved lysine residues. Gray shading indicates lysine residues conserved throughout flaviviruses.
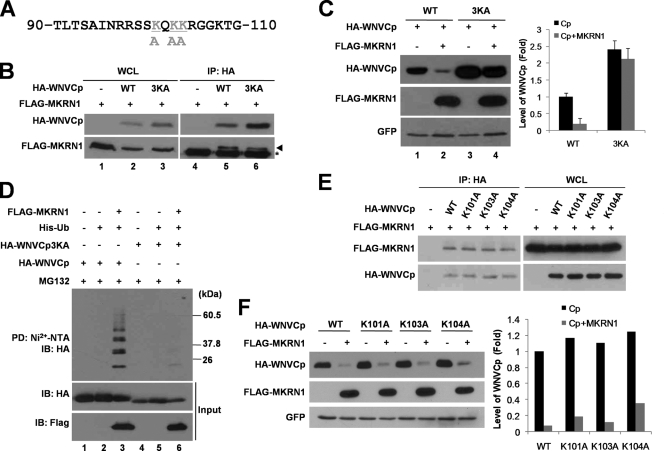
When the region between positions 90 and 105 was analyzed, three lysine residues (101, 103, and 104) were found to be conserved in flaviviruses, including Murray Valley encephalitis virus, Kunjin virus, and St. Louis encephalitis virus, while they were not conserved in dengue virus or yellow fever virus (Fig. 6C). To further investigate whether these sites were involved in MKRN1-dependent ubiquitination and degradation, the lysine residues were replaced with alanine (Fig. 7A). The WNVCp point mutant 3KA was expressible and able to bind to MKRN1 like wild-type WNVCp in 293T cells (Fig. 7B, lanes 3 and 6). When 3KA was expressed in H1299, the expression level was increased up to 2.4-fold compared to that of the wild type, suggesting that this mutant might be more resistant to degradation within the cells (Fig. 7C, lanes 1 and 3). When MKRN1 was cotransfected with wild-type WNVCp or 3KA, 3KA was observed to be quite resistant to MKRN1-dependent degradation (Fig. 7C, lanes 3 and 4). Finally, ubiquitination analysis showed that WNVCp 3KA was not as readily ubiquitinated by MKRN1 as the wild type, implicating the three lysines as ubiquitination sites (Fig. 7D, lane 6). We further constructed three point mutants of WNVCp, K101A, K103A, and K104A, to identify which lysine site was the most responsible for MKRN1-mediated ubiquitination of WNVCp. All three point mutants were able to bind to MKRN1 like the wild type (Fig. 7E). The mutants were not resistant to MKRN1-mediated degradation (Fig. 7F). These observations suggest that all three or at least two lysine sites are required for ubiquitination of WNVCp. Overall, our data suggest that MKRN1 specifically targets three lysine sites located at positions 101, 103, and 104 for polyubiquitination and subsequent degradation.
FIG. 7.
Lysines at residues 101, 103, and 104 are important for ubiquitination labeling and nuclear localization. (A) Sequence of WNVCp between positions 90 and 110. Gray shading indicates lysine residues replaced by alanine. (B) Interaction between 3KA and MKRN1. A plasmid expressing FLAG-MKRN1 was cotransfected with or without a plasmid expressing HA-WNVCp or HA-WNVCp 3KA in 293T cells. The whole-cell lysates (WCL) were immunoprecipitated (IP) with anti-HA antibodies. WCL and IP samples were detected with anti-HA and anti-FLAG antibodies. WT, wild type. (C) MKRN1-mediated degradation of 3KA. H1299 cells were transfected with a plasmid expressing FLAG-MKRN1 in the presence or absence of HA-WNVCp and 3KA. pEGFP-C2 expressing enhanced GFP was cotransfected as a transfection control. The cell lysates were detected with anti-HA, anti-FLAG, and anti-GFP antibodies. Protein quantification was performed as described for Fig. 1. Error bars indicate standard deviations. (D) Ubiquitination of WNVCp 3KA by MKRN1. Plasmids expressing HA-WNVCp, HA-WNVCp 3KA, FLAG-MKRN1, and His-Ub were transfected into H1299 cells. Cells were treated MG132 for 3 h before harvest. The cell lysates were pulled down (PD) with Ni2+-NTA resin. WCL and PD samples were detected with anti-HA and anti-FLAG antibodies. (E) Interaction between WNVCp point mutants and MKRN1. A plasmid expressing FLAG-MKRN1 was transfected with or without HA-WNVCp or its point mutants in 293T cells. WCL were immunoprecipitated with anti-HA antibodies. WCL and IP were detected with anti-HA and anti-MKRN1 antibodies. (F) Degradation of WNVCp point mutants by MKRN1. Mock vector or plasmids expressing FLAG-MKRN1, HA-WNVCp, or HA-WNVCp point mutants were transfected into H1299 cells. pEGFP-C2 expressing enhanced GFP was cotransfected as a transfection control. The lysates were detected with anti-HA antibodies, anti-FLAG antibodies, and anti-GFP antibodies.
MKRN1 prevents WNV-induced cell death.
To further identify the protective effects of MKRN1 on WNV, we tested whether MKRN1 expression could affect WNV-mediated cell death of 293T cells. As a control, we included MKRN1 H307E mutants and Hdm2. Upon infection of WNV on mock vector-transfected 293T cells, 18% cell death was observed. In contrast, the expression of MKRN1 almost completely suppressed the cytotoxic effects of WNVCp. The H307E mutant or Hdm2 did not have any significant effects on WNV cytotoxic effects (Fig. 8A). Proliferation of WNV upon infection of 293T cells showed an exponential virus proliferation after 2 days of infection. On the other hand, overexpression of MKRN1 in these cells suppressed the exponential virus growth after 2 days (Fig. 8B). As shown in Fig. 8A, neither the H307E mutant nor Hdm2 displayed any inhibitory effects on viral proliferation. When MKRN1 was ablated in 293T cells (Fig. 8C), there was a significant increase of cell death from 10% to 45%, indicative of a negative effect of MKRN1 on WNV cytotoxicity (Fig. 8D). Corroborating these observations, a lack of MKRN1 actually increased the viral replication compared to that for the control (Fig. 8E). The data indicate that MKRN1 protected cells against WNV infection. Moreover, MKRN1 was able to suppress virus proliferation, suggesting that MKRN1 could repress viral growth within human cells.
FIG. 8.
MKRN1 protects cells against WNV-induced cell death and inhibits WNV replication. (A) Effects of MKRN1 overexpression on WNV cytotoxicity. 293T cells transiently expressing MKRN1, MKRN1 H307E, or Hdm2 were seeded at 8 × 105 in a 60-mm-diameter plate and infected with WNV at a multiplicity of infection of 1 PFU/cell. After 72 h, the cells were fixed, followed by PI staining. The stained cells were detected and analyzed via flow cytometry as described for Fig. 1C. (B) WNV replication in MKRN1-overexpressing 293T cells. MKRN1-, MKRN1 H307E-, or Hdm2-overexpressing 293T cells were seeded at 3.9 × 105 cells in six-well tissue culture plates and then infected with WNV at a multiplicity of infection of 100 PFU/cell. Virus replication titers at 24, 36, 40, 44, and 64 h postinfection were determined using plaque assay on Vero cells. This experiment was repeated three times. WT, wild type. Error bars indicate standard deviations. (C) Ablation of MKRN1. 293T cells were transfected using MKRN1 siRNA. After 72 h, the levels of the endogenous MKRN1 were measured using anti-MKRN1 antibodies. (D) Effects of MKRN1 depletion against WNV cytotoxicity. 293T cells transfected with MKRN1 or control siRNA were seeded at 8 × 105 in a 60-mm-diameter plate and infected with WNV at a multiplicity of infection of 1 PFU/cell. After 72 h, the cells were fixed, followed by PI staining. The stained cells were detected and analyzed by flow cytometry. (E) Effect of MKRN1 depletion on WNV replication. 293T cells transfected with control siRNA or MKRN1 siRNA were seeded at 3.9 × 105 cells in six-well tissue culture plates and then infected with WNV at a multiplicity of infection of 100 PFU/cell. Virus replication titers at 24, 36, 40, 44, and 64 h postinfection were determined using plaque assay on Vero cells. This experiment was repeated three times. Error bars indicate standard deviations.
DISCUSSION
Flaviviruses, including WNV, commence their replicative cycle by attaching to the surface of host cells; attachment is followed by receptor-mediated endocytosis (11). Once in the cells, the positive-sense RNAs are translated and further processed to the structural and nonstructural proteins (2). The assembly of the virus, which takes place on the ER membrane, requires the initial formation of the membrane-associated nucleocapsid followed by the association of PrM-E heterodimers (12, 42). Thus, in the process of viral assembly, capsid proteins localize on the ER. However, the capsid proteins of flaviviruses, including WNV, Dengue virus, Kunjin virus, JEV, and HCV, can also localize in the nucleus and nucleolus (4, 24, 26, 33, 39, 44). The effects of capsid protein localized to the nucleus and the nucleolus have been implicated in the enhancement of viral replication or induction of apoptotic cell death. In JEV, a mutant capsid protein defective in the nuclear localization signal displays a low level of viral replication (24). Previously, we reported that the nucleolar localization of WNVCp induced relocalization of Hdm2, an E3 ligase of apoptotic protein p53, and resulted in the accumulation of p53 and induction of apoptosis in cancer and neuron cells (47). Thus, it seems that capsid proteins of flaviviruses have a role in the nucleus to enhance viral propagation in addition to its main role in viral genome assembly.
The present study has identified a novel WNVCp E3 ligase, MKRN1. MKRN1 is known to function as an E3 ligase of hTERT, p21, and p53 (16, 20). However, other substrates of MKRN1 are largely unknown and require further investigation. We established U2OS cell lines constitutively overexpressing MKRN1 to identify the role of MKRN1 against various cellular stresses. Since U2OS cells are susceptible to the cytotoxic effects of WNVCp (47), we transfected WNVCp into U2OS cells. The results demonstrate that cell lines with constitutive expression of MKRN1 are quite resistant to the cytotoxic effects of WNVCp (Fig. 1C). These results corroborate observations of reduced exogenous levels of WNVCp in the MKRN1 stable cell line (Fig. 1B). Furthermore, MKRN1 was able to reduce the levels of WNVCp by catalyzing ubiquitination followed by a 20S proteasome-dependent degradation (Fig. 4). On the other hand, MKRN1 H307E, a mutant defective in ligase activity, was not able to induce degradation of WNVCp, while it interacted with the capsid protein (Fig. 5). The identification of three lysine sites of WNVCp responsible for MKRN1-dependent ubiquitination indicates that MKRN1 is indeed an E3 ligase of WNVCp protein (Fig. 7). MKRN1 is not the first flavivirus E3 ligase to be identified, suggesting that a protective mechanism of host cells against viral proteins includes posttranslational modification leading to proteasomal degradation. For example, E6AP, an activator of E6 protein from human papillomavirus types 16 and 18, mediates degradation of HCV capsid through ubiquitination (35). Upon expression of this protein, viral propagation is suppressed, indicating that this protein functions as a host defense factor. Several other proteins, such as E2, NS3, and F in HCV, also undergo ubiquitination (9, 29, 45). A recent RNA interference screening of human genes responsible for interaction with WNV proteins identified the Ub ligase CBLL1 as being critical for WNV internalization (19). Thus, it seems that flaviviruses can also employ host E3 ligases in favor of their propagation.
Elucidation of the protective mechanisms against WNV infection has largely focused on studies at the systemic level, which include studies of immune cells and signals (7, 8, 36, 41). The cytotoxicity of WNV for individual cells has also been studied recently. For example, WNV is known to induce Bax-dependent or -independent apoptosis in cancer and brain cells (28). Factors such as NS2B-NS3 and capsid also enforce the cytotoxic effects of WNV through the apoptotic process (31). On the other hand, cellular components that protect host cells from WNV are not well known. Recent studies concerning the comprehensive interactions between host and WNV proteins (19) are beginning to reveal the complex nature of the communications between host cells and virus. In this context, the characterization of MKRN1 as the first E3 ligase of WNVCp will be meaningful for greater understanding of host-virus interaction. It is not absolutely clear presently whether E3 ligase activity of MKRN1 is required for protection against viral replication and cytotoxicity unless WNV with a 3KA mutant capsid is employed and tested for cytotoxicity and viral propagation. Moreover, studies have to be carried out to identify whether MKRN1 can affect other flavivirus capsid proteins. Due to the high homology of capsid protein in the ubiquitination site (Fig. 6C), it is very probable that MKRN1 might participate in similar degradation pathways in other flaviviruses. Currently, the possible effects of MKRN1 on capsid proteins in the same Falvividae virus are being studied. Overexpressed MKRN1 protected cells from the cytotoxic effects of WNVCp (Fig. 8A), while depletion of MKRN1 by siRNA made cells more susceptible to WNVCp cytotoxicity (Fig. 8D). Furthermore, viral propagation was prevented by the overexpressed MKRN1 (Fig. 8B) but promoted by depletion of MKRN1 by siRNA (Fig. 8E). Based on these observations, we suggest that MKRN1 may function as a host cell protector against flaviviral infection by depleting capsid proteins through ubiquitination and degradation.
This is the first report, to our knowledge, to identify an E3 ligase for WNVCp. Since MKRN1 is a novel E3 ligase with no clear regulatory mechanisms observed so far, further mechanistic studies of regulation of MKRN1 could help to elucidate the cellular response against WNV infection.
Acknowledgments
This work was supported by a grant (A080333) from the Korea Healthcare Technology R&D Project, Ministry of Health & Welfare, Republic of Korea; a grant (Z-AD16-2007-07-05) from the National Veterinary Research & Quarantine Service, Ministry for Food, Agriculture, Forestry and Fisheries; and a grant (R01-2007-000-20327-0) from the Korea Science and Engineering Foundation (KOSEF) funded by the Korea government (MOST).
Footnotes
Published ahead of print on 21 October 2009.
REFERENCES
- 1.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, and F. X. Heinz. 2001. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J. Virol. 75:4268-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brinton, M. A. 2002. The molecular biology of West Nile Virus: a new invader of the western hemisphere. Annu. Rev. Microbiol. 56:371-402. [DOI] [PubMed] [Google Scholar]
- 3.Chambers, T. J., A. Nestorowicz, S. M. Amberg, and C. M. Rice. 1993. Mutagenesis of the yellow fever virus NS2B protein: effects on proteolytic processing, NS2B-NS3 complex formation, and viral replication. J. Virol. 67:6797-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, S. C., J. H. Yen, H. Y. Kang, M. H. Jang, and M. F. Chang. 1994. Nuclear localization signals in the core protein of hepatitis C virus. Biochem. Biophys. Res. Commun. 205:1284-1290. [DOI] [PubMed] [Google Scholar]
- 5.Chang, Y. S., C. L. Liao, C. H. Tsao, M. C. Chen, C. I. Liu, L. K. Chen, and Y. L. Lin. 1999. Membrane permeabilization by small hydrophobic nonstructural proteins of Japanese encephalitis virus. J. Virol. 73:6257-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clum, S., K. E. Ebner, and R. Padmanabhan. 1997. Cotranslational membrane insertion of the serine proteinase precursor NS2B-NS3(Pro) of dengue virus type 2 is required for efficient in vitro processing and is mediated through the hydrophobic regions of NS2B. J. Biol. Chem. 272:30715-30723. [DOI] [PubMed] [Google Scholar]
- 7.Diamond, M. S., B. Shrestha, A. Marri, D. Mahan, and M. Engle. 2003. B cells and antibody play critical roles in the immediate defense of disseminated infection by West Nile encephalitis virus. J. Virol. 77:2578-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diamond, M. S., E. M. Sitati, L. D. Friend, S. Higgs, B. Shrestha, and M. Engle. 2003. A critical role for induced IgM in the protection against West Nile virus infection. J. Exp. Med. 198:1853-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franck, N., J. Le Seyec, C. Guguen-Guillouzo, and L. Erdtmann. 2005. Hepatitis C virus NS2 protein is phosphorylated by the protein kinase CK2 and targeted for degradation to the proteasome. J. Virol. 79:2700-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray, T. A., L. Hernandez, A. H. Carey, M. A. Schaldach, M. J. Smithwick, K. Rus, J. A. Marshall Graves, C. L. Stewart, and R. D. Nicholls. 2000. The ancient source of a distinct gene family encoding proteins featuring RING and C(3)H zinc-finger motifs with abundant expression in developing brain and nervous system. Genomics 66:76-86. [DOI] [PubMed] [Google Scholar]
- 11.Heinz, F. X., and S. L. Allison. 2000. Structures and mechanisms in flavivirus fusion. Adv. Virus Res. 55:231-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heinz, F. X., K. Stiasny, G. Puschner-Auer, H. Holzmann, S. L. Allison, C. W. Mandl, and C. Kunz. 1994. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology 198:109-117. [DOI] [PubMed] [Google Scholar]
- 13.Khromykh, A. A., P. L. Sedlak, and E. G. Westaway. 2000. cis- and trans-acting elements in flavivirus RNA replication. J. Virol. 74:3253-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khromykh, A. A., P. L. Sedlak, and E. G. Westaway. 1999. trans-Complementation analysis of the flavivirus Kunjin ns5 gene reveals an essential role for translation of its N-terminal half in RNA replication. J. Virol. 73:9247-9255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khromykh, A. A., and E. G. Westaway. 1996. RNA binding properties of core protein of the flavivirus Kunjin. Arch. Virol. 141:685-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, J. H., S. M. Park, M. R. Kang, S. Y. Oh, T. H. Lee, M. T. Muller, and I. K. Chung. 2005. Ubiquitin ligase MKRN1 modulates telomere length homeostasis through a proteolysis of hTERT. Genes Dev. 19:776-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koonin, E. V. 1993. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and lambda 2 protein of reovirus. J. Gen. Virol. 74:733-740. [DOI] [PubMed] [Google Scholar]
- 18.Koonin, E. V. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72:2197-2206. [DOI] [PubMed] [Google Scholar]
- 19.Krishnan, M. N., A. Ng, B. Sukumaran, F. D. Gilfoy, P. D. Uchil, H. Sultana, A. L. Brass, R. Adametz, M. Tsui, F. Qian, R. R. Montgomery, S. Lev, P. W. Mason, R. A. Koski, S. J. Elledge, R. J. Xavier, H. Agaisse, and E. Fikrig. 2008. RNA interference screen for human genes associated with West Nile virus infection. Nature 455:242-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, E. W., M. S. Lee, S. Camus, J. Ghim, M. R. Yang, W. Oh, N. C. Ha, D. P. Lane, and J. Song. 2009. Differential regulation of p53 and p21 by MKRN1 E3 ligase controls cell cycle arrest and apoptosis. EMBO J. 28:2100-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindenbach, B. D., and C. M. Rice. 2003. Molecular biology of flaviviruses. Adv. Virus Res. 59:23-61. [DOI] [PubMed] [Google Scholar]
- 22.Lindenbach, B. D., and C. M. Rice. 1997. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J. Virol. 71:9608-9617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mackenzie, J. M., and E. G. Westaway. 2001. Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J. Virol. 75:10787-10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mori, Y., T. Okabayashi, T. Yamashita, Z. Zhao, T. Wakita, K. Yasui, F. Hasebe, M. Tadano, E. Konishi, K. Moriishi, and Y. Matsuura. 2005. Nuclear localization of Japanese encephalitis virus core protein enhances viral replication. J. Virol. 79:3448-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muylaert, I. R., R. Galler, and C. M. Rice. 1997. Genetic analysis of the yellow fever virus NS1 protein: identification of a temperature-sensitive mutation which blocks RNA accumulation. J. Virol. 71:291-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oh, W., M. R. Yang, E. W. Lee, K. M. Park, S. Pyo, J. S. Yang, H. W. Lee, and J. Song. 2006. Jab1 mediates cytoplasmic localization and degradation of West Nile virus capsid protein. J. Biol. Chem. 281:30166-30174. [DOI] [PubMed] [Google Scholar]
- 27.Omwancha, J., X. F. Zhou, S. Y. Chen, T. Baslan, C. J. Fisher, Z. Zheng, C. Cai, and L. Shemshedini. 2006. Makorin RING finger protein 1 (MKRN1) has negative and positive effects on RNA polymerase II-dependent transcription. Endocrine 29:363-373. [DOI] [PubMed] [Google Scholar]
- 28.Parquet, M. C., A. Kumatori, F. Hasebe, K. Morita, and A. Igarashi. 2001. West Nile virus-induced bax-dependent apoptosis. FEBS Lett. 500:17-24. [DOI] [PubMed] [Google Scholar]
- 29.Pavio, N., D. R. Taylor, and M. M. Lai. 2002. Detection of a novel unglycosylated form of hepatitis C virus E2 envelope protein that is located in the cytosol and interacts with PKR. J. Virol. 76:1265-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Preugschat, F., C. W. Yao, and J. H. Strauss. 1990. In vitro processing of dengue virus type 2 nonstructural proteins NS2A, NS2B, and NS3. J. Virol. 64:4364-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramanathan, M. P., J. A. Chambers, P. Pankhong, M. Chattergoon, W. Attatippaholkun, K. Dang, N. Shah, and D. B. Weiner. 2006. Host cell killing by the West Nile Virus NS2B-NS3 proteolytic complex: NS3 alone is sufficient to recruit caspase-8-based apoptotic pathway. Virology 345:56-72. [DOI] [PubMed] [Google Scholar]
- 32.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 33.Sacco, R., T. Tsutsumi, R. Suzuki, M. Otsuka, H. Aizaki, S. Sakamoto, M. Matsuda, N. Seki, Y. Matsuura, T. Miyamura, and T. Suzuki. 2003. Antiapoptotic regulation by hepatitis C virus core protein through up-regulation of inhibitor of caspase-activated DNase. Virology 317:24-35. [DOI] [PubMed] [Google Scholar]
- 34.Sampson, B. A., C. Ambrosi, A. Charlot, K. Reiber, J. F. Veress, and V. Armbrustmacher. 2000. The pathology of human West Nile Virus infection. Hum. Pathol. 31:527-531. [DOI] [PubMed] [Google Scholar]
- 35.Shirakura, M., K. Murakami, T. Ichimura, R. Suzuki, T. Shimoji, K. Fukuda, K. Abe, S. Sato, M. Fukasawa, Y. Yamakawa, M. Nishijima, K. Moriishi, Y. Matsuura, T. Wakita, T. Suzuki, P. M. Howley, T. Miyamura, and I. Shoji. 2007. E6AP ubiquitin ligase mediates ubiquitylation and degradation of hepatitis C virus core protein. J. Virol. 81:1174-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shrestha, B., and M. S. Diamond. 2004. Role of CD8+ T cells in control of West Nile virus infection. J. Virol. 78:8312-8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Speight, G., and E. G. Westaway. 1989. Positive identification of NS4A, the last of the hypothetical nonstructural proteins of flaviviruses. Virology 170:299-301. [DOI] [PubMed] [Google Scholar]
- 38.Steele, K. E., M. J. Linn, R. J. Schoepp, N. Komar, T. W. Geisbert, R. M. Manduca, P. P. Calle, B. L. Raphael, T. L. Clippinger, T. Larsen, J. Smith, R. S. Lanciotti, N. A. Panella, and T. S. McNamara. 2000. Pathology of fatal West Nile virus infections in native and exotic birds during the 1999 outbreak in New York City, New York. Vet. Pathol. 37:208-224. [DOI] [PubMed] [Google Scholar]
- 39.Wang, S. H., W. J. Syu, K. J. Huang, H. Y. Lei, C. W. Yao, C. C. King, and S. T. Hu. 2002. Intracellular localization and determination of a nuclear localization signal of the core protein of dengue virus. J. Gen. Virol. 83:3093-3102. [DOI] [PubMed] [Google Scholar]
- 40.Wang, T., T. Town, L. Alexopoulou, J. F. Anderson, E. Fikrig, and R. A. Flavell. 2004. Toll-like receptor 3 mediates West Nile virus entry into the brain causing lethal encephalitis. Nat. Med. 10:1366-1373. [DOI] [PubMed] [Google Scholar]
- 41.Wang, Y., M. Lobigs, E. Lee, and A. Mullbacher. 2003. CD8+ T cells mediate recovery and immunopathology in West Nile virus encephalitis. J. Virol. 77:13323-13334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wengler, G., and G. Wengler. 1989. Cell-associated West Nile flavivirus is covered with E+pre-M protein heterodimers which are destroyed and reorganized by proteolytic cleavage during virus release. J. Virol. 63:2521-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Westaway, E. G., M. A. Brinton, S. Gaidamovich, M. C. Horzinek, A. Igarashi, L. Kaariainen, D. K. Lvov, J. S. Porterfield, P. K. Russell, and D. W. Trent. 1985. Flaviviridae. Intervirology 24:183-192. [DOI] [PubMed] [Google Scholar]
- 44.Westaway, E. G., A. A. Khromykh, M. T. Kenney, J. M. Mackenzie, and M. K. Jones. 1997. Proteins C and NS4B of the flavivirus Kunjin translocate independently into the nucleus. Virology 234:31-41. [DOI] [PubMed] [Google Scholar]
- 45.Xu, Z., J. Choi, W. Lu, and J. H. Ou. 2003. Hepatitis C virus f protein is a short-lived protein associated with the endoplasmic reticulum. J. Virol. 77:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang, J. S., M. P. Ramanathan, K. Muthumani, A. Y. Choo, S. H. Jin, Q. C. Yu, D. S. Hwang, D. K. Choo, M. D. Lee, K. Dang, W. Tang, J. J. Kim, and D. B. Weiner. 2002. Induction of inflammation by West Nile virus capsid through the caspase-9 apoptotic pathway. Emerg. Infect. Dis. 8:1379-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang, M. R., S. R. Lee, W. Oh, E. W. Lee, J. Y. Yeh, J. J. Nah, Y. S. Joo, J. Shin, H. W. Lee, S. Pyo, and J. Song. 2008. West Nile virus capsid protein induces p53-mediated apoptosis via the sequestration of HDM2 to the nucleolus. Cell. Microbiol. 10:165-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yusof, R., S. Clum, M. Wetzel, H. M. Murthy, and R. Padmanabhan. 2000. Purified NS2B/NS3 serine protease of dengue virus type 2 exhibits cofactor NS2B dependence for cleavage of substrates with dibasic amino acids in vitro. J. Biol. Chem. 275:9963-9969. [DOI] [PubMed] [Google Scholar]