Abstract
MicroRNAs (miRNAs) have emerged as key regulators in many biological processes, from development to defense, at almost all organismal levels. Recently, their role has been highlighted in pathogen-host interactions. Emerging evidence from a variety of virus-host systems indicates that cellular as well as virally encoded miRNAs influence viral replication. Here, we report changes in expression levels of host miRNAs upon ascovirus infection in an insect cell line and investigated the role of a host miRNA, Hz-miR24, in the host-virus system. We found that Hz-miR24 is differentially expressed following virus infection, with an increase in its expression levels late in infection. Experimental evidence demonstrated that Hz-miR24 downregulates ascoviral DNA-dependent RNA polymerase and its β subunit transcript levels late in infection. The specific miRNA-target interactions were investigated and confirmed using the ectopic expression of Hz-miR24 and a green fluorescent protein-based reporter system. Further, the expression of the target gene was substantially enhanced in cells transfected with a synthesized inhibitor of Hz-miR24. These findings suggest that ascoviruses manipulate host miRNAs that in turn regulate the expression of their genes at specific time points after infection. To our knowledge, this is the first cellular miRNA reported to interact with an insect virus.
MicroRNAs (miRNAs) are 18- to 25-nucleotide (nt) small noncoding RNAs that are involved in the regulation of gene expression. During this decade, there has been a tremendous increase in the number of miRNAs discovered in organisms ranging from unicellular organisms to humans to some viruses. Their roles have been revealed in development, defense, and apoptosis, and they have been estimated to regulate more than one-third of the human genome (14, 29). The first step in miRNA biogenesis includes the transcription of a primary RNA transcript (pri-miRNA) of various sizes by RNA polymerase II or III (7). Pri-miRNA is cleaved by drosha in the nucleus to a 60- to 80-nt precursor miRNA (pre-miRNA) with multiple mismatches and bulges, which then is exported to the cytoplasm and cleaved by dicer 1, generating an approximately 22-nt double-stranded miRNA. Usually, only one strand with weaker 5′ base pairing participates in binding to mRNA after it is recruited by RNA-induced silencing complex proteins (32, 34). Recent findings have shown multiple binding sites of mRNA-miRNA interaction, namely the 5′-untranslated region, 3′-untranslated region, and the coding region (5, 21). The fate of mRNA in this interaction, either mRNA cleavage or translation inhibition, depends on several factors, including complementarity in the seed region (2 to 8 nt from the 5′ end) and the rest of the miRNA sequence. At present, in addition to virus-encoded miRNAs, cellular miRNAs in relation to virus biology are being investigated as another level of virus-host interaction. The expression level of cellular miRNAs may be induced or inhibited upon virus infection and may help their replication or inhibit them through targeting viral mRNAs (6). The tissue tropism of viruses also can be explained by their impact on the level of certain cellular miRNAs (29). There are published examples that highlight the interaction of cellular miRNAs with virus infection. A cellular miRNA (miR-32) inhibits the accumulation of primate foamy virus type 1 (20), miR-122 facilitates viral replication by binding to the 5′ end of the hepatitis C virus genome (18), human immunodeficiency virus type 1 downregulates cellular miRNAs that have negative impacts on viral growth (28), and Epstein-Barr virus mediates the dysregulation of cellular miRNAs (13).
Ascoviruses (AV) cause lethal infection in lepidopteran insects, mostly those belonging to the family Noctuidae (25). These viruses possess a single circular double-stranded DNA genome ranging from 119 to 186 kbp and is encapsidated in virions of allantoid to bacilliform shapes (10). There are four recognized AV species, Spodoptera frugiperda AV (SfAV-1), Trichoplusia ni AV (TnAV-2), Heliothis virescens AV (HvAV-3), and Diadromus pulchellus AV (DpAV-4), and their complete genome sequences now are available (1-3, 31). AVs are transmitted by wasp vectors, and their association with the vector could either be antagonistic (SfAV-1, TnAV-2, and HvAV-3) or mutualistic (DpAV-4) (4). The salient changes in cell morphology upon AV infection start with the enlargement and breakdown of nuclei, leading to the fragmentation of cells and vesicle formation. These vesicles are disseminated in the hemocoel and produce milky white hemolymph, which results in the death of the host insect (10). They encode three predicted proteins with homology to eukaryotic RNA polymerase RPC1, RPC2, and β subunits. The regulation of two of these subunits (RPC2 and β) is the subject of this report.
Previously, we reported an miRNA (HvAV-miR1) encoded by HvAV-3e that specifically targets transcripts of the virus-encoded DNA polymerase (ORF1) regulating viral replication (17). Here, we investigated the differential expression of cellular miRNAs from an insect cell line derived from Helicoverpa zea fat bodies (HzFB) upon infection with HvAV-3e and further analyzed, in detail, the role of a cellular miRNA (Hz-miR24) in host-virus interactions. The expression of Hz-miR24 drops to a very low level between 24 and 48 h postinfection (hpi) and then increases to significantly higher levels after 72 h of virus infection. We analyzed Hz-miR24 at the functional level and found that it regulates the expression of viral DNA-dependent RNA polymerase II RPC2 (DdRP; ORF64) and DdRP β subunits (DdRPβ; ORF82) by specifically interacting with the targets.
MATERIALS AND METHODS
Cell cultures and virus infection.
HzFB cells (22) were maintained in SF900-II serum-free medium (Invitrogen) as a monolayer in cell culture flasks at 27°C.
The AV isolate HvAV-3e, extracted from infected Helicoverpa armigera larvae and amplified in HzFB cells, were used throughout the experiments (1). For virus infection, cells were transferred into 35- by 10-mm petri dishes (Falcon) and allowed to settle for 1 h. Medium was removed and cells were inoculated with 106 HvAV-3e PFU as determined by plaque assay (19). An hour after incubation at room temperature with shaking, fresh medium was added to the cells and incubated further at 27°C.
Cloning of cellular miRNAs.
miRNAs were cloned according to the mRAP protocol (26). In short, 30 μg of small RNA prep (from the PureLink miRNA isolation kit; Invitrogen) was fractionated on a 15% urea polyacrylamide gel, excising the gel in the size range of 18 to 25 nt. We followed the same procedure as that described in the protocols for generating a cDNA library of small RNAs and for concatamerization. Concatemers then were cloned in pGEM-T Easy vector (Promega) and sequenced. We used the GenBank database at the NCBI to find homologues of small RNA sequences.
miRNA microarray analysis.
Total RNA extracted from mock-infected HzFB cells and cells infected with HvAV-3e for 24 and 48 h, using TRIzol reagent, were sent to LC Sciences for miRNA microarray analysis. Available insect miRNAs (a total of 288 insect miRNAs; miRBase, release 12.0) in addition to custom miRNA probes from HzFB cells and Bombyx mori (a total of 94 miRNAs) were printed on chips in nine replicates and hybridized with Cy3-labeled small RNAs isolated from inoculated larvae. Labeled small RNAs from mock-infected cells were used as controls. In addition, multiple control probes were included on each chip. The control probes were used for quality controls of chip production, sample labeling, and assay conditions. Data were statistically analyzed using analysis of variance and a t test.
RT-qPCR expression analysis.
Reverse transcription-quantitative PCR (RT-qPCR) was performed using an Invitrogen miRNA first-strand synthesis and qPCR kit (NCode miRNA qRT-PCR; Invitrogen) according to the manufacturer's instructions. Total RNAs from virus-infected HzFB cells were extracted, and miRNAs were isolated according to the manufacturer's instructions (PureLink miRNA isolation kit; Invitrogen). For polyadenylation, 250 ng of miRNA from each sample was used. cDNA from polyadenylated RNA was prepared by using an Hz-miR24-specific primer as the forward sequence and a universal RT primer as the reverse sequence, while U6 was used as a control. RT-qPCR was performed in a Rotor-Gene 6000 (Qiagen). Real-time PCR conditions were 50°C for 2 min, 95°C for 2 min, and 40 cycles of 95°C for 15 s and 57°C for 30 s. Expression levels from each sample were analyzed with the Qgene template program. Reactions were repeated three times for two biological replicates.
qPCR.
Total genomic DNA was extracted from cells and subjected to qPCR using specific primers to ORF19 from the HvAV-3e genome. DNA concentrations were measured with Nanodrop, and 50 ng total genomic DNA was used for each qPCR reaction using SYBR green (Invitrogen) with a Rotor-Gene 6000. Real-time PCR conditions were 50°C for 2 min and 95°C for 2 min, followed by 40 cycles of 95°C for 10 s, 60°C for 10 s, 72°C for 20 s, and a final extension of 72°C for 20 s. Actin replication was used for normalizing the data. Relative DNA levels from each sample were analyzed in the Qgene template program. Reactions were repeated three times.
Northern blot analyses.
For the detection of miRNAs, we followed Bartel's laboratory protocol (http://web.wi.mit.edu/bartel/pub/protocols.html). In brief, 20-μg aliquots of small RNA prep (from the PureLink miRNA isolation kit; Invitrogen) from each sample were run on 15% urea denaturing polyacrylamide gels, electroblotted to nylon membranes, and UV cross-linked. Reverse complementary DNA oligonucleotides (21 mer) to specific miRNA sequences were labeled with [α-32P]dCTP using terminal nucleotide transferase. All of the probe hybridizations and washings were done at 50°C using solutions indicated in the protocol. The blots then were exposed to a phosphorimager screen overnight, and radioactive signals were detected using a phosphorimager scanner.
The transcripts of target and nontarget genes were detected by Northern analyses of total RNA (10 μg) run on 1.2% agarose formaldehyde gels as described previously (17).
Western blot analysis.
Cells were resuspended in 1× phosphate-buffered saline (PBS) and 4× sodium dodecyl sulfate (SDS) loading buffer. Proteins were resolved on SDS-polyacrylamide gel electrophoresis (10%) and transferred to a nitrocellulose membrane in transfer buffer. Antiserum raised against HvAV-3e major capsid protein (MCP) was used for hybridization, along with polyclonal anti-rabbit as the secondary antiserum. Blots were developed using nitroblue tetrazolium and 5-bromo-4-chloro-3-indoyl phosphate.
miRNA target studies.
We used RNAHybrid (24) software to find potential targets of Hz-miR24 in the HvAV-3e genome. To verify these targets, we cloned all of the constructs in the pIZ/V5-His vector (Invitrogen), and all of the primer sequences used are listed in Table 1. First, we constructed a green fluorescent protein (GFP)-based target replacement construct. A GFP fragment of 700 bp was amplified and cloned using SacI and XbaI sites in the pIZ vector. Sequences from target genes DdRP (783 bp) and DdRPβ (347 bp) as well as a nontarget gene, RNase III (200 bp; ORF27), were amplified and cloned downstream of GFP using XbaI and SacII restriction sites. These three gene constructs were named pIZ/GFP-DdRP, pIZ/GFP-β, and pIZ/GFP-RNaseIII. For the overexpression of Hz-miR24, a fragment of 250 bp that includes pre-miRNA sequence of Hz-miR24 was amplified from HzFB genomic DNA (using pre-24 forward and reverse primers) and cloned into the pIZ vector at HindIII and SacII sites to generate the pIZ/miR24 construct. For cotransfection experiments, two more gene constructs based on target and nontarget sequences were cloned into the pIZ vector. The DdRP gene was amplified in two halves, DdRP-A (the first 1,701 nt, which contains a target sequence) and DdRP-B (from bp 1701 to 3468, which contains a nontarget sequence), that were cloned at BamHI and EcoRI restriction sites, resulting in pIZ/DdRP-A and pIZ/DdRP-B, respectively.
TABLE 1.
Primer sequences
| Name | Sequence |
|---|---|
| Hz-miR24 sense | GTCCGGATACTCTTTGCGGAC |
| Hz-miR24 antisense | GTCCGCAAAGAGTATCCGGAC |
| Pre-24 F | CCAAGCTTGGCGCGACATCTCAACGAAAC |
| Pre-24 R | GCCCGCGGCCCAAAGTGGCCTGAAGACGC |
| AV RNA polymerase II F | ATGGATTACGACAGCTACCGCGCCGTG |
| AV RNA polymerase II R | CAGTGATCGTGTCTCTTTACGACGTAT |
| AV RNA Poly beta F | ATGCAGACGGCACTGATTACGCTCCGG |
| AV RNA Poly beta R | CAAAAGACCGTCATCAATCAAAGCGGT |
| BRO20 F | ATGTGTATTTCAAAACACAAGAGGGCT |
| BRO20 R | GCAGTCGTCGTTGGTGGCGCGTTCGAA |
| RNA polymerase target F | GGTCTAGATGCCGACGTGGTGAATG |
| RNA polymerase target R | GGCCGCGGTAACATTCCGGTACG |
| Beta subunit F target | GGTCTAGATACAACTGTTGTCGATG |
| Beta subunit R target | GGCCGCGGTTACAAAAGACCGTCAT |
| DdRPII pIZ F1 | GCGGATCCGCCATGGATTACGACAGCTAC |
| DdRPII pIZ R1 | GGGAATTCGGTGCCTTCTGTGGTCTC |
| DdRPII PIZ F2 | GCGGATCCGCCATGGACGTTGGTATAAGGTTA |
| DdRPII PIZ R2 | GGGAATTCGTGATCGTGTCTCTTTA |
| DdRP RNAi F | TAATACGACTCACTATAGGGGACATTGTCGTAGCGG |
| DdRP RNAi R | TAATACGACTCACTATAGGGGCCTGTAACTTCATC |
| Beta subunit RNAi F | TAATACGACTCACTATAGGGGTACAACTGTTGTCGA |
| Beta subunit RNAi R | TAATACGACTCACTATAGGGGTTACAAAAGACCGTC |
| RNaseIII targetF | GCGCTCTAGAACGATACTGCTTCGCAACGT |
| RNaseIII targetR | GCCCGCGGATTGGTTACATTTCGTGAA |
Hz-miR24 (5′-GUCCGCAAAGAGUAUCCGGAC-3′) and control (an irrelevant sequence; 5′-UCUACUCUUUCUAGGAGGUUGUGA-3′) inhibitors were synthesized by Genepharma Co., Ltd. One hundred nanograms of the Hz-miR24 inhibitor or the control inhibitor was transfected into 106 HzFB cells using Cellfectin transfection reagent (Invitrogen) 24 h prior to transfection with pIZ/DdRP-A and pIZ/DdRP-B constructs. Other control cells were mock transfected without the inhibitor. Cells were analyzed 72 h after transfection by Northern blotting using corresponding DNA constructs (DdRP-A and DdRP-B) as probes.
RNAi-mediated gene silencing.
For RNA interference (RNAi), we used double-stranded RNA (dsRNA) synthesized in vitro by using a Megascript transcription kit (Ambion) according to the manufacturer's instructions. T7 promoter sequences were incorporated in both forward and reverse primers designed to amplify 470 and 350 bp of DdRP and DdRPβ genes, respectively. For dsRNA synthesis, 1 μg PCR product was used in a 16-h incubation at 37°C, and DNase was treated and precipitated by the lithium chloride method. We used 0.5 μg dsRNA for transfection in HzFB and Sf9 cells 72 h prior to virus inoculation. Cells were collected for RNA and protein extraction at 48 and 72 hpi.
RESULTS
HvAV-3e infection affects cellular miRNA expression.
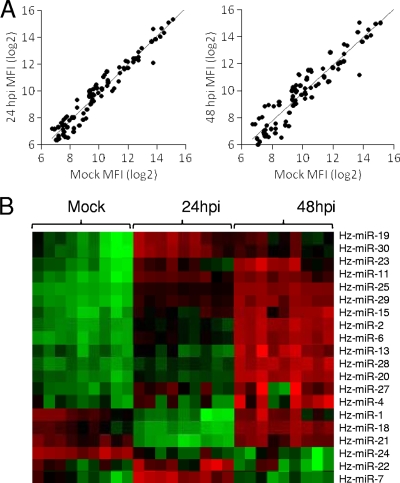
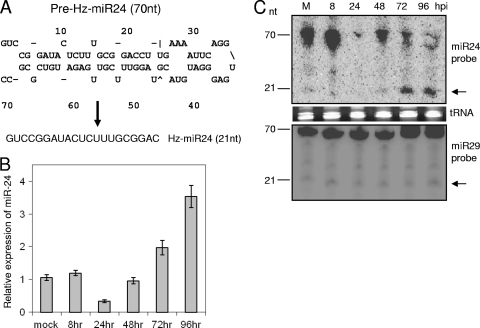
Initially, we generated a small RNA cDNA library from HzFB cells infected with HvAV-3e (96 to 120 hpi) and cloned 30 cellular miRNA sequences (designated Hz-miRs). We used antisense probes of all of these HzFB miRNAs for microarray analyses in mock-infected as well as virus-infected cells at 24 and 48 hpi. Moreover, 352 known insect miRNAs from Bombyx mori, Anophelese gambiae, Apis mellifera, Drosophila melanogaster, D. ananassae, and D. grimshawi (sequences are available from miRBase) were used in this microarray analysis. Significant changes in miRNA expression levels were found in virus-infected cells compared to those of mock-infected cells (Fig. 1A). Figure 1B shows the outcome for 20 Hz-miRs that were significantly up- or downregulated. Most of the Hz-miRs were upregulated at 48 hpi, while three miRNAs, Hz-miR-24, Hz-miR-22, and Hz-miR-7, were downregulated. Here, we concentrated on the functional analysis of one of these differentially expressed miRNAs (Hz-miR24) upon HvAV-3e infection. The mature Hz-miR24 sequence cloned and putative pre-miRNA loop structure are shown in Fig. 2A.
FIG. 1.
HvAV-3e infection affects cellular miRNA expression profiles. (A) Comparison of results from miRNA microarray experiments. The expression profiles of 92 miRNAs that were significantly up- or downregulated at 24 and 48 h of virus infection in HzFB cells compared to results for mock infection are shown. MFI, mean fluorescent intensity. (B) miRNA microarray color map showing expression levels of Hz-miRNAs in mock-infected cells and cells infected for 24 and 48 h. The expression of most miRNAs was significantly upregulated at 48 hpi, while Hz-miR-24 and Hz-miR-22 showed downregulation. Each miRNA was replicated nine times on each chip. Red indicates the upregulation of expression, and green indicates the downregulation of expression.
FIG. 2.
Temporal expression of Hz-miR24 after HvAV-3e infection. (A) Predicted stem-loop secondary structure of pre-Hz-miR24 and mature Hz-miR24 sequences. (B) RT-qPCR analysis of Hz-miR24 expression at different time intervals following virus infection in HzFB cells. qPCR forward primers specific to Hz-miR24 and U6 were used, and the normalized expression of Hz-miR24 was calculated relative to that of U6. Error bars represent standard deviations from three replicates. (C) Northern blot analysis showing pre-miRNA and mature miRNA bands (arrows) using a reverse complementary probe to mature Hz-miR24 (upper) or Hz-miR29 (lower) as a control. tRNA is shown as a loading control. M, molecular size marker.
Differential expression of Hz-miR24 upon virus infection.
For the confirmation of microarray results, we analyzed Hz-miR24 expression levels following HvAV-3e infection with RT-qPCR, which showed a decrease at 24 hpi compared to levels for mock-infected cells or cells infected for 8 h and then gradually increased to significantly higher levels at 72 and 96 hpi (Fig. 2B). These results were confirmed by the Northern hybridization of pre-miRNA and mature miRNA during the same time course of HvAV-3e infection (Fig. 2C, upper). Levels of another cellular mature miRNA (Hz-miR29), which was used as a control in the Northern hybridization, was unaffected by viral infection (Fig. 2C, lower). This suggested that HvAV-3e transiently suppresses Hz-miR24 expression during 24 to 48 hpi.
Hz-miR24 regulates viral gene expression.
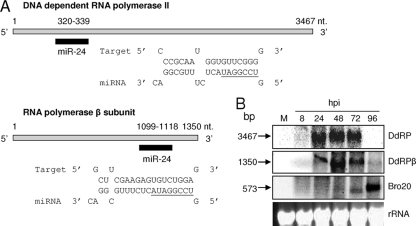
Potential targets of Hz-miR24 were predicted in the HvAV-3e genome using RNAhybrid based on the seed region complementarity and minimum free energy (24). Two viral genes, DdRP (ORF64) and DdRPβ (ORF82), were identified as likely targets (Fig. 3A). There is a complete complementarity in the seed region for both of these targets, with minimum free energy of −28 and −29.7 kcal/mol for DdRP and DdRPβ, respectively. Target sequences for both of these genes were found in their coding regions, at nucleotide coordinates 70175 to 70196 for DdRP and at 89766 to 89786 for DdRPβ. Considering that DdRP and DdRPβ are potential targets of Hz-miR24, we analyzed their expression levels in virus-infected HzFB cells at different time intervals. Northern hybridizations showed higher transcript levels for both genes at 48 hpi and then a sharp decrease after 72 hpi (Fig. 3B), which coincided with higher expression levels of Hz-miR24 (Fig. 2B and C). However, when the same blot was hybridized with Bro20 (an ORF154-specific probe that is not a target for Hz-miR24), unlike the case for DdRP and DdRPβ, its transcript levels were found to increase after 72 hpi (Fig. 3B). This suggested that the downregulation of transcripts at later hours is not general for all viral genes.
FIG. 3.
Hz-miR24 targets viral transcripts. (A) RNAhybrid prediction for Hz-miR24 target sites and sequences in DdRP and DdRPβ genes of HvAV-3e. Seed regions are underlined. (B) Northern blot detection of DdRP, DdRPβ (targets), and Bro20 (nontarget) at various time points after viral infection. The equal loading of samples is shown by rRNA bands.
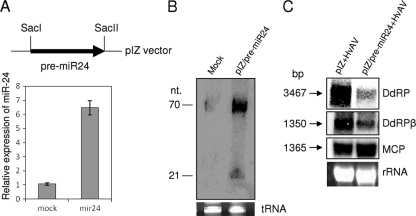
To achieve higher expression levels of Hz-miR24 in HzFB cells before virus infection, we ectopically expressed the miRNA using an insect expression plasmid that contained the pre-miRNA sequence for Hz-miR24 (pIZ/pre-miR24). The significantly higher expression levels of Hz-miR24 in transfected cells with pIZ/pre-miR24 compared to those of mock-transfected cells were confirmed by RT-qPCR (Fig. 4A) and Northern hybridization (Fig. 4B) at 72 h posttransfection. After 72 h, the transfected cells were inoculated with HvAV-3e, and samples were collected after 48 hpi. While significant reductions in transcript levels of DdRP and DdRPβ were detected compared to the levels of control transfection (pIZ empty vector; pIZ+HvAV), transcript levels of a nontarget gene coding for the MCP (ORF56) was found to be unaffected, indicating the specific targeting of DdRP and DdRPβ mRNAs by Hz-miR24 (Fig. 4C).
FIG. 4.
Ectopic expression of Hz-miR24 and downregulation of target transcripts. (A) Cloning map of pre-Hz-miR24 in the pIZ vector (pre-miR24) for the ectopic expression of the miRNA and RT-qPCR results showing significantly higher expression levels of the miRNA (mir24) in pIZ/pre-miR24 compared to those of mock-transfected cells at 72 h posttransfection. Error bars represent standard deviations from three replicates. (B) Northern blot analysis of cells from panel A probed with a reverse complementary oligonucleotide to mature Hz-miR24. (C) Northern blots showing significant decreases in transcript levels of DdRP and DdRPβ when Hz-miR24 was ectopically expressed using pIZ/pre-miR24, while MCP (nontarget) transcript levels remained the same. pIZ+HvAV indicates cells transfected with the pIZ empty vector. Cells were analyzed at 48 hpi. The equal loading of samples is shown by rRNA bands.
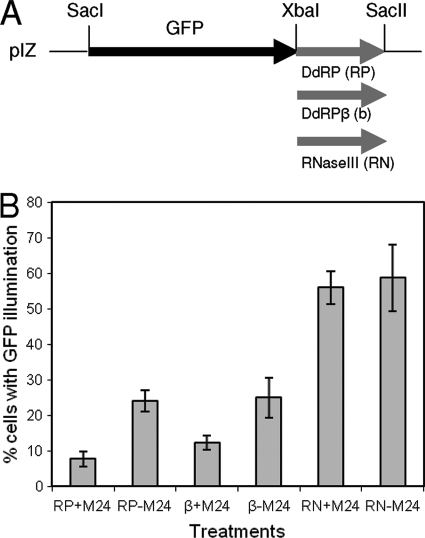
Further, to confirm target sequence specificity, we designed gene constructs based on the expression of GFP as a reporter gene. The target sequences of DdRP and DdRPβ and a nontarget DNA fragment from the HvAV-3e RNase III gene (ORF27) were amplified along with their flanking sequences and cloned downstream of a GFP construct (Fig. 5A). All three constructs (pIZ/GFP-DdRP, pIZ/GFP-β, and pIZ/GFP-RNaseIII) were cotransfected with pIZ/pre-miR24 individually into HzFB cells. After 72 h, cells were observed under a UV microscope to analyze the number of cells expressing the reporter gene in various treatments. The number of cells expressing GFP was significantly reduced in cells transfected with pIZ/pre-miR24 and target constructs compared to that of cells that were transfected only with the target constructs without pIZ/pre-miR24 (Fig. 5B). However, the number of GFP-expressing cells was significantly higher in the case of the nontarget construct (pIZ-GFP-RNaseIII), which was not affected by the presence of pIZ/pre-miR24 (Fig. 5B). A lower number of GFP-expressing cells transfected with the target sequences in the absence of pIZ/pre-miR24 than with the nontarget construct is because of endogenous Hz-miR24 base expression levels that affect the GFP constructs with target sequences and not the one with nontarget sequences. These results confirmed the target specificity of Hz-miR24 for DdRP and DdRPβ.
FIG. 5.
(A) Cloning strategy to produce GFP-based reporter constructs with target and nontarget sequences. DdRP (RP), DdRPβ (β), and RNase III (RN; nontarget) were cloned downstream of the GFP open reading frame. (B) All three constructs were cotransfected individually with (+M24) or without (−M24) pIZ/pre-miR24 in HzFB cells, and the numbers of GFP-expressing cells were determined 72 h after the transfections. Error bars represent standard deviations from three independent determinations.
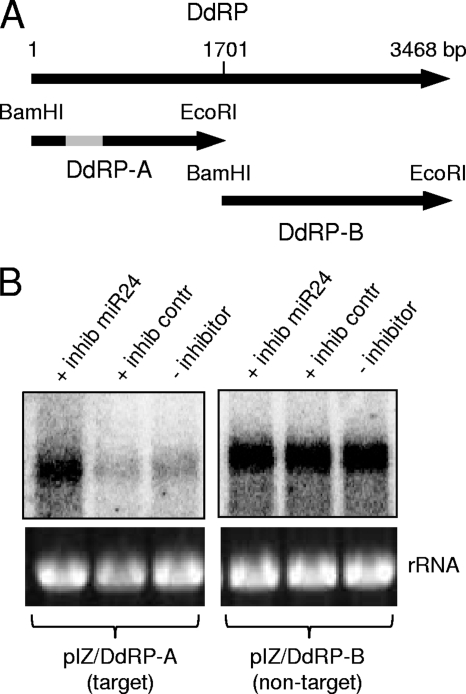
In addition, to further confirm miRNA target specificity, we conducted another set of experiments by expressing DdRP target and nontarget sequences in the absence or presence of an Hz-miR24 inhibitor. We cloned two gene constructs for DdRP, one fragment containing the target sequence (pIZ/DdRP-A) and another lacking the target (pIZ/DdRP-B) (Fig. 6A). We used an Hz-miR24-specific synthetic inhibitor to block Hz-miR24 function. The two constructs were expressed in the presence or absence of Hz-miR24 inhibitor in HzFB cells. Northern blot analyses using specific probes for each construct showed higher expression levels of the DdRP-A target gene in cells transfected with Hz-miR24 inhibitor than in those without the inhibitor or with the control inhibitor, while DdRP-B expression was the same in all treatments (Fig. 6B). This further confirmed that Hz-miR24 specifically affects the expression of DdRP by interacting with the target sequence, and that the inhibition of the miRNA leads to higher expression levels of the gene.
FIG. 6.
Upregulation of DdRP expression in the presence of the inhibitor of Hz-miR24. (A) Two DdRP constructs with (DdRP-A) and without (DdRP-B) DdRP target sequences (gray box) were produced and cloned into the pIZ vector. (B) pIZ/DdRP-A and pIZ/DdRP-B were transfected into HzFB cells previously transfected with (+ inhib miR24) or without Hz-miR24 synthetic inhibitor (− inhibitor) and a control inhibitor (+ inhib contr). Cells were analyzed 72 h after transfection by Northern blotting using corresponding DNA constructs (DdRP-A and DdRP-B) as probes. The equal loading of samples is shown by rRNA bands.
Hz-miR24 and virus replication.
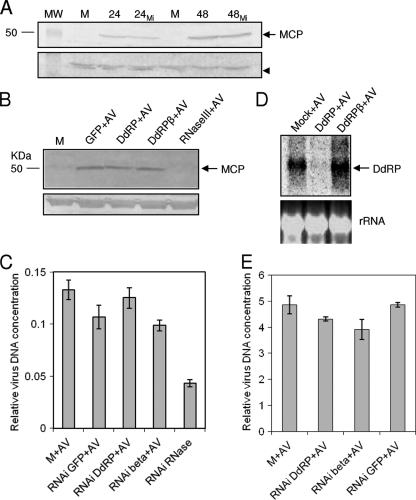
Considering the negative regulation of viral DdRP and DdRPβ by Hz-miR24, we looked for any effect on virus replication. Cells transfected with pIZ/pre-miR24 were inoculated with HvAV-3e, and cells were collected at 24 and 48 h after infection. Virus replication was analyzed in a Western blot by the detection of the viral MCP using a specific antiserum to the protein. The same levels of the 50-kDa MCP were detected in samples with or without Hz-miR24, which suggested that this miRNA has no effect on virus replication (Fig. 7A). This raised the possibility that the two target genes are not essential for virus replication. To look further into the role of DdRP and DdRPβ in virus replication, we applied RNAi-mediated silencing for both genes. We generated dsRNA specific to DdRP and DdRPβ and transfected those into HzFB cells, which then were inoculated with HvAV-3e. The silencing of DdRP and DdRPβ (data not shown) was confirmed by Northern hybridization (Fig. 7D). As a negative control, dsRNA specific to GFP was used, and as a positive control dsRNA specific to the viral RNase III gene (ORF27) was used. RNase III is an essential gene for virus replication and directly or indirectly affects DNA replication as well (unpublished data). Virus replication was analyzed by Western hybridization with the antiserum against MCP at 48 hpi. MCP expression was the same in RNAi DdRP and DdRPβ as that in RNAi GFP treatment, while it was absent in RNAi RNase III treatment (Fig. 7B). However, these results should be interpreted cautiously, as the silencing of DdRP and DdRPβ may not have affected the expression of MCP gene but the expression of other late genes and, as a consequence, the maturation of viral particles.
FIG. 7.
Hz-miR24 does not affect virus replication. (A) Western blot analysis of virus-infected cells at 24 and 48 hpi in the absence or presence (24MI and 48MI) of pIZ/pre-miR24 probed with an MCP-specific antiserum. Mock-infected cells (M) were used as a control. A nonspecifically reacted protein with the antibody is shown as a control for the equal loading of samples (arrowhead). MW, molecular weight in thousands. (B) RNAi experiment to investigate the role of the target genes DdRP and DdRPβ in virus replication. dsRNAs specific to these genes were synthesized in vitro and transfected into HzFB cells, and 72 h after transfection cells were inoculated with HvAV-3e. Western blot analysis with samples taken at 48 hpi showed the same level of MCP detection in negative control RNAi (GFP RNAi; designated GFP+AV), DdRP RNAi (DdRP+AV), and DdRPβ RNAi (DdRPβ+AV), while there is no band detected in cells with a positive control (RNase III RNAi; designated RNase III+AV). (C) Confirmation of the RNAi-mediated silencing of DdRP using a gene-specific probe. The equal loading of samples is shown by rRNA bands. (D) qPCR of samples treated as described for panel B showing that the silencing of DdRP and DdRPβ did not have a significant effect on virus DNA replication. (E) qPCR analysis of Sf9 cells at 72 hpi previously transfected with dsRNA specific to DdRP, DdRPβ, and GFP. Error bars represent standard deviations from three replicates.
qPCR results on replicates of the RNAi experiment using specific primers to ORF19 indicated that the silencing of DdRP and DdRPβ genes did not have a significant effect on virus DNA replication, whereas in the positive control (ORF27) viral DNA replication was significantly reduced (Fig. 7C). Relative virus DNA levels in the RNAi of DdRP and AV were similar to that of mock infection, and levels of the RNAi of DdRPβ were similar to those of the RNAi of GFP and AV. The silencing of the genes in Sf9 cells reduced viral DNA replication compared to that of controls, but it was not a severalfold reduction (Fig. 7E). This suggested that DdRP and DdRPβ are not essential genes for virus DNA replication in vitro but could be required for the expression of late genes that are involved in the production of mature virions or pathogenicity in vivo.
DISCUSSION
In this study, we examined changes in the expression profile of cellular miRNAs in connection with an AV (HvAV-3e) infection in host cells and further analyzed viral genes targeted by one of the regulated miRNAs (Hz-miR24). The expression analysis using an miRNA-microarray approach showed both the down- and upregulation of subsets of miRNAs, which is consistent with observations made in other host-virus interactions (e.g., those of human immunodeficiency virus and human cytomegalovirus [16, 30]). Such information on global changes in cellular miRNAs with respect to insect viruses still is lacking. In our overall microarray analysis, some of the highly regulated miRNAs were found to have homologs that have been reported to control certain apoptotic pathways and insect development (33). In addition, we observed significant changes in the expression levels of miRNAs that we cloned from virus-infected HzFB cells (Hz-miRs) and propose that they play significant roles in the regulation of virus infection or host cell defense against viral infection.
Following our target predictions in the HvAV-3e genome for the differentially regulated Hz-miRs upon virus infection, we focused our efforts on Hz-miR24. The expression levels of Hz-miR24 were found to decrease at early stages of virus infection compared to those of mock-infected cells but significantly increased late in infection. Logically, it benefits the virus to downregulate cellular miRNAs during the period in which they may target viral transcripts (see below). The possible mechanism(s) behind the early suppression of Hz-miR24 has yet to be explored, but Northern hybridization results demonstrated that the expression of the primary transcript is consistent at all times following virus infection, while pre-miRNA expression is downregulated at 24 and 48 h (data not shown). This could mean that HvAV-3e infection transiently has a direct or indirect influence on miRNA biogenesis elements in the processing of pri-miRNA to pre-miRNA (e.g., drosha). The temporal regulation of cellular miRNAs following viral infection also has been observed in other interactions. For example, an immunomodulatory miRNA, miR-146a, was reported to be downregulated earlier and upregulated severalfold later in Epstein-Barr virus-infected cells (13).
Our functional analysis of Hz-miR24 suggested that it plays an important role in interactions with HvAV-3e by downregulating viral DdRP and DdRPβ transcripts later in infection. Both of these genes encode multiple domains that are conserved in the RNA polymerase superfamily across various organisms that are involved in DNA-dependent polymerization for transcription. It is likely that both of these proteins work together for transcribing some HvAV-3e genes. Target sequences for Hz-miR24 were found in coding regions of DdRP and DdRPβ, resulting in a sharp decrease in their transcript levels late after virus infection. We confirmed our target predictions through Northern analyses that showed the downregulation of both target gene transcripts at the time of virus infection (96 hpi) when Hz-miR24 level is higher and by the ectopic expression of the miRNA, while the expression of nontarget genes was unaffected. Further, using GFP target sequence constructs, our results showed a significant decrease in GFP expression in the presence of Hz-miR24 compared to that of a GFP-nontarget sequence construct. Further, we made use of a synthetic miRNA inhibitor (21-nt RNA sequence antisense to Hz-miR24) to block Hz-miR24 function. The expression levels of a cloned fragment of DdRP in the pIZ vector with the target sequence were noticeably lower in the absence of Hz-miR24 inhibitor.
Recent reports have shown functional target sites in the coding regions of genes for certain miRNAs. Let7 target sites in the coding region of human dicer were experimentally validated (11). Similarly, a human miRNA, miR-148, was reported to target the coding sequence of DNA methyltransferase 3 (9). Target sites in coding regions also were found in mouse transcripts that encode the pluripotency factors nanog, OCT4, and SOX2 (27). A sharp decline in DdRP and DdRPβ transcript levels later in virus infection and in all target specificity experiments could be due to perfect complementarity at the central nt 10 and 11 in mRNA-miRNA interactions. It is now well documented that centrally matched pairs lead to mRNA slicing, ultimately resulting in the degradation of mRNA. For example, Let7 was made capable of cleaving its target mRNA by changing its central nucleotides complementary to its target (5). In Arabidopsis thaliana, the Cu/Zn superoxide dismutase gene is regulated by miR-398a through slicing as well as translational inhibition. After introducing mismatches at the central nt 10 and 11 in this mRNA-miRNA interaction, mRNA degradation was prevented (8). This is mechanistically logical, because mRNA is exposed to the Ago RNase H active site 10 nt from the start of the mRNA-miRNA duplex.
Our RNAi results suggested that DdRP and DdRPβ are not essential for viral DNA replication in vitro. However, they could be involved in the expression of late viral genes and affect the maturation of the virus. This explains why the ectopic expression of Hz-miR24 had no effect on virus replication. In baculoviruses, late expression factors LEF4, LEF8, LEF9, and p47 constitute the four subunits of the viral RNA polymerase. Various reports have shown that the four subunits are strictly involved in transcription, and their deletion/mutation had no effect on viral DNA synthesis (reviewed in reference 23). Similarly, DdRP and DdRPβ do not seem to be required for viral DNA replication, but they could be essential for the expression of late genes and the maturation of virions, which requires further investigation. Complete genome sequences of AVs have been available only in the last 2 years, and therefore the majority of genes (including DdRP and DdRPβ) and their roles in virus biology have not been investigated. In addition to DdRP and DdRPβ, the HvAV-3e genome encodes another protein that shows similarity to RNA polymerases (ORF11). The function of these three genes, which may operate independently or as subunits of an RNA polymerase, remains to be elucidated. Immediate-early and early genes usually are transcribed by the host RNA polymerase II, whereas late genes are transcribed by a virus-encoded RNA polymerase (e.g., baculoviruses) (12, 15). Similarly, higher expression levels of DdRP and DdRPβ from 24 to 72 hpi, due to HvAV-induced lower levels of Hz-miR24, may be sufficient to transcribe late viral genes. However, very late in infection they may not be required and hence may be downregulated by the host Hz-miR24, which increases in concentration very late in infection. To our knowledge, this is the first study showing the effect of an insect virus infection on the cellular miRNA profile and targeting of the viral genes by a cellular miRNA late in infection.
Acknowledgments
This project was funded partly by Horticulture Australia Ltd. (VG06044) to S.A. and a UQ Ph.D. scholarship to M.H.
Footnotes
Published ahead of print on 21 October 2009.
REFERENCES
- 1.Asgari, S., J. Davis, D. Wood, P. Wilson, and A. McGrath. 2007. Sequence and organization of the Heliothis virescens ascovirus genome J. Gen. Virol. 88:1120-1132. [DOI] [PubMed] [Google Scholar]
- 2.Bideshi, D. K., M.-V. Demattei, F. Rouleux-Bonnin, K. Stasiak, Y. Tan, S. Bigot, Y. Bigot, and B. A. Federici. 2006. Genomic sequence of Spodoptera frugiperda ascovirus 1a, an enveloped, double-stranded DNA insect virus that manipulates apoptosis for viral reproduction. J. Virol. 80:11791-11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bigot, Y., S. Renault, J. Nicolas, C. Moundras, M.-V. Demattei, S. Samain, D. K. Bideshi, and B. A. Federici. 2009. Symbiotic virus at the evolutionary intersection of three types of large DNA viruses; iridoviruses, ascoviruses, and ichnoviruses. PLoS One 4:e6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bigot, Y., S. Samain, C. Augé-Gouillou, and B. A. Federici. 2008. Molecular evidence for the evolution of ichnoviruses from ascoviruses by symbiogenesis. BMC Evol. Biol. 8:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodersen, P., and O. Voinnet. 2009. Revisiting the principles of microRNA target recognition and mode of action. Nat. Rev. Mol. Cell Biol. 10:141-148. [DOI] [PubMed] [Google Scholar]
- 6.Cullen, B. R. 2009. Viral and cellular messenger RNA targets of viral microRNAs. Nature 457:421-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cullen, B. R. 2006. Viruses and microRNAs. Nat. Gen. 38:S25-S30. [DOI] [PubMed] [Google Scholar]
- 8.Dugas, D. V., and B. Bartel. 2008. Sucrose induction of Arabidopsis miR398 represses two Cu/Zn superoxide dismutases. Plant Mol. Biol. 67:403-417. [DOI] [PubMed] [Google Scholar]
- 9.Duursma, A. M., M. Kedde, M. Schrier, C. Le Sage, and R. Agami. 2008. miR-148 targets human DNMT3b protein coding region. RNA 14:872-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Federici, B. A., D. K. Bideshi, Y. Tan, T. Spears, and Y. Bigot. 2009. Ascoviruses: superb manipulators of apoptosis for viral replication and transmission. Curr. Top. Microbiol. Immunol. 328:171-196. [DOI] [PubMed] [Google Scholar]
- 11.Forman, J. J., A. Legesse-Miller, and H. A. Coller. 2008. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc. Natl. Acad. Sci. USA 105:14879-14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friesen, P. D. 1997. Regulation of baculovirus early gene expression, p. 141-170. In L. K. Miller (ed.), The baculoviruses. Plenum Press, New York, NY.
- 13.Godshalk, S. E., S. Bhaduri-McIntosh, and F. J. Slack. 2008. Epstein-Barr virus-mediated dysregulation of human microRNA expression. Cell Cycle 7:3595-3600. [DOI] [PubMed] [Google Scholar]
- 14.Griffiths-Jones, S., H. K. Saini, S. V. Dongen, and A. J. Enright. 2008. miRBase: tools for microRNA genomics. Nucleic Acids Res. 36:D154-D158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guarino, L. A., B. Xu, J. Jin, and W. Dong. 1998. A virus-encoded RNA polymerase purified from baculovirus-infected cells. J. Virol. 72:7985-7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, Y., and X. Gu. 2007. A bootstrap based analysis pipeline for efficient classification of phylogenetically related animal miRNAs. BMC Genomics 8:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hussain, M., R. J. Taft, and S. Asgari. 2008. An insect virus-encoded microRNA regulates viral replication. J. Virol. 82:9164-9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jopling, C. L. 2008. Regulation of hepatitis C virus by microRNA-122. Biochem. Soc. Trans. 36:1220-1223. [DOI] [PubMed] [Google Scholar]
- 19.King, L. A., and R. D. Possee. 1992. The baculovirus expression system. A laboratory guide. Chapman & Hall, New York, NY.
- 20.Lecellier, C. H., P. Dunoyer, K. Arar, J. Lehmann-Che, S. Eyquem, C. Himber, A. Saïb, and O. Voinnet. 2005. A cellular microRNA mediates antiviral defense in human cells. Science 308:557-560. [DOI] [PubMed] [Google Scholar]
- 21.Lytle, J. R., T. A. Yario, and J. A. Steitz. 2007. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc. Natl. Acad. Sci. U. S. A. 104:9667-9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McIntosh, A. H., and C. M. Ignoffo. 1981. Replication and infectivity of the single-embedded nuclear polyhedrosis virus, baculovirus heliothis, in homologous cell lines. J. Invertebr. Pathol. 37:258-264. [Google Scholar]
- 23.Passarelli, A. L., and L. A. Guarino. 2007. Baculovirus late and very late gene regulation. Curr. Drug Targets 8:1103-1115. [DOI] [PubMed] [Google Scholar]
- 24.Rehmsmeier, M., P. Steffen, M. Höchsmann, and R. Giegerich. 2004. Fast and effective prediction of microRNA/target duplexes. RNA 10:1507-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stasiak, K., S. Renault, M.-V. Demattei, Y. Bigot, and B. A. Federici. 2003. Evidence for the evolution of ascoviruses from iridoviruses. J. Gen. Virol. 84:2999-3009. [DOI] [PubMed] [Google Scholar]
- 26.Takada, S., and H. Mano. 2007. Profiling of microRNA expression by mRAP. Nat. Protoc. 2:3136-3145. [DOI] [PubMed] [Google Scholar]
- 27.Tay, Y., J. Zhang, A. M. Thomson, B. Lim, and I. Rigoutsos. 2008. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455:1124-1128. [DOI] [PubMed] [Google Scholar]
- 28.Triboulet, R., B. Mari, Y.-L. Lin, C. Chable-Bessia, Y. Bennasser, K. Lebrigand, B. Cardinaud, T. Maurin, P. Barbry, V. Baillat, J. Reynes, P. Corbeau, K.-T. Jeang, and M. Benkirane. 2007. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 315:1579-1582. [DOI] [PubMed] [Google Scholar]
- 29.Umbach, J. L., and B. R. Cullen. 2009. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 23:1151-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, F.-Z., F. Weber, C. Croce, C.-G. Liu, X. Liao, and P. E. Pellett. 2008. Human cytomegalovirus infection alters the expression of cellular microRNA species that affect its replication. J. Virol. 82:9065-9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, L., J. Xue, C. P. Seaborn, B. M. Arif, and X.-W. Cheng. 2006. Sequence and organization of the Trichoplusia ni ascovirus 2C (Ascoviridae) genome. Virology 354:167-177. [DOI] [PubMed] [Google Scholar]
- 32.Winter, J., S. Jung, S. Keller, R. I. Gregory, and S. Diederichs. 2009. Many roads to maturity:microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 11:228-234. [DOI] [PubMed] [Google Scholar]
- 33.Yu, X., Q. Zhou, S.-C. Li, Q. Luo, Y. Cai, W.-C. Lin, H. Chen, Y. Yang, S. Hu, and J. Yu. 2008. The silkworm (Bombyx mori) microRNAs and their expressions in multiple developmental stages. PLoS One 3:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng, Y., R. Yi, and B. R. Cullen. 2005. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 24:138-148. [DOI] [PMC free article] [PubMed] [Google Scholar]