Abstract
The rubella virus (RV) capsid is an RNA-binding protein that functions in nucleocapsid assembly at the Golgi complex, the site of virus budding. In addition to its role in virus assembly, pools of capsid associate with mitochondria, a localization that is not consistent with virus assembly. Here we examined the interaction of capsid with mitochondria and showed that this viral protein inhibits the import and processing of mitochondrial precursor proteins in vitro. Moreover, RV-infected cells were found to contain lower intramitochondrial levels of matrix protein p32. In addition to inhibiting the translocation of substrates into mammalian mitochondria, capsid efficiently blocked import into yeast mitochondria, thereby suggesting that it acts by targeting a highly conserved component of the translocation apparatus. Finally, mutation of a cluster of five arginine residues in the amino terminus of capsid, though not interfering with its binding to mitochondria, abrogated its ability to block protein import into mitochondria. This is the first report of a viral protein that affects the import of proteins into mitochondria.
Rubella virus (RV) is a human pathogen that causes severe birth defects (reviewed in reference 17). Teratogenicity undoubtedly results from deleterious interactions between virus proteins and host cell proteins, but little is known about this phenomenon. The viral genome encodes two nonstructural proteins (p150 and p90) and three structural proteins, the capsid protein, E2, and E1. The capsid protein is a multifunctional RNA-binding protein and is the focus of our studies. The primary function of the capsid protein is to package the viral genome into nucleocapsids, a process that appears to be regulated by phosphorylation (27, 29). Recent evidence suggests that, in addition to their structural roles in virus assembly, capsid proteins may be key determinants in virus-host interactions. For example, the hepatitis C virus capsid may affect disease development by modulating apoptotic and innate immune pathways (5, 36, 42). Moreover, localization of capsids appears to be an important factor in viral pathogenesis. Specifically, it has been reported that nuclear localization of the Japanese encephalitis virus capsid is necessary for neuroinvasion (38). The RV capsid also localizes to subcellular compartments that have no obvious relationship to the virus budding site (Golgi complex). For example, a pool of capsid colocalizes with the nonstructural protein p150 on virus-induced tubular structures (26). Later it was demonstrated that capsid binds p150 and modulates the transcription of viral RNA (8, 47-49). In addition to its role as a replicase cofactor, a number of studies indicate that a large pool of capsid localizes to mitochondria (2, 22, 32).
Among togaviruses, localization of capsid proteins to mitochondria is unique to RV (31). The significance of this phenomenon is not known, but we hypothesize that the mitochondrial pool of capsid is engaged in functions not directly related to virus budding. We and others have shown that the RV capsid binds to mitochondrial protein p32 (3, 37). p32 was originally identified as a factor that copurified with alternative splicing factors (13, 25), but subsequent studies showed that it interacts with a wide variety of cellular and viral proteins that are not involved in splicing (reviewed in reference 14). Among its various cellular functions, p32 is known to function in a number of apoptotic pathways (9, 23, 46). Ablation of the p32 binding site in capsid does not abrogate targeting to mitochondria, but virus replication is severely impaired (2). The observation that capsid expression has a dramatic effect upon the distribution and morphology of mitochondria (2, 3) prompted us to investigate whether this viral protein affects mitochondrial physiology. In the study described here, we show that the RV capsid protein associates with the surface of mitochondria and impairs the import of newly synthesized proteins into mitochondria. Given the linkage between apoptosis and the translocation machinery of mitochondria, it is tempting to speculate that the ability of capsid to block the import of proteins into this organelle has functional implications for apoptosis.
MATERIALS AND METHODS
Reagents.
The following reagents were purchased from the respective suppliers: protein G-Sepharose from GE Healthcare Bio-Sciences Corp. (Princeton, NJ), general lab chemicals from Sigma Chemical Co. (St. Louis, MO), TnT Quick Coupled transcription/translation systems from Promega (Madison, WI), Redivue l-[35S]methionine aqueous solution from Perkin-Elmer Life Sciences (Boston, MA), medium and fetal bovine serum for cell culture from Life Technologies-Invitrogen, Inc. (Carlsbad, CA), and Vero cells from the American Type Culture Collection (Manassas, VA).
Mammalian cell culture and virus infection.
Vero cells were cultured in Dulbecco's minimal essential medium (high glucose) containing 5% fetal bovine serum, 2 mM glutamine, 1 mM HEPES, and antibiotics. Cells were incubated at 37°C in a humidified atmosphere with 5% CO2. RV stocks were diluted with cell culture medium and then added to Vero cells that had been washed with phosphate-buffered saline (PBS). Cells were incubated with the virus inoculum (1 ml/35-mm dish) for 4 h at 35°C, after which time the inoculum was replaced with normal growth medium. Infected cultures were kept at 35°C until experimental analyses.
Purification of recombinant proteins.
The RV capsid was purified from Escherichia coli by two different methods. For import assays using rat liver mitochondria, the purification of capsid was exactly as described previously by this laboratory (22). Purified capsid and the negative control protein green fluorescent protein (GFP) were stored in buffer containing 20 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 7.4) and 10 mM KCl. When yeast mitochondria were employed, capsid proteins lacking the E2 signal peptide (wild-type and C5RA mutant versions) were expressed in transformed BL21(DE3) E. coli cells at 16°C for 16 h using the pET23 expression vector (Novagen, EMD Chemicals, Inc., Gibbstown, NJ). Expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were harvested and resuspended in a lysis buffer containing 50 mM sodium phosphate buffer (pH 8.0), 8 M urea, and 1 mM phenylmethylsulfonyl fluoride (PMSF). After the lysate was clarified, the supernatant was applied to an Ni-nitrilotriacetic acid-Sepharose matrix which was then washed with 50 mM sodium phosphate buffer (pH 6.3)-8 M urea-1 mM PMSF. Elution of the capsid protein was performed with 50 mM sodium phosphate buffer (pH 4.5)-8 M urea-1 mM PMSF-50 mM EDTA. For storage of the protein, MgCl2 was added to a final concentration of 50 mM.
Purification of maltose-binding protein (MBP) from bacteria was performed as follows. MBP was expressed in BL21(DE3) E. coli cells from a pMALcRI expression vector. Expression was induced by 1 mM IPTG. Cells were harvested after growth at 30°C for 3 h. Cell lysis was performed in 20 mM HEPES (pH 7.4)-1% Triton X-100-150 mM KCl-1 mM PMSF. The lysate was clarified by centrifugation and applied to an amylose matrix (New England BioLabs). The matrix was washed with lysis buffer. MBP was eluted with 20 mM HEPES (pH 7.4)-150 mM KCl-1 mM PMSF-10 mM maltose. The protein was stored in the same buffer. Glutathione S-transferase (GST) purification was performed essentially as described previously (15). GST was eluted by incubation of the resin with 50 mM Tris/HCl (pH 7.4)-100 mM NaCl-20 mM glutathione.
Expression of RV capsid in yeast.
Full-length capsid was cloned into the pGREG536 plasmid by the drag-and-drop cloning technique in Saccharomyces cerevisiae (24). A cDNA encoding the entire capsid protein, including the E2 signal peptide, was amplified by PCR using forward and reverse primers 5′-GAATTCGATATCAAGCTTATCGATACCGTCGACAATGGCTTCCACTACCCCCATCACC-3′ and 5′-GCGTGACATAACTAATTACATGACTCGAGGTCGACCTACGGCGCGCGCGG-3′, respectively, where the restriction sites are in boldface italics. The resulting PCR product was cotransformed with SalI-digested plasmid pGREG536 into S. cerevisiae wild-type strain B44742, and transformants were selected on complete synthetic medium (Bio 101, Inc., Carlsbad, CA) lacking uracil. Capsid expression was induced by addition of galactose (2%) to the culture medium.
To test the binding of Mam33p to capsid, yeast cells were constructed which expressed a capsid variant that lacked the E2 signal peptide (capsidΔSP). The capsidΔSP cDNA (28) was subcloned into the EcoRI and NotI sites of pYES2/NT (Invitrogen). The plasmid was transformed into a yeast strain that was genomically tagged with a GFP cassette at the 3′ end of the MAM33 locus (21). Capsid expression was induced by addition of 2% galactose. To assess the influence of endogenously expressed capsid on protein import, full-length capsid was expressed from a pYES2/NT plasmid in wild-type yeast cells.
Immunoprecipitation and immunoblotting.
Yeast cells expressing capsidΔSP were harvested by centrifugation and then resuspended in 1% NP-40-0.5% sodium deoxycholate-0.1% sodium dodecyl sulfate (SDS)-150 mM NaCl-50 mM Tris-HCl (pH 8.0) containing Complete EDTA-free protease inhibitors (Roche). An equal volume of acid-washed 425- to 600-μm glass beads (Sigma-Aldrich) was added, and cells were disrupted with four cycles of homogenization (30 s) at 4°C by bead beating (Mini Beadbeater 8; BioSpec Products Inc., Bartlesville, OK). The cell slurries were clarified by centrifugation (1,000 × g, 5 min), after which the resulting supernatants were precleared with protein G-Sepharose for 1 h at 4°C. Mam33p-GFP was recovered by immunoprecipitation with goat anti-GFP (Eusera, Edmonton, AB, Canada) that had been prebound to protein G-Sepharose. Samples were washed three times with PBS containing 0.1% Triton X-100, and bound proteins were eluted by boiling in protein gel sample buffer. Proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride membranes (Immobilon-P; Millipore, Bedford, MA), which were incubated for 1 h at room temperature with 1:1,000 rabbit anti-RV capsid (7W7 [3]) or 1:5,000 rabbit anti-GFP (Eusera, Edmonton, AB) antibodies. After three washes with Tris-buffered saline-Tween, the membranes were incubated with goat anti-rabbit horseradish peroxidase-conjugated immunoglobulin G (IgG; Bio-Rad, Hercules, CA) for 1 h. Membranes were washed four times with Tris-buffered saline-Tween, and immunoreactive proteins were detected using Supersignal West Pico chemiluminescent substrate (Pierce Biotechnology, Rockford, IL), followed by exposure to X-ray film (Fuji Photo Film Co., Ltd., Tokyo, Japan).
Indirect immunofluorescence.
Vero cells cultured on glass coverslips were infected with RV (multiplicity of infection [MOI] = 1). Cells were processed for indirect immunofluorescence 48 h after infection (22). For localization of capsid in yeast, strains expressing full-length capsid were cultured in YES (yeast extract at 0.5%, glucose at 3%, 225 mg/liter adenine, histidine, leucine, uracil, and lysine hydrochloride) at 30°C. Cells from log-phase cultures were fixed for 1 h by adding 3.7% (vol/vol) formaldehyde directly to the culture. After fixation, cells were washed once with 0.1 M potassium phosphate (pH 6.5) and then twice with 0.1 M potassium phosphate (pH 6.5) containing 1.2 M sorbitol. Dithiothreitol was added to a final concentration of 25 mM, and samples were incubated for 30 min at 30°C. Cell walls were digested by adjusting the samples to 0.25 mg/ml zymicase I and β-mercaptoethanol (0.5%) and then incubating them at 37°C for 20 min. Cell suspensions were then placed onto microscope slides that had been coated with 0.1% poly-l-lysine and then fixed with ice-cold methanol for 6 min and then ice-cold acetone for 30 s. All of the washes were done in PBS supplemented with 0.1 mM CaCl2 and 1 mM MgCl2.
RV proteins were detected by using previously described rabbit anti-capsid (7W7), rabbit anti-p150, or mouse-anti E1 (H2C213) antibodies (3, 26). Mitochondria in mammalian cells were detected by using rabbit anti-cytochrome c antibodies (from L. Berthiaume, University of Alberta). Yeast mitochondria were labeled with a monoclonal antibody to subunit II of complex IV (MitoSciences, Eugene, OR). Primary antibodies were detected with Alexa Fluor 594-conjugated chicken anti-mouse, Alexa Fluor 488-conjugated donkey anti-rabbit, and/or Alexa Fluor 488-conjugated donkey anti-mouse antibodies (Molecular Probes, Invitrogen, Carlsbad, CA). Coverslips were mounted onto microscope slides using ProLong Gold antifade reagent with 4′-6-diamidino-2-phenylindole (DAPI; Molecular Probes, Invitrogen), and samples were examined using a Zeiss 510 confocal microscope.
Isolation of mitochondria.
Mitochondria isolated from a single rat liver were used for the import assays on the same day. Livers were homogenized using a Dounce homogenizer with a loose-fitting pestle in ice-cold mitochondrial isolation buffer containing 200 mM mannitol, 70 mM sucrose, 10 mM HEPES, 1 mM EGTA, and 0.1% (wt/vol) bovine serum albumin (pH 7.5) supplemented with Complete protease inhibitors. Unbroken cells and nuclei were pelleted by centrifugation at 500 × g for 5 min at 4°C. The supernatants were then centrifuged at 7,800 × g for 10 min at 4°C to pellet the crude mitochondrial fraction. Mitochondrial pellets were washed once with mitochondrial isolation buffer without bovine serum albumin. The final mitochondrial pellet was resuspended in 500 μl of 250 mM sucrose-10 mM HEPES-1 mM ATP-5 mM sodium succinate-0.08 mM ADP-2 mM K2HPO4-1 mM dithiothreitol (pH 7.4). Protein concentrations were determined using the BCA protein assay kit (Pierce Biotechnology, Rockford, IL) using bovine serum albumin as the standard.
Mitochondria were isolated from S. cerevisiae precisely as described previously (16) and then stored at −80°C until required.
Import assays.
The substrate used for mammalian mitochondrial import assays was Su9(1-69)DHFR, a hybrid protein consisting of the presequence of subunit 9 of the mitochondrial ATP synthase (residues 1 to 69) of Neurospora crassa fused to murine dihydrofolate reductase (45). For import into yeast mitochondria, a slightly longer derivative of this fusion protein was used [Su9(1-112)DHFR]. We refer to both as Su9-DHFR here. 35S-labeled Su9-DHFR was synthesized using the TNT Quick Coupled transcription/translation rabbit reticulocyte system (Promega).
Protein import into rat liver mitochondria.
Ten micrograms of freshly isolated mitochondria was incubated with radiolabeled Su9-DHFR in a buffer containing 20 mM PIPES (pH 7.4) and 10 mM KCl. To maintain the coupling of mitochondria, reaction mixtures were supplemented with ATP (2 mM) and sodium succinate (10 mM). Where indicated, C-terminally histidine-tagged capsidΔSP-His or GFP was added to the reaction mixtures. Import reaction mixtures were incubated at 30°C for 20 min, after which import was stopped by adding valinomycin (Sigma Chemical Co., St. Louis, MO) to a final concentration of 1 μM, followed by immediate transfer to ice.
Samples were resolved by 12% SDS-PAGE, and gels were fixed in a 50% (vol/vol) methanol-10% (vol/vol) acetic acid solution for 45 min at room temperature. Next, the gels were treated with 1 M sodium salicylate-0.01% (vol/vol) β-mercaptoethanol for 45 min to intensify the 35S signal before drying on filter paper. Radiolabeled proteins were detected by exposure to X-ray film or using a Storm 840 PhosphorImager (GE Healthcare Life Sciences). Quantitation of relative amounts of mature proteins compared to the amount of imported protein in the positive control reaction mixture was performed using ImageQuant software (Molecular Dynamics).
Protein import into yeast mitochondria.
Import of Su9-DHFR into yeast mitochondria was assayed essentially as described by us previously (6). Mitochondria (5 μg) were incubated in buffer containing 2 mM NADH and 2 mM ATP, and import reaction mixtures were incubated at 16°C for 5 to 20 min. Where indicated, purified capsid, GST, or MBP was added to the reaction mixtures. Nonimported precursor proteins were removed by incubation with proteinase K (100 μg/ml) for 30 min on ice. The protease was inactivated by addition of 2 mM phenylmethylsulfonyl fluoride. Samples were then subject to SDS-PAGE and autoradiography. Radioactive signals were quantified using an AIDA software package (Raytest).
Where indicated, mitochondria were pretreated with trypsin to remove surface-exposed transport receptors before the import assays were conducted. Destruction of the receptors was verified by immunoblotting with rabbit antibodies to Tom22 or mitochondrial ribosomal protein Mrpl40, which served as an internal control.
Immunoelectron microscopy.
Mock- and RV-infected cells were fixed in a mixture of 0.5% glutaraldehyde and 2% formaldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) for 1 h at 4°C. Samples then were rinsed in 0.075 M sodium cacodylate buffer (pH 7.4), dehydrated with a graded alcohol series (30%, 50%, 70%, and 80% ethanol), and then infiltrated with LR White (London Resin Co., Berkshire, United Kingdom). Infiltrated samples were embedded in gelatin capsules and polymerized under UV light for 24 h at 4°C. Following polymerization, ultrathin sections (60 nm) were cut and loaded onto a 300-mesh nickel grid without coating.
Prior to incubation with antibodies, the dried ultrathin sections were blocked overnight at 4°C with 8% bovine serum albumin in Tris-buffered saline (pH 7.4). Antibodies to capsid and p32 have been described previously (2, 3). Sequential labeling of each primary antibody, rabbit anti-capsid (1:200) and goat anti-p32 (1:200), was performed for 90 min. Incubations with the 12-nm colloidal gold-conjugated donkey anti-rabbit IgG (1:20) and 6 nm-colloidal gold-conjugated donkey anti-goat IgG (1:40) secondary antibodies were performed for 40 min each at room temperature. All antibodies were diluted with Tris-buffered saline containing 1% bovine serum albumin. After incubations with the primary and secondary antibodies, the sections were contrasted with 2% aqueous uranyl acetate for 15 min before viewing with a Philips 410 transmission electron microscope at 80 kV equipped with a charge-coupled device camera (MegaView III Soft Imaging System; Olympus).
For the quantitation of p32 in the mitochondria of mock- and RV-infected samples, mitochondria with good contrast and clearly delimited membranes and cristae were selected and segmented using iTEM software (Olympus Soft Imaging Solutions Co., Markham, Ontario, Canada). The segmentations were manually drawn along the mitochondrial membranes, and the areas (nm2) were measured and displayed in Adobe Photoshop 7.0 files. Prior to counting, the contrast of the 6-nm colloidal gold particles was increased to aid in identification. For each sample, between six and eight sections with more than 1,000 gold particles (6 nm) were counted.
RESULTS
RV infection results in lower levels of p32 in mitochondria.
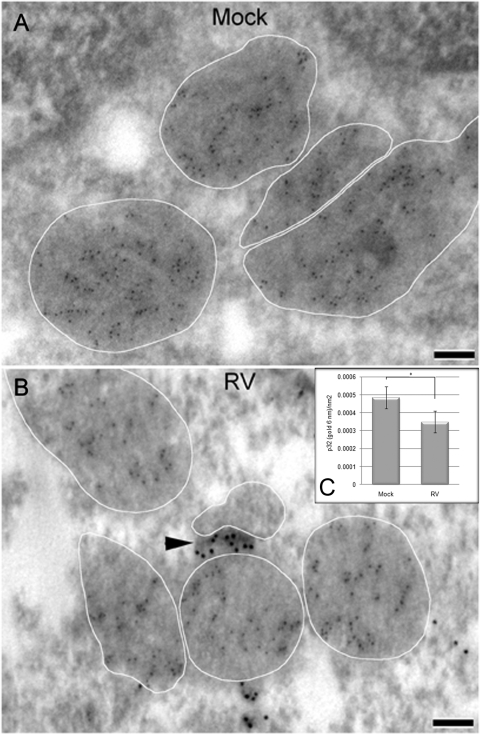
Mitochondria have long been thought to play an important role in RV replication (1), and one of the hallmarks of viral infection is drastic rearrangement of the mitochondrial network. Specifically, mitochondria often take on a club shape and tend to cluster in the perinuclear region of infected cells (31). Loss of cristae has also been reported in RV-infected cells (30). In light of previous data showing that blocking import into mitochondria leads to progressive loss of cristae (40), we questioned whether viral infection affects the import of proteins into this organelle. We used immunoelectron microscopy to examine the relative density and distribution of matrix protein p32 in mitochondria of mock- and RV-infected cells. Figure 1 shows that p32-specific labeling is confined almost exclusively to the matrix and is not detectable in the intermitochondrial plaques. Conversely, a pool of capsid (12-nm gold) localizes to the cytoplasmic surface of mitochondria, including the plaques (Fig. 1B, arrowhead). Qualitative analyses of micrographs such as those shown in Fig. 1A and B suggested that the concentration of p32 in the mitochondria of infected cells was lower than in mock-treated cells. Indeed, quantitation of data from multiple micrographs revealed that the mitochondria in RV-infected cells contained, on average, 30% less p32 than the mitochondria in mock-treated cells (Fig. 1C).
FIG. 1.
Mitochondrial levels of matrix protein p32 are reduced by RV infection. Vero cells were mock treated or infected with RV (MOI = 2) for 40 h, after which they were processed for immunoelectron microscopy. Samples were double labeled with antibodies specific for capsid (12-nm gold) and p32 (6-nm gold). White traces show the limiting membranes of the mitochondria. (A) Mock-infected cells. (B) RV-infected cells. A pool of capsid is visible in a plaque between two apposing mitochondria (arrowhead). Bar = 100 nm. (C) Quantitation of p32-specific gold particles (6 nm) within mitochondria. *, P < 0.001.
Capsid is the only RV antigen that associates with mitochondria.
To date, capsid is the only RV-encoded protein known to associate with mitochondria (2, 3, 32). However, a systematic comparison of the localization of other viral antigens in relation to this organelle has not been reported. Accordingly, Vero cells were infected with RV for 48 h and then processed for indirect immunofluorescence. Samples were incubated with antibodies to RV antigens, and mitochondria were detected using antibodies to cytochrome c or p32. Similar to our previous findings, the bulk of glycoprotein E1 was concentrated in the Golgi region, which is the major viral budding site (19). There was no apparent overlap between E1 and the mitochondrial marker cytochrome c (Fig. 2). The E2 glycoprotein, which is the binding partner of E1, exhibits a localization nearly identical to that of E1 at the Golgi complex (18) and was not observed to associate with mitochondria (data not shown). Next, we endeavored to compare the distribution of the nonstructural proteins with mitochondrial markers. Unfortunately, the only p90-specific antibodies that have been described are not suitable for indirect immunofluorescence (11). Therefore, our studies were limited to the localization of p150. As reported by Kujala et al. (26), p150 associates with unusual tubular structures that emanate from the perinuclear region (Fig. 2). In some cases, the p150-positive tubules were closely apposed to mitochondrial cisternae; however, very little overlap with the mitochondrial marker was evident. In contrast, there were large pools of mitochondrion-associated capsid protein in many of the infected cells (Fig. 2).
FIG. 2.
Capsid is the only RV antigen that localizes to mitochondria. Vero cells were infected with RV (MOI = 1), and at 48 h postinfection, samples were processed for indirect immunofluorescence using antibodies to RV proteins (E1, capsid, and p150) and mitochondria (cytochrome c and p32). Viral antigens are red, and mitochondrial marker proteins are green. Rabbit anti-cytochrome c antibody was used to stain mitochondria in all of the samples, except where p150 was detected. In this case, mitochondria were stained with goat anti-p32 antibody. Arrowheads indicate the tubular structures that are decorated with anti-p150 antibody. The insets in the bottom right of the panels are enlargements of the areas within the white squares. Nuclei were stained with DAPI. Bar = 10 μm.
Capsid protein blocks import into mitochondria.
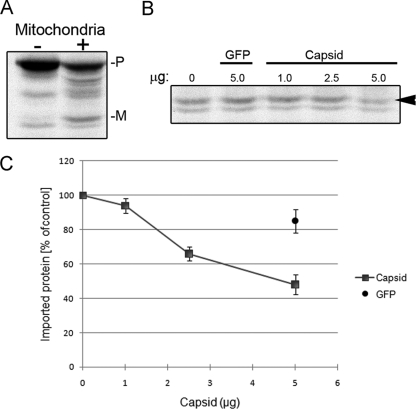
The data in Fig. 1 are consistent with a scenario in which RV infection blocks the import of proteins into mitochondria, and based upon the observation that capsid appears to be the only virus antigen that localizes to this organelle (Fig. 2), we hypothesized that capsid is responsible for this effect. To address this theory directly, mitochondria were isolated from rat liver and in vitro import assays were performed. For these experiments, we used Su9-DHFR as an import substrate. Su9-DHFR is a chimera that contains an N-terminal 69-amino-acid mitochondrial targeting signal from ATPase subunit 9 fused to the mouse dihydrofolate reductase protein, and it has been used extensively in mitochondrial import assays (45). 35S-labeled Su9-DHFR was incubated with mitochondria in the presence or absence of recombinant RV capsid protein. Following its translocation into mitochondria, Su9-DHFR is processed by the matrix-processing peptidase, which cleaves the N-terminal mitochondrial targeting sequence, leading to a concomitant decrease in the size of the mature protein (Fig. 3A). Addition of capsid to reaction mixtures resulted in dose-dependent inhibition of import, as evidenced by the reduction in the appearance of the mature processed form of Su9-DHFR (Fig. 3B, arrowhead). When 5 μg of capsid protein was included in the reaction mixtures, a twofold reduction in import was observed (Fig. 3C). Conversely, addition of the same amount of a similarly sized protein, GFP, did not significantly affect the import process.
FIG. 3.
Capsid inhibits import of substrates into mitochondria isolated from rat liver. (A) 35S-labeled Su9-DHFR was incubated with or without mitochondria for 20 min at 30°C, after which samples were subjected to SDS-PAGE and fluorography. Cleavage of the Su9-DHFR precursor occurs following translocation into mitochondria. Precursor (P) and mature (M) forms of Su9-DHFR are indicated. (B) Capsid inhibits mitochondrion-dependent maturation of Su9-DHFR in a dose-dependent manner. Increasing amounts of capsid or GFP were added to in vitro import assays, after which samples were analyzed by SDS-PAGE and fluorography. Regions of the gel showing the mature form of Su9-DHFR (arrowhead) are shown. (C) Data from three independent import experiments were quantitated and plotted.
Mitochondria isolated from yeast expressing capsid are impaired for import.
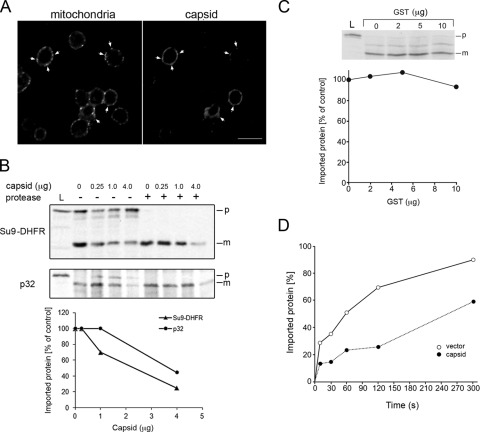
As a first step toward understanding the mechanism by which capsid inhibits the translocation of mitochondrial precursor proteins, we investigated whether import of precursor proteins into yeast mitochondria was affected by expression of the capsid protein. Mitochondrial import and export processes are very similar between animals and fungi (20), and the budding yeast S. cerevisiae has been used extensively to study these processes. Accordingly, if capsid also blocks import into yeast mitochondria, it would suggest that the viral protein interacts with a highly conserved component(s) of the import machinery. We first confirmed that the capsid protein localizes to the mitochondria in yeast. Similar to what we have observed in mammalian cells infected with RV, the majority of the capsid expressed in yeast cells was associated with mitochondria (Fig. 4A, arrowheads).
FIG. 4.
Capsid blocks protein import into yeast mitochondria. (A) Yeast cells were transformed with a yeast expression vector encoding RV capsid or the vector alone. Log-phase cultures were fixed and processed for indirect immunofluorescence. Capsid was detected with a rabbit polyclonal antibody, and mitochondria were stained with a mouse monoclonal antibody to subunit II of complex IV. Secondary antibodies were Alexa Fluor 488 donkey anti-rabbit IgG and Alexa Fluor 594 chicken anti-mouse IgG. Arrowheads denote colocalization between capsid and mitochondria. Bar = 5 μm. (B) Mitochondria isolated from wild-type yeast were incubated with 35S-lableled Su9-DHFR in the presence of capsid protein for 20 min at 16°C. In parallel, import assays were conducted with a second substrate, human p32. Prior to SDS-PAGE and autoradiography, samples were treated with proteinase K for 30 min at 0°C to remove nonimported precursor (p). The mature (m) imported forms of Su9-DHFR and p32 are indicated. Ten percent of the reticulocyte lysates (L) without mitochondria was loaded. The relative amounts of mature Su9-DHFR and p32 as a function of capsid concentration were determined and then plotted. (C) Here, import assays were conducted in the presence of variable amounts of a control protein (GST). Only the protease-treated samples are shown in this panel. The relative amounts of mature Su9-DHFR as a function of GST concentration were determined and then plotted. (D) Mitochondria were isolated from yeast strains that express capsid protein or vector alone. The mitochondria were incubated with 35S-lableled Su9-DHFR at 16°C for 0 to 300 s. Prior to SDS-PAGE and autoradiography, samples were treated with proteinase K for 30 min at 0°C to remove nonimported precursor. The relative amounts of mature Su9-DHFR as a function of time were determined and then plotted.
Next, import assays were performed using mitochondria isolated from wild-type yeast. As shown in Fig. 4B, import of Su9-DHFR was drastically reduced by the capsid protein in a dose-dependent manner. The translocation of a second radiolabeled substrate, human p32, was similarly affected. In contrast, the control protein GST had very little effect on the import process (Fig. 4C). Similar results were obtained when MBP was used as the negative control (data not shown). Together, these results suggest that targeting of capsid to mitochondria and the subsequent negative effect on mitochondrial import involve interaction with conserved components of this organelle. Moreover, using the yeast system offers a number of technical advantages, as well as access to a genome-wide set of null mutants with which to dissect the mechanism of the capsid-dependent block in translocation.
While in vitro import assays are invaluable for identifying factors that are required for translocation into mitochondria, there are a number of caveats to this system. To mimic the situation in RV-infected cells more closely, we expressed the RV capsid in living yeast cells. Mitochondria were isolated from these capsid-expressing cells or from yeast mutants harboring the empty vector as a control. The import of radiolabeled Su9-DHFR into these mitochondria was measured as a function of time. Compared to wild-type mitochondria, the rate of Su9-DHFR import into mitochondria isolated from yeast cells expressing capsid protein was markedly lower (Fig. 4D). Moreover, the efficiency of substrate import was reduced by approximately 33%. Therefore, as well as validating the in vitro import assays which employed soluble recombinant capsid protein, these results are in agreement with the data in Fig. 1, which suggest that capsid blocks mitochondrial import in vivo.
The capsid-binding components p32 and cardiolipin are not required for capsid-induced import inhibition.
After having established budding yeast as a valid model to study the effects of capsid on import into mitochondria, we next focused on the potential importance of mitochondrion-localized proteins and lipids that have been linked to RV replication and assembly. First, we examined the requirement for expression of matrix protein p32, which has been shown to interact with RV capsid protein p32 (2, 3, 37). Unfortunately, mammalian cell lines that are both null for p32 and permissive for RV infection are not available. Therefore, we employed a yeast null mutant that lacks the gene for the p32 ortholog Mam33p (43). Mam33p shares 22% identity and 34% similarity with human p32 at the amino acid level (Fig. 5A). The most-conserved region between p32 and Mam33p is the carboxyl terminus, which is where the capsid-binding site is localized (2). Before conducting the import experiments, it was important to verify that Mam33p interacts with capsid. Yeast lysates were subjected to immunoprecipitation with anti-GFP antibody, followed by immunoblot analyses. As shown in Fig. 5B, the capsid protein forms a stable complex with the yeast ortholog of p32. As a negative control for binding to GFP-tagged yeast proteins, capsid was expressed in a yeast strain containing GFP integrated at the HCR1 locus and the immunoprecipitation and immunoblot analyses were repeated. Hcr1p (∼29.6 kDa) is similar in size to Mam33p (30.1 kDa) but, as shown in Fig. 5B, does not form a stable complex with capsid.
FIG. 5.
Capsid-dependent inhibition of import does not require Mam33p. (A) Alignment of human p32 and its yeast ortholog Mam33p. Regions of identify (black shading) and similarity (gray shading) are indicated. Arrowheads indicate predicted sites of cleavage by mitochondrial proteases. (B) Yeast strains genomically tagged at the MAM33 or HRC1 locus with GFP were transformed with a plasmid encoding RV capsid. Expression of capsid protein was induced with galactose (Gal). Cell lysates were subjected to immunoprecipitation with anti-GFP, followed by SDS-PAGE and immunoblotting with anti-capsid antibodies. The asterisk indicates capsid that copurifies with Mam33p. CO-IP, coimmunoprecipitation. (C) Mitochondria isolated from wild-type (WT) and Δmam33 mutant yeast strains were incubated with 35S-lableled Su9-DHFR in the presence or absence of RV capsid for 20 min at 16°C. Prior to SDS-PAGE and autoradiography, samples were treated with proteinase K for 30 min at 0°C to remove nonimported precursor. The relative amounts of mature Su9-DHFR in the samples were determined and then plotted.
Next, we investigated whether expression of Mam33p in mitochondria is required for the ability of capsid to block translocation into this organelle. Mitochondria were isolated from wild-type and Δmam33 mutant yeast cells, and import assays were conducted as described above. Data in Fig. 5C show that capsid had similar effects on import into wild-type and Δmam33 mutant mitochondria. In both cases, import of Su9-DHFR was reduced approximately eightfold when 10 μg of capsid was included in the reaction mixtures. These results indicate that expression of Mam33p and, by extrapolation, p32 in mammalian mitochondria is not required for the ability of capsid to inhibit the translocation of proteins into this organelle.
Recently, it was reported that mitochondrion-specific anionic phospholipids such as cardiolipin may be important host determinants that regulate the targeting of viral polymerase complexes (44). For this reason and the fact that it has been known for more than 30 years that RV virions are enriched in cardiolipin (1), we next investigated whether this phospholipid is required for capsid to block mitochondrial import. Using an in vitro lipid-binding assay, we showed that capsid specifically interacts with cardiolipin (Fig. 6A). To determine whether binding of capsid to cardiolipin in the outer or inner mitochondrial membrane is required for blocking mitochondrial import, mitochondria were isolated from a yeast strain lacking the gene for cardiolipin synthase (Δcrd1 mutant) and in vitro import assays were performed as described above. Results shown in Fig. 6B and C clearly show that capsid efficiently blocks the import of Su9-DHFR into cardiolipin-deficient mitochondria. Together, these results indicate that cardiolipin is not required for the ability of capsid to block mitochondrial import.
FIG. 6.
Capsid-dependent inhibition of import does not require cardiolipin. (A) Purified capsid protein was incubated with a membrane that had been spotted with various lipids. Bound capsid was detected by enhanced chemiluminescence with a secondary antibody conjugated to horseradish peroxidase. (B) Mitochondria were isolated from a yeast strain (Δcrd1 mutant) lacking the gene for cardiolipin synthase. The mitochondria were incubated with 35S-labeled Su9-DHFR in the presence or absence of RV capsid for 20 min at 16°C. Prior to SDS-PAGE and autoradiography, samples were treated with proteinase K for 30 min at 0°C to remove nonimported precursor. The mature (m) form of Su9-DHFR is indicated. (C) The relative amounts of mature Su9-DHFR in the samples were determined and then plotted. WT, wild type.
cis- and trans-acting factors that affect the import-blocking ability of capsid.
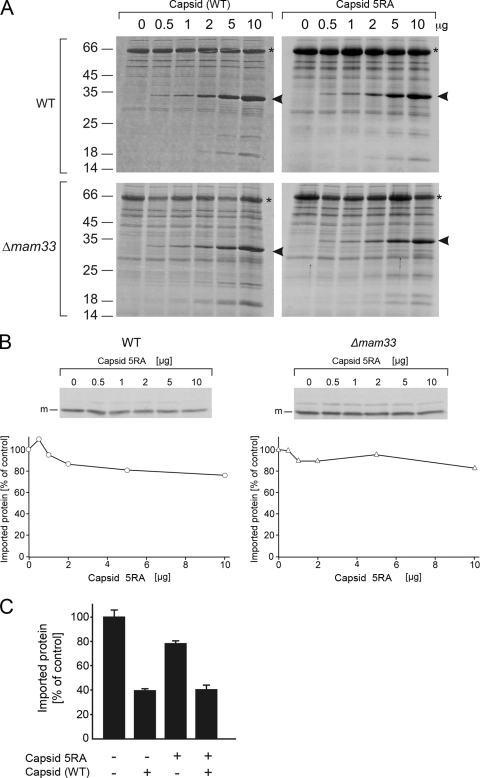
To gain further insight into how capsid inhibits the translocation of proteins into mitochondria, we took advantage of a well-characterized capsid mutant in which a cluster of five arginine residues present in the amino-terminal region of the protein were changed to alanine residues (2). This motif is clearly important for capsid function, as RV strains that contain the C5RA capsid gene produce 1,000-fold less infectious virus than does the wild-type virus. Using an in vitro binding assay, we showed that the C5RA mutant still associates with mitochondria (Fig. 7A). However, results from the import assays revealed that, unlike wild-type capsid, the C5RA capsid did not efficiently block the translocation of Su9-DHFR into mitochondria isolated from wild-type or Δmam33 mutant yeast (Fig. 7B). In order to examine whether the C5RA mutant capsid could act in a dominant negative manner to prevent wild-type capsid from blocking import, we treated mitochondria first with the C5RA mutant capsid before wild-type capsid was added. As shown in Fig. 7C, the presence of the mutant capsid did not reduce the inhibitory effect of capsid on the import of Su9-DHFR. This suggests that the binding of the C5RA mutant capsid does not saturate potential capsid-binding sites on the mitochondria. Hence, the positively charged region in the wild-type capsid can still block import, presumably by its interaction with negatively charged presequence receptors on the mitochondrial surface.
FIG. 7.
(A) The binding of capsid to mitochondria is independent of Mam33p and a cluster of amino-terminal arginine residues in capsid. Mitochondria were isolated from wild-type (WT) and Δmam33 mutant cells and incubated for 10 min at 16°C with increasing amounts of purified WT capsid or C5RA mutant capsid protein. The mitochondria were reisolated, washed, and subjected to SDS-PAGE. Proteins were stained with Coomassie brilliant blue. The arrowheads depict the capsid proteins. Due to its altered isoelectric point, the C5RA mutant protein migrates slightly slower. Bovine serum albumin, which is present in the storage buffer of the mitochondria, is indicated by asterisks. (B) A cluster of arginine residues in the amino terminus of capsid is critical for the translocation-inhibiting activity of capsid. Mitochondria isolated from wild-type and Δmam33 mutant yeast strains were incubated with 35S-lableled Su9-DHFR in the presence or absence of C5RA mutant capsid for 20 min at 16°C. Prior to SDS-PAGE and autoradiography, samples were treated with proteinase K for 30 min at 0°C to remove nonimported precursor. The mature (m) imported form of Su9-DHFR is indicated. The relative amounts of mature Su9-DHFR in the samples were determined and then plotted. (C) Wild-type mitochondria (5 μg) were incubated in a first reaction mixture with or without 5 μg of C5RA mutant capsid at 16°C for 10 min. Then, capsid (5 μg) was added as indicated and the import reaction mixtures were started by addition of 35S-lableled Su9-DHFR. After 5 min, the samples were treated with proteinase K for 30 min at 0°C to remove nonimported precursor. Proteins were analyzed by SDS-PAGE and autoradiography. The relative amounts of imported protein in the samples were determined. Shown are mean values from three independent experiments. The values on the left of panel A are molecular sizes in kilodaltons.
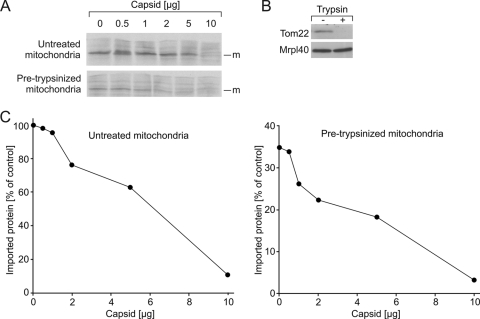
Lastly, we examined whether protease-sensitive components of the TOM (translocase of the outer membrane) complex are required for capsid to block translocation. For these experiments, mitochondria were preincubated with trypsin to remove the surface-exposed import receptors that comprise the TOM complex. As shown in Fig. 8, removal of the receptors strongly diminished import efficiency. However, the addition of capsid still interferes with the import of Su9-DHFR into mitochondria to an extent that is comparable to that in untreated mitochondria. This indicates that also in the absence of surface-exposed TOM receptors, capsid blocks the translocation of mitochondrial substrates.
FIG. 8.
A functional TOM complex is not required for capsid to block mitochondrial import. (A) Mitochondria isolated from wild-type yeast were incubated with 35S-lableled Su9-DHFR in the presence or absence of capsid for 20 min at 16°C. Where indicated, mitochondria were pretreated with trypsin to remove components of the TOM complex. Prior to SDS-PAGE and autoradiography, samples were treated with proteinase K for 30 min at 0°C to remove nonimported precursor. The mature (m) imported form of Su9-DHFR is indicated. (B) Western blot against TOM receptor Tom22 with untreated and pretrypsinized mitochondria. Mitochondrial ribosomal protein Mrpl40 served as a loading control. (C) The amounts of mature Su9-DHFR in the samples were determined and then plotted relative to the amount of protein imported into untreated mitochondria in the absence of capsid.
DISCUSSION
Mitochondria play important roles in regulating the outcome of viral infections (reviewed in reference 12). In addition to manufacturing ATP, which is required for numerous aspects of viral replication and assembly, for some viruses, mitochondrial membranes serve as platforms for viral replicase complexes. Flock house virus is probably the most well characterized virus that replicates on this organelle (35). In this case, the viral RNA polymerase is targeted to mitochondria by an amino-terminal targeting motif (34). Interestingly, protease-accessible components of the TOM complex are not necessary for association of the polymerase with mitochondria, but anionic phospholipids such as cardiolipin are required (44). Aside from the flock house virus RNA polymerase, there are very few examples of RNA virus-encoded proteins that are known to associate with mitochondria. Furthermore, with the exception of the RV and hepatitis C virus capsid proteins, all known viral mitochondrial proteins are nonstructural or accessory proteins.
Here we report for the first time that a virus-encoded protein blocks the import of precursor proteins into mitochondria. The mechanism is not substrate specific, as evidenced by the fact that the translocation and processing of two different mitochondrially targeted proteins, p32 and Su9-DHFR, were both impaired by capsid. Moreover, because import into mammalian and yeast mitochondria was similarly affected, the capsid presumably targets a highly conserved component(s) of the translocation machinery. Our data suggest that binding of capsid to mitochondria is not sufficient for its ability to interfere with import. This is based on the observation that the C5RA mutant capsid does not block import even though it is targeted to mitochondria (2). Because capsid does not possess enzymatic activity, we believe that its inhibitory effect on translocation is stoichiometric in nature. We originally hypothesized that the replication defects of the CR5A mutant strain were due to lack of binding between capsid and p32; however, it is now tempting to speculate that the inability of the CR5A mutant protein to block mitochondrial import may also affect replication.
Capsid was still able to block the import of substrates into mitochondria in which receptors of the TOM complex had been removed by trypsin digestion. Accordingly, interference with surface-exposed transport receptors is not the main or only mechanism by which capsid interferes with import. An alternative scenario is that the capsid protein, which is rich in basic amino acids, interacts with negatively charged mitochondrial lipids and/or protease-resistant components of the translocation machinery such as the general insertion pore (41). Consistent with this theory is the observation that mutagenesis of the arginine cluster in the amino terminus of capsid ablates the import-blocking property. Similar to the flock house virus polymerase (44), capsid binds to cardiolipin; however, this anionic phospholipid is not strictly required in order for capsid to impair translocation.
The obvious question is why is it advantageous for a virus to interfere with the translocation of host proteins into mitochondria. We hypothesize that this process is important to inhibit virus-induced apoptosis. Indeed, recent studies suggest that mitochondrial import may be an integral part of some apoptotic mechanisms (reviewed in reference 39). It is well established that targeting of some proapoptotic factors to the mitochondria requires the TOM complex. For example, a number of recent studies suggest that the mitochondrial receptor for the pore-forming protein Bax is Tom22 (4, 7), and indeed, genetic evidence indicates that multiple subunits of the TOM complex are required for efficient Bax-dependent apoptosis (10). Because the replication cycle of RV is quite long compared to that of related viruses, it would be highly beneficial if apoptosis were inhibited or delayed until replication and assembly occurred. Moreover, blocking programmed cell death may facilitate the establishment and/or maintenance of persistent infections.
Preliminary studies from our laboratory, indeed, suggest that the RV capsid inhibits apoptosis in a variety of cultured cell lines (C. S. Ilkow and T. C. Hobman, unpublished data). If the antiapoptotic function of capsid is related to its effects on mitochondrial import, there are at least three non-mutually exclusive potential mechanisms by which this can occur. First, by interfering with the TOM complex through steric hindrance, the pool of mitochondrion-associated capsid could reduce the amount of Bax that is recruited to mitochondria. Second, binding of capsid to cardiolipin, which is important for targeting of the proapoptotic protein tBid to mitochondria (33), could decrease the amount of tBid recruitment. Lastly, capsid could block apoptosis by reducing the levels of p32 in mitochondria. p32 contributes to programmed cell death through multiple mechanisms (9, 23, 46), all of which require localization of p32 to mitochondria.
Acknowledgments
We thank Luc Berthiaume (University of Alberta), Dejana Mokranjac, and Kai Hell (Institute for Physiological Chemistry, Munich, Germany) for reagents.
C.I. and M.D.B. were supported by graduate studentships from the Alberta Heritage Foundation for Medical Research (AHFMR). T.C.H. is the recipient of a Scientist award from AHFMR. This work was supported by grants from the Canadian Institutes of Health Research to T.C.H. and from the Deutsche Forschungsgemeinschaft and the Stiftung für Innovation Rheinland-Pfalz to J.M.H. I.S.G. is the recipient of a Recruitment Award from the Alberta Cancer Research Institute.
Footnotes
Published ahead of print on 21 October 2009.
REFERENCES
- 1.Bardeletti, G., and D. C. Gautheron. 1976. Phospholipid and cholesterol composition of rubella virus and its host cell BHK 21 grown in suspension cultures. Arch. Virol. 52:19-27. [DOI] [PubMed] [Google Scholar]
- 2.Beatch, M. D., J. C. Everitt, L. J. Law, and T. C. Hobman. 2005. Interactions between rubella virus capsid and host protein p32 are important for virus replication. J. Virol. 79:10807-10820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beatch, M. D., and T. C. Hobman. 2000. Rubella virus capsid associates with host cell protein p32 and localizes to mitochondria. J. Virol. 74:5569-5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellot, G., P. F. Cartron, E. Er, L. Oliver, P. Juin, L. C. Armstrong, P. Bornstein, K. Mihara, S. Manon, and F. M. Vallette. 2007. TOM22, a core component of the mitochondria outer membrane protein translocation pore, is a mitochondrial receptor for the proapoptotic protein Bax. Cell Death Differ. 14:785-794. [DOI] [PubMed] [Google Scholar]
- 5.Benali-Furet, N. L., M. Chami, L. Houel, F. De Giorgi, F. Vernejoul, D. Lagorce, L. Buscail, R. Bartenschlager, F. Ichas, R. Rizzuto, and P. Paterlini-Brechot. 2005. Hepatitis C virus core triggers apoptosis in liver cells by inducing ER stress and ER calcium depletion. Oncogene 24:4921-4933. [DOI] [PubMed] [Google Scholar]
- 6.Bihlmaier, K., M. Bien, and J. M. Herrmann. 2008. In vitro import of proteins into isolated mitochondria. Methods Mol. Biol. 457:85-94. [DOI] [PubMed] [Google Scholar]
- 7.Cartron, P. F., G. Bellot, L. Oliver, X. Grandier-Vazeille, S. Manon, and F. M. Vallette. 2008. Bax inserts into the mitochondrial outer membrane by different mechanisms. FEBS Lett. 582:3045-3051. [DOI] [PubMed] [Google Scholar]
- 8.Chen, M. H., and J. P. Icenogle. 2004. Rubella virus capsid protein modulates viral genome replication and virus infectivity. J. Virol. 78:4314-4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chowdhury, A. R., I. Ghosh, and K. Datta. 2008. Excessive reactive oxygen species induces apoptosis in fibroblasts: role of mitochondrially accumulated hyaluronic acid binding protein 1 (HABP1/p32/gC1qR). Exp. Cell Res. 314:651-667. [DOI] [PubMed] [Google Scholar]
- 10.Colin, J., J. Garibal, B. Mignotte, and I. Guenal. 2009. The mitochondrial TOM complex modulates bax-induced apoptosis in Drosophila. Biochem. Biophys. Res. Commun. 379:939-943. [DOI] [PubMed] [Google Scholar]
- 11.Forng, R. Y., and T. K. Frey. 1995. Identification of the rubella virus nonstructural proteins. Virology 206:843-853. [DOI] [PubMed] [Google Scholar]
- 12.Galluzzi, L., C. Brenner, E. Morselli, Z. Touat, and G. Kroemer. 2008. Viral control of mitochondrial apoptosis. PLoS Pathog. 4:e1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ge, H., P. Zuo, and J. L. Manley. 1991. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell 66:373-382. [DOI] [PubMed] [Google Scholar]
- 14.Ghebrehiwet, B., B. L. Lim, R. Kumar, X. Feng, and E. I. Peerschke. 2001. gC1q-R/p33, a member of a new class of multifunctional and multicompartmental cellular proteins, is involved in inflammation and infection. Immunol. Rev. 180:65-77. [DOI] [PubMed] [Google Scholar]
- 15.Grumbt, B., V. Stroobant, N. Terziyska, L. Israel, and K. Hell. 2007. Functional characterization of Mia40p, the central component of the disulfide relay system of the mitochondrial intermembrane space. J. Biol. Chem. 282:37461-37470. [DOI] [PubMed] [Google Scholar]
- 16.Herrmann, J., H. Fölsch, W. Neupert, and R. Stuart. 1994. Isolation of yeast mitochondria and study of mitochondrial protein translation, p. 538-544. In J. Celis (ed.), Cell biology: a laboratory handbook. Academic Press, Inc., San Diego, CA.
- 17.Hobman, T., and J. K. Chantler. 2007. Rubella virus, p. 1069-1100. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, fifth ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 18.Hobman, T. C. 1993. Targeting of viral glycoproteins to the Golgi complex. Trends Microbiol. 1:124-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hobman, T. C., L. Woodward, and M. G. Farquhar. 1993. The rubella virus E2 and E1 spike glycoproteins are targeted to the Golgi complex. J. Cell Biol. 121:269-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoogenraad, N. J., L. A. Ward, and M. T. Ryan. 2002. Import and assembly of proteins into mitochondria of mammalian cells. Biochim. Biophys. Acta 1592:97-105. [DOI] [PubMed] [Google Scholar]
- 21.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 22.Ilkow, C. S., V. Mancinelli, M. D. Beatch, and T. C. Hobman. 2008. Rubella virus capsid protein interacts with poly(A)-binding protein and inhibits translation. J. Virol. 82:4284-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itahana, K., and Y. Zhang. 2008. Mitochondrial p32 is a critical mediator of ARF-induced apoptosis. Cancer Cell 13:542-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jansen, G., C. Wu, B. Schade, D. Y. Thomas, and M. Whiteway. 2005. Drag&Drop cloning in yeast. Gene 344:43-51. [DOI] [PubMed] [Google Scholar]
- 25.Krainer, A. R., A. Mayeda, D. Kozak, and G. Binns. 1991. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell 66:383-394. [DOI] [PubMed] [Google Scholar]
- 26.Kujala, P., T. Ahola, N. Ehsani, P. Auvinen, H. Vihinen, and L. Kaariainen. 1999. Intracellular distribution of rubella virus nonstructural protein P150. J. Virol. 73:7805-7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Law, L. J., C. S. Ilkow, W. P. Tzeng, M. Rawluk, D. T. Stuart, T. K. Frey, and T. C. Hobman. 2006. Analyses of phosphorylation events in the rubella virus capsid protein: role in early replication events. J. Virol. 80:6917-6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Law, L. M., R. Duncan, A. Esmaili, H. L. Nakhasi, and T. C. Hobman. 2001. Rubella virus E2 signal peptide is required for perinuclear localization of capsid protein and virus assembly. J. Virol. 75:1978-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Law, L. M., J. C. Everitt, M. D. Beatch, C. F. Holmes, and T. C. Hobman. 2003. Phosphorylation of rubella virus capsid regulates its RNA binding activity and virus replication. J. Virol. 77:1764-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, J. Y., and D. S. Bowden. 2000. Rubella virus replication and links to teratogenicity. Clin. Microbiol. Rev. 13:571-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, J. Y., D. S. Bowden, and J. A. Marshall. 1996. Membrane junctions associated with rubella virus infected cells. J. Submicrosc. Cytol. Pathol. 28:101-108. [PubMed] [Google Scholar]
- 32.Lee, J. Y., J. A. Marshall, and D. S. Bowden. 1999. Localization of rubella virus core particles in Vero cells. Virology 265:110-119. [DOI] [PubMed] [Google Scholar]
- 33.Lutter, M., M. Fang, X. Luo, M. Nishijima, X. Xie, and X. Wang. 2000. Cardiolipin provides specificity for targeting of tBid to mitochondria. Nat. Cell Biol. 2:754-761. [DOI] [PubMed] [Google Scholar]
- 34.Miller, D. J., and P. Ahlquist. 2002. Flock house virus RNA polymerase is a transmembrane protein with amino-terminal sequences sufficient for mitochondrial localization and membrane insertion. J. Virol. 76:9856-9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, D. J., M. D. Schwartz, and P. Ahlquist. 2001. Flock house virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J. Virol. 75:11664-11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, K., S. McArdle, M. J. Gale, Jr., D. A. Geller, B. Tenoever, J. Hiscott, D. R. Gretch, and S. J. Polyak. 2004. Effects of the hepatitis C virus core protein on innate cellular defense pathways. J. Interferon Cytokine Res. 24:391-402. [DOI] [PubMed] [Google Scholar]
- 37.Mohan, K. V., B. Ghebrehiwet, and C. D. Atreya. 2002. The N-terminal conserved domain of rubella virus capsid interacts with the C-terminal region of cellular p32 and overexpression of p32 enhances the viral infectivity. Virus Res. 85:151-161. [DOI] [PubMed] [Google Scholar]
- 38.Mori, Y., T. Okabayashi, T. Yamashita, Z. Zhao, T. Wakita, K. Yasui, F. Hasebe, M. Tadano, E. Konishi, K. Moriishi, and Y. Matsuura. 2005. Nuclear localization of Japanese encephalitis virus core protein enhances viral replication. J. Virol. 79:3448-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paschen, S. A., A. Weber, and G. Hacker. 2007. Mitochondrial protein import: a matter of death? Cell Cycle 6:2434-2439. [DOI] [PubMed] [Google Scholar]
- 40.Perkins, G. A., C. W. Renken, I. J. van der Klei, M. H. Ellisman, W. Neupert, and T. G. Frey. 2001. Electron tomography of mitochondria after the arrest of protein import associated with Tom19 depletion. Eur. J. Cell Biol. 80:139-150. [DOI] [PubMed] [Google Scholar]
- 41.Pfaller, R., N. Pfanner, and W. Neupert. 1989. Mitochondrial protein import. Bypass of proteinaceous surface receptors can occur with low specificity and efficiency. J. Biol. Chem. 264:34-39. [PubMed] [Google Scholar]
- 42.Sacco, R., T. Tsutsumi, R. Suzuki, M. Otsuka, H. Aizaki, S. Sakamoto, M. Matsuda, N. Seki, Y. Matsuura, T. Miyamura, and T. Suzuki. 2003. Antiapoptotic regulation by hepatitis C virus core protein through up-regulation of inhibitor of caspase-activated DNase. Virology 317:24-35. [DOI] [PubMed] [Google Scholar]
- 43.Seytter, T., F. Lottspeich, W. Neupert, and E. Schwarz. 1998. Mam33p, an oligomeric, acidic protein in the mitochondrial matrix of Saccharomyces cerevisiae is related to the human complement receptor gC1q-R. Yeast 14:303-310. [DOI] [PubMed] [Google Scholar]
- 44.Stapleford, K. A., D. Rapaport, and D. J. Miller. 2009. Mitochondrion-enriched anionic phospholipids facilitate flock house virus RNA polymerase membrane association. J. Virol. 83:4498-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stojanovski, D., N. Pfanner, and N. Wiedemann. 2007. Import of proteins into mitochondria. Methods Cell Biol. 80:783-806. [DOI] [PubMed] [Google Scholar]
- 46.Sunayama, J., Y. Ando, N. Itoh, A. Tomiyama, K. Sakurada, A. Sugiyama, D. Kang, F. Tashiro, Y. Gotoh, Y. Kuchino, and C. Kitanaka. 2004. Physical and functional interaction between BH3-only protein Hrk and mitochondrial pore-forming protein p32. Cell Death Differ. 11:771-781. [DOI] [PubMed] [Google Scholar]
- 47.Tzeng, W. P., and T. K. Frey. 2003. Complementation of a deletion in the rubella virus p150 nonstructural protein by the viral capsid protein. J. Virol. 77:9502-9510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tzeng, W. P., and T. K. Frey. 2005. Rubella virus capsid protein modulation of viral genomic and subgenomic RNA synthesis. Virology 337:327-334. [DOI] [PubMed] [Google Scholar]
- 49.Tzeng, W. P., J. D. Matthews, and T. K. Frey. 2006. Analysis of rubella virus capsid protein-mediated enhancement of replicon replication and mutant rescue. J. Virol. 80:3966-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]