Abstract
It has been firmly established that many interphase nuclear functions, including transcriptional regulation, are regulated by chromatin and histones. How mitotic progression and quality control might be influenced by histones is less well characterized. We show that histone H3 plays a crucial role in activating the spindle assembly checkpoint in response to a defect in mitosis. Prior to anaphase, all chromosomes must attach to spindles emanating from the opposite spindle pole bodies. The tension between sister chromatids generated by the poleward pulling force is an integral part of chromosome biorientation. Lack of tension due to erroneous attachment activates the spindle assembly checkpoint, which corrects the mistakes and ensures segregation fidelity. A histone H3 mutation impairs the ability of yeast cells to activate the checkpoint in a tensionless crisis, leading to missegregation and aneuploidy. The defects in tension sensing result directly from an attenuated H3-Sgo1p interaction essential for pericentric recruitment of Sgo1p. Reinstating the pericentric enrichment of Sgo1p alleviates the mitotic defects. Histone H3, and hence the chromatin, is thus a key factor transmitting the tension status to the spindle assembly checkpoint.
During mitosis, chromatin goes through significant compaction and condensation to form metaphase chromosomes for segregation. While there is a wealth of information on the crucial roles played by chromatin structures and histone modifications in controlling transcription, replication, repair, and recombination (30), much less is known about how individual histones contribute mechanistically to mitotic progression and regulation.
Forward and reverse genetic studies have suggested that histones, rather than being merely a part of the cargo during mitotic segregation, may play key roles in cell cycle progression and regulation. A histone H4 allele, hhf1-20, compromises the interaction between H4 and the centromere-specific H3 variant Cse4p, thus impeding centromeric functions and mitosis at the restrictive temperature (50). Two alleles of histone H2A (44) cause cold-sensitive growth defects and a significant increase in ploidy. This hyperploidy phenotype can be suppressed by mutations affecting a histone deacetylase, HDA1 (19). Similarly, the Gcn5p histone acetyltransferase genetically interacts with several inner kinetochore components and is physically mapped to the centromeric regions (57). Deleting the flexible tail domain of H3 and H4 results in mitotic delay (36) via a mechanism that can be suppressed by inhibiting the spindle assembly checkpoint activity (J.L. and M.H.K., unpublished data). Together, these data warrant a more thorough examination of how chromatin may proactively regulate the process of mitotic segregation.
The center stage for mitotic segregation and control is the kinetochore, a large proteinaceous complex assembled on centromeres. The ultimate function of the kinetochore is to capture the spindle microtubules during mitosis. The kinetochore-spindle attachment drives the movement of chromatids to daughter cells, and error-free attachment is essential for even partitioning of the entire genetic complement. To prepare for segregation, S-phase cells first establish sister chromatid cohesion by loading the cohesin complex to centromeres, pericentromeres, and selective regions in the chromatin arms (8, 38). Cohesion prevents precocious segregation before alignment of chromosomes at the metaphase midplate. When cells enter the prophase, spindles are assembled in and emanate from the two spindle pole bodies and are captured by kinetochores. The opposing poleward pulling force then generates tension between sisters and causes the congression of bioriented chromosomes toward the midplane (11). Equatorial alignment of all chromosomes leads to activation of the anaphase-promoting complex/cyclosome, which catalyzes polyubiquitylation and degradation of the securin protein Pds1p and cyclins (23, 60), and activation of the separase Esp1p, which cleaves the Mcd1p/Scc1p subunit of the cohesin complex (37) and hence permits sister chromatid segregation.
One of the most critical mitotic control mechanisms for segregation is the spindle assembly checkpoint, SAC (31), which monitors both the kinetochore-spindle attachment and the resultant tension between sisters (43, 61). The tension-sensing function of the SAC is critical for cells to detect and correct the so-called syntelic attachment; that is, both sister kinetochores capture spindles emanating from the same spindle pole body. This condition meets the attachment requirement but does not generate tension. Aneuploidy will result if this type of error is not eradicated. One of the factors essential for cells to detect the tensionless crisis is the Shugoshin protein (Sgo) (7, 15, 21). Downregulation of human Sgo1 expression is linked to about half of the colorectal cancer cases analyzed in one study (16). Yeast Sgo1p is localized to the centromeres and pericentromeres (22, 46). Centromeres and pericentromeres are the most likely loci in which tension is generated and monitored (3, 14). Here we show that histone H3 plays a key role in the pericentric recruitment of Sgo1p for the tension-sensing function in mitosis.
MATERIALS AND METHODS
Yeast strains and plasmid constructs.
The yeast strains, plasmids, and oligonucleotide primers used in this work are listed in Tables 1 and 2. The sequences of the oligonucleotides used in this study are available upon request.
TABLE 1.
Yeast strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| A10652 | W303 MATaSGO1-6×HA | 22 |
| JHY200 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 hht1-hhf1::KAN hht2-hhf2::KAN hta1-htb1::Nat hta2-htb2::HPH/pJH33 [ARS CEN URA3 HTA1-HTB1 HHT2-HHF2] | 1 |
| SBY214 | MATaade2-1 bar1Δ can1-100 his3-11::pCUP1-GFP12-lacI12::HIS3 leu2,3-112 lys2Δ trp1-1::lacO::TRP1 ura3-1 | 2 |
| yJL118 | MATaade2-1 bar1Δ can1-100 his3-11::pCUP1-GFP12-lacI12::HIS3 leu2,3-112 lys2Δ trp1-1::lacO::TRP1 ura3-1 hht1-hhf1::KAN | This study |
| yJL145 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 hht1-hhf1::KAN hht2-hhf2::KAN hta1-htb1::Nat hta2-htb2::HPH/pMK439G44S [ARS CEN LEU2 HTA1-HTB1 hht2-G44S-HHF2] | This study |
| yJL171 | MATaade2-1 bar1Δ can1-100 his3-11,15::pGAL-MCD1::HIS3 leu2-3,112 trp1-1::PDS1-Myc13::TRP1 ura3-1 hht1-hhf1::KAN hht2-hhf2::KAN hta1-htb1::Nat hta2-htb2::HPH/pQQ18 [ARS CEN LEU2 HTA1-HTB1 HHT2-HHF2] | This study |
| yJL172 | MATaade2-1 bar1Δ can1-100 his3-11,15::pGAL-MCD1::HIS3 leu2-3,112 trp1-1::PDS1-Myc13::TRP1 ura3-1 hht1-hhf1::KAN hht2-hhf2::KAN hta1-htb1::Nat hta2-htb2::HPH/pMK439G44S [ARS CEN LEU2 HTA1-HTB1 hht2-G44S-HHF2] | This study |
| yJL292 | MATaade2-1 bar1Δ can1-100 his3-11::pCUP1-GFP12-lacI12::HIS3 leu2,3-112 lys2Δ trp1-1::lacO::TRP1 ura3-1::hht2-hhf2::HHT2-HHF2::URA3 hht1-hhf1::KAN | This study |
| yJL293 | MATaade2-1 bar1Δ can1-100 his3-11::pCUP1-GFP12-lacI12::HIS3 leu2,3-112 lys2Δ trp1-1::lacO::TRP1 ura3-1::hht2-hhf2::hht2-G44S-HHF2::URA3 hht1-hhf1::KAN | This study |
| yJL343 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1::SGO1-6×HA::TRP1 ura3-1 hht1-hhf1::KAN hht2-hhf2::KAN hta1-htb1::Nat hta2-htb2::HPH/pQQ18 [ARS CEN LEU2 HTA1-HTB1 HHT2-HHF2] | This study |
| yJL344 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1::SGO1-6×HA::TRP1 ura3-1 hht1-hhf1::KAN hht2-hhf2::KAN hta1-htb1::Nat hta2-htb2::HPH/pMK439G44S [ARS CEN LEU2 HTA1-HTB1 hht2-G44S-HHF2] | This study |
| yMK1243 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 hht1-hhf1::KAN hht2-hhf2::KAN hta1-htb1::Nat hta2-htb2::HPH/pQQ18 [ARS CEN LEU2 HTA1-HTB1 HHT2-HHF2] | This study |
| yMK1329 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1::PDS1-Myc13::TRP1 ura3-1 hht1-hhf1::KAN hht2-hhf2::KAN hta1-htb1::Nat hta2-htb2::HPH/pJH33 [ARS CEN URA3 HTA1-HTB1 HHT2-HHF2] | This study |
| yMK839 | MATaleu2-3 trp1 ura3-52 | 32 |
TABLE 2.
Plasmid constructs used in this study
| Plasmid | Main features | Source or reference |
|---|---|---|
| pJH33 | pRS316-HTA1-HTB1 HHT2-HHF2 | 1 |
| pJL51 | 2μm 3×HA-SGO1 | This study |
| pJL53 | ARS1 CEN4 URA3/pADH1-3×HA-SGO1-tADH1 | This study |
| pJL55 | pGEX-4T-1 3×HA-SGO1 | This study |
| pJL76 | ARS1 CEN4 URA3/pADH1-3×HA-SGO1-BD-tADH1 | This study |
| pMK439G44S | pRS315-HTA1-HTB1 hht2 G44S-HHF2 | This study |
| pMK572 | 2μm URA3 vector with ADH1 promoter and terminator | This study |
| pMK573 | 2μm URA3 SGO1 with ADH1 promoter and terminator | This study |
| pMK621 | pJJ244 URA3-HHF2-KTR5 insert | This study |
| pMK622G44S | pJJ244 hht2 G44S-HHF2-URA3-HHF2-KTR5 | This study |
| pMK622WT | pJJ244 HHT2-HHF2-URA3-HHF2-KTR5 | This study |
| pQQ18 | pRS315-HTA1-HTB1 HHT2-HHF2 | 1 |
| pYCF1/CEN3.L | YRp14/TEL cassette (pYCF1) with a CEN3 insert | 51 |
Most of the strains were derived from the W303 background (JHY200) (1). Initially, phenotypic characterization, including cellular responses to various stresses (e.g., see Fig. 1A) and suppression by 2μm SGO1, was conducted in parallel with both JHY200 and its S288C mutant counterpart JHY205 (1). Both strains exhibited literally identical phenotypes. Subsequent studies were thus focused on the W303 background strains.
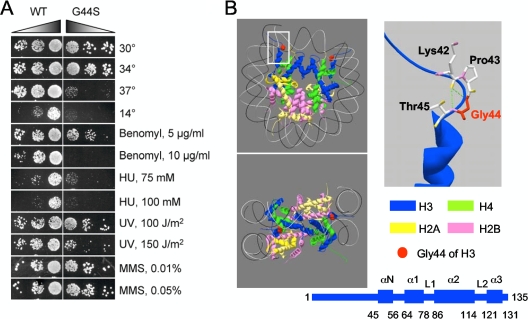
FIG. 1.
The G44S mutation confers pleiotropic phenotypes. (A) Yeast cells bearing the G44S allele as the sole copy of H3 were tested on YPD medium under the indicated conditions. Left panel, sixfold serially diluted log-phase cells were spotted for growth tests. All drug tests were conducted at 30°C. MMS, methyl methanesulfonate. (B) Position of G44 of H3 within a nucleosomal core particle. Left panels: two views of the crystal structure (Protein Data Bank entry 1ID3) based on White et al. (58). Right panels: closeup view of the 42Lys-Pro-Gly-Thr β turn (top) and the secondary structural domains (bottom) of histone H3. Numbers below the secondary structure are amino acid residues at the junctions of the indicated domains. The green dotted lines in the closeup represent possible hydrogen bonds between the carbonyl oxygen of K42 and the amide groups of G44 and Thr45. Hydrogen is not included. DNA is omitted for clarity.
To delete SGO1, primers O361 and O362 were used to PCR amplify the Kluyveromyces lactis TRP1 selective marker from plasmid pBS1479 (47), and the PCR product was transformed into yeast cells for tryptophan prototroph selection.
To tag endogenous Pds1p with 13×Myc, O323 and O324 were used to PCR amplify pFA6a-13Myc-TRP1 for yeast integrative transformation. Integration was verified with PDS1-specific primers O325 and O326 and TRP1-specific primers O375 and O376. Mcd1p-13Myc was created in a scheme identical to that used for Pds1p-13Myc, except for the use of MCD1-specific primers OJL21, OJL22 (for integration), and OJL23 (for verification). To introduce a carboxyl-terminal six-hemagglutinin (6×HA) tag into Sgo1p, yeast strain A10652 (22) was used for a genomic PCR (primers O363 and O364) that amplified the 6×HA-TRP1 fragment flanked by SGO1 sequences. The resultant PCR product was transformed into yMK1243 and yJL145 to knock in the HA tag and the TRP1 marker, creating yJL343 and yJL344, respectively. Contrary to Kitajima et al. (25), who analyzed Myc-tagged SGO1, we did not observe discernible phenotypes associated with carboxyl-terminal HA tagging.
To place the MCD1 gene under the control of the GAL1,10 promoter (i.e., yJL171 and yJL172), the His3MX6-PGAL1 sequence (42) was amplified with primers O388 and O389. The PCR fragment was agarose gel purified and transformed into yMK1329 for histidine prototroph selection. Yeast genomic PCR was conducted using primers O390 and O391 (derived from the MCD1 locus) against O392 or O393 (derived from the His3MX6 sequence) for verification. The correct integration of the GAL promoter into the MCD1 5′ untranslated region was further verified by the inability of cells to grow in the presence of glucose.
To introduce the G44S mutation into a green fluorescent protein (GFP)-marked strain, SBY214, the (hht1-hhf1)Δ::KanMX allele from JHY200 was PCR amplified using primers O396 and O397 and transformed into SBY214 to knock out HHT1-HHF1 (creating yJL118). Correct integration was verified by genomic PCR using primers O398, O399 (from the HHT1/HHF1 locus), and mk133 (from KanMX). To replace the remaining copy of H3 and H4, i.e., HHF2-HHT2, with either wild-type (WT) H3 or the G44S mutant with the URA3 selective marker, plasmid pMK622 (WT or G44S mutant) was cleaved with SnaBI and EcoRI for integrative transformation. Ura+ colonies were isolated for genomic PCR to verify the integration. Genomic PCR using primers O404 and O405 was done to amplify an HHT2 fragment for ScaI digestion (indicative of the G44S mutation) and sequencing to rule out the existence of any unwanted mutations. To create congenic strains that differ only at the HHT2 locus, both the WT and G44S mutant versions of pMK622 were digested for yJL118 transformation, resulting in yJL292 (HHT2-HHT2::URA3-KTR5) and yJL293 (hht2 G44S-HHF2::bpURA3-KTR5).
The yeast chromosomal copy of HHT2 was mutated by integrating pMK622 bearing the desired mutation. To create pMK622, pMK621 was first generated by PCR to amplify from yeast genomic DNA part of HHF2 and KTR5 with primers O400 and O401. The PCR fragment was digested with EcoRI and SphI and cloned into the same sites of pJJ244 (18). pMK622 was made by inserting the SpeI (blunted) and AatII fragment from pQQ18 bearing the G44S mutation into the NarI (blunted) and AatII sites of pMK621, resulting in pMK622 with two 140-bp direct repeats spanning the URA3 gene.
Histone mutations were generated by two-step PCR site-directed mutagenesis. Briefly, the desired mutations were incorporated into two complementary oligonucleotides, and each was used for PCR against O17 (downstream of the HHT2 open reading frame [ORF]) or O19 (downstream of the HHF2 ORF) that hybridized outside the HHT2/HHF2 genes in pQQ18. The two PCR fragments (amplified using pQQ18 as the template by Pfu Turbo DNA polymerase [Stratagene]) were agarose gel purified and subjected to a second round of PCR. The complementary sequence at the 3′ ends of these two PCR fragments allowed annealing and extension. Primers O17 and O19 were included in the reaction mixture to exponentially amplify the entire HHT2/HHF2 gene with the mutation. The final PCR fragments were then digested with SalI and SpeI and used to replace the WT SalI/SpeI sequence in pQQ18. The entire H3 and H4 genes were sequenced for verification. This second-step PCR usually did not work well with Pfu Turbo polymerase. Taq polymerase was used to circumvent this problem. However, Taq polymerase frequently introduced unwanted mutations that had to be revealed by sequencing and, if detected, set aside. The original G44S mutation was obtain fortuitously in this manner.
pMK573, a 2μm URA3 SGO1 plasmid, was created by PCR amplifying the SGO1 ORF from yeast genomic DNA with primers O329 and O330 that included 42 bp of homology to the vector pMK572 (see below) at the 5′ ends. The PCR fragment was cotransformed with HindIII- and EcoRI-digested pMK572 into yeast strain yMK839 (32). Ura+ colonies were subjected to DNA isolation and bacterial transformation. Miniprep DNA was analyzed by restriction digestion and sequencing across the insertion junctions for confirmation of a correct insert.
To create pMK572 (a multicloning sequence flanked by the ADH1 promoter and terminator), ADH-Ras-ΔBamHI (4) was digested with SmaI and self-ligated to remove the Ras sequence. The resultant plasmid, pMK322, then was used as the template for a PCR using primers O327 and O328 to amplify the ADH1 sequence and the multicloning sites. The PCR fragment was cotransformed with HindIII- and EcoRI-digested YEplac195 (a 2μm URA3 plasmid [10]), resulting in pMK572. The multicloning sequence contains unique HindIII, SmaI, SalI, BssHII, MluI, SacI, NotI, EagI, SfiI, BalI, and EcoRI restriction sites.
To create a construct for glutathione S-transferase (GST)-HA-Sgo1p in Escherichia coli, the SGO1 ORF was PCR amplified with primers OJL25 and OJL26. The PCR fragment obtained was digested with NotI and ligated to NotI-linearized pMK595, resulting in in-frame fusion of 3×HA and SGO1 (pJL51). To further generate a GST fusion of HA-SGO1 for bacterial production, 3×HA-SGO1 was isolated from pJL51 and inserted into pSP72 (Promega) at the HindIII and XhoI sites. The BamHI-XhoI fragment was then isolated and ligated to the BamHI and XhoI sites of pGEX4T-1, generating pJL55.
Yeast methods.
Yeast growth media, conditions, and transformation were based on standard procedures (49). When appropriate, a 5% concentration of Casamino Acids (CAA) was used to substitute for synthetic amino acid mixtures as a selective medium for a uracil, tryptophan, or adenine prototroph. Yeast transformation was done by the lithium acetate method (9).
Chromosome stability of the WT and G44S mutant strains was examined according to Spencer et al. (51), using plasmid pYCF1/CEN3.L cut with BglII and transformed into yMK1243 and yJL145. Ura+ transformants were grown in CAA-Ura medium overnight and then plated directly onto yeast extract-peptone-dextrose (YPD) plates to allow colony formation and scoring. A tension-sensing test using the pGAL-MCD1 mutant strains was done according precisely to reference 15, using strains yJL171 and yJL172.
For Western analyses of yeast proteins, yeast extracts were prepared by directly boiling cell pellets in appropriate volumes of 2× sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) loading dye for 5 min, followed by vortexing with 1 lysate volume of glass beads (0.45 μm in diameter; Sigma) at room temperature for 5 min. The mixtures were boiled again for 5 min and then centrifuged at 14,000 × g at room temperature for 1 min. The supernatant was transferred to another tube for SDS-PAGE.
Suppressor screening.
To screen for a multicopy suppressor of the G44S mutation, a YEplac195-based library was constructed by ligating 1 to 3 kb of DpnI-digested yeast genomic DNA (enriched by sucrose gradient ultracentrifugation) to the BamHI site of YEplac195 (10). G44S mutant cells transformed with 0.3 μg of library DNA in each of 20 transformation reactions were plated onto CAA-Ura, and Ura+ colonies (estimated to be about 60,000 in total) were then replica plated onto YPD plates supplemented with 20 μg/ml benomyl and incubated at 30°C. Initial benomyl-resistant colonies were regrown in YPD and streaked onto 5-fluoroorotic acid plates to select for clones that had lost the URA3 plasmid. Benomyl sensitivity tests were repeated to screen for clones displaying plasmid-dependent benomyl resistance. Candidates were subjected to DNA isolation for E. coli transformation. Plasmid DNAs were isolated for restriction digestion and retransformation into the G44S mutant strain to verify the suppression phenotypes, including hypersensitivity to benomyl, heat shock, and cold. The identity of the insert was revealed by DNA sequencing using universal M13 primers.
Chromatin immunoprecipitation (ChIP).
ChIP was conducted as previously described (28). To quantify the ChIP results, the semiquantitative PCR products were purified and resolved by 9% PAGE and stained with ethidium bromide. The captured gel images were then quantified by NIH Image. Intensities of the CEN/pericentric fragments were compared to that of a common internal control (the DED1 or, in some cases, the PGK1 locus). The ratio was further normalized to 0.1% input DNA (set at 1.0) for PCR amplifications carried out in parallel with all of the reactions. The ChIP data were obtained from at least three independent yeast cultures.
Recombinant Sgo1p preparation.
To express and purify GST-HA-Sgo1p from E. coli, 125 ml of BL21 codon+ cells (Stratagene) were subjected to induction (optical density at 600 nm of 0.5 to 0.6 in LB-ampicillin medium) with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 37°C for 4 h. Cells were pelleted (5,000 × g for 5 min) at 4°C and sonicated in 5 ml of HEMGT buffer (25 mM HEPES, pH 7.9, 12.5 mM MgCl2, 150 mM KCl, 0.1 mM EDTA, 0.1% Tween 20, 10% glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride) six times for 20 s each time with 1-min chilling intervals. The soluble fraction was obtained by centrifugation at 10,000 × g for 15 min at 4°C. Sgo1p was purified by incubating the cytosol with 200 μl of reduced glutathione Sepharose 4B beads (Amersham) at 4°C for 1 h. Bound Sgo1p was washed twice with 5 ml of binding buffer for 5 min each time, followed by another wash with 1.5 ml of binding buffer, and transferred to a microcentrifuge tube. The elution was done by gently rocking beads in 200 μl of 50 mM glutathione in the binding buffer for 30 min at 4°C. Eluate was collected, and the elution was repeated once under the same conditions. Two batches of eluate were separately dispensed and stored at −70°C. The yield and purity of GST-HA-Sgo1p were estimated by SDS-PAGE.
Sgo1p interaction with H3, nucleosomal particles, and oligonucleosomes.
Histones were prepared according to reference 6. Recombinant yeast histone H3 and reconstituted nucleosomal particles were gifts from K. Luger. For pulldown assays, approximately 5.5 μg of soluble recombinant Sgo1p was incubated with about 3.4 μg of yeast histones or oligonucleosomes liberated by micrococcal nuclease digestion from a nuclear preparation with an A260 of 200 (WT and G44S mutant) in 150 μl of HEMGT buffer at 4°C for 1 h. Six microliters of glutathione beads and 150 μl of the HEMGT buffer were added to the reaction mixtures, which were rocked gently at 4°C for another hour. Beads were washed with 500 μl of HEMGT buffer three times for 5 min each time, followed by boiling in 2× SDS-PAGE loading buffer for 5 min. Eluate was resolved by 15% SDS-PAGE and blotted for anti-H3 Western analyses. The anti-H3 antibodies were rabbit polyclonal serum made in house and raised against the peptide H2N-CIQLARRIRGERA-COOH. The Western assay used a 1:2,000 dilution of the primary antibody. For far-Western assays, 0.3 μg of yeast histones was first resolved by 15% SDS-PAGE and then blotted onto a polyvinylidene difluoride (PVDF) membrane. Small strips of the membrane were blocked with 10% milk in TTBS (0.9% NaCl, 0.1% [vol/vol] Tween 20, 50 mM Tris, pH 7.4) at room temperature for 30 min, followed by two 10-min TTBS washes. For the Sgo1p-histone interaction, the membrane was incubated with 0.3 μg of GST-HA-SgoI in 3 ml of TTBS buffer supplemented with 0.1% gelatin. The binding was conducted at 4°C with gentle rocking for 3 to 5 h. The membrane was then subjected to regular Western blotting procedures using anti-HA monoclonal antibody 12CA5 (Roche) at a 1:750 dilution.
RESULTS
An H3 mutation causes mitotic chromosome instability.
To identify histone H3 mutations that affect chromatin metabolism in the budding yeast Saccharomyces cerevisiae, we fortuitously encountered a Gly-to-Ser mutation of histone H3 at position 44 (referred to as G44S henceforth) that caused hypersensitivity to benomyl, cold temperature (14°C), heat shock (37°C), and hydroxyurea (HU; Fig. 1A). Mild growth defects were also caused by UV light; however, methyl methanesulfonate did not have an obvious effect. At 26°C, the doubling time of log-phase cultures of the WT and G44S mutant strains are 110.9 ± 7.6 and 129.1 ± 8.8 min/generation, respectively (data not shown). Fluorescence-activated cell sorter analyses of cultures grown at 14°C, 26°C, or 37°C did not reveal obvious defects in ploidy or cell cycle control (data not shown). These results demonstrated that the H3 G44S mutation conferred pleiotropic phenotypes and that cells were defective in dealing with certain stresses. It is worth noting that some of the observed phenotypes, in particular, those related to mitosis (see below), were not specific to the original serine substitution, as an alanine mutation introduced at Gly44 resulted in literally identical phenotypes (data not shown). All of the analyses henceforth were conducted using the G44S allele.
Residue G44 resides at the end of a unique region of H3 (K37-G44) that connects the flexible tail domain (residues 1 to 36) and the well-structured core (residues 45 to about 130) (Fig. 1B) (34). The small region of K37-G44 inserts through the aligned minor grooves of two DNA gyres and then makes a sharp β turn into the nucleosome core. This conserved β turn structure brings the K42 carbonyl oxygen to a topographically favorable position to form a hydrogen (H) bond with the amide group of T45 (Fig. 1B, right) (58). In addition, the amide group of G44 may also H bond with the DNA phosphate backbone (34). By contrast, P43 does not appear to participate in H bond formation with any nearby atoms of DNA or histones, suggesting that G44 plays unique roles in determining the architecture of H3 as it transitions from the extended flexible tail domain into a rigidly structured nucleosomal core. Given the unique position of G44 and the cellular hypersensitivity of the mutant to such genotoxic insults as benomyl, HU, and UV light, we suspected that some of the observed phenotypes were results of aberrations of chromatin metabolism. This work focused on the mitotic functions of G44.
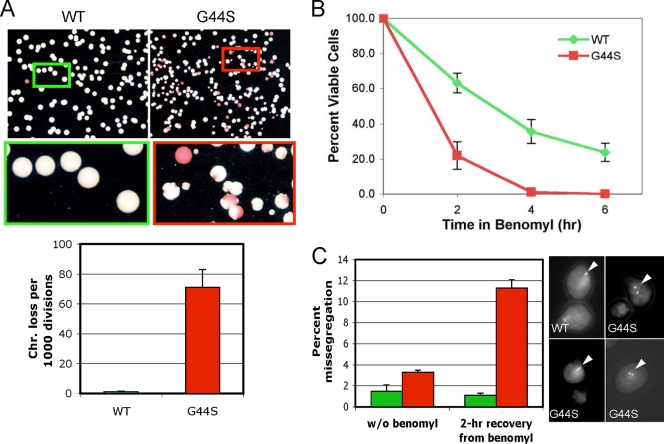
To examine the role of H3 G44 in mitotic chromosome metabolism, we first measured chromosome stability by introducing an artificial, nonessential chromosome into WT and G44S mutant cells bearing the ade2-1 ochre mutant allele (51). ade2− yeast cells accumulate a red pigment and produce red colonies, whereas Ade+ colonies are white. The artificial chromosome used in this study contained a mutant SUP11 tRNA gene that suppresses the ochre mutation. Recipient cells thus formed white colonies. The frequency of first-division chromosome loss of WT and G44S mutant cells was measured (Fig. 2A). Compared with WT cells, G44S mutant cells lost the indicator chromosome at a much greater rate (71 losses versus <1 loss per 1,000 divisions). In addition, red sectoring of G44S mutant cells was commonly accompanied by indentation of the otherwise round, smooth colonies (see closeups). This phenomenon may be due to slower cellular growth rates arising from aneuploidy of native chromosomes and is consistent with the notion that the G44S mutation causes stochastic mitotic errors that retard growth (data not shown).
FIG. 2.
The G44S mutation causes chromosome instability. (A) Mitotic chromosome stability tests. Representative pictures of colonies on YPD plates are shown at the top. The bar graph was prepared from three independent experiments with the standard error of the mean. Colonies with an at least 50% continuous red sector were counted as the first-division chromosome (Chr.) loss. Colonies that were totally red, as a result of chromosome loss prior to cell plating on YPD, were excluded. (B) G44S mutant cells lose viability faster after short benomyl exposure. Log-phase cells were treated with 60 μg/ml benomyl for the indicated time, washed, counted, and spread onto benomyl-free YPD plates. Percent viability was calculated by dividing the total number of colonies by the number of cells inoculated (counted microscopically) and was normalized to that of the T0′ samples. Results are from at least three independent experiments. (C) Higher missegregation is associated with the G44S mutation. WT and G44S mutant cells with the TRP1 locus marked by GFP were treated with 30 μg/ml benomyl for 2 h, collected, and regrown in YPD medium containing α-factor for 2 h before fixation for microscopy. At least 200 unbudded cells with GFP dots were scored in four independent cell cultures of each strain. Green and red bars represent WT and G44S mutant cells, respectively. Error bars show standard deviations. Randomly selected images of two-dotted WT and G44S mutant cells (marked by white triangles) are shown on the right. w/o, without.
We next investigated the molecular defects underlying the benomyl hypersensitivity of G44S mutant cells. Eukaryotes respond to benomyl-triggered microtubule depolymerization by activating the SAC, which stabilizes the securin protein (Pds1p in yeast) and arrests cells at G2/M phase (31). When G44S mutant and WT cells were examined for the budding index and the Pds1p level after benomyl treatment, we observed a similar efficiency of G2/M arrest (Fig. 3A) and upregulation of Pds1p for up to 180 min (Fig. 3B), indicating that dissolving microtubules by benomyl was sufficient to trigger the normal spindle checkpoint responses. However, if yeast cells were plated on drug-free YPD medium to assess viability after 2, 4, or 6 h of benomyl treatment, mutant cells lost viability more quickly (Fig. 2B). These results suggested that a lethal error, most likely missegregation (47), occurred when cells resumed mitosis after benomyl removal. To examine whether the segregation fidelity of G44S mutant cells was impaired after the brief benomyl shock, we used a GFP-marked haploid strain in which a lac operator array was integrated into the TRP1 locus 12 kb from CEN4 (53). In this background, one of the two copies of H3 was deleted and the remaining one was replaced with the G44S allele. This new GFP-marked G44S mutant strain exhibited phenotypes comparable to those seen with the original, non-GFP-marked strain, including sensitivity to benomyl, cold, and heat (data not shown).
FIG. 3.
G44S mutant cells activate the spindle checkpoint in response to benomyl toxicity. (A) Comparable budding indices were obtained from WT and mutant cells. Benomyl (60 μg/ml) was added to log-phase cells. The percentage of large-budded cells (with daughter cells at least half the diameter of their mothers) was determined microscopically. (B) Pds1p was activated and stabilized in both normal and G44S mutant cells in the presence of benomyl. Comparable numbers of α-factor-arrested G1 cells were released at T0′ into YPD medium containing 0 or 30 μg/ml benomyl. The same volume of cell suspension was taken at the indicated times for boiling and whole-cell extract preparation, followed by anti-Myc Western blotting to quantify the abundance of Pds1p.
When GFP-marked yeast cells were allowed to recover from the 2-h benomyl shock, the WT strain exhibited a low rate of missegregation (1.1% ± 0.2% of the G1-phase cells showed two GFP dots), whereas the G44S mutant cells showed 11.3% ± 0.7% cosegregation of both chromosome IV sister chromatids (Fig. 2C). Without benomyl treatment, both strains had a low incidence of missegregation, although there seemed to be a mild elevation in the G44S background (1.5% ± 0.6% versus 3.3% ± 0.2%). These data suggested that the G44S mutant cells were unable to either detect or correct the erroneous kinetochore-spindle attachment that frequently happens during recovery from benomyl toxicity (56). Together, the results in Fig. 2 strongly suggested that the G44S mutation caused chromosome instability by impairing a cellular control mechanism that ensured the bipolar microtubule-kinetochore attachment before committing to anaphase.
G44S mutant cells are defective in tension sensing.
Prior to anaphase onset, sister chromatids are held together by the cohesin complex (27, 39). Cohered chromatids resist the poleward pulling force of bipolar spindles, hence generating tension. The SAC can be activated by attachment errors or by the lack of tension. Given that G44S mutant cells exhibited normal SAC activation upon microtubule depolymerization (Fig. 3) and that attachment errors likely eluded cellular scrutiny (Fig. 2C), we tested whether the SAC was activated in G44S mutant cells under a tensionless condition.
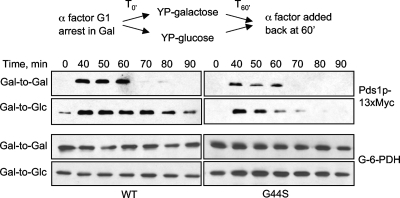
To examine the tension-sensing function, we used a method (15) whereby the MCD1 gene encoding an essential cohesin component was placed under the repressible control of the GAL1 promoter (pGAL). Shifting cells from galactose to glucose medium represses MCD1 expression and hence perturbs the formation of sister cohesion, which consequently prevents tension establishment but does not influence the spindle-kinetochore attachment (2). This tensionless crisis activates the SAC. Using this approach, we compared the SAC activation of the WT and G44S mutant strains. Yeast cells bearing the pGAL-MCD1 gene were grown to log phase in YPgal medium and synchronized at G1 by α-factor. Cells were then released into either YPgal (Gal to Gal) or YPD (Gal to Glc) and harvested at different time points before the cells were again arrested at the next G1 stage by α-factor. Western blotting was conducted to compare the level of Pds1p to see whether the tensionless crisis triggered the activation of SAC in G44S mutant cells (Fig. 4). Gal-to-Gal treatment allowed yeast cells to continue dividing, as evidenced by the degradation of Pds1p when cells finished the first round of mitosis. In contrast, shifting WT cells from galactose to glucose (Gal to Glc) stabilized Pds1p, demonstrating the activation of the SAC. On the other hand, G44S mutant cells degraded Pds1p about 60 min after release from the first G1 block under both Gal-to-Gal and Gal-to-Glc conditions, indicating that the G44S mutation impaired the ability of cells to detect or to respond correctly to the tensionless condition.
FIG. 4.
The G44S mutation impairs the tension-sensing function. MCD1 was placed under the control of the GAL1 promoter for glucose repression. The abundance of Myc-tagged Pds1p was analyzed by Western blotting in the presence (Gal to Gal) or absence (Gal to Glc) of Mcd1p. Experimental schemes are shown above the Western blot assay results. G-6-PDH, glucose-6-phosphate dehydrogenase.
Shugoshin is a high-copy-number suppressor of G44S tension-sensing defects.
The above results revealed that the G44S mutation of H3 caused a defect in the tension-sensing function in mitosis. To further delineate the molecular mechanism underlying this new function of histone H3, we conducted a high-copy-number suppressor screen for genes that could rescue the benomyl hypersensitivity phenotype. From about 60,000 transformants representing ninefold coverage of the S. cerevisiae genome, we obtained 20 independent clones. Sequencing results revealed five nonoverlapping genomic DNA inserts (data not shown). Histone H3 was one of the clones, thus validating the screening. Two multifunctional transcriptional activators, YAP1 and CAD1, were found multiple times in the screen. However, these two proteins have been shown to confer resistance to a variety of stresses, including benomyl (40, 59), and hence were considered unlikely to be relevant to the tension-sensing defects. Another candidate contained the intragenic region between the BIO1 and YGR287C genes without a discernible gene in this fragment. No further work was conducted on this clone. The last and most likely candidate contained the SGO1 gene missing the first 34 amino acids and the 5′ promoter element. Sgo1p plays a key role in mitotic tension sensing (15, 21). The identification of this gene, albeit slightly shorter, as a suppressor agreed well with the observed mitotic phenotypes of the G44S mutant. We posited that a cryptic promoter in the vector activated the expression of a slightly truncated but functional Sgo1p that was responsible for the observed suppression.
To verify that Sgo1p was a bona fide suppressor of G44S, we cloned the SGO1 ORF, with or without the first 34 amino acids, downstream of the constitutive ADH1 promoter in a 2μm plasmid. G44S mutant cells transformed with different SGO1-overexpressing plasmids were tested for responses to benomyl, cold, HU, and UV light. Overexpressing Sgo1p alone rescued hypersensitivities to benomyl and cold but not to HU or UV light (Fig. 5A), indicating that the suppression was specific to mitotic defects caused by this mutation. Deleting the first 34 amino acids did not affect the suppression (data not shown), consistent with the notion that the original 34-amino-acid truncation did not alter the normal function of Sgo1p. Critically, overproducing Sgo1p alleviated the phenotypes of benomyl-induced lethality, chromosome missegregation (Fig. 5B), and the inability to stabilize Pds1p under a tensionless condition (Fig. 5C). The expression of endogenous SGO1 was normal at both transcription and translation levels (Fig. 5D), arguing against the possibility that the G44S mutation downregulated the abundance of Sgo1p. Together, these genetic data strongly suggested that Sgo1p acted downstream of H3 and that the G44S mutation undermined the function of Sgo1p in mitosis.
FIG. 5.
G44S mutant mitotic phenotypes are suppressed by overexpressing Sgo1p. (A) Sgo1p overproduction specifically rescues the mitotic phenotypes of G44S mutant cells. The SGO1 ORF was cloned into a 2μm plasmid bearing the promoter and terminator sequences of ADH1. WT and G44S mutant cells transformed with SGO1 or the corresponding vector were tested under the indicated conditions. (B) Left panel, cell viability test after benomyl treatment. This plot was generated from three independent experiments, and the error bars depict the standard deviations. Right panel, chromosome missegregation assay. Shown are percentages of two-GFP-dotted G1-phase cells. Data were collected from three or four independent cultures, and error bars represent standard deviations. See the legend to Fig. 2B and C for details. (C) Tension-sensing defects conferred by the G44S mutation are alleviated by SGO1 overexpression. G44S mutant cells receiving a 2μm plasmid with or without the ADH1-driven SGO1 gene were tested for the molecular response following MCD1 shutdown. Experiments were done exactly as those shown in Fig. 4. (D) Neither SGO1 transcription nor protein abundance is affected by the G44S mutation. Left, reverse transcription (RT)-PCR shows normal expression of SGO1 in the WT and G44S mutant strains. Right, Western blotting of C′-6×HA-tagged Sgo1p demonstrates the equal abundance of Sgo1p in these two strains. The loading control is a cross-reacting band from both yeast lysates.
G44S mutation attenuates Sgo1p interaction and localization.
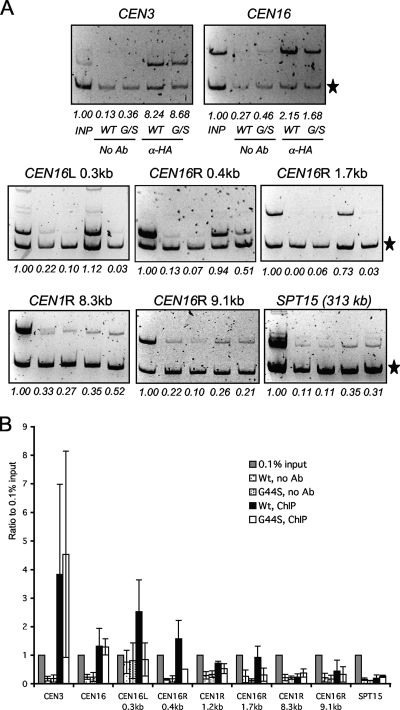
Based on the genetic interaction between SGO1 and H3 revealed by the high-copy-number suppressor screening, we suspected that the G44S mutation might affect a function of Sgo1p. Sgo1p is enriched at centromeres and pericentric regions, where tension is most likely monitored by the hitherto unspecified machinery (22). Mutations that eliminate the Sgo1p recruitment to these loci also cause tension-sensing defects (5). We thus used ChIP to examine whether the recruitment of Sgo1p was impaired by the G44S mutation. To this end, we tagged chromosomal SGO1 with HA at the 3′ end. The HA tag did not alter the benomyl (hyper)sensitivity of either WT or G44S mutant cells (data not shown). Mitotic cells were harvested for ChIP, and immunoprecipitation efficiency was compared by semiquantitative PCR. The ChIP results showed that Sgo1p was present at both centromeres and pericentromeres in WT cells (Fig. 6). However, pericentric Sgo1p was significantly reduced in G44S mutant cells, while the centromeric localization of Sgo1p was normal. The pericentric Sgo1p domain was narrow, for PCR fragments 9.1 kb from CEN16 and 8.3 kb from CEN1 were so weak that it did not show a significant difference between the two strains (Fig. 6B). At the SPT15 locus 313.3 kb from CEN5, there was essentially no Sgo1p detected in either background. The differential effects on centromeric and pericentric recruitment of Sgo1p were consistent with the fact that the canonical H3 is replaced by Cse4p at centromeres (35). Mutating H3 thus had no effect on Sgo1p-centromere association. In conclusion, the ChIP data demonstrated that the G44S mutation specifically diminished the pericentric localization of Sgo1p during mitosis.
FIG. 6.
The G44S mutation selectively downregulates the pericentric recruitment of Sgo1p in vivo. (A) ChIP analysis of HA-tagged Sgo1p. Samples in all of the panels are arranged in the same order. The common internal control (marked by the star on the right) for multiplex PCRs is from within the ORF of the DED1 gene 386.2 kb to the right of CEN15. Targets of the PCR fragments and their distance to the cognate centromeres are listed at the top of each gel image. R, right; L, left. SPT15 is 313.3 kb to the right of CEN5. (B) Quantification of ChIP results. Ethidium bromide-stained DNA gel images were quantified by NIH Image. The intensity of each CEN or pericentric fragment was compared to that of the DED1 internal control (star). The ratio was then normalized to 0.1% input (INP) DNA (set at 1.0). Error bars represent standard deviations from at least three independent cell cultures for ChIP. Ab, antibody.
The genetic and ChIP data shown above predicted that histone H3 and Sgo1p interacted for mitotic tension surveillance and that this association was attenuated by the G44S mutation. To test these predictions biochemically, we expressed GST and HA double-tagged Sgo1p in E. coli and subjected it to in vitro binding assays using core histones or oligonucleosomes purified from WT and G44S mutant yeast cells. In pulldown assays, histone H3 was incubated with Sgo1p and trapped by glutathione beads. Anti-H3 Western blotting then was used to compare the relative abundance of WT and G44S mutant H3 trapped by recombinant Sgo1p. In a parallel far-Western approach, the purified yeast histones were first resolved by SDS-PAGE and then blotted onto a membrane. After incubating the blot with HA-tagged recombinant Sgo1p, anti-HA antibodies were used to probe the trapped Sgo1p by each histone. Figure 7A (pulldown) shows that WT free and nucleosomal H3 bound Sgo1p efficiently, whereas G44S mutant H3 bound to Sgo1p with a diminished affinity. In the far-Western assay (Fig. 7B), HA-Sgo1p also bound more effectively to WT H3. Importantly, the H3-Sgo1p interaction was independent of other histones, and Sgo1p only interacted with H3 (Fig. 7B). Furthermore, chicken H3, recombinant yeast H3, and reconstituted nucleosomal core particles all bound Sgo1p (data not shown). It is noteworthy that the G44S mutation did not completely eliminate Sgo1p binding (compare the 0.6× and 1× histone doses in the left panel of Fig. 7A). The remnant affinity for Sgo1p provided a molecular explanation for the dosage-dependent suppression seen in vivo.
FIG. 7.

Physical interaction between H3 and Sgo1p is attenuated by the G44S mutation. (A) Pulldown assays assessing the interactions between bacterially expressed GST-3×HA-Sgo1p and histones and oligonucleosomes purified from WT or G44S mutant yeast cells. Trapped H3 was quantified by anti-H3 antibodies. (B) Far-Western assays detecting direct binding between H3 and Sgo1p. Yeast core histones were resolved, blotted onto a PVDF membrane, and incubated with GST-3×HA-Sgo1p. Sgo1p, trapped via association with H3, was probed by anti-HA antibodies. A parallel gel was stained with Coomassie blue R250 (CBR) to reveal the relative mobility of yeast histones.
Together, these results demonstrated that the molecular reason for the G44S mitotic defects was an attenuated interaction between histone H3 and Sgo1p, resulting in the loss of Sgo1p from pericentromeres. While the centromeric enrichment of Sgo1p was not affected by the H3 mutation, the downregulation of pericentric Sgo1p enrichment was likely sufficient to cause the tension-sensing phenotypes associated with the G44S mutation.
Reinstating pericentric Sgo1p partially rescues G44S mitotic defects.
If the only purpose of the observed H3-Sgo1p interaction was to bring the latter to pericentromeres for tension surveillance, artificially tethering Sgo1p back to the pericentric loci should effectively eliminate the mitotic phenotypes of G44S mutant cells. On the other hand, if the interaction between H3 and Sgo1p was also important for mitotic control, forcing Sgo1p to pericentromeres by an approach other than the natural H3-Sgo1p association may, at best, only partially suppress the mitotic defects caused by the G44S mutation. The challenges of artificially tethering a protein to the pericentric regions of the budding yeast are that the pericentromeres do not form heterochromatin like other eukaryotic systems and that there is no known epigenetic mark specific to this region. Nonetheless, the ChIP data in Fig. 7A suggested that the establishment of an Sgo1p domain was likely nucleated from the H3-independent recruitment at the centromere, where the canonical histone H3 was absent. The physical association of H3 and Sgo1p then allowed the latter to spread outward to generate a pericentric domain (see Discussion for details). If this is so, fusing Sgo1p to a general chromatin-binding motif may reestablish the pericentric retention of Sgo1p and would allow us to assess the effect of pericentric recruitment of Sgo1p in the G44S mutant background.
To regenerate the pericentric enrichment of Sgo1p in the G44S mutant background, we fused a bromodomain (BD) from Bdf1p to the carboxyl end of Sgo1p and a trimeric HA tag to the amino terminus. Bdf1p binds both histones H3 and H4 (41), and the BD in different proteins has been shown to interact with acetyllysine (5, 17). Genome-wide ChIP analyses detected comparable histone acetylation between pericentromeres and the bulk of the yeast genome (29), suggesting that a general (acetylated) histone-binding module such as the BD may complement the weakened affinity of Sgo1p for G44S mutant H3. As shown in Fig. 8A, this indeed was the case. When Sgo1p and Sgo1p fused to the BD (Sgo1p-BD) were introduced into sgo1Δ mutant cells expressing the WT or G44S mutant allele of H3, Sgo1p-BD exhibited efficient pericentric enrichment (Fig. 8A and C), whereas Sgo1p without the BD remained underrepresented at pericentromeres. These two alleles of Sgo1p were expressed at comparable levels (Fig. 8B), indicating that the BD fusion increased the affinity for a histone at the pericentromeres. Importantly, the reinstatement of pericentric Sgo1p was accompanied by partial suppression of benomyl hypersensitivity, loss of viability following benomyl shock, and stabilization of Pds1p during MCD1 shutdown (Fig. 9A to C). Together, these results strongly suggested that dislodging Sgo1p from pericentromeres was the major underlying cause of the G44S mutant mitotic defects.
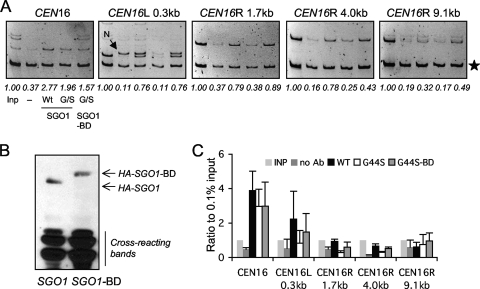
FIG. 8.
Reestablishing pericentric Sgo1p recruitment by BD fusion. (A) ChIP assays assessing the distribution of Sgo1p and Sgo1p-BD at selective loci. G44S sgo1Δ mutant cells were transformed with an ARS CEN plasmid containing 3×HA-Sgo1p or 3×HA-Sgo1p-BD genes and analyzed by anti-HA ChIP. The assay conditions were identical to those described in the legend to Fig. 6. A nonspecific PCR fragment (N) amplified along with CEN16L 0.3 kb is marked with an arrow. (B) Comparable expression of 3×HA-Sgo1p and 3×HA-Sgo1p-BD. G44S sgo1Δ mutant cells bearing the indicated recombinant SGO1 construct were examined by anti-HA Western blotting. Equal loading was evidenced by Coomassie blue R250 staining (not shown) and by proteins that cross-reacted with the anti-HA antibodies. (C) Quantification of the ChIP results was done as detailed in the legend to Fig. 6B.
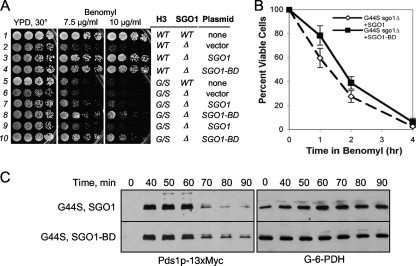
FIG. 9.
BD fusion partially rescues the tension-sensing defects of G44S mutant cells. (A) The indicated yeast strains expressing Sgo1p with or without the BD were tested for resistance to benomyl (A), for viability after benomyl exposure (B), and for the Pds1p level after MCD1 shutdown. See the legends to Fig. 2 and 4 for details. In panels B and C, only data from G44S mutant cells are shown.
In summary, histone H3 plays a critical role in Sgo1p recruitment to the mitotic pericentromeres, where the tension between sister chromatids is likely monitored. Genetic data strongly suggest an intimate association between H3 and Sgo1p, a notion supported by ChIP and biochemical assays. We thus conclude that the mitotic tension-sensing function depends critically on the appropriate association between histone H3 and Sgo1p in the pericentric regions.
DISCUSSION
Establishment of a pericentric Sgo1p domain.
This work uncovered a novel function of histone H3 in mitotic checkpoint control. By recruiting Sgo1p specifically to the pericentric region, histone H3 was shown for the first time to play a role key in segregation fidelity. As the G44S mutation selectively compromises the pericentric enrichment of Sgo1p but apparently leaves centromeric Sgo1p unaltered (Fig. 6), we suspect that Sgo1p localization is nucleated by an H3-independent mechanism at the centromere/kinetochore, followed by spreading toward the pericentric regions via direct association with histone H3. This scenario is analogous to the mechanism of establishing the telomeric heterochromatin in yeast (12), in which Rap1p nucleates the heterochromatin formation by binding to the telomeric C1-3A repeats. Silent information regulator, or Sir, proteins are recruited by Rap1p. Via the direct association between Sir3p/Sir4p and the amino-terminal tails of histones H3 and H4 (6), Sir proteins spread from the nucleation site to the neighboring region (45). Inserting the telomeric C1-3A repeats into an ectopic location is sufficient to create a new transcription silent domain, conclusively ruling out the need for a specific cis-acting element for Sir protein spread or retention (52). Similarly, relocating the centromere to an ectopic site establishes a new Sgo1p domain (22).
The notion that the H3-Sgo1p association is critical for mitotic regulation is also supported by the observation that the G44S allele caused mild benomyl sensitivity in the presence of the WT counterpart (data not shown). It is conceivable that the concomitant incorporation of WT and G44S mutant H3 into nucleosomes in the pericentric region results in intermediate affinity for Sgo1p, thus resulting in visible but not as severe defects in coping with benomyl toxicity.
The protein directly responsible for bringing Sgo1p to the centromere remains unidentified. Among the 30-plus kinetochore proteins and the SAC components, one of the likely Sgo1p recruiters is Bub1p. Although deleting BUB1 causes loss of Sgo1p in centromeres and pericentromeres (7, 22), the physical evidence for Bub1p-Sgo1p association is lacking to date. Moreover, Bub1p is a kinase that is essential for SAC assembly at the kinetochore (33). It is thus also possible that Bub1p regulates the protein that physically brings Sgo1p to the centromere.
Given the diverse posttranslational modifications of histones, it is tempting to speculate about an alternative model in which a novel epigenetic mark at the pericentromeres facilitates Sgo1p retention. For example, Gcn5p and Hda1p have been linked genetically to mitotic segregation (19, 57), and Ser31 phosphorylation of the human H3.3 variant is enriched at the pericentric heterochromatin (13). However, we do not believe this notion is very likely because Sgo1p interacts with bulk H3 from yeast and chicken, as well as recombinant H3 synthesized in E. coli (data not shown), arguing against an H3 modification essential for Sgo1p binding. Nonetheless, we do not rule out the possibility of an auxiliary function of an elusive, pericentromere-specific modification. Nor do we disregard a notion that a chromosome arm-specific epigenetic mark prevents the H3-Sgo1p interaction outside the pericentric region. Due to the absence of functionally and structurally distinct pericentromeres in the budding yeast, the identification of this epigenetic mark presents a major technical challenge.
Tension sensing-specific relationship between H3 and Sgo1p.
While the G44S mutation causes pleiotropic phenotypes (Fig. 1A), the genetic and physical interactions between H3 and Sgo1p are specific for tension sensing based on multiple lines of evidence. First, overexpressing Sgo1p does not have an effect on a histone H4 allele, hhf1-20, that impairs centromere/kinetochore functions (50; data not shown). The hta1-300 allele of histone H2A, which causes defects in ploidy control, is also insensitive to SGO1 overexpression (I. Pinto, personal communication). Second, we have yet another H4 mutant allele, R35S, that also causes benomyl hypersensitivity (J. Luo, X. Xu, and M.-H. Kuo, unpublished data). 2μm SGO1 constructs fail to suppress this mutant. In contrast, other candidates fished out of the original suppressor screens, such as CAD1 and YAP1, which have been linked to multidrug resistance, rescued the benomyl hypersensitivity associated with either the H3 G44S or the H4 R35S mutation (data not shown). Third, although G44S mutant cells are sensitive to UV light and HU, neither defect is responsive to Sgo1p overproduction (Fig. 5A), indicating that, whereas the G44S mutation may also impair the control of DNA metabolism or damage repair, this is an SGO1-independent function. Indeed, deleting SGO1 does not render cells sensitive to UV light or HU (data not shown). Lastly, the G44S mutation does not affect the pericentric recruitment of the cohesin component Mcd1p (data not shown), indicating that pericentric H3 is a specific interaction target for Sgo1p, whereas Mcd1p (i.e., the cohesin complex) is recruited to the pericentric region via a different pathway (8). The last notion is consistent with the fact that cohesin also forms clusters in the chromosome arms whereas Sgo1p is absent therein (19).
One might argue that the G44 mutation causes a general transcriptional defect that indirectly impairs Sgo1p recruitment or acts in a different pathway that leads to the tension-sensing defects observed in this work. We do not think this notion likely because when we examined the transcription of a variety of genes representing functions in mitosis, checkpoint control, nutrient responses, mating, and transcriptional silencing, we did not detect any discernible differences between WT and G44S mutant cells (data not shown). The only transcription defect that we found associated with G44 mutations is the deregulation of transcription driven by cryptic promoter elements within certain ORFs (20; E. M. Hyland, and J. D. Boeke, submitted for publication). However, this phenotype was not affected by SGO1 overexpression (data not shown), further strengthening the mitosis-specific relationship between H3 and Sgo1p.
While the present work focused on the G44 allele of H3, which was obtained fortuitously in an attempt to create other mutants with mitotic phenotypes, our preliminary data indicated that residues surrounding Gly44 of H3 perform similar mitotic functions and that targeted mutations thereof lead to tension-sensing defects similar to those of the G44S mutant (J. Luo and M.-H. Kuo, unpublished data). We suspect that G44 is part of a “tension-sensing motif” that makes physical contact with Sgo1p. Biochemical and molecular studies are in progress to test this hypothesis.
Other factors related to Sgo1p and tension sensing.
Sgo1p belongs to the Shugoshin family proteins found in many eukaryotes. Several conserved proteins, including Bub1p, PP2A, and microtubules, are linked to Shugoshin functions. For example, Bub1 homologues are critical for centromeric and pericentric localization of Shugoshin in different organisms (7, 22, 24, 55). Human and yeast Shugoshin proteins collaborate with a specific form of protein phosphatase 2A (PP2A) to protect meiotic cohesin (26, 46, 54). The Sgo1p-PP2A cooperation appears to be independent of the Sgo1p-H3 interaction, for deleting CDC55 or RTS1, the key B and B′ regulatory subunits of PP2A complexes, does not affect the ability of 2μm SGO1 to suppress the benomyl hypersensitivity phenotype of the G44S mutant (J. Luo and M.-H. Kuo, unpublished data). The human and Xenopus Shugoshin proteins bind and stabilize mitotic kinetochore microtubules (48). It is unclear whether budding yeast Sgo1p does so. However, this microtubule stabilization function is clearly a postbiorientation activity preceded by the tension surveillance function. Therefore, we believe that histone H3 is the first protein shown to act directly upstream of Sgo1p in mitotic tension sensing.
A key player in correcting tension defects is the Aurora B kinase encoded by IPL1 in budding yeast. Ipl1p destabilizes spindle-kinetochore attachment before anaphase nucleation, hence permitting correction of attachment errors (2). We observed synthetic phenotypes when Ipl1p or one of its partners, Sli15p, was overexpressed in an sgo1Δ or H3 G44S mutant background (H. Hall and T. Hazbun, data not shown). These genetic interactions suggest that increasing the detachment activity of Ipl1p/Sli15p brings about a molecular defect (i.e., spindle detachment) similar to that caused by benomyl treatment. Lack of functional Sgo1p or intact H3 causes growth defects.
It is critical that the BD-Sgo1p fusion protein can be recruited to the pericentromeres. However, close examination of the efficacy of suppression revealed that Sgo1p-BD only partially corrected the mitotic defects (Fig. 8). The enrichment of recombinant Sgo1p is consistent with the “nucleation-and-spread model” depicted above for establishing the Sgo1p domain. However, the inability of the Sgo1p-BD chimeric protein to completely restore benomyl tolerance suggests two possibilities. First, the BD may somehow interfere with the tension-sensing function of Sgo1p but not with the ability to interact with the nucleating protein for recruitment. Alternatively, the physical interaction between Sgo1p and H3 at G44 might be critical for the tension-sensing function. For example, if Sgo1p docks directly to G44 and nearby residues, the BD may place Sgo1p toward the far end of the amino-terminal tail domain of H3 or even at a different spot on the nucleosomal particle where the H4 tail resides. If this is so, the activity of Sgo1p in tension sensing may be attenuated even though it has been directed back to the pericentromeres. Structural and biochemical experiments are required to examine these questions.
Acknowledgments
We are grateful for C. David Allis, Angelika Amon, Sue Biggins, Jennifer Gerton, Phil Hieter, Karolin Luger, Mitchell Smith, and Fred Winston for generous supply of materials; Sue Biggins for advice; and Inés Pinto for sharing unpublished results. We thank Sue Biggins, Sharon Dent, and John Wang for critical reading of the manuscript; Yang Liu for creating the original G44S allele; and David Almy, No-Ya Hung, and Andy Lin for technical assistance.
This work was partly supported by a grant (CMB 0315542) from the National Science Foundation to M.H.-K.
J.L. contributed to all of the experimental data, except the following. X.X. conducted suppressor screening and initial characterization of 2μm SGO1. Genetic interactions among IPL1, H3, and SGO1 were contributed by H.H. and T.H. Work related to cryptic promoter regulation was contributed by E.M.D. and J.D.B. M.-H.K. coordinated the project.
There is no conflict of interest for this work.
Footnotes
Published ahead of print on 16 November 2009.
REFERENCES
- 1.Ahn, S. H., W. L. Cheung, J. Y. Hsu, R. L. Diaz, M. M. Smith, and C. D. Allis. 2005. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell 120:25-36. [DOI] [PubMed] [Google Scholar]
- 2.Biggins, S., and A. W. Murray. 2001. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 15:3118-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloom, K., S. Sharma, and N. V. Dokholyan. 2006. The path of DNA in the kinetochore. Curr. Biol. 16:R276-R278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broder, Y. C., S. Katz, and A. Aronheim. 1998. The ras recruitment system, a novel approach to the study of protein-protein interactions. Curr. Biol. 8:1121-1124. [DOI] [PubMed] [Google Scholar]
- 5.Dhalluin, C., J. E. Carlson, L. Zeng, C. He, A. K. Aggarwal, and M. M. Zhou. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491-496. [DOI] [PubMed] [Google Scholar]
- 6.Edmondson, D. G., M. M. Smith, and S. Y. Roth. 1996. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 10:1247-1259. [DOI] [PubMed] [Google Scholar]
- 7.Fernius, J., and K. G. Hardwick. 2007. Bub1 kinase targets SgoI to ensure efficient chromosome biorientation in budding yeast mitosis. PLoS Genet. 3:2312-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gartenberg, M. 2009. Heterochromatin and the cohesion of sister chromatids. Chromosome Res. 17:229-238. [DOI] [PubMed] [Google Scholar]
- 9.Gietz, D., A. St Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 11.Goshima, G., and M. Yanagida. 2000. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell 100:619-633. [DOI] [PubMed] [Google Scholar]
- 12.Grunstein, M. 1997. Molecular model for telomeric heterochromatin in yeast. Curr. Opin. Cell Biol. 9:383-387. [DOI] [PubMed] [Google Scholar]
- 13.Hake, S. B., B. A. Garcia, M. Kauer, S. P. Baker, J. Shabanowitz, D. F. Hunt, and C. D. Allis. 2005. Serine 31 phosphorylation of histone variant H3.3 is specific to regions bordering centromeres in metaphase chromosomes. Proc. Natl. Acad. Sci. U. S. A. 102:6344-6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He, X., S. Asthana, and P. K. Sorger. 2000. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell 101:763-775. [DOI] [PubMed] [Google Scholar]
- 15.Indjeian, V. B., B. M. Stern, and A. W. Murray. 2005. The centromeric protein SgoI is required to sense lack of tension on mitotic chromosomes. Science 307:130-133. [DOI] [PubMed] [Google Scholar]
- 16.Iwaizumi, M., K. Shinmura, H. Mori, H. Yamada, M. Suzuki, Y. Kitayama, H. Igarashi, T. Nakamura, H. Suzuki, Y. Watanabe, A. Hishida, M. Ikuma, and H. Sugimura. 2009. Human SgoI downregulation leads to chromosomal instability in colorectal cancer. Gut 58:249-260. [DOI] [PubMed] [Google Scholar]
- 17.Jacobson, R. H., A. G. Ladurner, D. S. King, and R. Tjian. 2000. Structure and function of a human TAFII250 double bromodomain module. Science 288:1422-1425. [DOI] [PubMed] [Google Scholar]
- 18.Jones, J. S., and L. Prakash. 1990. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast 6:363-366. [DOI] [PubMed] [Google Scholar]
- 19.Kanta, H., L. Laprade, A. Almutairi, and I. Pinto. 2006. Suppressor analysis of a histone defect identifies a new function for the hda1 complex in chromosome segregation. Genetics 173:435-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaplan, C. D., L. Laprade, and F. Winston. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301:1096-1099. [DOI] [PubMed] [Google Scholar]
- 21.Kawashima, S. A., T. Tsukahara, M. Langegger, S. Hauf, T. S. Kitajima, and Y. Watanabe. 2007. Shugoshin enables tension-generating attachment of kinetochores by loading Aurora to centromeres. Genes Dev. 21:420-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiburz, B. M., D. B. Reynolds, P. C. Megee, A. L. Marston, B. H. Lee, T. I. Lee, S. S. Levine, R. A. Young, and A. Amon. 2005. The core centromere and SgoI establish a 50-kb cohesin-protected domain around centromeres during meiosis I. Genes Dev. 19:3017-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King, R. W., J. M. Peters, S. Tugendreich, M. Rolfe, P. Hieter, and M. W. Kirschner. 1995. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell 81:279-288. [DOI] [PubMed] [Google Scholar]
- 24.Kitajima, T. S., S. Hauf, M. Ohsugi, T. Yamamoto, and Y. Watanabe. 2005. Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting Shugoshin localization. Curr. Biol. 15:353-359. [DOI] [PubMed] [Google Scholar]
- 25.Kitajima, T. S., S. A. Kawashima, and Y. Watanabe. 2004. The conserved kinetochore protein Shugoshin protects centromeric cohesion during meiosis. Nature 427:510-517. [DOI] [PubMed] [Google Scholar]
- 26.Kitajima, T. S., T. Sakuno, K. Ishiguro, S. Iemura, T. Natsume, S. A. Kawashima, and Y. Watanabe. 2006. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature 441:46-52. [DOI] [PubMed] [Google Scholar]
- 27.Koshland, D. E., and V. Guacci. 2000. Sister chromatid cohesion: the beginning of a long and beautiful relationship. Curr. Opin. Cell Biol. 12:297-301. [DOI] [PubMed] [Google Scholar]
- 28.Kuo, M. H., and C. D. Allis. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19:425-433. [DOI] [PubMed] [Google Scholar]
- 29.Kurdistani, S. K., S. Tavazoie, and M. Grunstein. 2004. Mapping global histone acetylation patterns to gene expression. Cell 117:721-733. [DOI] [PubMed] [Google Scholar]
- 30.Latham, J. A., and S. Y. Dent. 2007. Cross-regulation of histone modifications. Nat. Struct. Mol. Biol. 14:1017-1024. [DOI] [PubMed] [Google Scholar]
- 31.Lew, D. J., and D. J. Burke. 2003. The spindle assembly and spindle position checkpoints. Annu. Rev. Genet. 37:251-282. [DOI] [PubMed] [Google Scholar]
- 32.Liu, Y., X. Xu, S. Singh-Rodriguez, Y. Zhao, and M.-H. Kuo. 2005. Histone H3 Ser10 phosphorylation-independent function of Snf1 and Reg1 proteins rescues a gcn5− mutant in HIS3 expression. Mol. Cell. Biol. 25:10566-10579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Logarinho, E., and H. Bousbaa. 2008. Kinetochore-microtubule interactions “in check” by Bub1, Bub3 and BubR1: the dual task of attaching and signalling. Cell Cycle 7:1763-1768. [DOI] [PubMed] [Google Scholar]
- 34.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 35.Meluh, P. B., P. Yang, L. Glowczewski, D. Koshland, and M. M. Smith. 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94:607-613. [DOI] [PubMed] [Google Scholar]
- 36.Morgan, B. A., B. A. Mittman, and M. M. Smith. 1991. The highly conserved N-terminal domains of histones H3 and H4 are required for normal cell cycle progression. Mol. Cell. Biol. 11:4111-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nasmyth, K. 2005. How do so few control so many? Cell 120:739-746. [DOI] [PubMed] [Google Scholar]
- 38.Nasmyth, K. 2002. Segregating sister genomes: the molecular biology of chromosome separation. Science 297:559-565. [DOI] [PubMed] [Google Scholar]
- 39.Nasmyth, K., J. M. Peters, and F. Uhlmann. 2000. Splitting the chromosome: cutting the ties that bind sister chromatids. Science 288:1379-1385. [DOI] [PubMed] [Google Scholar]
- 40.Nguyên, D. T., A. M. Alarco, and M. Raymond. 2001. Multiple Yap1p-binding sites mediate induction of the yeast major facilitator FLR1 gene in response to drugs, oxidants, and alkylating agents. J. Biol. Chem. 276:1138-1145. [DOI] [PubMed] [Google Scholar]
- 41.Pamblanco, M., A. Poveda, R. Sendra, S. Rodriguez-Navarro, J. E. Perez-Ortin, and V. Tordera. 2001. Bromodomain factor 1 (Bdf1) protein interacts with histones. FEBS Lett. 496:31-35. [DOI] [PubMed] [Google Scholar]
- 42.Petracek, M. E., and M. S. Longtine. 2002. PCR-based engineering of yeast genome. Methods Enzymol. 350:445-469. [DOI] [PubMed] [Google Scholar]
- 43.Pinsky, B. A., and S. Biggins. 2005. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 15:486-493. [DOI] [PubMed] [Google Scholar]
- 44.Pinto, I., and F. Winston. 2000. Histone H2A is required for normal centromere function in Saccharomyces cerevisiae. EMBO J. 19:1598-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Renauld, H., O. M. Aparicio, P. D. Zierath, B. L. Billington, S. K. Chhablani, and D. E. Gottschling. 1993. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 7:1133-1145. [DOI] [PubMed] [Google Scholar]
- 46.Riedel, C. G., V. L. Katis, Y. Katou, S. Mori, T. Itoh, W. Helmhart, M. Galova, M. Petronczki, J. Gregan, B. Cetin, I. Mudrak, E. Ogris, K. Mechtler, L. Pelletier, F. Buchholz, K. Shirahige, and K. Nasmyth. 2006. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature 441:53-61. [DOI] [PubMed] [Google Scholar]
- 47.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 48.Salic, A., J. C. Waters, and T. J. Mitchison. 2004. Vertebrate Shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell 118:567-578. [DOI] [PubMed] [Google Scholar]
- 49.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 50.Smith, M. M., P. Yang, M. S. Santisteban, P. W. Boone, A. T. Goldstein, and P. C. Megee. 1996. A novel histone H4 mutant defective in nuclear division and mitotic chromosome transmission. Mol. Cell. Biol. 16:1017-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spencer, F., S. L. Gerring, C. Connelly, and P. Hieter. 1990. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics 124:237-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stavenhagen, J. B., and V. A. Zakian. 1994. Internal tracts of telomeric DNA act as silencers in Saccharomyces cerevisiae. Genes Dev. 8:1411-1422. [DOI] [PubMed] [Google Scholar]
- 53.Straight, A. F., A. S. Belmont, C. C. Robinett, and A. W. Murray. 1996. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6:1599-1608. [DOI] [PubMed] [Google Scholar]
- 54.Tang, Z., H. Shu, W. Qi, N. A. Mahmood, M. C. Mumby, and H. Yu. 2006. PP2A is required for centromeric localization of SgoI and proper chromosome segregation. Dev. Cell 10:575-585. [DOI] [PubMed] [Google Scholar]
- 55.Tang, Z., Y. Sun, S. E. Harley, H. Zou, and H. Yu. 2004. Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis. Proc. Natl. Acad. Sci. U. S. A. 101:18012-18017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trautmann, S., S. Rajagopalan, and D. McCollum. 2004. The S. pombe Cdc14-like phosphatase Clp1p regulates chromosome biorientation and interacts with Aurora kinase. Dev. Cell 7:755-762. [DOI] [PubMed] [Google Scholar]
- 57.Vernarecci, S., P. Ornaghi, A. Bagu, E. Cundari, P. Ballario, and P. Filetici. 2008. Gcn5p plays an important role in centromere kinetochore function in budding yeast. Mol. Cell. Biol. 28:988-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.White, C. L., R. K. Suto, and K. Luger. 2001. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 20:5207-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu, A., J. A. Wemmie, N. P. Edgington, M. Goebl, J. L. Guevara, and W. S. Moye-Rowley. 1993. Yeast bZip proteins mediate pleiotropic drug and metal resistance. J. Biol. Chem. 268:18850-18858. [PubMed] [Google Scholar]
- 60.Yu, H. 2002. Regulation of APC-Cdc20 by the spindle checkpoint. Curr. Opin. Cell Biol. 14:706-714. [DOI] [PubMed] [Google Scholar]
- 61.Zhou, J., J. Yao, and H. C. Joshi. 2002. Attachment and tension in the spindle assembly checkpoint. J. Cell Sci. 115:3547-3555. [DOI] [PubMed] [Google Scholar]