Abstract
Functional coordination between DNA replication helicases and DNA polymerases at replication forks, achieved through physical linkages, has been demonstrated in prokaryotes but not in eukaryotes. In Saccharomyces cerevisiae, we showed that mutations that compromise the activity of the MCM helicase enhance the physical stability of DNA polymerase α in the absence of their presumed linker, Mcm10. Mcm10 is an essential DNA replication protein implicated in the stable assembly of the replisome by virtue of its interaction with the MCM2-7 helicase and Polα. Dominant mcm2 suppressors of mcm10 mutants restore viability by restoring the stability of Polα without restoring the stability of Mcm10, in a Mec1-dependent manner. In this process, the single-stranded DNA accumulation observed in the mcm10 mutant is suppressed. The activities of key checkpoint regulators known to be important for replication fork stabilization contribute to the efficiency of suppression. These results suggest that Mcm10 plays two important roles as a linker of the MCM helicase and Polα at the elongating replication fork—first, to coordinate the activities of these two molecular motors, and second, to ensure their physical stability and the integrity of the replication fork.
The key players of the replication machinery are the DNA polymerases that synthesize the leading and lagging daughter strands and the replicative helicase that unwinds the parental strands ahead of the polymerases. Coordination between the helicase and the polymerases is critical during replication. Uncoupling of these two molecular machines, especially during lagging strand synthesis, may result in an unrestrained helicase and the exposure of extensive single-stranded DNA (ssDNA), as observed in checkpoint mutants treated with hydroxyurea (HU) (37). Although there is no direct evidence, the implication is that the replicative helicase would be moving at a faster pace than would the DNA polymerase if synchrony were destroyed. In Escherichia coli, the replicative helicase (DnaB) and the primase (DnaG) are coupled by direct contact to form a tight complex (3). In T7, processivity of the gp5 polymerase in lagging strand synthesis requires coupling to the gp4 helicase (16). Recent studies of the budding yeast Saccharomyces cerevisiae suggest that Mrc1 may couple DNA polymerase ɛ and the MCM helicase on the leading strand as well as activate the checkpoint response under replication stress (1, 22, 28). A candidate for coupling DNA polymerase α primase and the MCM helicase on the lagging strand is Mcm10, because Mcm10 interacts with subunits of the Mcm2-7 helicase (26, 29) as well as Polα (14, 33) and the stability of Polα requires Mcm10 in both budding yeast and human cells (8, 33). Mcm10 is an essential protein known to be involved in various aspects of the replication process. It is required during both initiation and elongation steps of DNA replication and interacts with a wide range of replication factors, such as ORC (17, 23, 29), MCM helicase, DNA polymerases ɛ and δ (23), Cdc45 (34), and Polα (33). Therefore, Mcm10 is important for the overall stability of the elongation complex, but its essential function remains unknown.
Accumulating evidence suggests that the major function of many checkpoint proteins is the stabilization of the replication machinery at the fork (9, 22, 39), in addition to regulation of the temporal and spatial firing of origins and prevention of premature mitosis (31, 35, 39). The main signal that leads to checkpoint activation is believed to be the exposure of RPA-coated ssDNA (42). In Xenopus, ssDNA exposure has been shown to be mediated by a functional uncoupling between the polymerase and the helicase (7), and it has been shown that the level of checkpoint activation depended on the extent of ssDNA accumulation. This observation suggests that uncoupling of the polymerase and the helicase activity would result in ssDNA accumulation that in turn would activate the checkpoint pathway to stabilize the fork.
In our study, we carried out a random and a gene-targeted mutagenesis screen to identify mutations that suppress the conditional lethality of mcm10 caused by the lability of Mcm10 in budding yeast (27). We found suppressor mutations in MCM2, which encodes one of the six distinct subunits of the MCM helicase. These mcm2 mutations correct the fork defects of mcm10, particularly that which leads to Polα instability. The altered helicase activity and activation of the checkpoint pathway of the mcm2 mutants appeared to be required for viability of mcm10 mcm2. We showed that uncoupling the MCM helicase and DNA polymerase α by destabilizing Mcm10 leads to accumulation of ssDNA, which is suppressed by reducing the MCM helicase activity. Our findings suggest that the physical coupling of Polα and the helicase by Mcm10 may be replaced by an alternative stabilization mechanism that involves slowing down the helicase and activating the checkpoint proteins.
MATERIALS AND METHODS
Strains and plasmids.
Strains used in this study are listed in Table 1. All strains were isogenic derivatives of W303-1A, unless otherwise indicated. Strains carrying various deletions were made by crossing the mcm10-1 mcm2 strain with the appropriate deletion strain and selecting desired segregants by their conditional phenotypes and/or auxotrophy and by sequencing. Genotypes were confirmed by PCR, sequencing, or by plasmid complementation, where applicable. Plasmids used in this study are listed in Table 2. Plasmids used for yeast two-hybrid analysis were constructed by the Gateway system (Invitrogen, San Diego, CA). The Gateway recombination cassette was inserted into the BamHI site of pGAD2F and pBTM116 plasmids (13) for conversion into destination vectors.
TABLE 1.
Strains used in this study
| Strain isogenic to W303 | Description | Source |
|---|---|---|
| W303-1A | MATaade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-1 | R. Rothstein |
| W303-1B | MATα ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-1 | R. Rothstein |
| BTY100 | W303 MATamcm10-1 | This lab |
| BTY101 | W303 MATα mcm10-1 | This lab |
| BTY103 | W303 MATamcm10-43 | This lab |
| BTY102 | W303 MATα mcm10-43 | This lab |
| ILY230 | MATa13myc-MCM10 TRP1 | This lab |
| ILY232 | MATa13myc-mcm10-43 TRP1 | This lab |
| SSY84 | MATa13myc-mcm10-1 HIS3MX | This lab |
| CLY88 | MATa13myc-mcm10-43 TRP1 mcm2-G400D | This study |
| CLY90 | MATa13myc-mcm10-1 HIS3MX mcm2-G400D | This study |
| CLY91 | W303 MATamcm2-P399L | This lab |
| CLY92 | W303 MATamcm2-G400D | This lab |
| CLY93 | W303 MATα mcm2-D472G | This lab |
| CLY94 | W303 MATamcm2-R617H | This lab |
| ILY215 | W303 MATamcm2-S619F | This lab |
| CLY95 | W303 MATamcm2-P399L mcm10-1 | This study |
| CLY96 | W303 MATα mcm2-G400D mcm10-1 | This study |
| CLY97 | W303 MATα mcm2-D472G mcm10-1 | This study |
| CLY98 | W303 MATα mcm2-R617H mcm10-1 | This study |
| ILY245 | W303 MATamcm2-S619F mcm10-1 | This study |
| XL16 | W303 MATarad53::URA3 sml1::HIS3 | This lab |
| XL18 | W303 MATamec1::LEU2 sml1::URA3 | This lab |
| XL232 | W303 MATα sgs1::URA3 | This lab |
| XL158 | W303 MATα srs2::HIS3 | This lab |
| CLY99 | W303 MATaexo1::URA3 | This study |
| CLY84 | W303 MATamre11::LEU2 | This lab |
| CLY102 | W303 MATamcm10-1 rad53::URA3 sml1::HIS3 | This study |
| CLY103 | W303 MATamcm10-1 mec1::LEU2 sml1::URA3 | This study |
| CLY105 | W303 MATα mcm10-1 sgs1::URA3 | This study |
| CLY108 | W303 MATα mcm10-1 exo1::URA3 | This study |
| CLY85 | W303 MATamcm10-1 mre11::LEU2 | This study |
| CLY113 | W303 MATamcm2-G400D rad53::URA3 sml1::HIS3 | This study |
| CLY114 | W303 MATα mcm2-G400D mec1::LEU2 sml1::URA3 | This study |
| CLY116 | W303 MATamcm2-G400D sgs1::URA3 | This study |
| CLY119 | W303 MATamcm2-G400D exo1::URA3 | This study |
| CLY120 | W303 MATamcm2-G400D srs2::HIS3 | This study |
| CLY86 | W303 MATamcm2-G400D mre11::LEU2 | This study |
| CLY125 | W303 MATamcm2-G400D mcm10-1 rad53::URA3 sml1::HIS3 | This study |
| CLY126 | W303 MATα mcm2-G400D mcm10-1 mec1::LEU2 sml1::URA3 | This study |
| CLY128 | W303 MATα mcm2-G400D mcm10-1 sgs1::URA3 | This study |
| CLY131 | W303 MATamcm2-G400D mcm10-1 exo1::URA3 | This study |
| CLY132 | W303 MATamcm2-G400D mcm10-1 srs2::HIS3 | This study |
| CLY87 | W303 MATamcm2-G400D mcm10-1 mre11::LEU2 | This study |
| CLY152 | W303 MATa3×HA-Cdc17 HIS3 | This study |
| CLY144 | W303 MATα 3×HA-Cdc17 HIS3 mcm10-1 | This study |
| CLY145 | W303 MATα 3×HA-Cdc17 HIS3 mcm10-1 mcm2-G400D | This study |
| CLY148 | W303 MATa3×HA-Cdc17 HIS3 mec1 | This study |
| CLY149 | W303 MATa3×HA-Cdc17 HIS3 mec1 mcm10-1 mcm2-G400D | This study |
| CLY140 | W303 MATa3×HA-Rad53 kanMX | This study |
| CLY141 | W303 MATα 3×HA-Rad53 kanMX mcm10-1 | This study |
| CLY142 | W303 MATa3×HA-Rad53 kanMX mcm2-G400D | This study |
| CLY143 | W303 MATα 3×HA-Rad53 kanMX mcm10-1 mcm2-G400D | This study |
| CLY150 | W303 MATa3×HA-Cdc17 HIS3 rad53 | This study |
| CLY151 | W303 MATa3×HA-Cdc17 HIS3 rad53 mcm10-1 mcm2-G400D | This study |
| CLY152 | W303 MATaADE2 RFA1-8ala-YFP | R. Rothstein |
| CLY153 | W303 MATα ADE2 RFA1-8ala-YFP mcm10-1 | This study |
| CLY154 | W303 MATaADE2 RFA1-8ala-YFP mcm2-G400D | This study |
| CLY155 | W303 MATα ADE2 RFA1-8ala-YFP mcm10-1 mcm2-G400D | This study |
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| pRS315 | YCP LEU2 | New England Biolabs |
| pRS315MCM10 | YCP LEU2 MCM10 | This lab |
| pRS315mcm2-G400D | YCP LEU2 mcm2-G400D | This lab |
| pRS316MCM10 | YCP URA3 MCM10 | This lab |
| pGAD2F | 2μm LEU2 GAD4-AD | S. Fields |
| pBTM116 | 2μm TRP1 LEXA-DBD | S. Fields |
| pSH18-34 | URA3 LacZ with LEXA binding sites | S. Fields |
| pGADgw | pGAD2F with Gateway cassette | This lab |
| pBTMgw | pBTM116 with Gateway cassette | This lab |
| pGBKgw | pGBKT7 with Gateway cassette; Ampr | This lab |
| pBTMMCM10 | pBTMgw MCM10 | This lab |
| pBTMmcm10-1 | pBTMgw mcm10-1 | This lab |
| pBTMMCM2 | pBTMgw MCM2 | This lab |
| pBTMmcm2-G400D | pBTMgw mcm2-G400D | This lab |
| pBTMmcm2-S619F | pBTMgw mcm2-S619F | This lab |
| pGADMCM10 | pGADgw MCM10 | This lab |
| pGADmcm10-1 | pGADgw mcm10-1 | This lab |
| pGADMCM2 | pGADgw MCM2 | This lab |
| pGADmcm2-G400D | pGADgw mcm2-G400D | This lab |
| pGADmcm2-S619F | pGADgw mcm2-S619F | This lab |
| YCp1 | LEU2 CENV ARS1 | This lab |
Suppressor screen.
Suppressor screenings for random suppressor mutations of mcm10-1 were carried out as described previously (27). Plasmid-based mutagenesis of MCM2 was subsequently carried out to screen for more suppressor mutations. MCM2 was cloned into the pRS316 plasmid and mutagenized in Escherichia coli by using XL1-red competent cells (Stratagene). Mutagenized plasmids were obtained from E. coli, transformed into mcm10-1, and plated at 37°C to select for suppressors. The plasmids were sequenced to determine the nature of the mutations.
Expression and purification of MCM mutant proteins.
All Methanothermobacter thermautotrophicus MCM mutant proteins used in this study are derivatives of the full-length enzyme and were generated using the QuikChange site-directed mutagenesis kit (Stratagene), using the full-length MCM in the pET-21a vector (Novagen). All constructs contain a C-terminal His6 tag. The oligonucleotides used for the mutagenesis are G190D forward, 5′-AACCTTTCCGGTGATGAACAGCCCCGG-3′; G190D reverse, 3′-CCGGGGCTGTTCATCACCGGAAAGGTT-5′; R392H forward, 5′-CGTGAGGAGGACCACTCAGCCATACAC-3′; and R392H reverse, 3′-GTGTATGGCTGAGTGGTCCTCCTCACG-5′. The wild-type and mutant proteins were overexpressed in codon plus cells (Stratagene) at 16°C and purified as previously described (21).
DNA helicase assay.
Substrates for the helicase assay were generated as previously described (36) by hybridizing two oligonucleotides (DF50, 5′-GGGACGCGTCGGCCTGGCACGTCGGCCGCTGCGGCCAGGCACCCGATGGC-3′, and DF25F, 5′-CCGACGTGCCAGGCCGACGCGTCCC-3′). DF25F was labeled using [γ-32P]ATP (PerkinElmer) and T4 polynucleotide kinase (Fermentas) and hybridized to DF50, and the substrate was purified as previously described (36). Helicase assays were performed as previously described in reaction mixtures (15 μl) containing 20 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 2 mM dithiothreitol (DTT), 100 μg/ml bovine serum albumin (BSA), 3.33 mM ATP, 10 fmol of 32P-labeled DNA substrate, and MCM proteins, as indicated in the figure legends. Mixtures were incubated at 60°C for 30 min. Reactions were stopped by adding 5 μl of buffer containing 1% sodium dodecyl sulfate (SDS), 100 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue, and 50% glycerol, and then placed on ice. Aliquots were fractionated on an 8% native polyacrylamide gel in 0.5× TBE (45 mM Tris, 45 mM boric acid, 1 mM EDTA) and electrophoresed for 1 h at 150 V at 25°C. The helicase activity was visualized and quantitated by phosphorimaging.
Protein-protein interactions.
The wild-type W303 strain with the pSH18-34 reporter plasmid was transformed with pGAD2F and pBTM116 constructs for the two-hybrid assay (13). Transformants were selected on the appropriate dropout plates. Interactions were assessed by the appearance of blue colonies on plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; Sigma). Relevant strains were inoculated for saturated cultures and spotted onto X-Gal plates and photographed after 2 to 4 days of growth at 30°C.
Plasmid stability assays.
MCM assays were carried out as described in reference 12. Wild-type and mutant strains were transformed with the plasmid YCp1 that contains an origin of replication (ARS1), a centromere, and the LEU2 selectable marker. Assessment of plasmid loss rate in the mutants was done as described.
2D DNA gel electrophoresis.
Protocols for two-dimensional (2D) DNA gel electrophoresis were adapted from the method of Huberman (18) and the rapid DNA purification method (40). Cells were broken and spheroplasts were collected by centrifugation for 10 min at 8,000 rpm (4°C) according to the neutral-neutral method. The spheroplasts were resuspended in G2 buffer (Qiagen), and subsequent steps were carried out according to the rapid DNA purification method.
For visualization of replication intermediates at the ARS1 region, purified DNA was digested to completion with NcoI to produce a 5-kb fragment. To enrich the sample for replicating DNA, digested DNA was passed through BND cellulose (Sigma-Aldrich) columns as described in reference 11. ARS1 probes were made by amplifying a 1.5-kb region centered at ARS1 by PCR. The probes were radiolabeled with [α-32P]dATP by using the Prime-It II random primer labeling kit from Stratagene.
Western blot analysis.
Cdc17 was tagged with 3×HA at the C terminus. Mcm10 in wild-type, mcm10-1, and mcm10-43 strains was tagged with 13×myc and introduced into mcm2-G400D or mcm2-S619F strains. The strains were grown to log phase at 30°C and subsequently shifted to 37°C. Samples were collected at various time points for Western blot analysis. Proteins were extracted either by treating the cells briefly with mild alkali and then boiling in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer as described in reference 25 or by glass bead lysis in the presence of protease inhibitors. Extraction of phosphorylated Rad53 also contained phosphatase inhibitors. The mild-alkali-treatment (0.2 M NaOH) method produced a protein extraction yield similar to that of glass bead lysis. Mouse anti-myc (Santa Cruz) and mouse anti-hemagglutinin (HA; Roche) antibodies were used to probe for the appropriate myc-tagged and HA-tagged proteins. Goat anti-mouse horseradish peroxidase-conjugated secondary antibodies were obtained from Bio-Rad. Blots were visualized by chemiluminescence reagents (Promega).
FACS.
For overall ratio of cells in G1, S, or G2 phase, log-phase cells of wild-type, mcm10-1, mcm2-G400D, and mcm10 mcm2 strains were collected without α-factor arrest. For cell cycle progression, log-phase cultures were arrested in G1 phase by α-factor for 2 h. Cells were spun down and resuspended in fresh yeast extract-peptone-dextrose (YPD) media containing 100 μg/ml of pronase (Sigma) for rapid α-factor degradation and release into S phase. The G1-arrested cells were released at either 30°C or 37°C. For the latter, the cells were preincubated at 37°C for 1 h before release to allow Mcm10 degradation to occur before the onset of S phase. Samples at different time points were collected for fluorescence-activated cell sorting (FACS) analysis at the Cornell FACS facility.
Fluorescence microscopy.
Visualization of yellow fluorescent protein (YFP)-conjugated RPA was carried out in live cells under a conventional fluorescence microscope with a 100× objective. The images were obtained with a charge-coupled-device (CCD) detector by using Openlab (Improvision). Log-phase cells were prepared by growing in synthetic media at 30°C or subsequent exposure at 37°C for 2 h.
RESULTS
Mutations in MCM2 that suppress mcm10 TS affect the helicase activity.
Two temperature-sensitive (TS) mutants with MCM10, mcm10-1(P269L), and mcm10-43(C320Y) mutations share many of the same phenotypes (17). Both mutants show reduced replication initiation activity and fork pausing at unfired replication origins at the permissive temperature but arrest in S phase at the restrictive temperature. The protein products of both mutant alleles are heat labile (33, 34), suggesting that the instability may be the cause of these phenotypes. To determine the essential role of Mcm10 that was compromised at the restrictive temperature, two suppressor screens for mcm10 TS were carried out (27). In the first screen, spontaneous TS suppressors that have simultaneously acquired cold sensitivity (CS) were isolated. They all lie in MCM2 at two positions, R617 and S619, and they are all dominant suppressors (27). To identify other mutations in MCM2 that suppress the TS phenotype of mcm10, but did not necessarily have a CS phenotype, a CEN plasmid carrying MCM2 was randomly mutagenized and transformed into mcm10 cells. The transformation reaction mixture was plated at 37°C for identification of dominant suppressors. The resultant suppressor alleles were sequenced and integrated into the genomes of both wild-type and mcm10 cells. In all, 10 dominant suppressors comprising six alleles were isolated. Nine out of the 10 mutations clustered in two regions of MCM2 at either the region from P399 to R401 or that from R617 to S619 (Fig. 1A, panel i). With the exception of the P399L mutant, these mcm2 mutants do not display TS on their own (Fig. 1A, panel ii). Spot dilution of the mutants shows that the outlier D472G mutation is least able to suppress mcm10-1 TS. The mcm2 mutants are all allele-nonspecific suppressors, as they suppress both mcm10-1 and mcm10-43 (Fig. 1A, panel i). As both mcm10-1 and mcm10-43 express unstable forms of the protein that degrades at the restrictive temperature, we speculated that suppression by the mcm2 mutants may involve either restoration of Mcm10 stability or compensatory changes which lead to increased affinity between the proteins or bypass of function.
FIG. 1.
Suppression of mcm10 TS by mcm2 mutants. (A) (i) Fivefold serial dilutions of wild-type, mcm10-1, and mcm10-43 cells and the different mcm2 suppressors in the mcm10-1 or mcm10-43 background were spotted onto YPD plates and incubated for 1 to 2 days at either 30°C or 37°C. The mcm2 mutants are non-allele-specific suppressors of both mcm10-1 and mcm10-43. (ii) mcm2 suppressors, except mcm2-P399L, do not display TS. (iii) mcm2 mutants are located in two specific regions of the gene. Numbers in parentheses indicate the number of times a specific mutation was independently isolated. The mutations and their corresponding positions in the three-dimensional (3D) structure shown in panel B are color coded in shades of yellow, red and blue. Asterisks mark the mutants that display cold sensitivity. (B) The corresponding residues for the well-conserved ScG400, ScD472, and ScR617 in the archaeal Sulfolobus solfataricus are G207, D270, and R415, respectively. The red arrows indicate the locations of the three residues within the primary structure. The residues are located in the SsMCM structure by Brewster et al. (5). G207 and R415 of adjacent subunits are localized close in space at the subunit interface.
Most of the mcm2 suppressor mutations are located in regions of Mcm2 that are conserved throughout archaeal and eukaryotic MCM helicase (Fig. 1A, panel iii). In particular, the residues G400 and R617 in eukaryotic MCM2 are highly conserved in all eukaryotic MCM2-7 subunits and the archaeal MCM protein. Based on a recent study of the archaeal MCM helicase crystal structure from Sulfolobus solfataricus (5), these two regions are at the interface of adjacent subunits of the MCM helicase with the residue corresponding to S. cerevisiae G400 (ScG400) of one subunit juxtaposed to the residue corresponding to ScR617 of the neighboring subunit (Fig. 1B). The positions of the mutated residues suggest that suppression of mcm10 TS by the different mcm2 mutations may occur through a common mechanism and may involve altered interaction between the subunits at that particular interface.
The corresponding ScG400D and ScR617H mutations were individually introduced into the archaeal MCM protein for in vitro helicase assays to determine how they would affect the helicase activity. The helicase activity was measured by the extent of strand displacement when the substrate double-stranded DNA (dsDNA) was incubated with the purified proteins. The mutant helicases displayed weaker helicase activity than did the wild type (Fig. 2A and B). Keeping in mind that the archaeal MCM helicase is a homohexamer, the effect of an mcm2 equivalent mutation in the archaeal MCM is likely amplified. Therefore, the helicase defect of these two mcm2 suppressors in yeast is likely to be subtle, as the suppressors showed no obvious growth defects (Fig. 1A, panel i).
FIG. 2.
Suppressor mutations affect the helicase activity. (A) Wild-type and mutant M. thermautotrophicus (mt) MCM proteins were purified, and helicase assays were performed. A partial duplex DNA substrate was made by hybridization of 50-mer and 25-mer ssDNAs. The extent of helicase activity was determined by measuring the displacement of the radiolabeled 25-mer from the 50-mer. Lanes 3 to 5, wild-type MCM protein; lanes 6 to 8, G190D (ScG400D) mtMCM protein; lanes 9 to 11, R392H (ScR617H) mtMCM protein; lane 1, substrate only; lane 2, boiled substrate; lanes 3, 6, and 9, 10 ng (8.7 nM as monomers) MCM protein; lanes 4, 7, and 10, 30 ng (26 nM as monomers) MCM protein; lanes 5, 8, and 11, 90 ng (78 nM as monomers) MCM protein. S, substrate; P, product. (B) Average of the results for three independent experiments. (C) mcm assay to measure plasmid loss rate of mcm2 mutants at 30°C. The mcm2 suppressors show a minichromosome maintenance defect independent of the mcm10 mutation.
The archaeal MCM helicase result supports the in vivo minichromosome maintenance (mcm) assay result of the budding yeast MCM helicase. The mcm assay is used to assess the general replication proficiency of yeast cells and measures how well the cells are able to replicate and maintain plasmids in the absence of selective pressure. Mutants defective in replication display higher levels of plasmid loss. The mcm2 mutants displayed various degrees of mcm defect, independent of mcm10-1 (Fig. 2C). The result shows that the corresponding mutations that decreased helicase activity in the archaeal MCM helicase also showed a modest reduction in replication proficiency in the budding yeast. These observations suggest that although the decreased helicase activity compromises the replication efficiency of the mcm10 mutant at the permissive temperature, it is important for rescuing the lethal effects of mcm10 at the nonpermissive temperature.
mcm2 suppressors do not restore Mcm10-1 protein-protein interactions or stability.
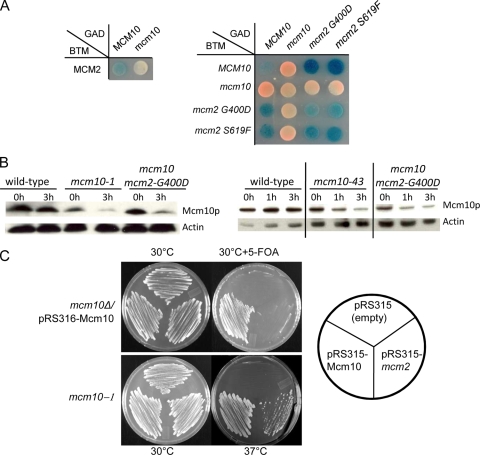
Mcm10 interacts with the Mcm2-7 subunits (17, 29), but this interaction is disrupted in the mcm10-1 strain. To investigate whether the mcm2 suppressors restore this interaction, we performed a yeast two-hybrid analysis of the Mcm2 suppressors, mcm2-G400D and mcm2-S619F, with the Mcm10-1 protein (Fig. 3A). We found that the level of interaction between the mutant Mcm2 construct and wild-type Mcm10 construct was similar to that of wild-type Mcm2 and wild-type Mcm10 interaction. However, we could not detect interaction between the mcm10-1 and mcm2 constructs. This result suggests that the protein interaction between Mcm10 and Mcm2 is not restored by the mcm2 mutations.
FIG. 3.
Mutations in Mcm2 do not restore interaction with Mcm10-1 or stabilize the mutant Mcm10 protein. (A) Two-hybrid reporter constructs with mcm10 and mcm2 alleles were transformed into a wild-type W303 strain. Two-hybrid interactions are detected by blue color. The loss of interaction between Mcm10-1 and Mcm2 is not restored by Mcm2-G400D or Mcm2-S619F proteins. (B) Western blot analysis of myc-tagged Mcm10, Mcm10-1, or Mcm10-43 from wild-type and mcm2-G400D log-phase cells at 37°C. (C) Plasmid shuffling to determine whether the mcm2-G400D suppressor can substitute for the wild-type MCM10 gene in an mcm10 null strain. mcm10Δ/pRS316-MCM10 (URA3) was transformed with an empty pRS315-LEU2, pRS315-Mcm10, and pRS315-mcm2-G400D and plated onto 5-fluoroorotic acid (5-FOA) plates. A control experiment shows that pRS315-mcm2-G400D is functional and is able to suppress mcm10-1 TS.
Although the mcm2 suppressor mutations did not restore physical interactions with Mcm10-1, we wanted to know if they restored the stability of the mutant Mcm10 protein at 37°C, a suspected cause of the TS phenotype of mcm10 cells. Mcm10 protein levels in the wild-type, mcm10, and mcm10 mcm2 suppressor strains were visualized by Western blots. Both Mcm10-1 and Mcm10-43 proteins were labile in the presence or absence of the mcm2 suppressor mutations (Fig. 3B). Therefore, the mcm2 suppressors do not prevent the degradation of either Mcm10-1 or Mcm10-43 proteins.
Since the mcm2 suppressors did not restore their interactions with the mutant Mcm10 protein or prevent its degradation, we asked if Mcm10 is dispensable in the mcm2 suppressor strains. We used an mcm10 knockout strain that was kept viable by a wild-type copy of MCM10 on a plasmid and determined whether we could replace the plasmid carrying MCM10 URA3 with one carrying mcm2-G400D LEU2 by plasmid shuffling (Fig. 3C). We found that mcm10 knockout strains required the MCM10 plasmid regardless of the presence or absence of mcm2-G400D, suggesting that the mcm2 suppressor could not bypass all of the essential functions of MCM10 but only the essential function of mcm10-1 and mcm10-43 compromised at the restrictive temperature of 37°C.
mcm2 suppressors suppress multiple replication fork defects of mcm10.
Replication forks in mcm10-1 pause at unfired origins as shown by the accumulation of DNA replication intermediates near the origin sequences of ARS1 or ARS121 in 2D gel electrophoresis analysis (2, 29). The locations of the pauses suggest that a defect in the elongation machinery may have compromised the fork's ability to move past bound prereplication complexes (pre-RCs) at unfired origins. The accumulation of the pause structures in mcm10-1 is more striking at 30°C than at 25°C (Fig. 4A), suggesting that the severity of fork pausing at the restrictive temperature may be the cause of death.
FIG. 4.
Suppression of replication elongation defects of mcm10 by mcm2. (A) (Top) Schematic of Southern blot of 2D gel probed with ARS1 DNA. (Bottom) DNA from log-phase cultures of mcm10-1 cells grown at either 25°C or 30°C was analyzed by 2D gel. The arrow points to pause signal corresponding to replication intermediates accumulated in mcm10-1 at unfired ARS1 at 30°C. This pause signal is not observed at 25°C. (B) DNAs from wild-type, mcm10-1, mcm2-G400D, and mcm10 mcm2 cells grown at 30°C were analyzed by 2D gel. mcm2-G400D alleviates the pause signal, but not the initiation defect of mcm10-1. (C) HU and MMS sensitivity of mcm10-1. Fivefold serial dilutions of wild-type and mutant strains were spotted onto YPD, YPD with 100 mM HU, and YPD with 0.02% MMS. mcm10-1 displays HU and MMS sensitivity, which is rescued by mcm2. (D) Cells expressing mcm10-1 display synthetic growth defects with genes in DSB and fork repair pathway. Those expressing sgs1, exo1, mre11, and rad50 display synthetic growth defects with mcm10-1. Deletion of srs2 is synthetically lethal with mcm10-1, as mcm10 srs2 is viable only when it carries a plasmid containing the wild-type MCM10 gene. The strain is unable to grow on 5-FOA when the plasmid is lost due to the URA3 marker. mcm10 sgs1, mcm10 exo1, and mcm10 srs2 synthetic growth defects are suppressed by mcm2-G400D, whereas those caused by mcm10 mre11 and mcm10 rad50 are not suppressed.
If fork pausing were indeed associated with mcm10 TS, it would be suppressed by mcm2. Therefore, we asked whether the mcm2 mutants are able to suppress the pause phenotype. Replication intermediates of wild-type, mcm10-1, mcm2-G400D, and mcm10 mcm2 strains grown at 30°C were analyzed by 2D gel electrophoresis (Fig. 4B). The pause signals observed in mcm10-1 strains were no longer observed in the mcm10 mcm2 strains, suggesting that the mcm2 suppressor has alleviated the fork pausing at unfired pre-RC. Furthermore, the enhancement rather than the suppression of the replication initiation defect (reduced bubble signal intensity) in the double mutant suggests that the lethality of mcm10-1 at the restrictive temperature is not due to replication initiation at the origins. Failure to suppress the replication initiation defect and suppression of the pause phenotype were also observed with mcm2-S619F (data not shown). This result suggests that the defect of mcm10 that leads to replication fork pausing and TS is in replication elongation.
HU depletes nucleotide pools and causes replication forks to stall. Methylmethane sulfonate (MMS) is a DNA alkylating reagent that causes DNA damage. Defects in replication fork stabilization and DNA repair have been associated with sensitivity to these chemicals (10, 38). Sensitivity to HU reflects defects in the replication fork, and sensitivity to MMS may be due to defects either in the fork or in DNA repair. We found that mcm10-1 is sensitive to both HU and MMS and the mcm2-G400D suppressor alleviates this sensitivity to both reagents (Fig. 4C). The sensitivity to these reagents is more likely to be associated with the defect at the fork rather than with DNA repair because mcm10-1 does not display increased spontaneous mutation rate by the canavanine assay (data not shown). These results further suggest that mcm10 compromises the replication fork and that this fork defect is compensated by the mcm2 mutation.
The observation that mcm10 loses viability as the cells go through S phase at the restrictive temperature (2) suggests that DNA damage accumulates as the defective replication fork progresses. Therefore, the defective fork in mcm10 may be creating damage and mcm2 may be preventing such damage from being formed. The damage could be in the form of double-strand breaks or altered fork structures. If so, proteins that function in dsDNA break (DSB) repair or resolution of aberrant fork structures should be required in mcm10.
In searching for gene deletions in strains that showed synthetic growth defect or lethality with mcm10-1, we found mre11, rad50, sgs1, exo1, and srs2 (Fig. 4D). Even at 30°C, strains with mre11, rad50, sgs1, and exo1 deleted displayed synthetic growth defects with mcm10-1. Also, mcm10 srs2 is synthetically lethal; the strain is viable only when it carries a plasmid expressing the wild-type MCM10 gene. MRE11 and RAD50 are required during the initial processing of DSB repair (32). DNA helicases SGS1, SRS2 and nuclease EXO1 process ssDNA overhangs during DSB repair (19, 20). It has been previously reported that mcm10-1 is synthetically lethal with yet another DNA helicase/nuclease dna2-2 (2) that also functions in DSB repair.
However, Sgs1, Exo1, and Srs2 also function in fork repair as their helicase or nuclease activities are involved in promoting progression and/or resolution of reversed forks and Holliday junction structures. Srs2 is known to disrupt Rad51 binding to ssDNA to prevent aberrant recombination (24) and most srs2 synthetic lethal mutants are rescued by deletion of rad51 (15). Indeed, we observed that rad51Δ suppresses the mcm10 srs2 synthetic lethality as well (data not shown). Interestingly, mcm2-G400D also rescues this synthetic lethality (Fig. 4D). If rescue of mcm10 srs2 synthetic lethality by rad51Δ is due to disruption of Rad51 filament formation and prevention of aberrant recombination events, then mcm2 may be preventing mcm10 from producing substrates for Srs2 and/or Rad51.
We observed that mcm2-G400D also suppresses mcm10 sgs1 and mcm10 exo1 growth defects (Fig. 4D), suggesting that the DNA helicases/nucleases that are important for the viability of mcm10 are no longer vital in mcm2-G400D cells. However, mcm2-G400D does not suppress mcm10 mre11 or mcm10 rad50 synthetic defect (Fig. 4D), which indicates that DSBs are still occurring in mcm10 mcm2. In summary, these results suggest that the role of Sgs1, Exo1, and Srs2 in mcm10 is different from that of Mre11 and Rad50, implying that different types of damage are occurring at the replication fork due to mcm10 defect.
Checkpoint proteins are required for the suppression of mcm10-1.
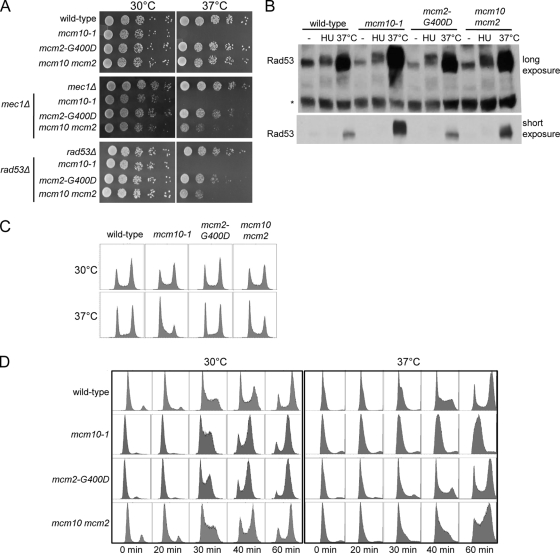
The nature of the various mcm10 phenotypes that are suppressed by mcm2 strongly suggests that the defect in the replication fork is the cause of cell death at the restrictive temperature. However, the suppressors do not suppress the TS by restoring physical interaction between the mutant Mcm10 and Mcm2 proteins or by preventing degradation of the mutant Mcm10 protein (Fig. 3). Therefore, the mechanism by which mutations in mcm2 restore viability of mcm10 cells at the restrictive temperature must involve mechanisms that compensate for the function of Mcm10 at the fork. One possible scenario is that Mcm10 is an important fork stabilizer. Mutations in factors that can stabilize the fork independently of Mcm10 would appear as suppressors of mcm10-1. Another is that Mcm10 may be essential for fork repair and the suppressor has gained the function to facilitate fork repair by alternative mechanisms. These hypotheses may be tested by candidate mutations from the different DNA repair and checkpoint pathways that negate or weaken the suppression of mcm10 TS by the mcm2 mutants. We introduced deletions of mec1, rad53, rad51, mrc1, tof1, rad6, dnl4, rad9, exo1, mre11, sgs1, srs2, and ddc1 into the wild-type, mcm10-1, mcm2-G400D, and mcm10 mcm2 strains (Fig. 5A) (results not shown for double mutants with no effect). In addition to mre11 and rad50, gene deletions that have negative effects on suppression were rad53 and mec1. Since both rad53 and mec1 also require sml1 deletion for viability, we confirmed that sml1 is not responsible for the negative effect on suppression (data not shown).
FIG. 5.
Checkpoint functions are required for viability of mcm10 mcm2. (A) Serial dilutions of cells are spotted onto YPD and incubated at 30°C or 37°C. Deletion of MEC1 and RAD53 has a negative effect on suppression. (B) Rad53 phosphorylation in mcm10-1 and mcm10 mcm2. 3×HA-tagged Rad53 strains with mcm10-1 and mcm2-G400D mutations were grown to log phase, arrested by α-factor for 1.5 h, and released into fresh media with or without HU at 30°C for 1 h or without HU at 37°C for 1 h. Samples were collected for Western blot analysis to assay the phosphorylation state of Rad53. Exposure of mcm10-1 to 37°C leads to hyperphosphorylation of Rad53. Suppression of TS by mcm2 is accompanied by a decrease in Rad53 phosphorylation. (C) FACS analysis of wild-type, mcm10-1, mcm2-G400D, and mcm10 mcm2 cells. For overall ratio of cells in G1, S, or G2 phase, log-phase cells of wild-type, mcm10-1, mcm2-G400D, and mcm10 mcm2 strains were collected without α-factor arrest. (D) Analysis of cell cycle progresses was carried out by arresting the cells at G1 and releasing into S phase at either 30°C or 37°C. Samples analyzed at 37°C were preincubated at 37°C during α-factor arrest to allow time for Mcm10 protein degradation. At 37°C, with Mcm10 depletion, significant delay in S-phase entry and progression was observed as published previously (29). While mcm2 cells do not show any difference in cell cycle progression from that of wild-type cells, a slight delay of S-phase progression in mcm10 mcm2 cells was observed.
Rad53 and Mec1 are both key players in the checkpoint signaling pathway that have a role in stabilizing stalled forks as well as in transducing signals to downstream effectors (for a review, see reference 4). Deletion of rad53 and mec1 greatly diminished the ability of the mcm2 mutants to suppress mcm10 TS. These results suggest that the checkpoint pathway is activated in mcm10 mcm2. Phosphorylation of Rad53 is required for replication fork stabilization.
To determine if Rad53 is indeed activated, we examined the phosphorylation state of Rad53 in mcm10-1, mcm2-G400D, and mcm10 mcm2 mutants. Log-phase cells were grown at 37°C for 2 h and collected for protein extraction and Western blot analysis (Fig. 5B). We found that Rad53 is hyperphosphorylated in the mcm10 mutant at 37°C. Previous work showed that a shift to 37°C causes an irreversible loss of viability upon return to permissive temperature in mcm10 cells (2). Therefore, degradation of Mcm10p, which causes irreparable DNA damage, must have activated Rad53. In the mcm10 mcm2 mutant, Rad53 is also phosphorylated, though the shift due to phosphorylation is much weaker. While Rad53 is activated in both mcm10 and mcm10 mcm2 mutants, the consequences of its activation are drastically different, as the mcm10 mutant loses viability, while the mcm10 mcm2 mutant is phenotypically similar to the wild type. Rad53 phosphorylation is not enhanced in the mcm2 mutant, ruling out the possibility that the mutation in the helicase alone activates the checkpoint pathway. It appears that the mcm2 mutant helicase in combination with mcm10 causes activation of Rad53 that prevents or corrects the damage by mcm10. Our results suggest that the unstable replication fork in mcm10 is stabilized by the mcm2 suppressor and a mechanism that involves activation of the checkpoint pathway.
Since activated Rad53 can slow down S phase to provide more time for repair of damages, we determined whether Rad53 activation in mcm10 mcm2 cells is accompanied by a slower S phase by FACS analysis. To obtain the overall ratio of cells in G1, S, or G2 phase, log-phase cells of wild-type, mcm10-1, mcm2-G400D, and mcm10 mcm2 strains were collected without α-factor arrest. At 30°C, the overall ratios of G1/S/G2-phase cells are similar for all strains, with two peaks at G1 and G2 phases with the G2-phase peak being slightly stronger (Fig. 5C). However, at 37°C, in the mcm10 cells, the G1 peak is much greater than the G2 peak, suggesting that the cells have difficulty entering S phase. This problem seems to be corrected in mcm2-G400D cells, though not completely, as the G1 peak is still stronger than the G2 peak. A closer examination of how the cell cycle progresses was carried out by arresting the cells at G1 phase and releasing into S phase at either 30°C or 37°C (Fig. 5D). At 37°C, with Mcm10 depletion, significant delay in S-phase entry and progression was observed as reported previously (29). While mcm2 cells do not show any significant difference in cell cycle progression from the wild type, a slight delay of S-phase progression in mcm10 mcm2 cells was observed. Entry into S phase seemed to be similar for all strains, as they entered S phase at the 30-min time point. However, the delay in progression was evident, because mcm10 mcm2 cells were still in S phase while wild-type or mcm2 cells were already into G2 phase at the 60-min time point. Therefore, the S-phase delay in mcm10 mcm2 cells is consistent with the checkpoint activation and ongoing DNA repair.
Mutations in MCM2 stabilize Cdc17p in mcm10 cells in a checkpoint-dependent manner.
Cdc17 is the catalytic subunit of the Polα primase, the only DNA polymerase that has the capability of de novo DNA synthesis (6). The primase is required for priming Okazaki fragments on the lagging strand throughout elongation. In budding yeast, it is suggested that Mcm10 functions as a linker between the Polα primase and the helicase, because Mcm10 is required for Cdc17 stabilization and its association with chromatin (33). In both mcm10-1 and mcm10 temperature-degron (TD) mutants, Mcm10 protein degradation at 37°C was accompanied by Cdc17 degradation with similar kinetics (33). We found that the mcm2 suppressors did not suppress degradation of the mutant Mcm10 protein (Fig. 3B). However, since the primase activity is indispensable for DNA replication, we reasoned that suppression of mcm10 TS by mcm2 must be accompanied by restoration of Cdc17 function, either directly or indirectly. Therefore, we performed Western blot experiments with Cdc17-3xHA-tagged wild-type, mcm10-1, and mcm10-1 mcm2-G400D strains to determine the stability of Cdc17p. We found that while the mcm2 suppressor failed to stabilize Mcm10 in mcm10-1 cells, it was able to stabilize Cdc17 (Fig. 6A).
FIG. 6.
mcm2-G400D stabilizes Cdc17 in a Mec1-dependent manner. (A) Log-phase cells were incubated at either room temperature or 37°C for 90 min and collected for Western blot analysis. Cdc17 is unstable in mcm10-1 cells at 37°C but stabilized in mcm2-G400D mcm10-1 cells. (B) Log-phase cells of mec1 and mec1 mcm10-43 mcm2-G400D strains were incubated at 37°C for 0, 1, and 3 h and harvested for Western blot analysis. Loss of Mec1 function in mcm10-43 mcm2-G400D cells leads to degradation of Cdc17. Log-phase cells of rad53 and rad53 mcm10-43 mcm2-G400D strains were incubated at 37°C for 0 and 3 h and before being harvested for Western blot analysis. Loss of Rad53 function has no effect on Cdc17 stability. Asterisk indicates the cross-reacting band, which serves as a loading control. (C) (i) Log-phase cells of RFA1-YFP wild-type, mcm10-1, mcm2-G400D, and mcm10 mcm2 strains were grown at either 30°C or 37°C and subjected to microscopy analysis. The extent of RPA focus formation in mcm10-1 cells increased with the temperature, and mcm2-G400D suppressed this. (ii) The percentage of cells with RPA foci was quantified by averaging three independent counts of >100 cells. The level of RPA focus formation in mcm10 mcm2 cells at 37°C was similar to that in mcm10-1 cells at 30°C.
Since Mcm10 is suggested to be a chaperone for Cdc17 stability, it was of interest how Cdc17 is stabilized despite Mcm10 instability. Two-hybrid analysis did not show interaction between Mcm2-G400D and Cdc17, suggesting that the Mcm2 suppressor is not directly involved in the stabilization of Cdc17 by establishing new interactions. We had noticed that suppression of mcm10-1 TS by mcm2-G400D was greatly diminished in a mec1 or rad53 null background, suggesting that the checkpoint function may be required for Cdc17 stability. We tested this idea by examining Cdc17 stability in mec1 and in rad53 cells. We carried out Western blot analysis of Cdc17-3xHA to determine protein stability in mec1Δ mcm10 mcm2 and rad53Δ mcm10 mcm2 cells, respectively (Fig. 6B). We found that Cdc17 was no longer stable when Mec1 function was lost. However, loss of Rad53 did not affect Cdc17 stability. This result suggests that stability of Cdc17 in mcm10 mcm2 cells depends on a specific function of Mec1 in stabilizing the replication fork.
As degradation of Cdc17 would lead to cessation of DNA synthesis on the lagging strand, ssDNA is expected to accumulate at the fork in mcm10-1 at 37°C. To examine ssDNA at replication forks, we visualized RPA-YFP localization in log-phase cells of wild-type, mcm10-1, mcm2-G400D, and mcm10 mcm2 strains at either 30°C or 37°C (Fig. 6C, panel i). The frequencies of RPA foci observed in S-phase cells in these different strains are compared in a bar graph (Fig. 6C, panel ii). In mcm10-1 cells, we observed intense RPA focus formation, and the number of cells with RPA foci increased with temperature. Importantly, mcm2-G400D, which does not display RPA focus formation on its own, suppresses RPA focus formation in mcm10-1 cells, as the level of focus formation in mcm10 mcm2 cells at 37°C is similar to that of mcm10-1 cells at 30°C. This decrease in RPA focus formation suggests that the mutant helicase is preventing ssDNA accumulation that results from the loss of Mcm10.
DISCUSSION
The properties of Mcm10, its role in Cdc17 stability and association at the fork (33), and its interaction with the MCM helicase all suggest that Mcm10 plays a pivotal role in physically linking and coordinating the activities of polymerase α primase and the MCM helicase. We reasoned that suppression of the loss of this linker function would involve either restoration of Mcm10 function or recruiting another pathway to coordinate the polymerase and helicase activities. The finding that mcm2 suppressors do not restore the interaction of Mcm2 with the mutant Mcm10 protein or stabilize the mutant Mcm10 protein suggests that the latter is more likely. In achieving this end, the mutant helicase has to play a critical role. Although no detectable physiological defect is observed in the mcm2-G400D suppressor other than the mild mcm defect, the archaeal MCM helicase bearing the suppressor mutations invariably showed a compromised helicase activity. This result suggests that the altered helicase activity is critical for the suppression of the conditional lethality of mcm10.
What are the phenotypes associated with the mcm10 conditional lethality? They should be phenotypes that are also suppressed by the mcm2 suppressors. We showed that mcm2 suppresses the replication fork pausing phenotype as well as HU and MMS sensitivity of mcm10. Furthermore, it suppresses the synthetic growth defects of the loss of Sgs1, Exo1, or Srs2, a cohort of DNA helicases/nucleases, in mcm10 strains. These helicases/nucleases are involved in DSB repair (DSBR) in two capacities: (i) through resolution of aberrant fork structures to prevent DSB formation, and (ii) through resection of DSBs after their formation (19, 30, 41). In contrast, mcm2-G400D fails to suppress the synthetic growth defects of mcm10 mre11 or mcm10 rad50 strains. Mre11 and Rad50 are the major proteins involved in all DSB repair. The differences in requirement of these DSBR proteins in the suppression of mcm10 by mcm2 suggest that mcm2 is able to either substitute for Sgs1, Exo1, and Srs2 in the repair of their DSB substrates or prevent the formation of these substrates. The roles of Sgs1 and Exo1 are well defined in the DSB repair pathway, so it is doubtful that a defective MCM helicase could carry out their functions, especially those involving nuclease activities. A more likely scenario is that mcm2 is preventing the formation of aberrant fork structures and thereby abrogates the need for these helicases/nucleases. Under this scenario, we imagine that reduced activity of the helicase prevents the formation of a subset of DNA damage due to aberrant fork structures caused by the instability of Mcm10-1.
Mec1 is important for preventing dissociation of fork components, polymerase α in particular, when replication forks stall under replication stress (9). It was shown that Mec1, rather than Rad53, plays a key role in maintaining the association of Polα with the replication fork when forks stall. Therefore, the Mec1-dependent stabilization of Cdc17 in mcm10 mcm2 cells suggests that preventing fork collapse is a key factor in preserving the viability of cells despite loss of Mcm10 function. Though it is possible that the Mcm2 suppressor stabilizes Cdc17 by acquiring the ability to interact directly between Polα and the helicase, two-hybrid analysis of Cdc17 with either wild-type or mutant Mcm2 does not support this hypothesis (data not shown). We believe that normally Mcm10 may stabilize Polα by direct interaction, but in the event that Mcm10 fails to carry out this function, alternative pathways may be evoked to substitute for this critical activity. An alternative explanation is that the stability of Polα depends on fork integrity as a whole rather than interaction with any particular protein and that cells are multifaceted in maintaining the integrity of the replication fork under normal or stress conditions.
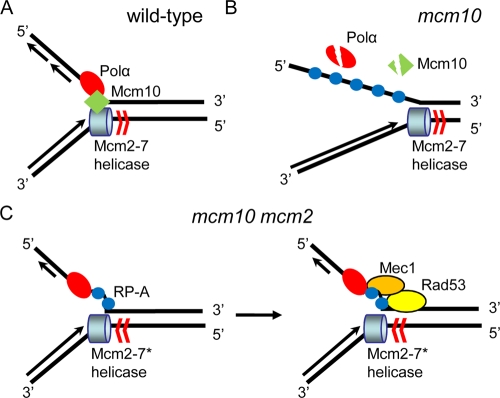
In summary, our study suggests that reduced MCM helicase activity rendered by the mcm2 suppressor mutation is able to mediate fork stabilization by activating the checkpoint pathway and coordinating the helicase and polymerase activities in the absence of Mcm10. A model of how mcm2 may suppress mcm10 is shown in Fig. 7. In a normal replication fork, Mcm10, by interaction with both Polα and the MCM helicase, coordinates the polymerizing and unwinding activities on the lagging strand (Fig. 7A). In the mcm10 mutant, Mcm10 is unstable at 37°C, resulting in the decoupling of Polα from the helicase. Polα released from chromatin is destabilized. The uncoordinated unwinding and polymerizing activities expose extensive ssDNA (Fig. 6C), especially on the lagging strand, resulting in fork collapse and other damage that cannot be rescued by checkpoint-activated repair (Fig. 7B). We imagine that mcm2 suppresses the mcm10 conditional lethality by preventing such irreversible damage. The suppressor mutations may alter the rate of helicase unwinding to the extent that ssDNA accumulation is reduced and fork collapse is diverted; however, coordination between the unwinding and polymerizing activities may still be imperfect. As a result, chronic activation of checkpoint response in mcm10 mcm2 (Fig. 5B) by persistent low-level ssDNA exposure works in the favor of the faulty replication fork by stabilizing it (Fig. 7C). In other words, we propose that the loss of physical stabilization at the fork caused by the unstable Mcm10 can be compensated for by a mechanistic stabilization that results from the compromised helicase and the activated checkpoint proteins to coordinate the lagging strand synthesis. This hypothesis points to the dynamics of fork components in adapting to the defects of one another and the integration of different cellular pathways, such as replication, repair, and checkpoints, to maintain the integrity of the genome.
FIG. 7.
Model of mechanism by which mcm2 suppresses mcm10 conditional lethality. (A) Mcm10 couples helicase to Polα primase under normal replication conditions. (B) Degradation of Mcm10 causes dissociation of Polα from chromatin and failure to coordinate the unwinding activity with the polymerizing activity result in the exposure of ssDNA, fork collapse, and checkpoint activation. (C) Mutation in the helicase compromises the unwinding activity, allowing the polymerase to keep up without a coupling factor. The imperfect synchrony results in chronic exposure of ssDNA that renders the fork “prone to checkpoint activation” but able to remain intact. The checkpoint proteins recruited to the fork help stabilize the components of the fork and replace the function of Mcm10 as a coupler.
Acknowledgments
We thank a very talented group of undergraduate and rotation students, Alice Leung, Mike Singer, Jany Chan, and Stephanie Yazinski, for identifying many of the suppressor alleles. We thank Nozomi Sakakibara for her help in the construction of the Mth mutants. We also thank Marcus Smolka and Francisco M. Bastos de Oliviera for their intellectual and technical input, especially with regard to checkpoint effects. Lastly, we thank the Huberman lab for their patience and time in teaching C.L. the 2D gel technique.
This work was supported by NSF (MBG-0453773) and NIH GM072557 awarded to B.K.T. and NSF (MCB-0815646) awarded to Z.K.
Footnotes
Published ahead of print on 16 November 2009.
REFERENCES
- 1.Alcasabas, A. A., A. J. Osborn, J. Bachant, F. Hu, P. J. H. Werler, K. Bousset, K. Furuya, J. F. X. Diffley, A. M. Carr, and S. J. Elledge. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3:958-965. [DOI] [PubMed] [Google Scholar]
- 2.Araki, Y., Y. Kawasaki, H. Sasanuma, B. K. Tye, and A. Sugino. 2003. Budding yeast mcm10/dna43 mutant requires a novel repair pathway for viability. Genes Cells 8:465-480. [DOI] [PubMed] [Google Scholar]
- 3.Bailey, S., W. K. Eliason, and T. A. Steitz. 2007. Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science 318:459-463. [DOI] [PubMed] [Google Scholar]
- 4.Branzei, D., and M. Foiani. 2008. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 9:297-308. [DOI] [PubMed] [Google Scholar]
- 5.Brewster, A. S., G. Wang, X. Yu, W. B. Greenleaf, J. M. Carazo, M. Tjajadi, M. G. Klein, and X. S. Chen. 2008. Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc. Natl. Acad. Sci. USA 105:20191-20196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burgers, P. M. J. 1998. Eukaryotic DNA polymerases in DNA replication and DNA repair. Chromosoma 107:218-227. [DOI] [PubMed] [Google Scholar]
- 7.Byun, T. S., M. Pacek, M.-C. Yee, J. C. Walter, and K. A. Cimprich. 2005. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19:1040-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chattopadhyay, S., and A.-K. Bielinsky. 2007. Human Mcm10 regulates the catalytic subunit of DNA polymerase α and prevents DNA damage during replication. Mol. Biol. Cell 18:4085-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cobb, J. A., L. Bjergbaek, K. Shimada, C. Frei, and S. M. Gasser. 2003. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 22:4325-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desany, B. A., A. A. Alcasabas, J. B. Bachant, and S. J. Elledge. 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 12:2956-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dijkwel, P. A., J. P. Vaughn, and J. L. Hamlin. 1991. Mapping of replication initiation sites in mammalian genomes by two-dimensional gel analysis: stabilization and enrichment of replication intermediates by isolation on the nuclear matrix. Mol. Cell. Biol. 11:3850-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donato, J. J., S. C. C. Chung, and B. K. Tye. 2006. Genome-wide hierarchy of replication origin usage in Saccharomyces cerevisiae. PLoS Genet. 2:e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245-256. [DOI] [PubMed] [Google Scholar]
- 14.Fien, K., Y.-S. Cho, J.-K. Lee, S. Raychaudhuri, I. Tappin, and J. Hurwitz. 2004. Primer utilization by DNA polymerase {alpha}-primase is influenced by its interaction with Mcm10p. J. Biol. Chem. 279:16144-16153. [DOI] [PubMed] [Google Scholar]
- 15.Gangloff, S., C. Soustelle, and F. Fabre. 2000. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 25:192-194. [DOI] [PubMed] [Google Scholar]
- 16.Hamdan, S. M., D. E. Johnson, N. A. Tanner, J.-B. Lee, U. Qimron, S. Tabor, A. M. van Oijen, and C. C. Richardson. 2007. Dynamic DNA helicase-DNA polymerase interactions assure processive replication fork movement. Mol. Cell 27:539-549. [DOI] [PubMed] [Google Scholar]
- 17.Homesley, L., M. Lei, Y. Kawasaki, S. Sawyer, T. Christensen, and B. K. Tye. 2000. Mcm10 and the MCM2-7 complex interact to initiate DNA synthesis and to release replication factors from origins. Genes Dev. 14:913-926. [PMC free article] [PubMed] [Google Scholar]
- 18.Huberman, J. A. 1997. Mapping replication origins, pause sites, and termini by neutral/alkaline two-dimensional gel electrophoresis. Methods 13:247-257. [DOI] [PubMed] [Google Scholar]
- 19.Ira, G., A. Malkova, G. Liberi, M. Foiani, and J. E. Haber. 2003. Srs2 and Sgs1Top3 suppress crossovers during double-strand break repair in yeast. Cell 115:401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ira, G., A. Pellicioli, A. Balijja, X. Wang, S. Fiorani, W. Carotenuto, G. Liberi, D. Bressan, L. Wan, N. M. Hollingsworth, J. E. Haber, and M. Foiani. 2004. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431:1011-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasiviswanathan, R., J.-H. Shin, E. Melamud, and Z. Kelman. 2004. Biochemical characterization of the Methanothermobacter thermautotrophicus minichromosome maintenance (MCM) helicase N-terminal domains. J. Biol. Chem. 279:28358-28366. [DOI] [PubMed] [Google Scholar]
- 22.Katou, Y., Y. Kanoh, M. Bando, H. Noguchi, H. Tanaka, T. Ashikari, K. Sugimoto, and K. Shirahige. 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424:1078-1083. [DOI] [PubMed] [Google Scholar]
- 23.Kawasaki, Y., S.-I. Hiraga, and A. Sugino. 2000. Interactions between Mcm10p and other replication factors are required for proper initiation and elongation of chromosomal DNA replication in Saccharomyces cerevisiae. Genes Cells 5:975-989. [DOI] [PubMed] [Google Scholar]
- 24.Krejci, L., S. Van Komen, Y. Li, J. Villemain, M. S. Reddy, H. Klein, T. Ellenberger, and P. Sung. 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423:305-309. [DOI] [PubMed] [Google Scholar]
- 25.Kushnirov, V. V. 2000. Rapid and reliable protein extraction from yeast. Yeast 16:857-860. [DOI] [PubMed] [Google Scholar]
- 26.Lee, J.-K., Y.-S. Seo, and J. Hurwitz. 2003. The Cdc23 (Mcm10) protein is required for the phosphorylation of minichromosome maintenance complex by the Dfp1-Hsk1 kinase. Proc. Natl. Acad. Sci. USA 100:2334-2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liachko, I., and B. K. Tye. 2005. Mcm10 is required for the maintenance of transcriptional silencing in Saccharomyces cerevisiae. Genetics 171:503-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lou, H., M. Komata, Y. Katou, Z. Guan, C. C. Reis, M. Budd, K. Shirahige, and J. L. Campbell. 2008. Mrc1 and DNA polymerase [var epsilon] function together in linking DNA replication and the S phase checkpoint. Mol. Cell 32:106-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merchant, A., Y. Kawasaki, Y. Chen, M. Lei, and B. Tye. 1997. A lesion in the DNA replication initiation factor Mcm10 induces pausing of elongation forks through chromosomal replication origins in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:3261-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mimitou, E. P., and L. S. Symington. 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455:770-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nyberg, K. A., R. J. Michelson, C. W. Putnam, and T. A. Weinert. 2002. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36:617-656. [DOI] [PubMed] [Google Scholar]
- 32.Paull, T. T., and M. Gellert. 1998. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol. Cell 1:969-979. [DOI] [PubMed] [Google Scholar]
- 33.Ricke, R. M., and A.-K. Bielinsky. 2004. Mcm10 regulates the stability and chromatin association of DNA polymerase-α. Mol. Cell 16:173-185. [DOI] [PubMed] [Google Scholar]
- 34.Sawyer, S. L., I. H. Cheng, W. Chai, and B. K. Tye. 2004. Mcm10 and Cdc45 cooperate in origin activation in Saccharomyces cerevisiae. J. Mol. Biol. 340:195-202. [DOI] [PubMed] [Google Scholar]
- 35.Shechter, D., V. Costanzo, and J. Gautier. 2004. Regulation of DNA replication by ATR: signaling in response to DNA intermediates. DNA Repair 3:901-908. [DOI] [PubMed] [Google Scholar]
- 36.Shin, J.-H., B. Grabowski, R. Kasiviswanathan, S. D. Bell, and Z. Kelman. 2003. Regulation of minichromosome maintenance (MCM) helicase activity by Cdc6. J. Biol. Chem. 278:38059-38067. [DOI] [PubMed] [Google Scholar]
- 37.Sogo, J. M., M. Lopes, and M. Foiani. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297:599-602. [DOI] [PubMed] [Google Scholar]
- 38.Tercero, J. A., and J. F. X. Diffley. 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412:553-557. [DOI] [PubMed] [Google Scholar]
- 39.Tercero, J. A., M. P. Longhese, and J. F. X. Diffley. 2003. A central role for DNA replication forks in checkpoint activation and response. Mol. Cell 11:1323-1336. [DOI] [PubMed] [Google Scholar]
- 40.Wu, J., and D. Gilbert. 1995. Rapid DNA preparation for 2D gel analysis of replication intermediates. Nucleic Acids Res. 23:3997-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu, Z., W.-H. Chung, E. Y. Shim, S. E. Lee, and G. Ira. 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134:981-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zou, L., and S. J. Elledge. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542-1548. [DOI] [PubMed] [Google Scholar]