Abstract
Ty1 is a retrovirus-like retrotransposon whose replication is influenced by diverse cellular processes in Saccharomyces cerevisiae. We have identified cytoplasmic P-body components encoded by DHH1, KEM1, LSM1, and PAT1 as cofactors that posttranscriptionally enhance Ty1 retrotransposition. Using fluorescent in situ hybridization and immunofluorescence microscopy, we found that Ty1 mRNA and Gag colocalize to discrete cytoplasmic foci in wild-type cells. These foci, which are distinct from P-bodies, do not form in P-body component mutants or under conditions suboptimal for retrotransposition. Our immunoelectron microscopy (IEM) data suggest that mRNA/Gag foci are sites where virus-like particles (VLPs) cluster. Overexpression of Ty1 leads to a large increase in retrotransposition in wild-type cells, which allows VLPs to be detected by IEM. However, retrotransposition is still reduced in P-body component mutants under these conditions. Moreover, the percentage of Ty1 mRNA/Gag foci and VLP clusters and levels of integrase and reverse transcriptase are reduced in these mutants. Ty1 antisense RNAs, which have been reported to inhibit Ty1 transposition, are more abundant in the kem1Δ mutant and colocalize with Ty1 mRNA in the cytoplasm. Therefore, Kem1p may prevent the aggregation of Ty1 antisense and mRNAs. Overall, our results suggest that P-body components enhance the formation of retrotransposition-competent Ty1 VLPs.
Ty elements comprise five related families of long terminal repeat (LTR) retrotransposons present in the genome of the budding yeast Saccharomyces. Ty1 is the most abundant retrotransposon in Saccharomyces cerevisiae and resembles retroviruses in genome organization and replication (59). The Ty1 genome consists of two overlapping open reading frames, GAG and POL, which are flanked by LTRs. Ty1 is transcribed by RNA polymerase II to yield a high level of a 5.9-kb genomic transcript, which accounts for 5 to 10% of the total poly(A) RNA. Like retroviruses, the genomic RNA can be translated or packaged as a dimer into virus-like particles (VLPs). The primary translation products are Gag (p49) and Gag-Pol (p199) precursors, the latter resulting from a +1 ribosomal frameshift during translation. Gag is the main structural component of VLPs. Pol contains protease (PR), integrase (IN), and reverse transcriptase (RT), which are enzymes required for retrotransposition. Once VLPs undergo proteolytic processing of Gag and Gag-Pol precursor proteins, Ty1 genomic RNA is transcribed into cDNA within VLPs. Ty1 cDNA and IN are imported into the nucleus, and transposition is completed by integration into the host genome.
Despite high levels of Ty1 mRNA and many active elements present in the yeast genome, retrotransposition occurs at a low rate (16, 18-20). Endogenous VLPs are rarely detected, proteolytic processing of Pol is extremely inefficient, and the level of cDNA is less than one copy per cell (13, 17, 24, 44). These observations suggest that “transpositional dormancy” occurs posttranscriptionally, prior to or during VLP assembly. However, overexpression of a competent Ty1 element from the strong inducible GAL1 promoter carried on a multicopy pGTy1 plasmid overcomes transpositional dormancy and results in a >10,000-fold increase in retrotransposition, even though the level of Ty1 mRNA increases only 5- to 15-fold (17). Thus, the utilization of Ty1 mRNA in normal and transposition-induced cells changes; however, the factors responsible for the trafficking of Ty1 mRNA and transpositional dormancy are not well understood.
The results of several genetic screens have shown that cytoplasmic processing body (P-body) components in budding yeast facilitate the replication of retrotransposons (Ty1 and Ty3) and brome mosaic virus (BMV), a positive-strand RNA plant virus (2, 5, 22, 27, 28, 33, 47). P-bodies are discrete cytoplasmic foci that contain messenger ribonucleoprotein complexes (mRNPs), which can be stored or degraded (10, 50, 55, 57). In wild-type cells grown to mid-log phase, P-bodies are difficult to visualize by microscopy unless cells are placed under stress, such as glucose deprivation, which leads to an increase in the pool of nontranslated mRNPs (57). Proteins encoded by the essential genes DCP1 and DCP2, along with those encoded by EDC3, DHH1, LSM1-7 complex, and PAT1, form the conserved core of a P-body. Dhh1p, Lsm1p, and Pat1p facilitate the recruitment of repressed mRNA into P-bodies and stimulate mRNA decapping, which is carried out by Dcp1p and Dcp2p (12, 55). KEM1/XRN1 encodes the 5′-3′ exonuclease that degrades mRNA in P-bodies. Dcp2 and Pat1p contribute to P-body assembly, although no single P-body component is absolutely required (56). The formation of P-bodies is a dynamic process which affects the level of transcripts available for translation (12, 57). Thus, competition between P-bodies and the translational apparatus impacts the regulation of gene expression. P-bodies are also sites of microRNA-mediated repression (50); however, S. cerevisiae lacks the homologous RNA interference pathways conserved in many other eukaryotes.
In addition, Ty1 possesses intrinsic mechanisms for regulating its propagation via the expression of cis-encoded antisense RNAs. These antisense RNAs have been reported to decrease retrotransposition by repressing transcription (7) or inhibiting accumulation of RT and IN (43). Antisense RNA degradation is regulated by cellular RNA surveillance pathways, which include the P-bodies. Therefore, determining where Ty1 mRNA and antisense RNAs localize in the cell and whether the lack of individual P-body components affects their localization may shed light on the inhibition of transposition by antisense RNAs and the molecular basis of transpositional dormancy. In this study, we show that several P-body components are required for the accumulation of Ty1 mRNA and Gag in discrete cytoplasmic foci, which enhances efficient proteolytic processing of Gag, accumulation of IN and RT, and formation of VLP clusters that contain retrotransposition-competent VLPs. Interestingly, Kem1p may affect retrotransposition by controlling the interactions between Ty1 mRNA and antisense transcripts.
MATERIALS AND METHODS
Yeast strains, plasmids, and growth conditions.
The yeast strains used in this study are listed in Table 1. The haploid MATα deletion strains (26) derived from BY4742 (8) were obtained from Open Biosystems (Huntsville, AL). SPT3, DHH1, and KEM1 were deleted in BY4742 to create DG2247, MAC532, and MAC103, respectively. Gene disruptions were carried out using the KanMX4 cassette (60). The KanMX disruption cassettes were PCR amplified using template DNA extracted from deletion mutants with gene-specific primers A and D (Saccharomyces Genome Database, http://www-sequence.stanford.edu/group/yeast_deletion_project/Deletion_primers_PCR_sizes.txt). Correct gene disruptions by KanMX for all of the deletion mutant strains used in this study were confirmed by PCR using locus-specific primers. BY4742 and isogenic mutants were transformed with pBJC573 (11, 53), a URA3-based integrating plasmid carrying a complete Ty1 element marked with a modified indicator gene, his3-AId1 (abbreviated as his3-AI) (18, 48), to create DG2122 and isogenic mutants MAC535, MAC353, MAC450, MAC453, and DG2277. P-body proteins encoded by DCP2, DHH1, KEM1, and PAT1 were tagged with three hemagglutinin (HA) epitopes at the C terminus using plasmid pFA6a-3HA-KanMX6 (kindly provided by M. Longtine) (40). Correct tagging with HA was verified by PCR analysis using gene-specific primers and Western blot analysis. A Ty1-less S. paradoxus strain (DG1768) which is devoid of Ty1, Ty2, Ty4, and Ty5 elements (25, 45) served as a negative control. For overexpression of Ty1 driven by the GAL1 promoter, BY4742, MAC103, MAC532, YSC1021-551146, and YSC1021-549835 were transformed with pBDG945 (pGTy1-H3his3-AId1), which is compatible with the his3-Δ1 allele present in BY4742 (48, 53), to create DG2027, MAC350, MAC584, MAC587, and MAC590, respectively. Strains DG2027, MAC350, MAC587, MAC590, and DG1781, which contains plasmid pBLR96 (pGTy1-H3his3AI) (52), were used for analysis of VLPs by immunoelectron microscopy (IEM). For in vitro transcription of strand-specific riboprobes, pMACB22-1 was constructed by amplification of the 5′ end of Ty1H3 GAG with primers TyA-316For (CCGGAATTCAACATTCACCCAATTCTCATGG [EcoRI site underlined]) and TyA-646Rev (GGACAGATCTAGGTGGAAAGTACATAGGCG [BglII site underlined]), followed by ligation into pSP70 (Promega, Madison, WI). Yeast strains were grown in synthetic complete (SC) or SC-ura medium (54) supplemented with 2% glucose at 20°C unless otherwise noted. For induction of a Ty1 element driven by the GAL1 promoter, strains containing pBDG945 or pBLR96 were grown in SC-ura medium containing 2% raffinose (pH 6.5) at 30°C until early log phase and then placed in SC-ura medium containing 2% galactose and grown at 20°C for 16 h. For glucose deprivation, cells were grown in SC medium containing 2% glucose at 20°C until log phase, washed with SC containing glucose (controls) or without glucose, and then resuspended in SC medium with or without glucose and incubated for 25 min.
TABLE 1.
Yeast strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| BY4742 (wild type) | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0 | 8 |
| DG1768 (Ty1-less) | MATα his3-Δ200 hisG ura3 gal3 Ty1-less Spo− (S. paradoxus) | 25 |
| DG1781 (Ty1-less) | DG1768(pBLR96)a | 25 |
| DG2027 | BY4742(pBDG945)b | This study |
| DG2122 | BY4742(pJC573)c | 48 |
| DG2247 | BY4742 spt3Δ::KanMX | 48 |
| DG2277 | DG2247(pJC573) | 48 |
| MAC532 | BY4742 (YDL160C) dhh1Δ::KanMX | This study |
| YSC1021-551146 | BY4742 (YJL124C) lsm1Δ::KanMX | Open Biosystems (26) |
| YSC1021-549835 | BY4742 (YCR077C) pat1Δ::KanMX | Open Biosystems (26) |
| MAC103 | BY4742 (YGL173C) kem1Δ::KanMX | This study |
| MAC353 | BY4742 (YGL173C) kem1Δ::KanMX(pJC573) | This study |
| MAC535 | BY4742 (YDL160C) dhh1Δ::KanMX(pJC573) | This study |
| MAC450 | BY4742 (YJL124C) lsm1Δ::KanMX(pJC573) | This study |
| MAC453 | BY4742 (YCR077C) pat1Δ::KanMX(pJC573) | This study |
| MAC421 | BY4742 DCP2-3HA:KanMX6 | This study |
| MAC424 | BY4742 DHH1-3HA:KanMX6 | This study |
| MAC428 | BY4742 KEM1-3HA:KanMX6 | This study |
| MAC432 | BY4742 PAT1-3HA:KanMX6 | This study |
| MAC466 | MAC421(pJC573) | This study |
| MAC468 | MAC424(pJC573) | This study |
| MAC471 | MAC428(pJC573) | This study |
| MAC473 | MAC432(pJC573) | This study |
| MAC350 | MAC103(pBDG945) | This study |
| MAC584 | MAC532(pBDG945) | This study |
| MAC587 | BY4742 (YJL124C) lsm1Δ::KanMX(pBDG945) | This study |
| MAC590 | BY4742 (YCR077C) pat1Δ::KanMX(pBDG945) | This study |
Rate of Ty1 mobility.
Ty1his3-AI mobility assays were determined as previously described (48). To determine the rate of Ty1 mobility from endogenous elements, ∼103 cells were inoculated into five individual 1-ml SC-ura liquid cultures containing 2% glucose and grown at 20°C until saturation. For strains containing pGTy1, cells were grown in SC-ura medium containing 2% galactose at 20°C until saturation. Dilutions of each cell suspension were spread on SC-ura and SC-ura-his plates to obtain Ura+ and His+ CFU titers, respectively. The total Ura+ CFU titer and the His+ CFU titers were calculated for each 1-ml culture. The rate of Ty1his3AI mobility was calculated as previously described using the median frequency from the Drake equation (51). Standard deviations were calculated from at least three independent experiments.
Analysis of Ty1 integration events upstream of SUF16.
Wild-type strain DG2122 and isogenic deletion mutant strains, grown to late-log phase, were analyzed for de novo Ty1 integration events upstream of SUF16 (tRNAGly) by PCR using 1.5 μg of genomic DNA as previously described (48). A PCR control for each DNA sample was performed using primers specific for CPR7.
Southern blot analysis of Ty1 cDNA.
The level of unintegrated Ty1 cDNA from wild-type strain DG2122 and isogenic deletion mutant strains, grown to late-log phase, was determined by Southern analysis using 2 μg of genomic DNA digested with PvuII (35, 48). The intensity of the unincorporated PvuII cDNA fragment was determined by analysis with a Typhoon Trio PhosphorImager (GE Healthcare, Piscataway, NJ) and ImageQuant software (GE Healthcare). The unincorporated cDNA fragment was normalized to four conserved Ty1 chromosomal junction fragments detected in the same blot. The relative percentage of unintegrated cDNA was defined as the ratio of the normalized intensity of the mutant cDNA to that of the wild type. Standard deviations were calculated from three different samples analyzed per strain.
Northern blot analysis of Ty1 transcripts.
Total RNA was extracted from wild-type strain BY4742 and isogenic deletion mutant strains grown to mid-to-late log phase and analyzed by Northern blotting as previously described (48). For the detection of Ty1 mRNA and antisense RNAs, strand-specific riboprobes were synthesized in vitro using SP6 or T7 RNA polymerase, linearized MACB22-1 plasmid, [α-32P]UTP, and the MAXIscript in vitro transcription kit (Applied Biosystems, Foster City, CA) by following the manufacturer's instructions. Strand-specific riboprobes were treated with DNase I and purified using a NucAway spin column (Applied Biosystems). Levels of Ty1 mRNA or antisense RNA bands were normalized to the 18S and 25S rRNAs visualized by ethidium bromide staining. Hybridization and ethidium bromide fluorescence signals were determined using a Typhoon Trio PhosphorImager and ImageQuant software. Relative levels of Ty1 RNA were defined as the ratio of the mutant RNA signal to that of the wild type. Standard deviations were calculated from at least three independent experiments.
Western blot analysis of Ty1 proteins.
To detect endogenous Ty1 Gag proteins, total protein was extracted from cells at an optical density at 600 nm (OD600) of 25 for wild-type strain BY4742 and isogenic mutants grown to mid-log phase as previously described (34). Total protein was extracted from cells of strains containing pGTy1 at an OD600 of 25 after ∼16 h of galactose induction at 20°C. Samples were separated electrophoretically on a 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel, followed by transfer to polyvinylidene difluoride membranes using an iBlot transfer apparatus (Invitrogen, Carlsbad, CA). Membranes were blocked in 5% milk-Tris-buffered saline containing 0.1% Tween 20 (TBS-T) for 1 h at room temperature and then incubated with primary antibody for 1 h. Gag was detected with a rabbit anti-Ty1 VLP antibody in TBS-T at a 1:15,000 dilution. Ty1 IN and RT were detected with rabbit anti-IN (B2) and rabbit anti-RT (B8) antibodies, respectively, at a 1:7,000 dilution in TBS-T. Blots were washed three times for 10 min each time in TBS-T. Primary antibodies were detected with horseradish peroxidase-conjugated donkey anti-rabbit immunoglobulin G (IgG; GE Healthcare, Pittsburgh, PA) at a 1:7,000 dilution in TBS-T for 1 h, washed as described above, visualized by enhanced chemiluminescence (Perkin-Elmer, Waltham, MA), and exposed to X-ray film (BioMax XAR; Kodak, Rochester, NY). The ratio of mature (p45) to precursor (p49) Gag was determined by densitometry using Quantity One software (Bio-Rad Laboratories, Hercules, CA). Standard deviations were calculated from three independent experiments.
Oligonucleotide probes used for fluorescence in situ hybridization (FISH).
Ty1 mRNA was detected with either the antisense TyA-322 (5′-GACTTCCTTAGAAGTAACCGAAGCACAGGCGCTACCATGAGAATTGGGTG-3′) or the antisense RT-4219 (5′-TGATGTACGGTATTGGATTGCATGCCTGAGTCGTAAGTGTCAGGATGCTG-3′) oligonucleotide. Ty1 antisense RNAs were detected using a sense TyA-340 oligonucleotide (5′-GCGCCTGTGCTTCGGTTACTTCTAAGGAAGTCCACACAAATCAAGATCCG-3′). ASH1 mRNA was detected using the oligonucleotides ASH1-433 antisense (5′-TGTTGTGGGCGCTCCGGTCTCTTAGATAAAGAGACGGACGATAGCCTGTC) and ASH1-884 antisense (5′-GGCCTCTACTCCTCTAAGCGTCGTGCGCTCCATGGTTCTATTGGTTGGTG).
All of the oligonucleotides were synthesized and high-performance liquid chromatography purified by Invitrogen. Oligonucleotides were 3′ end labeled with digoxigenin (DIG)-dUTP using the DIG Oligonucleotide Tailing Kit, 2nd Generation (Roche Applied Science, Indianapolis, IN) in accordance with the manufacturer's instructions. Labeled oligonucleotides were purified by lithium chloride precipitation and resuspended in a Tris-EDTA buffer (pH 8). A sense TyA-340 oligonucleotide labeled with DIG and an antisense RT-4219 oligonucleotide labeled at the 5′ end with Alexa Fluor 594 (Invitrogen) were used for simultaneous detection of both Ty1 antisense RNAs and mRNA.
Antibodies used for immunofluorescence microscopy (IF).
All antibodies used for IF were diluted in blocking reagent (Roche Applied Sciences). To detect Gag, a rabbit anti Ty1-VLP antibody (kindly provided by Alan Kingsman) was used at a 1:1,000 dilution (63). Proteins were visualized with Alexa Fluor 594-conjugated goat anti-rabbit IgG (2 mg/ml; Invitrogen) at a 1:100 dilution. To detect DIG-labeled probes, a fluorescein isothiocyanate (FITC)-conjugated sheep anti-DIG Fab fragment (Roche Applied Sciences) was used at 1 ng/μl.
FISH and IF conditions.
Procedures adapted for this study were described previously (38, 46). Briefly, log-phase cells were fixed in growth medium with 4% formaldehyde at growth temperature for 1 h and then washed with 0.1 M KPO4 (pH 6.5) and P solution (1.2 M sorbitol, 0.1 M KPO4, pH 6.5). About 1 × 108 cells in P solution were treated with 25 mM dithiothreitol at 24°C for 10 min and 200 μg/ml Zymolyase-100T (USBiological, Swampscott, MA) for 30 to 50 min. Spheroplasts were washed with P solution and transferred to slides coated with CLEARCELL Bio Adhesive (Thermo Scientific, Portsmouth, NH). Nonadherent cells were removed by aspiration. Cells were permeabilized with 0.5% Nonidet P-40 for 5 min, washed with P solution, and treated with 0.1 M triethanolamine hydrochloride (TEA), pH 8.0, for 2 min and with 0.25% acetic anhydride in 0.1 M TEA for 10 min. Cells were prehybridized in 50% formamide-4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M Na citrate)-1× Denhardt's solution-0.25 mg/ml S. cerevisiae tRNA-0.25 mg/ml denatured salmon sperm DNA-5 mM EDTA-0.1 mg/ml poly(A) for 1 h at 37°C in a humid chamber. Hybridization was carried out in the same solution containing 150 ng of Alexa Fluor 594-labeled oligonucleotide and/or 2.5 to 25 pmol of DIG-labeled oligonucleotide overnight at 40°C. Slides were washed once with 2× SSC (30 min at 40°C) and twice with 50% formamide in 2× SSC (15 min at 37°C), 1× SSC (5 min at 37°C), and 0.5× SSC (5 min at 23°C). Cells were treated with 0.1% SDS in 0.1 M sodium phosphate buffer for 2 min and rinsed with phosphate-buffered saline (PBS). For IF, cells were blocked with blocking reagent (Roche Applied Sciences) for 30 min at 23°C. To detect Gag, cells were incubated overnight at 4°C with dilutions of the appropriate antibody as described above. Cells were washed with PBS containing 0.2% Tween 20. Detection of DIG and protein was done with anti-DIG FITC-conjugated Fab fragment and Alexa Fluor 594-conjugated anti-rabbit IgG for 1 h at 23°C. Cells were washed twice with 8% formamide in 2× SSC (5 min) and PBS containing 0.2% Tween 20 (5 min) at 37°C and rinsed with PBS. Slides were mounted using VECTASHIELD mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA). About 100 cells per strain for each experiment were scored to obtain quantitative data for localization of RNA and protein signal. Percentages of Ty1 mRNA and Gag foci were calculated from cells that were stained with DAPI. Standard deviations were calculated from more than three experiments.
IF.
Fluorescence imaging was performed using a Nikon Eclipse E-1000 microscope equipped with a Nikon C-CU Universal condenser, an oil immersion Nikon Plan Fluor DIC 100× objective, a Semrock GFP-3035 brightline zero-bandpass filter cube, a Semrock tetramethyl rhodamine isocyanate filter cube, a UV-2E/C DAPI filter cube, and a Hamamatsu Orca-ER c4742-95 cooled charge-coupled device camera (Hamamatsu, Japan). Images were acquired using Openlab 5.0 software (Improvision, Lexington, MA). Fluorescent images were merged with Openlab to give the composite images shown. The exposure times used to capture fluorescent and DAPI images were kept consistent throughout each experiment. Figures were constructed with Adobe Photoshop software (Adobe Systems, Mountain View, CA).
Statistical analyses of colocalization of Ty1 mRNA and antisense RNAs.
The statistical significance of colocalization of Ty1 mRNA and antisense RNAs was assessed using a method based on a Monte Carlo simulation (14, 31, 32, 36). Briefly, the population of cells observed to have colocalization of Ty1 mRNA (foci) with Ty1 antisense RNAs (puncta) is defined as Nobs. If Nobs was significantly greater than the number of cells with colocalization expected to occur by chance, then it was deduced that there was a significant association of foci and puncta. Statistical significance was determined by calculating the probability, P, that Nobs could have occurred by chance assuming that the puncta were randomly located in each cell and that there was one focus per cell. To calculate P, the distribution of probabilities for the occurrence of each plausible value of Nobs (range, 0 to the number of cells in the population) was generated by computer simulation. Then, P was given by the integral under the distribution between the actual (empirical) value of Nobs and the number of cells in the population. A P value of <0.01 indicated that Nobs was significantly greater than the number of cells with colocalizations expected to occur by chance and implied that there was a specific association between foci and puncta in the cells.
Visual inspection of the microscopic images obtained led to three conclusions that simplified the subsequent analysis of significance. (i) About 30% of the cells had one mRNA focus, and only about 2% had more than one focus. Thus, analysis could be restricted to cells with exactly one focus. (ii) Ty1 antisense RNA puncta were significantly smaller than Ty1 mRNA foci and could be approximated as infinitely small points. Cells have variable numbers of puncta (range, 0 to 13). For each cell, the puncta were enumerated and it was determined whether a punctum colocalized with the focus. (iii) For each cell, the ratio of the area of the focus (Af) to the area of the cell (Ac) appeared similar across the cell population. To confirm this, the areas of individual cells and foci were measured from the microscope images using in-house image segmentation software, from which Af/Ac was calculated for each cell. The mean and standard deviation of Af/Ac across the population were determined to be 0.0213 ± 0.0022 using the method of van Kempen and van Vliet (58). The relatively low coefficient of variation of this mean ratio (10%) indicated that it was reasonable to approximate the ratio as the same for all of the cells.
Given simplifications ii and iii, the probability of chance colocalization of a focus and a punctum in a given cell was 0.0213 = p and, conversely, the probability of no colocalization was 1 − p = q. Therefore, for a cell, i, with Ni puncta, the probability of no colocalizations was qNi. A Matlab script generated the probability distribution for the number of cells with chance colocalization assuming that puncta are randomly located in each cell. Across all of the cells in the population, the script counted the number of times a uniformly distributed random number between 0 and 1, ri, was less than 1 − qNi, which equals the number of chance colocalizations. To generate a statistically accurate distribution, the script cycled through the population 10,000 times and accumulated the results. The probability, P, that the number of observed colocalizations, Nobs, could have occurred by chance was given by integral under the probability distribution between Nobs and the number of cells in the population. A P value of <1% indicated that Nobs was significantly greater than the number of cells with colocalizations expected to occur by chance and implied that there was a specific association between the foci and puncta in the cells. To ensure that P was not significantly influenced by the variation of Af/Ac across the population, P was recalculated by using a P value of 0.0279, which is the mean of Af/Ac plus 3 times the standard deviation.
IEM and transmission electron microscopy (TEM).
Strains were grown to log phase for detection of endogenous or pGTy1-induced VLPs. For IEM, cells were fixed with 4% formaldehyde-0.05% glutaraldehyde in PBS for 2 h at 4°C and washed three times with cold PBS, once with 0.1 M KPO4 (pH 6.5), and once with P solution. Cells were spheroplasted as described for FISH-IF and washed twice with PBS. Cell pellets were dehydrated in subsequent ethanol dilutions: twice for 10 min each with 35% ethanol, 50% ethanol, 70% ethanol, and 95% ethanol and three times with 100% ethanol. Pellets were incubated at 4°C in 2 volumes of 100% ethanol with 1 volume of LR White (Polysciences, Inc., Warrington, PA) for 1 h, in 1 volume of ethanol with 2 volumes of LR White for 1 h, and in 100% LR White overnight. Pellets were embedded in BEEM capsules with new LR White and cured in a 55°C oven for 24 h. For TEM, cells were fixed with 4% formaldehyde-2% glutaraldehyde in 0.1 M sodium cacodylate for 2 h at 4°C and washed three times with cold PBS, once with 0.1 M KPO4 (pH 6.5), and once with P solution. Cells were spheroplasted as described for FISH-IF, except that the Zymolyase treatment was carried out for at least 33 h at 35°C. Cells were then washed three times with PBS and postfixed with 1% osmium in 0.1 M sodium cacodylate, followed by en bloc staining with 0.5% uranyl acetate in 0.1 M acetate buffer. Cell pellets were dehydrated in subsequent ethanol dilutions as described for IEM and washed in 100% propylene oxide, and resin infiltration was carried out in an equal volume of propylene oxide and epoxy resin. The next day, pellets were embedded in BEEM capsules with pure resin and cured at 55°C for 48 h. Thin sections (80 to 90 nm) were made with an ultramicrotome (Leica, Inc., Bannockburn, IL) equipped with a diamond knife (Diatome, Inc., Hatfield, PA). Thin sections were mounted on 200-mesh nickel grids and blocked in blocking buffer (EMS, Inc., Hatfield, PA) for 30 min. Primary antibody and immunogold-conjugated secondary antibody (EMS, Inc.) incubations were carried out at room temperature in serial dilutions. Grids were stained in uranyl acetate, followed by lead citrate to enhance contrast. Sections were examined in an H7600 electron microscope (Hitachi, Inc., Tokyo, Japan), and digital images were taken with a 4K camera (AMT, Lawley, MA). For VLP analysis by TEM, ∼100 cells of similar size (transverse cross section) for each strain were examined for VLPs clustered (>10 VLPs), dispersed, or not present.
RESULTS
P-body components are required for Ty1 retrotransposition.
A genome-wide screen of haploid S. cerevisiae gene deletion mutants has identified factors involved in Ty1 mobility (48) (K. M. Nyswaner and D. J. Garfinkel, unpublished results). Among the host cofactor genes required for Ty1 mobility were DHH1, KEM1, LSM1, and PAT1, which encode P-body components. To assess the contribution that each P-body component makes to retrotransposition, we determined the rate of Ty1his3-AI mobility (18, 48) in the wild-type and isogenic deletion mutant P-body components. Ty1 mobility is measured by the number of His+ colonies that are formed, relative to the total number of viable cells, due to splicing of the artificial intron (AI) within his3-AI, followed by reverse transcription of a tagged mRNA. Deletion of DHH1, LSM1, and PAT1 decreased Ty1 mobility 8- to 16-fold (Table 2). Strikingly, His+ colonies were not detected in several independent Ty1 mobility assays of the kem1Δ mutant because mobility decreased beyond the limit of detection of this assay (>178-fold). An isogenic spt3Δ mutant that is defective for transcription of chromosomal Ty1 elements (62) served as a negative control.
TABLE 2.
Ty1his3-AI mobility in P-body component mutants
| Disrupted gene | Strain | Mean Ty1 mobility rate (107) ± SD | Relative mobility ratea (fold decrease) |
|---|---|---|---|
| None (wild type) | DG2122 | 1.23 ± 0.3 | 1 |
| dhh1Δ | MAC535 | 0.149 ± 0.05 | 8 |
| kem1Δ | MAC353 | <0.007 | >178 |
| lsm1Δb | MAC450 | 0.077 ± 0.05 | 16 |
| pat1Δb | MAC453 | 0.102 ± 0.07 | 12 |
| spt3Δ | DG2277 | 0.043 ± 0.01 | 29 |
Mobility rate relative to that in wild-type strain DG2122 (P < 0.0005).
LSM1 and PAT1 deletion mutations reintroduced into BY4742 do not produce a difference in Ty1 mobility compared to MAC450 and MAC453, respectively.
Ty1 mobility occurs by cDNA recombination or transpositional integration, which typically takes place upstream of genes transcribed by RNA polymerase III, such as tRNA genes (29). These integration events can be detected by PCR amplification of the junction between Ty1 and the tRNAGly gene (SUF16) on chromosome III (Fig. 1A) (35). In the wild-type strain, integration events are visualized as a ladder of PCR fragments between 0.5 and 1.5 kb. The intensity of these PCR products provides a qualitative measurement of de novo integration. Using this approach, all four P-body component mutants showed a reduced level of Ty1 insertions upstream of SUF16 (Fig. 1B). Insertions were not detected in the kem1Δ mutant. Therefore, P-body components are required for Ty1 retrotransposition.
FIG. 1.
Ty1 integration events upstream of the SUF16 tRNAGly gene are reduced in P-body component mutants. (A) Schematic representation depicting the region where Ty1 elements integrate upstream of SUF16 on chromosome III. LTRs are represented by hatched triangles. PCR using TyB Out and SNR33 Out primers (black horizontal arrows) yields products derived from the junctions of de novo Ty1 integration upstream of SUF16. For example, the vertical black arrow represents the location of a new Ty1 integration, which would generate a PCR product of 780 bp. PCR products range between 780 bp and ∼1,400 bp, with a periodicity of ∼80 bp (21). (B) Ty1 integration assay for wild-type (wt) strain DG2122 and isogenic P-body component mutants. An isogenic spt3Δ mutant, DG2277, served as a negative control. Three independent colonies were analyzed for each strain, and PCR products were resolved on 1.5% agarose gels, stained with ethidium bromide, and visualized by fluorescence imaging. Below are control PCR using primers specific for the CPR7 gene. Molecular size standards (kilobases) are alongside the gels.
P-body components affect Ty1 retrotransposition posttranscriptionally.
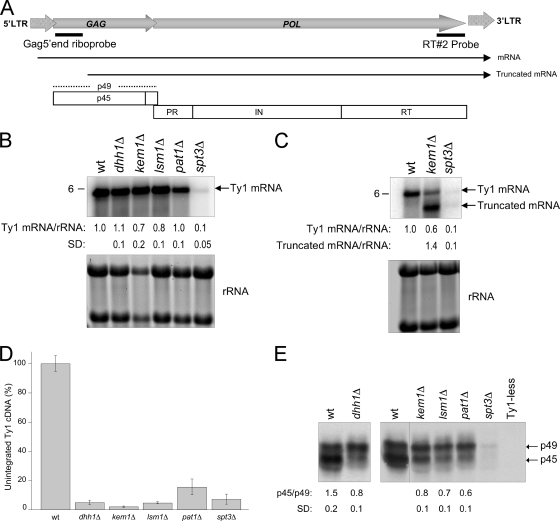
Northern blot analysis using total RNA and a 32P-labeled, strand-specific riboprobe corresponding to the 5′ end of GAG was used to determine Ty1 mRNA levels (Fig. 2A). RNA signals were normalized to the level of 18S and 25S rRNAs. The decrease in levels of endogenous full-length Ty1 mRNA was less than 1.4-fold in the dhh1Δ, kem1Δ, lsm1Δ, and pat1Δ mutants relative to that in the isogenic wild-type strain (Fig. 2B). Ty1 mRNA levels were reduced more than 10-fold in the spt3Δ mutant (62). A 5′-end-truncated form of Ty1 mRNA was previously reported in kem1Δ and spt3Δ mutants (7, 62). We used a probe corresponding to the 3′ end of RT to determine levels of the 5′-end-truncated mRNAs. In the kem1Δ mutant, the truncated mRNA level was 1.4-fold higher than that of the wild-type full-length Ty1 mRNA (Fig. 2C). The modest decrease in Ty1 mRNA levels in P-body component mutants was not consistent with the significant decrease in Ty1 mobility. To determine whether deletion of P-body components affects cDNA accumulation or PR function, we examined the levels of unintegrated Ty1 cDNA by Southern blot analysis and Gag processing by Western blotting. cDNA levels of all four P-body component mutants were significantly decreased (ranging from 2 to 15%) relative to those of the isogenic wild-type strain (Fig. 2D). The kem1Δ mutant displayed the lowest cDNA levels. Levels of precursor (p49) and mature (p45) Gag are a sensitive indicator of PR function, since Gag production and processing are critical for VLP maturation and reverse transcription (1, 63). Gag proteins were detected in all four P-body component mutants (Fig. 2E). The wild-type strain had more p45 than p49 Gag (p45/p49 ratio of 1.5), as determined by densitometric scans from three independent experiments. In contrast, a modest increase in the proportion of precursor p49 Gag was detected in the P-body component mutants, with a p45/p49 ratio of 0.6 to 0.8. These results suggest that Dhh1p, Kem1p, Lsm1p, and Pat1p act posttranscriptionally to enhance Ty1 cDNA accumulation and Gag processing.
FIG. 2.
P-body components are required for Ty1 retrotransposition posttranscriptionally. (A) Schematic representation of a Ty1 element showing the positions of mRNAs, hybridization probes, precursor (p49) and mature (p45) Gag proteins, and mature Pol proteins PR, IN, and RT. (B) Northern analysis of P-body component mutants using a 32P-labeled Gag 5′-end riboprobe. Below the blot is the amount of Ty1 mRNA relative to the 18S and 25S rRNAs after normalization to the wild-type (wt) transcript level. An spt3Δ mutant (DG2247) served as a negative control. SD, standard deviation from at least three independent experiments. (C) Northern analysis of Ty1 transcripts detected in a kem1Δ mutant using a radiolabeled RT#2 probe. Note that a 5′-truncated Ty1 transcript is detected in the kem1Δ mutant. Below the blot are the levels of full-length and 5′-truncated mRNAs relative to the level of full-length mRNA from the wild type. (D) Levels of unintegrated Ty1 cDNA are reduced in P-body component mutants. The graph summarizes the relative percentage of unintegrated Ty1 cDNA, as determined by Southern blot analysis. The decrease in Ty1 cDNA levels detected in the P-body mutants is expressed relative to the wild-type level. The error bars represent standard deviations from three independent cultures analyzed per strain. The level of unintegrated cDNA decreased at least 85% in the mutants compared with the wild-type level (P < 0.0001). (E) Levels of p49 and p45 in the P-body mutants. Total protein extracts from the wild type and P-body mutants, extracted from equivalent cell densities, were subjected to Western blot analysis using an anti-Ty1 VLP serum. Protein extracts from an spt3Δ mutant and a Ty1-less S. paradoxus strain (DG1768) were included as negative controls. The p45/p49 ratio was determined by densitometry. Standard deviations were calculated from three independent experiments (P < 0.005).
Colocalization of endogenous Ty1 mRNA and Gag requires P-body components and optimal conditions for retrotransposition.
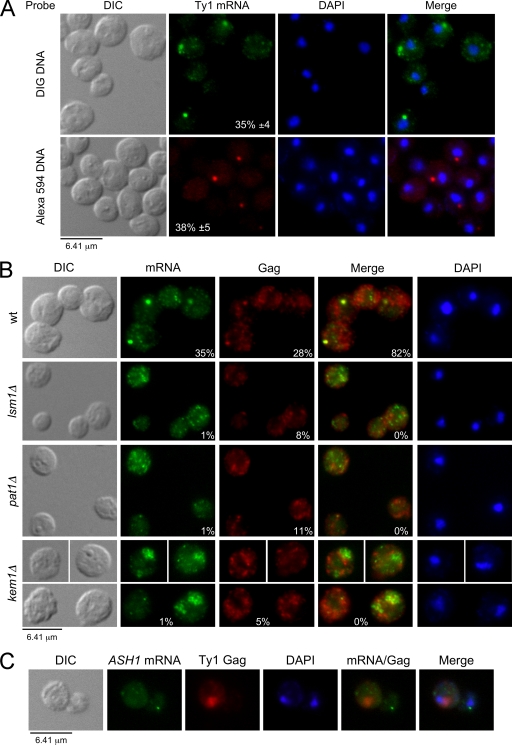
Given that P-body components are RNA binding proteins and are required for localization of VLP assembly intermediates of Ty3 (6), we determined their role in the localization of endogenous Ty1 mRNA and Gag by FISH and IF. Ty1 mRNA was detected using oligonucleotide probes labeled with DIG or Alexa Fluor 594, and Gag proteins were detected with an anti-VLP antibody. When the wild-type strain (BY4742) was grown at 20°C, which is the optimal temperature for Ty1 retrotransposition (49), endogenous Ty1 mRNA localized in a discrete cytoplasmic focus (>0.4 μm2) in ∼35% of the cells (Fig. 3A). The percentages of cells containing Ty1 mRNA foci detected with DIG- or Alexa Fluor-labeled probes were similar. However, the DIG detection method was more sensitive, since mRNA could also be detected in cytoplasmic puncta (<0.2 μm2) throughout most of the cells. Using the same conditions for detecting mRNA, Gag also localized in a discrete cytoplasmic focus in ∼28% of the cells (Fig. 3B). Gag foci colocalized with Ty1 mRNA in ∼82% of the cells that contained mRNA foci.
FIG. 3.
Localization of Ty1 mRNA and Gag in discrete cytoplasmic foci requires P-body components. Cells were grown in SC-glucose (2%) medium at 20°C and visualized by differential interference contrast (DIC) microscopy. Ty1 mRNA and Gag were detected by FISH-IF, and DNA was stained with DAPI. Representative images are shown. (A) Ty1 mRNA localizes in discrete cytoplasmic foci in wild-type (wt) BY4742 cells. Ty1 mRNA was detected using a probe corresponding to the RT region labeled with either DIG or Alexa Fluor 594. Merged Ty1 mRNA and DAPI staining is shown. Mean percentages of cells containing cytoplasmic Ty1 mRNA foci ± the standard error of the mean (SEM) calculated from more than six independent experiments are included. (B) P-body components are required for the formation of discrete Ty1 mRNA/Gag foci. Ty1 mRNA and Gag for the wild-type strain and isogenic P-body component mutants were detected using a DIG-labeled probe corresponding to the GAG sequence and an anti-Ty1 VLP serum, respectively. Merged Ty1 mRNA and Gag staining is shown. (C) Localization of ASH1 mRNA and Gag as a control for FISH-IF specificity. ASH1 mRNA was detected using ASH1-433 and -884 oligonucleotides labeled with DIG. Gag was detected as described for panel B. Merged ASH1 mRNA, Gag, and DAPI staining is shown.
In contrast, Ty1 mRNA/Gag localization was dramatically different in P-body component mutants (Fig. 3B). In the lsm1Δ and pat1Δ mutants, Ty1 mRNA localized in puncta dispersed throughout the cytoplasm. In these mutants, Gag was also detected throughout the cytoplasm in most of the cells and colocalization with mRNA foci was not detected. In the kem1Δ mutant, full-length and 5′-truncated mRNAs localized mainly in cytoplasmic aggregates (∼20% cells). As opposed to foci, these aggregates appeared to be a disorganized (nonspherical) accumulation of puncta with a nonuniform signal intensity. Gag rarely localized in foci but instead had a punctate distribution throughout the cytoplasm. As a control for specificity, Gag did not colocalize with ASH1 mRNA, which is found at the distal tip of buds (Fig. 3C) (39). Our results suggest that Lsm1p, Pat1p, and Kem1p are all required for the formation of discrete Ty1 mRNA/Gag cytoplasmic foci. Moreover, the kem1Δ mutant displays a distinct Ty1 mRNA/Gag localization phenotype compared to the other mutants analyzed. Kem1p appears to help form compact Ty1 mRNA/Gag foci.
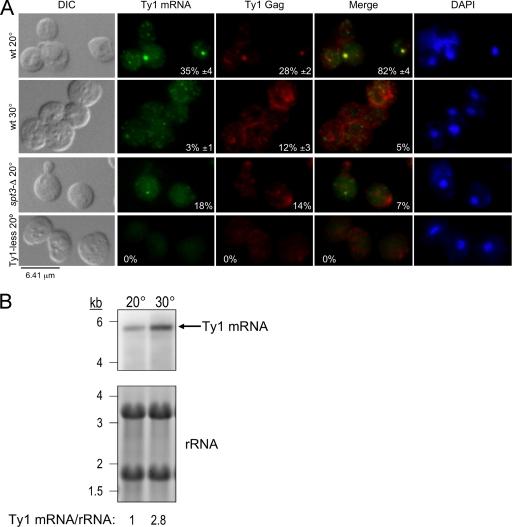
To determine whether the presence of Ty1 mRNA/Gag foci correlates with retrotransposition, FISH-IF was repeated with wild-type cells grown at a suboptimal temperature for retrotransposition (30°C). At this temperature, Ty1 mRNA and Gag could be detected; however, they were localized mainly in disperse cytoplasmic puncta (Fig. 4A). Colocalization of Ty1 mRNA with Gag was observed in only 5% of the cells despite the presence of threefold more mRNA (Fig. 4B) and detectable levels of Gag by Western blotting (M. A. Checkley and D. J. Garfinkel, unpublished data) in cells grown at 30°C than in cells grown at 20°C. As a negative control for retrotransposition, we examined an isogenic spt3Δ mutant where Ty1 mRNA (full length and 5′ truncated) and Gag localized in small faint foci with very little colocalization (7% of the cells; Fig. 4A), which may be due to reduced mRNA and Gag levels. Signals for mRNA or Gag were not detected in the Ty1-less S. paradoxus strain (DG1768), which served as another negative control. These results show a positive association between the presence of mRNA/Gag foci and retrotransposition.
FIG. 4.
Localization of Ty1 mRNA and Gag in discrete cytoplasmic foci requires optimal conditions for retrotransposition. Cells were grown in SC-glucose (2%) medium at 20°C, unless otherwise noted, and visualized by differential interference contrast (DIC) microscopy. Ty1 mRNA and Gag were detected by FISH-IF, and DNA was stained with DAPI. Representative images are shown. (A) Detection of Ty1 mRNA and Gag under conditions optimal and suboptimal for retrotransposition. Conditions optimal for retrotransposition are present when the wild type is grown at 20°C. Conditions suboptimal for retrotransposition are present when the wild type is grown at 30°C. An spt3Δ mutant (DG2247) and a Ty1-less S. paradoxus strain (DG1768) grown at 20°C served as negative controls. Ty1 mRNA and Gag were detected as described in the legend to Fig. 3B. Merged Ty1 mRNA and Gag staining is shown. Mean percentages of cells containing cytoplasmic Ty1 mRNA foci, Gag foci, and their respective colocalization ± the SEM are included. (B) Northern blot analysis of Ty1 mRNA of wild-type strain BY4742 grown at 20°C and 30°C. Equivalent amounts of total RNA were hybridized with a 32P-labeled riboprobe corresponding to the 5′ end of Gag (see Fig. 2A). The signal detected for Ty1 mRNA was normalized to rRNA levels. Below the blot is the relative Ty1 mRNA level.
Since P-body components influence the formation of Ty1 mRNA foci/Gag, we determined if these foci are distinct from P-bodies. P-bodies are difficult to visualize in exponentially growing cells; however, when cells are deprived of glucose for a short period of time, P-bodies become foci visible by fluorescence microscopy (9, 10). We used strains in which P-body components (Dcp2p, Dhh1p, Kem1p, and Pat1p) were tagged with HA, as detected by Western blotting (data no shown), but had no marked effect in Ty1 mobility (Table 3). After 25 min of glucose deprivation, Ty1 mRNA and Gag no longer localized as a discrete focus in most cells but instead were dispersed in a punctate pattern throughout the cytoplasm (Fig. 5A), while tagged P-body components were now clearly visible (Checkley and Garfinkel, unpublished). The levels of Ty1 mRNA remained unchanged when cells were deprived of glucose (Fig. 5B), suggesting that the mRNA foci were disassembling. Moreover, Ty1 mRNA/Gag foci remain visible in cells treated with the protein synthesis inhibitor cycloheximide (Checkley and Garfinkel, unpublished), while P-bodies disassemble (55). These results support the notion that Ty1 mRNA/Gag foci are different from P-bodies, which is in agreement with a previous report (42).
TABLE 3.
Ty1his3-AI mobility in strains containing P-body proteins tagged with HA
| Relevant genotype | Strain | Ty1 mobility rate (107) | Relative mobility ratea (fold change) |
|---|---|---|---|
| Wild type | DG2122 | 1.27 | 1.00 |
| DCP2-HA | MAC466 | 0.58 | 0.46 |
| DHH1-HA | MAC468 | 1.19 | 0.94 |
| KEM1-HA | MAC471 | 2.34 | 1.85 |
| PAT1-HA | MAC473 | 0.86 | 0.68 |
Mobility rate relative to that in strain DG2122.
FIG. 5.
Ty1 mRNA/Gag foci are distinct from P-bodies. (A) Ty1 mRNA/Gag foci disassemble when P-bodies are activated. Wild-type (wt) cells were grown at 20°C in SC medium in the presence of glucose (+Glu) and then deprived of glucose (−Glu) for 25 min, which activates the formation of P-bodies. Ty1 mRNA and Gag were detected as described in the legend to Fig. 3B. (B) Levels of Ty1 mRNA in cells deprived of glucose. Northern blot analysis of Ty1 mRNA for the wild-type strain (BY4742) and isogenic HA-tagged derivatives grown in the presence of glucose (+Glu) and then deprived of glucose (−Glu) for 25 min. Equivalent amounts of total RNA were hybridized with a 32P-labeled riboprobe corresponding to the 5′ end of GAG (see Fig. 2A). The signal detected for Ty1 mRNA was normalized to rRNA levels. Below the blot is the level of Ty1 mRNA without Glu relative to that of Ty1 mRNA with Glu.
P-body components are required for formation of retrotransposition-competent VLPs.
To determine whether mRNA/Gag foci are sites for VLP assembly, we examined Gag localization by IEM. Despite the high levels of Ty1 mRNA, retrotransposition from endogenous elements occurs in 1 out of ∼107 cells and endogenous VLPs are rarely observed in cells by electron microscopy (EM) (24, 44). However, when Ty1 is overexpressed from the GAL1 promoter on a multicopy pGTy1 plasmid, retrotransposition increases dramatically and numerous VLPs are detectable by EM. Consequently, Gag localization was analyzed in uninduced cells and after induction of pGTy1 tagged with his3-AI. Prior to IEM, we determined whether mRNA and Gag localization was altered when Ty1his3-AI was overexpressed after galactose induction. FISH experiments that include Ty1his3-AI detect both endogenous Ty1 and Ty1his3-AI mRNAs, hereafter referred to as Ty1 mRNA. Under transposition-inducing conditions, mRNA and Gag localized in multiple cytoplasmic foci in ∼36% and ∼31% of the cells, respectively, with ∼90% colocalization (Fig. 6A). The increased number and size of mRNA/Gag foci observed are most likely due to higher levels of Ty1 mRNA and Gag after galactose induction. Therefore, overexpression of Ty1 does not alter the localization of mRNA/Gag in cytoplasmic foci or the fraction of the cells containing discrete foci. Interestingly, foci are present 2.5-fold more often in the mothers of budding cells than in unbudded cells, which suggests that the presence of Ty1 mRNA/Gag foci may be associated with the cell cycle or cell age.
FIG. 6.
Gag colocalizes with VLP clusters. (A) Comparison of mRNA/Gag foci by FISH-IF in cells expressing endogenous levels of Ty1 (BY4742) versus cells overexpressing a Ty1 element from pGTy1 (DG2027). Cells containing pGTy1 were induced with galactose for 16 h. Ty1 mRNA and Gag were detected as described in the legend to Fig. 3B. Merged mRNA and Gag staining, DNA staining with DAPI, and corresponding DIC images are shown. The mean percentages of cells containing cytoplasmic Ty1 mRNA foci, Gag foci, and the propensity of Ty1 mRNA and Gag foci to colocalize ± the SEM are indicated. (B) Localization of Gag by IEM in cells expressing endogenous levels of Ty1 (BY4742) and cells overexpressing a pGTy1 plasmid (DG2027 and DG1781) after 16 h of galactose induction. A Ty1-less S. paradoxus strain (DG1768) served as a negative control. Gag was detected with anti-Ty1 VLP serum and visualized with 10-nm gold particles conjugated to anti-rabbit IgG. Enlargements of sections containing Gag are shown, and representative VLPs are indicated with arrows. A black bar shows the scale of each image. N, nucleus.
In IEM analyses of uninduced wild-type cells, endogenous Gag protein was detected in small cytoplasmic clusters (Fig. 6B). Morphologically distinct VLPs were not detected from cells expressing endogenous Ty1 elements, which is in agreement with previous reports (24, 44). In contrast, Gag localized with VLP clusters (containing >10 VLPs) in cells overexpressing Ty1his3-AI. The apparent number of Gag clusters also increased when distinct VLPs were not detected. Gag detection could be influenced by the different epitopes that are exposed in fully assembled VLPs compared to assembly intermediates, due to conformational changes or interaction with other proteins and/or RNA. Ty1 Gag clusters were not observed in the Ty1-less S. paradoxus strain. Therefore, Ty1 mRNA/Gag foci detected by FISH-IF may be preferred sites for VLP assembly, since we observed an association of VLP clusters with Gag detected by IEM.
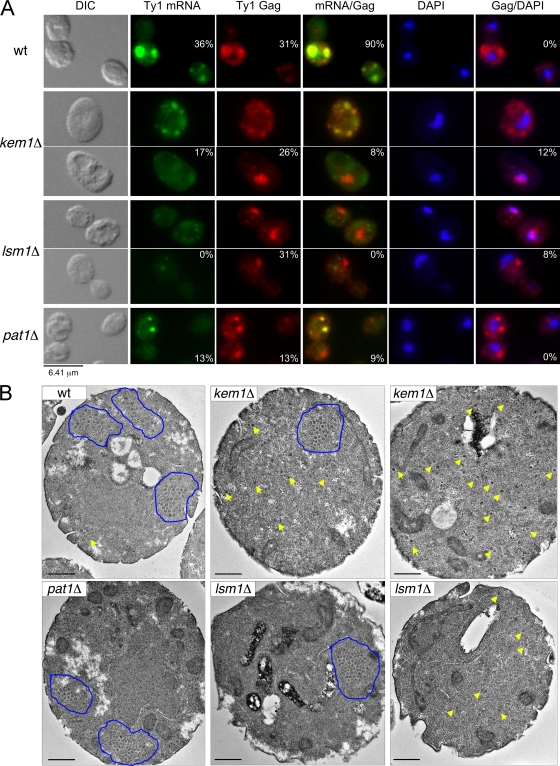
To address the role of P-body components in VLP assembly, we analyzed the effect of inducing pGTy1his3-AI in P-body deletion mutants with respect to Ty1 mobility, localization of mRNA and Gag, and formation of VLP clusters. First, we determined the rate of Ty1his3-AI mobility in P-body component mutants after galactose induction of pGTy1. Deletion of DHH1, KEM1, LSM1, and PAT1 decreased pGTy1his3-AI mobility 4- to 37-fold (Table 4). Both the kem1Δ and lsm1Δ mutants had the lowest levels of retrotransposition. Therefore, P-body components are required for mobility even when Ty1his3-AI is overexpressed. Next, we determined whether Ty1his3-AI overexpression alters mRNA and Gag localization by FISH-IF. Compared to the wild type, the kem1Δ, lsm1Δ, and pat1Δ mutants showed fewer or no detectable Ty1 mRNA foci or aggregates (Fig. 7A). Moreover, Ty1 mRNA localization was diffuse in most of the mutant cells. In each mutant, Gag was also detected diffusely throughout all of the cells, and the percentage of the cells showing Gag foci or aggregates (∼30%) was similar to that of the wild type (31%), except for the pat1Δ mutant (13%). The percentage of the cells where Ty1 mRNA and Gag foci or aggregates colocalized (0 to 9%) was significantly lower in all three mutants compared to the wild type (90%). Since Ty1 mRNA and Gag localized diffusely in all of the cells, it is likely that the apparent colocalization was more frequent and did not necessarily occur in foci or aggregates. Another phenotype observed in the kem1Δ and lsm1Δ mutants was the overlap of Gag foci with the nucleus, which was not observed in the wild type or the pat1Δ mutant. Analysis of Gag/nuclear overlap by confocal microscopy showed that Gag localized to the periphery of the nucleus (Checkley and Garfinkel, unpublished). Therefore, even when Ty1 was overexpressed, P-body components were still required for localization and/or transport of Ty1 mRNA and Gag to discrete cytoplasmic foci or aggregates.
TABLE 4.
Galactose-induced pGTy1his3-AI mobility in P-body component mutants
| Disrupted gene | Strain | Ty1 mobilityb rate (104) | Relative mobility ratea (fold decrease) |
|---|---|---|---|
| None (wild type) | DG2027 | 9.77 | 1 |
| dhh1Δ | MAC584 | 0.68 | 14 |
| kem1Δ | MAC350 | 0.32 | 31 |
| lsm1Δ | MAC587 | 0.26 | 37 |
| pat1Δ | MAC590 | 2.3 | 4 |
Mobility rate relative to that in wild-type strain DG2027.
Average from two independent mobility assays.
FIG. 7.
P-body components are required for the formation of retrotransposition-competent VLPs. (A) P-body components are required for colocalization of Ty1 mRNA/Gag in foci after galactose induction of a pGTy1 plasmid. Cells overexpressing pGTy1 (DG2027, MAC350, MAC587, and MAC590) were induced for 16 h. Ty1 mRNA and Gag were detected as described in the legend to Fig. 3B. Merged mRNA/Gag and Gag/DAPI staining, DNA staining with DAPI, and corresponding DIC images are shown. The percentages of cells containing cytoplasmic Ty1 mRNA and Gag foci and the propensity of Ty1 mRNA/Gag foci or aggregates to colocalize and Gag/DAPI overlap are indicated. (B) Localization of VLPs by TEM in P-body component mutants overexpressing a pGTy1 plasmid after 16 h of galactose induction. Representative cells containing VLP clusters outlined in blue (right and middle panels) and dispersed VLPs indicated by yellow arrows (right panel) are shown. The black bars represent 500 nm.
Since Gag was detected in association with VLP clusters by IEM in the wild type, we examined P-body component mutants. We used TEM to detect VLPs because it typically provides a sharper image than IEM. In the wild type and the P-body component mutants, VLPs were detected either in clusters or scattered throughout the cell (Fig. 7B). To determine the frequency of VLP cluster formation, we examined ∼100 cells for the presence of VLPs that were clustered, dispersed, or not detectable. Cells that contained both clustered and dispersed VLPs were scored as clustered. In the wild type, 18% of the cells contained clustered VLPs and 25% contained dispersed VLPs. In the kem1Δ and lsm1Δ mutants, fewer cells contained VLP clusters (9% and 4%, respectively), while 49% and 33% of the cells contained dispersed VLPs. In the pat1Δ mutant, 17% of the cells contained VLP clusters and 20% of the cells showed dispersed VLPs, similar to the wild type. The average diameter of VLPs expressed from the wild type (48.5 ± 6.1 nm) did not vary significantly from the kem1Δ (55.4 ± 6.7 nm), lsm1Δ (46.8 ± 7.6 nm), and pat1Δ (49.1 ± 6.1 nm) mutants. These results suggest that Kem1p and Lsm1p may play a role in maintaining VLP assembly within confined regions of the cell (clusters), rather than being dispersed. Furthermore, our data show that VLPs can assemble even in the absence of distinct Ty1 mRNA/Gag foci.
We measured IN and RT levels from total cell lysates of the kem1Δ and pat1Δ mutants by Western blotting, since these enzymes are critical for completing transposition after VLP assembly. These mutants were chosen because of their profound difference in Ty1 retrotransposition and Ty1 mRNA/Gag foci or VLP cluster formation. Strikingly, both the kem1Δ and pat1Δ mutants showed lower levels of mature IN and RT relative to the wild type when normalized to Gag (Fig. 8). In the kem1Δ mutant, the levels of IN were consistently at or below the limit of detection. Thus, although P-body component mutants can assemble VLPs, cellular levels of IN and RT were consistently diminished, which implies that these VLPs may be defective for Ty1 retrotransposition.
FIG. 8.
P-body components are required for accumulation of IN and RT. Levels of IN, RT, and Gag in P-body component mutants. Total protein extracts from the wild type (wt) and P-body component mutants overexpressing pGTy1 for 16 h, extracted from equivalent cell densities, were subjected to Western blot analysis using anti-IN, anti-RT, and anti-Ty1 VLP antibodies. Comparable levels of protein were loaded, as judged from Coomassie blue staining. Protein molecular size standards (kilodaltons) are indicated.
Localization of Ty1 antisense RNAs in P-body component mutants.
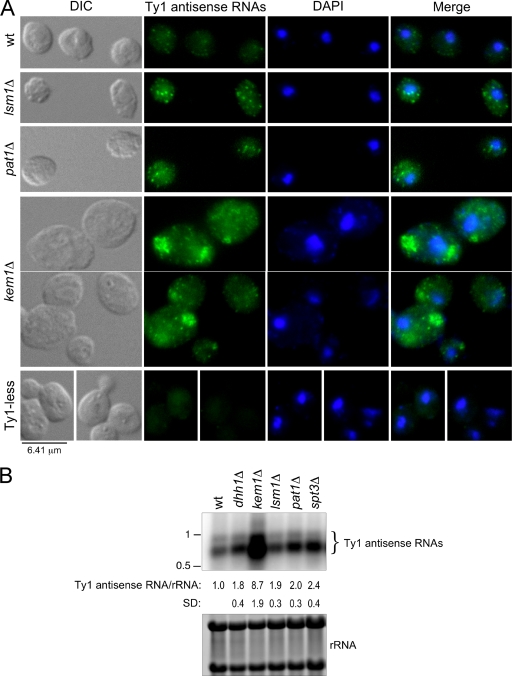
Since Lsm1p, Pat1p, and Kem1p are required for the formation of mRNA/Gag foci, we determined if deletion of P-body components affected the localization of Ty1 antisense RNAs by using a DIG-labeled probe corresponding to the 5′ end of GAG. A probe from this region detects multiple antisense RNAs (43). In ∼95% of the wild-type cells, antisense RNAs localized in puncta predominantly in the cytoplasm (Fig. 9A), with a range of 0 to 13 puncta/cell and an average of 5 puncta/cell. Ty1 antisense RNAs localized in more intense cytoplasmic puncta in the lsm1Δ and pat1Δ mutants, but there was no overall difference in their localization compared with that in wild-type cells. In contrast, antisense transcripts in the kem1Δ mutant were present in large cytoplasmic aggregates in about 56% of the cells and displayed a punctate distribution without aggregates in the remaining cells. The increased intensity of the puncta containing Ty1 antisense RNAs in the lsm1Δ and pat1Δ mutants is consistent with the increase in their levels, as shown by Northern blotting (Fig. 9B) (7). Furthermore, the about ninefold increase in the Ty1 antisense RNA level detected in a kem1Δ mutant helps explain the large aggregates observed by FISH.
FIG. 9.
Localization and levels of Ty1 antisense RNAs in wild-type (wt) cells and P-body component mutants. (A) Localization of Ty1 antisense RNAs in cytoplasmic puncta or aggregates in wild-type (BY4742) cells and isogenic P-body mutants by FISH-IF. Ty1 antisense RNAs were detected with a DIG-labeled probe corresponding to the GAG sequence (Gag340 probe). Merged antisense RNA and DAPI staining is shown. Ty1-less S. paradoxus strain DG1768 served as a negative control. (B) P-body component mutants have elevated levels of Ty1 antisense RNAs. Northern analysis of Ty1 antisense RNAs in the wild type and isogenic P-body component mutants. Equivalent amounts of total RNA were hybridized with a 32P-labeled riboprobe corresponding to the 5′ end of Ty1 (Gag 5′-end riboprobe). The hybridization signals for Ty1 antisense RNAs were normalized to the level of 18S and 25S rRNA, as determined by phosphorimaging. An isogenic spt3Δ mutant (DG2247) was included to verify that Ty1 mRNA synthesis and antisense RNA synthesis were independent. RNA molecular size standards (kilobases) are indicated. SD, standard deviation.
Ty1 mRNA and antisense RNAs colocalize, particularly in a kem1Δ mutant.
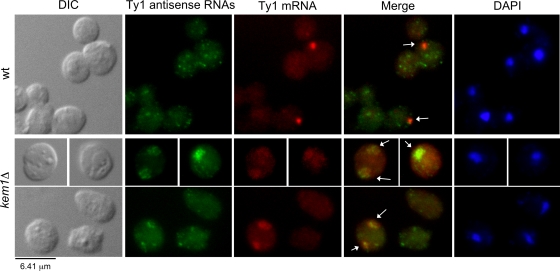
Ty1 antisense RNAs were detected using a DIG-labeled probe, and Ty1 mRNA was detected using an Alexa Fluor-labeled probe from the RT region of POL. In 27% of wild-type cells containing an mRNA focus, at least one antisense RNA punctum colocalized with the mRNA (Fig. 10). Colocalization of Ty1 antisense RNAs and mRNA was statistically significant (P < 0.001), as determined by analyzing the random distribution of colocalization events (see Materials and Methods). The colocalization of Ty1 mRNA/antisense RNAs was difficult to determine in the lsm1Δ and pat1Δ mutants, because the Alexa Fluor 594 detection method was not sensitive enough to detect the scattered mRNA signals. However, the colocalization of antisense RNAs with mRNA was much more prevalent in the kem1Δ mutant, since large cytoplasmic aggregates were detected for both transcripts (Fig. 10). Overall, our results suggest that Kem1p influences the localization of Ty1 mRNA and antisense RNAs, which may help maintain a low but detectable level of retrotransposition.
FIG. 10.
Colocalization of Ty1 mRNA and antisense RNAs in wild-type (wt) and kem1Δ mutant cells. Ty1 antisense RNAs were detected as described in the legend to Fig. 9A, and Ty1 mRNA was detected using a probe labeled with Alexa Fluor 594 corresponding to the RT region. Merged Ty1 antisense RNA and mRNA staining is shown, and colocalized foci and aggregates are indicated by arrows.
DISCUSSION
Here, we provide evidence that P-body components (Dhh1p, Kem1p, Lsm1p, and Pat1p) are important Ty1 cofactors that play multiple roles during retrotransposition after VLPs are assembled. First, P-body components are required for retrotransposition posttranscriptionally. Second, P-body components are necessary for the colocalization of Ty1 mRNA and Gag in cytoplasmic foci when expressed from either endogenous chromosomal Ty1 elements or a galactose-induced Ty1. mRNA/Gag localization in cytoplasmic foci may be required for retrotransposition, which is in agreement with a recent study on how transcription factors impact mRNA fate (42). Third, Kem1p and Lsm1p enhance VLP assembly in clusters. Our work suggests that mRNA/Gag foci detected by FISH-IF may be sites where VLPs clusters form, since these are found in association with Gag in wild-type cells. Fourth, P-body components are not required for VLP assembly per se but rather may be required for the formation of VLPs that are competent for retrotransposition, given that levels of IN and RT are reduced in kem1Δ and pat1Δ mutants. Fifth, Kem1p may be involved in degrading and trafficking Ty1 antisense RNAs, which may further help prevent their colocalization with Ty1 mRNA. Ty1 antisense RNAs associate with VLPs, where they also interfere with accumulation of RT and IN (43). Therefore, Kem1p may enhance the assembly of functional Ty1 VLPs by segregating mRNA/Gag foci from the antisense transcripts, which restrict retrotransposition. Although we cannot detect Ty1 antisense RNAs in the nucleus cytologically, and multiple studies failed to detect a marked decrease in the Ty1 mRNA level in a kem1Δ mutant (M. J. Curcio, personal communication), it has been reported that antisense transcripts repress Ty1 transcription in P-body mutants (7). The difference in Ty1 mRNA levels in P-body component mutants may be caused by strain variations or experimental conditions. Paradoxically, Ty1 and Saccharomyces have evolved a mechanism dependent on P-body components which allows high levels of Ty1 mRNA to accumulate in the cell but ensures that retrotransposition is a rare event.
P-body components facilitate multiple steps during Ty1 retrotransposition.
A complete cycle of Ty1 retrotransposition requires that mRNA, Gag, and Gag-Pol associate to form functional VLPs where reverse transcription occurs. Interestingly, Ty1 mRNA/Gag foci are observed in only ∼30% of the cells, even when overexpressed from a pGTy1 plasmid, which increases the level of mRNA severalfold (17, 19, 20). Ty1 expression may be associated with the cell cycle or cell age, since foci are observed mainly in the mothers of budding cells. Alternatively, the lack of Ty1 mRNA/Gag foci in ∼70% of the cells could be due to a transient transcriptional repression of endogenous Ty1 elements (30). However, this transcriptional repression does not occur in cells overexpressing Ty1 from a pGTy1 plasmid, yet the fraction of transposition-induced cells containing Ty1 mRNA/Gag foci remains unchanged.
Our IEM analyses show that Gag is present as clusters in defined zones when cells overexpress Ty1. These defined zones contain distinct VLPs and resemble the zones detected by FISH-IF. Ty1 VLPs are observed within the background of unassembled Gag complexes, suggesting that mRNA/Gag foci may be sites where VLPs cluster. Dispersed VLPs could also be detected in 25% of wild-type cells and in P-body component mutants, which contained fewer or no mRNA/Gag foci. These results suggest that accumulation of Gag above endogenous levels in either foci or puncta may be sufficient for VLP assembly, especially since Gag forms spherical particles when expressed in Escherichia coli (41). Despite the presence of high levels of endogenous mRNA, VLPs are not observed by IEM within clusters of Gag in cells expressing endogenous elements, which agrees with previous reports (24, 44). However, large clusters of Ty1 VLPs are observed in wild-type cells when levels of mRNA and Gag are increased by overexpression of a pGTy1 plasmid, which overcomes transpositional dormancy. Together, these results imply that the formation of endogenous VLPs is inefficient and may be under the control of cellular or Ty1-derived factors. This control could occur at the translational level when the concentration of Gag may be below the threshold required for efficient VLP formation, or at the posttranslational level where host- or Ty1-derived factors (such as antisense RNAs) could inhibit VLP assembly or function.
Deletion of P-body components leads to a modest decrease in the level of Ty1 mRNA but decreases the colocalization of mRNA and Gag in foci and the formation of VLP clusters. Perhaps Gag is transported along with mRNA by P-body components, as part of a Ty1 mRNP complex, from the nuclear periphery to a confined region(s) within the cytoplasm. By analogy, the localization of Oskar mRNA in Drosophila requires the Dcp1 subunit of the decapping complex Dcp1/Dcp2 present in P-bodies (37). Also, disruption of DHH1 in S. cerevisiae affects the subcellular distribution of Ty3 VLPs (6). The possibility that Gag is transported with Ty1 mRNA by P-body components from the nuclear periphery may help explain why Gag was observed to be perinuclear in a subset of kem1Δ and lsm1Δ mutant cells. Transport of Gag associated with genomic RNA to assembly sites has been shown for several retroviruses. For example, the genomic RNA from murine leukemia virus associates with Gag and Gag-Pol prior to trafficking to its assembly site at the plasma membrane (3). Also, it has been proposed that transit of Rous sarcoma virus Gag through the nucleus facilitates the packaging of genomic RNA (23).
There are several reasons to consider that the colocalization of Ty1 mRNA with Gag facilitates retrotransposition. First, our results suggest that P-body components and Ty1 mRNA/Gag foci may be required for optimal Gag processing, which is critical for generating functional VLPs (1, 63). Second, Ty1 mRNA and Gag do not colocalize in a discrete focus when cells are grown at 30°C, which is a suboptimal temperature for retrotransposition (49). Accordingly, Ty1 protein processing and reverse transcription decrease even more when cells are grown at temperatures above 30°C (34). Third, our results show that P-body components are necessary for cDNA accumulation. Fourth, colocalization of Ty1 mRNA and Gag and their dependence on P-body components may promote steps in VLP assembly necessary for retrotransposition. We show that total levels of IN and RT are greatly reduced in the kem1Δ and pat1Δ mutants, which has led us to speculate that the concentration of mRNA with Gag and Gag-Pol in foci may be required for the formation of retrotransposition-competent VLPs. This may further allow for Gag processing and reverse transcription. Close interaction of IN with RT is critical for the correct folding and activity of RT (61). This could explain why the concentration of Ty1 products in foci may support cDNA accumulation, which is reduced in P-body component mutants. Thus, P-body components may be required for the transport of mRNA and Gag/Gag-Pol as part of an mRNP to specific sites in the cell and may enhance the assembly of Ty1 components required to form retrotransposition-competent VLPs.
P-bodies are apparently sites of Ty3 VLP assembly, since they are required for retrotransposition, and Ty3 mRNA, proteins, and VLPs are found in association with P-body components (2, 6). P-bodies may also be assembly sites for a BMV replication complex that functions in S. cerevisiae, because P-body components are required for BMV genomic RNA translation (22, 33, 47) and sequester translationally repressed BMV RNA1 and RNA2 (4). The size of P-body foci changes according to the rate of mRNA decapping in the cell. Thus, deletion of P-body components affects the size or number of P-bodies (55). For example, the pat1Δ mutation leads to smaller P-bodies, while the lsm1Δ mutation leads to more abundant P-bodies. The kem1Δ mutation leads to larger and more abundant P-bodies. Similar phenotype changes are also observed with endogenous Ty1 mRNA/Gag foci. In the pat1Δ and lsm1Δ mutants, Ty1 mRNA/Gag foci are smaller and more abundant, while in the kem1Δ mutant they are larger but not more abundant. However, our results suggest that Ty1 mRNA/Gag foci are distinct from P-bodies, which is consistent with a recent report (42). For example, P-bodies can harbor translationally repressed mRNPs (55), but the apparent level of Ty1 Gag remains unchanged in mutants lacking the translational repressors DHH1 and PAT1. Moreover, Ty1 mRNA/Gag foci are visible in exponentially growing cells when P-bodies are difficult to detect, the mRNA/Gag foci disassemble in cells starved for glucose when P-bodies appear as distinct foci, and the mRNA/Gag foci remain visible in cells treated with the protein synthesis inhibitor cycloheximide, which causes P-bodies to disassemble. It is not known how glucose deprivation impacts Ty1 retrotransposition. Nonetheless, these results suggest that localization of Ty1 mRNA/Gag in foci is a dynamic process that may be dependent on cellular factors and translation.
Ty1 and S. cerevisiae have evolved a mechanism to limit retrotransposition that is dependent on P-body components.
How does S. cerevisiae cope with such high levels of Ty1 mRNA yet prevent Ty1 insertion events? The balance attained between Saccharomyces and Ty1 products keeps retrotransposition at a low level by sequestering the mRNA with Gag in subcellular compartments in transpositionally dormant cells. P-body components may ensure that some retrotransposition occurs by enhancing the formation of retrotransposition-competent VLPs. In the kem1Δ mutant, Ty1 antisense RNAs are very abundant and retrotransposition is virtually undetectable. Ty1 antisense RNAs produced in this mutant colocalized with Ty1 mRNA in large cytoplasmic aggregates. Therefore, Kem1p is responsible for maintaining low levels of antisense RNAs and may be involved in minimizing interactions between antisense RNAs and Ty1 products. Indeed, Ty1 antisense RNAs associate with VLPs and inhibit the accumulation of mature IN and RT (43), which is similar to the effect of the kem1Δ mutation observed in this study. However, the definitive mechanism underlying inhibition of Ty1 retrotransposition by antisense RNAs remains to be determined.
It is important to determine how Ty1 mRNA/Gag foci form since these foci appear to be an essential step in retrotransposition after VLP assembly. It is reasonable to assume that Gag may bind to Ty1 mRNA during or soon after translation, although Ty1 proteins do not preferentially act on mRNA in cis (16). Since little is known about the role of Ty1 Gag as an RNA chaperone (15), interactions between Gag and Ty1 mRNA may occur at any step in mRNA metabolism. Characterization of additional modulators involved in mRNA metabolism (27) will help define the pathway used to form Ty1 mRNA/Gag foci and may provide insight into the regulation of gene expression via RNA localization. Due to the common ancestry of Ty1 and retroviruses, further analysis of Ty1 RNA and Gag localization may shed light on how retroviruses replicate and help identify novel antiviral targets.
Acknowledgments
We thank Benjamin Luttge, Alison Rattray, and Dwight Nissley for critical review of the manuscript. We also thank Sharon Moore, Jeffrey Strathern, Hendrik Nielsen, and Brenda Youngren for helpful comments or technical suggestions during the course of this study. Anne Kamata provided technical assistance with EM. We thank Prabhakar Gudla, Kaustav Nandy, and Mathew Philip for building the image segmentation software.
This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. This project was also funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Published ahead of print on 9 November 2009.
REFERENCES
- 1.Adams, S. E., J. Mellor, K. Gull, R. B. Sim, M. F. Tuite, S. M. Kingsman, and A. J. Kingsman. 1987. The functions and relationships of Ty-VLP proteins in yeast reflect those of mammalian retroviral proteins. Cell 49:111-119. [DOI] [PubMed] [Google Scholar]
- 2.Aye, M., B. Irwin, N. Beliakova-Bethell, E. Chen, J. Garrus, and S. Sandmeyer. 2004. Host factors that affect Ty3 retrotransposition in Saccharomyces cerevisiae. Genetics 168:1159-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basyuk, E., T. Galli, M. Mougel, J. M. Blanchard, M. Sitbon, and E. Bertrand. 2003. Retroviral genomic RNAs are transported to the plasma membrane by endosomal vesicles. Dev. Cell 5:161-174. [DOI] [PubMed] [Google Scholar]
- 4.Beckham, C. J., H. R. Light, T. A. Nissan, P. Ahlquist, R. Parker, and A. Noueiry. 2007. Interactions between brome mosaic virus RNAs and cytoplasmic processing bodies. J. Virol. 81:9759-9768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beckham, C. J., and R. Parker. 2008. P bodies, stress granules, and viral life cycles. Cell Host Microbe 3:206-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beliakova-Bethell, N., C. Beckham, T. H. Giddings, Jr., M. Winey, R. Parker, and S. Sandmeyer. 2006. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA 12:94-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berretta, J., M. Pinskaya, and A. Morillon. 2008. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev. 22:615-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 9.Brengues, M., and R. Parker. 2007. Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol. Biol. Cell 18:2592-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brengues, M., D. Teixeira, and R. Parker. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310:486-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryk, M., M. Banerjee, D. Conte, Jr., and M. J. Curcio. 2001. The Sgs1 helicase of Saccharomyces cerevisiae inhibits retrotransposition of Ty1 multimeric arrays. Mol. Cell. Biol. 21:5374-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coller, J., and R. Parker. 2005. General translational repression by activators of mRNA decapping. Cell 122:875-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conte, D., Jr., E. Barber, M. Banerjee, D. J. Garfinkel, and M. J. Curcio. 1998. Posttranslational regulation of Ty1 retrotransposition by mitogen-activated protein kinase Fus3. Mol. Cell. Biol. 18:2502-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costes, S. V., D. Daelemans, E. H. Cho, Z. Dobbin, G. Pavlakis, and S. Lockett. 2004. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 86:3993-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cristofari, G., D. Ficheux, and J. L. Darlix. 2000. The GAG-like protein of the yeast Ty1 retrotransposon contains a nucleic acid chaperone domain analogous to retroviral nucleocapsid proteins. J. Biol. Chem. 275:19210-19217. [DOI] [PubMed] [Google Scholar]
- 16.Curcio, M. J., and D. J. Garfinkel. 1994. Heterogeneous functional Ty1 elements are abundant in the Saccharomyces cerevisiae genome. Genetics 136:1245-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curcio, M. J., and D. J. Garfinkel. 1992. Posttranslational control of Ty1 retrotransposition occurs at the level of protein processing. Mol. Cell. Biol. 12:2813-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Curcio, M. J., and D. J. Garfinkel. 1991. Single-step selection for Ty1 element retrotransposition. Proc. Natl. Acad. Sci. U. S. A. 88:936-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curcio, M. J., A. M. Hedge, J. D. Boeke, and D. J. Garfinkel. 1990. Ty RNA levels determine the spectrum of retrotransposition events that activate gene expression in Saccharomyces cerevisiae. Mol. Gen. Genet. 220:213-221. [DOI] [PubMed] [Google Scholar]
- 20.Curcio, M. J., N. J. Sanders, and D. J. Garfinkel. 1988. Transpositional competence and transcription of endogenous Ty elements in Saccharomyces cerevisiae: implications for regulation of transposition. Mol. Cell. Biol. 8:3571-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devine, S. E., and J. D. Boeke. 1996. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 10:620-633. [DOI] [PubMed] [Google Scholar]
- 22.Díez, J., M. Ishikawa, M. Kaido, and P. Ahlquist. 2000. Identification and characterization of a host protein required for efficient template selection in viral RNA replication. Proc. Natl. Acad. Sci. U. S. A. 97:3913-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garbitt-Hirst, R., S. P. Kenney, and L. J. Parent. 2009. Genetic evidence for a connection between Rous sarcoma virus Gag nuclear trafficking and genomic RNA packaging. J. Virol. 83:6790-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garfinkel, D. J., J. D. Boeke, and G. R. Fink. 1985. Ty element transposition: reverse transcriptase and virus-like particles. Cell 42:507-517. [DOI] [PubMed] [Google Scholar]
- 25.Garfinkel, D. J., K. Nyswaner, J. Wang, and J. Y. Cho. 2003. Post-transcriptional cosuppression of Ty1 retrotransposition. Genetics 165:83-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 27.Griffith, J. L., L. E. Coleman, A. S. Raymond, S. G. Goodson, W. S. Pittard, C. Tsui, and S. E. Devine. 2003. Functional genomics reveals relationships between the retrovirus-like Ty1 element and its host Saccharomyces cerevisiae. Genetics 164:867-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irwin, B., M. Aye, P. Baldi, N. Beliakova-Bethell, H. Cheng, Y. Dou, W. Liou, and S. Sandmeyer. 2005. Retroviruses and yeast retrotransposons use overlapping sets of host genes. Genome Res. 15:641-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji, H., D. P. Moore, M. A. Blomberg, L. T. Braiterman, D. F. Voytas, G. Natsoulis, and J. D. Boeke. 1993. Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell 73:1007-1018. [DOI] [PubMed] [Google Scholar]
- 30.Jiang, Y. W. 2002. Transcriptional cosuppression of yeast Ty1 retrotransposons. Genes Dev. 16:467-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson, C. S., C. P. Jerome, and R. Brommage. 2000. Unbiased determination of cytokine localization in bone: colocalization of interleukin-6 with osteoblasts in serial sections from monkey vertebrae. Bone 26:461-467. [DOI] [PubMed] [Google Scholar]
- 32.Knowles, D. W., C. Ortiz de Solarzano, A. Jones, and S. J. Lockett. 2000. Analysis of the 3D spatial organization of cells and sub cellular structures in tissue. Proc. SPIE 3921:66-73. [Google Scholar]
- 33.Kushner, D. B., B. D. Lindenbach, V. Z. Grdzelishvili, A. O. Noueiry, S. M. Paul, and P. Ahlquist. 2003. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc. Natl. Acad. Sci. U. S. A. 100:15764-15769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawler, J. F., Jr., D. P. Haeusser, A. Dull, J. D. Boeke, and J. B. Keeney. 2002. Ty1 defect in proteolysis at high temperature. J. Virol. 76:4233-4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, B. S., C. P. Lichtenstein, B. Faiola, L. A. Rinckel, W. Wysock, M. J. Curcio, and D. J. Garfinkel. 1998. Posttranslational inhibition of Ty1 retrotransposition by nucleotide excision repair/transcription factor TFIIH subunits Ssl2p and Rad3p. Genetics 148:1743-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lifshitz, L. M. 1998. Determining data independence on a digitized membrane in three dimensions. IEEE Trans Med. Imaging 17:299-303. [DOI] [PubMed] [Google Scholar]
- 37.Lin, M. D., S. J. Fan, W. S. Hsu, and T. B. Chou. 2006. Drosophila decapping protein 1, dDcp1, is a component of the Oskar mRNP complex and directs its posterior localization in the oocyte. Dev. Cell 10:601-613. [DOI] [PubMed] [Google Scholar]
- 38.Long, R. M., D. J. Elliott, F. Stutz, M. Rosbash, and R. H. Singer. 1995. Spatial consequences of defective processing of specific yeast mRNAs revealed by fluorescent in situ hybridization. RNA 1:1071-1078. [PMC free article] [PubMed] [Google Scholar]
- 39.Long, R. M., R. H. Singer, X. Meng, I. Gonzalez, K. Nasmyth, and R. P. Jansen. 1997. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science 277:383-387. [DOI] [PubMed] [Google Scholar]
- 40.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 41.Luschnig, C., M. Hess, O. Pusch, J. Brookman, and A. Bachmair. 1995. The gag homologue of retrotransposon Ty1 assembles into spherical particles in Escherichia coli. Eur. J. Biochem. 228:739-744. [DOI] [PubMed] [Google Scholar]
- 42.Malagon, F., and T. H. Jensen. 2008. The T-body, a new cytoplasmic RNA granule in Saccharomyces cerevisiae. Mol. Cell. Biol. 28:6022-6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsuda, E., and D. J. Garfinkel. 2009. Posttranslational interference of Ty1 retrotransposition by antisense RNAs. Proc. Natl. Acad. Sci. U. S. A. 106:15657-15662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mellor, J., S. M. Fulton, M. J. Dobson, W. Wilson, S. M. Kingsman, and A. J. Kingsman. 1985. A retrovirus-like strategy for expression of a fusion protein encoded by yeast transposon Ty1. Nature 313:243-246. [DOI] [PubMed] [Google Scholar]
- 45.Moore, S. P., G. Liti, K. M. Stefanisko, K. M. Nyswaner, C. Chang, E. J. Louis, and D. J. Garfinkel. 2004. Analysis of a Ty1-less variant of Saccharomyces paradoxus: the gain and loss of Ty1 elements. Yeast 21:649-660. [DOI] [PubMed] [Google Scholar]
- 46.Narayanan, A., J. Eifert, K. A. Marfatia, I. G. Macara, A. H. Corbett, R. M. Terns, and M. P. Terns. 2003. Nuclear RanGTP is not required for targeting small nucleolar RNAs to the nucleolus. J. Cell Sci. 116:177-186. [DOI] [PubMed] [Google Scholar]
- 47.Noueiry, A. O., J. Díez, S. P. Falk, J. Chen, and P. Ahlquist. 2003. Yeast Lsm1p-7p/Pat1p deadenylation-dependent mRNA-decapping factors are required for brome mosaic virus genomic RNA translation. Mol. Cell. Biol. 23:4094-4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nyswaner, K. M., M. A. Checkley, M. Yi, R. M. Stephens, and D. J. Garfinkel. 2008. Chromatin-associated genes protect the yeast genome from Ty1 insertional mutagenesis. Genetics 178:197-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paquin, C. E., and V. M. Williamson. 1984. Temperature effects on the rate of Ty transposition. Science 226:53-55. [DOI] [PubMed] [Google Scholar]
- 50.Parker, R., and U. Sheth. 2007. P bodies and the control of mRNA translation and degradation. Mol. Cell 25:635-646. [DOI] [PubMed] [Google Scholar]
- 51.Rattray, A. J., B. K. Shafer, and D. J. Garfinkel. 2000. The Saccharomyces cerevisiae DNA recombination and repair functions of the RAD52 epistasis group inhibit Ty1 transposition. Genetics 154:543-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rinckel, L. A., and D. J. Garfinkel. 1996. Influences of histone stoichiometry on the target site preference of retrotransposons Ty1 and Ty2 in Saccharomyces cerevisiae. Genetics 142:761-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scholes, D. T., M. Banerjee, B. Bowen, and M. J. Curcio. 2001. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159:1449-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- 55.Sheth, U., and R. Parker. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300:805-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teixeira, D., and R. Parker. 2007. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol. Biol. Cell 18:2274-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Teixeira, D., U. Sheth, M. A. Valencia-Sanchez, M. Brengues, and R. Parker. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11:371-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Kempen, G. M., and L. J. van Vliet. 2000. Mean and variance of ratio estimators used in fluorescence ratio imaging. Cytometry 39:300-305. [DOI] [PubMed] [Google Scholar]
- 59.Voytas, D. F., and J. B. Boeke. 2002. Ty1 and Ty5 of Saccharomyces cerevisiae, p. 614-630. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 60.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 61.Wilhelm, M., and F. X. Wilhelm. 2006. Cooperation between reverse transcriptase and integrase during reverse transcription and formation of the preintegrative complex of Ty1. Eukaryot. Cell 5:1760-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Winston, F., K. J. Durbin, and G. R. Fink. 1984. The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell 39:675-682. [DOI] [PubMed] [Google Scholar]
- 63.Youngren, S. D., J. D. Boeke, N. J. Sanders, and D. J. Garfinkel. 1988. Functional organization of the retrotransposon Ty from Saccharomyces cerevisiae: Ty protease is required for transposition. Mol. Cell. Biol. 8:1421-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]