Abstract
The small GTPase Ras, which transmits extracellular signals to the cell, and the kinase Aurora-A, which promotes proper mitosis, can both be inappropriately activated in human tumors. Here, we show that Aurora-A in conjunction with oncogenic Ras enhances transformed cell growth. Furthermore, such transformation and in some cases also tumorigenesis depend upon S194 of RalA, a known Aurora-A phosphorylation site. Aurora-A promotes not only RalA activation but also translocation from the plasma membrane and activation of the effector protein RalBP1. Taken together, these data suggest that Aurora-A may converge upon oncogenic Ras signaling through RalA.
Ras small GTPases (H-, N-, and K-Ras) function as regulated binary switches, typically at the plasma membrane, whereby extracellular signal-stimulated cell surface receptors stimulate guanine nucleotide exchange factors (GEFs) to promote GDP/GTP exchange to favor the formation of active, GTP-bound Ras. This, in turn, induces a conformational change in the effector binding domain in Ras, permitting the binding and activation of effector proteins, such as Raf proteins, phosphatidylinositol 3-kinase (PI3K), and RalGEF proteins, that mediate Ras signaling (19, 53). One-third of human cancers harbor point mutations in Ras that render the protein in a constitutively active GTP-bound state, promoting a host of cancer cell phenotypes (40).
Aurora-A belongs to a family of three related serine/threonine mitotic kinases critical for many stages of mitosis. Studies of a number of model systems indicate that Aurora-A phosphorylates a growing number of proteins in a spatially and temporally restricted manner to ensure proper centrosomal maturation and separation, mitotic entry, mitotic spindle assembly, chromosome alignment and separation, and subsequent cytokinesis (18, 42). Overexpression of Aurora-A is seen in human cancers (22, 31, 32) and can cause growth transformation of Rat1 and NIH 3T3 rodent fibroblast cell lines (5). That it can cause tumor formation in the mammary epithelia of mice only after a long latency (60, 68) and that alone it does not transform primary rodent cells (1) or induce pancreatic cancer formation in mice (61) argue that Aurora-A acts in concert with other changes to promote a transformed state.
Although the mechanism by which Aurora-A promotes oncogenesis remains to be understood, emerging evidence suggests that Aurora-A may cooperate with the Ras oncoprotein. First, activating mutations in KRAS occur in nearly all pancreatic cancers (28), and Aurora-A has been found to be overexpressed in this tumor type by gene amplification (21) or by elevated levels of mRNA or protein (21, 35). This overexpression likely fosters tumor growth, as suppression of Aurora-A expression by interfering RNA or treatment with Aurora-A inhibitors also impairs pancreatic cancer cell growth (26, 48). Second, overexpression of Aurora-A enhances Ras-induced transformation of murine 3T3A31-1 fibroblasts (59). Third, Aurora kinases physically interact with RasGAP in vitro (23), and inhibition of Aurora-B binding to RasGAP causes apoptosis (49). Fourth, two components of the RalGEF-Ral effector pathway of Ras, which is known to promote Ras oncogenesis (38), are substrates for Aurora-A (66). Specifically, active Ras binds to RalGEF proteins, a family of guanine nucleotide exchange factors (GEFs) and activators of the related small GTPases RalA and RalB. Both the RalGEF protein RalGDS and RalA were shown to be phosphorylated by Aurora-A. With regard to the latter, RalA is phosphorylated at S194 in its C-terminal membrane binding domain, leading to elevated levels of activated RalA-GTP. Constitutively active RalA (G23V) also cooperated with ectopically expressed Aurora-A to promote anchorage-independent growth of MDCK epithelial cells, whereas a RalA mutant that could not be phosphorylated by Aurora-A (RalAG23V,S194A) was impaired in this activity (66). Moreover, overexpression of both Aurora-A and RalA mRNA is associated with advanced human bladder cancer (58). Finally, as indirect evidence for the importance of the Aurora-A phosphorylation site in RalA, S194 and S183 were identified as sites of dephosphorylation by the phosphatase PP2A. Short hairpin RNA (shRNA) silencing of PP2A expression increased phosphorylation of S194 and S183 and formation of RalA-GTP, whereas replacing endogenous RalA with S194A or S183A mutants resulted in a loss of tumorigenic growth of human embryonic kidney (HEK) cells expressing oncogenic H-Ras, hTERT, and the early region of simian virus 40 (SV40) (56). Given these observations, we explored the molecular connection between Aurora-A and the Ras-RalGEF-RalA pathway.
MATERIALS AND METHODS
Plasmids.
pSuper-Retro-Puro plasmids encoding shRNA against RalA, RalB, RalBP1 (5′-GTAGAGAGGACCATGATGT), or a scramble sequence; pBabe-Neo plasmid encoding shRNA-resistant Myc-tagged RalA; pBabe-Puro plasmid encoding shRNA-resistant Flag-tagged RalA or Q72L; pBabe-Bleo plasmid encoding hemagglutinin (HA)-tagged Rlf-CAAX; pBabe-Puro RasG12V and derived effector mutants; small t antigen (t-Ag); DsRed-Rab11; pMT3-mycRalBP1; and pcDNA3-mycSec5 were previously described (16, 25, 36, 45). S194A and S194D point mutations in RalA were introduced to pBabe-Neo and pBabe-puro plasmids encoding shRNA-resistant RalA, respectively, by site-directed mutagenesis, and a Myc epitope tag (9E10) or a Flag epitope tag was added to the N terminus by PCR. Human Aurora-A was PCR amplified from a cDNA template (MGC-1605; ATCC) to add an N-terminal HA-epitope tag, after which K162R and T288D mutants were generated by site-directed mutagenesis. Resultant wild-type (WT), K162R, and T288D cDNAs were cloned into pBabe-Bleo or pBabe-Hygro for stable expression and pCGN for transient expression. shRNA directed against Aurora-A (5′-ATGCCCTGTCTTACTGTCA) was subcloned into pSuper-Retro-Puro-TET. Green fluorescent protein (GFP)-RalA fusion proteins were generated by subcloning wild-type RalA, RalAS194A, RalAS194D, and RalAD49N in frame to the C terminus of GFP in pEGFP-C2.
HEK-TtH cells, 293 cells, human pancreatic cancer cell lines and derived cell lines, inducible Aurora-A shRNA HPAC cells, and tissue samples.
HEK-TtH cells (human embryonic kidney cells stably expressing the SV40 early region and hTERT) were previously described (24). 293, 293T, AsPC-1, HPAC, HPAF-II, PANC-1, SW1990, Capan-1, Capan-2, CFPAC-1, and MIA PaCa-2 (38) were purchased and cultured as suggested by supplier (ATCC), and T3M4 cells were kindly provided by M. Korc (Dartmouth-Hitchcock Medical Center, Hanover, NH). Where noted, these cell lines were stably infected with retroviruses encoding the indicated shRNAs or transgenes as previously described (47). Generation of HPAC cells expressing inducible Aurora-A shRNA was done as described previously (37) except that a pSuper-Retro-Puro-TET plasmid (37) engineered to encode Aurora-A shRNA (26) was used and, at the indicated times, the cells were treated with either 60 mg/ml of doxycycline (Sigma) or a vehicle. Frozen surgical samples of normal human pancreatic tissues were kindly provided by A. D. Proia and D. Tyler (DUMC).
Immunoblotting.
Whole-cell lysates isolated in Triton X-100 lysis buffer from cultured cells (serum starved for 48 h for measurement of Ras effector pathways) or isolated in radioimmunoprecipitation assay (RIPA) buffer from tissue material were immunoblotted with the following antibodies, diluted according to the manufacturers' recommendations: antiactin, anti-RalBP1 (Santa Cruz), anti-RalA, anti-RalB (BD Transduction Laboratories), anti-Aurora-A (Cell Signaling Technology), and antitubulin (Sigma) (to detect endogenous actin, RalBP1, RalA, RalB, Aurora-A, and tubulin proteins, respectively) and anti-HA (Roche), anti-Myc 9E10 (Invitrogen), anti-RalA (BD Transduction Laboratories) and anti-pan-Ras (Santa Cruz) (to detect the ectopic proteins HA-Aurora-AWT, HA-Aurora-AT288D, HA-Aurora-AK162R, HA-Rlf-CAAX, shRNA-resistant RalA [wild type, S194A, or S194D], and H-Ras or effector domain mutants, respectively).
Immunoprecipitation.
Whole-cell lysates prepared in Triton X-100 buffer from 293T cells transiently transfected with plasmids encoding FLAG epitope-tagged RalA (wild type or S194D) and Myc-RalBP1 or Myc-Sec5 were subjected to immunoprecipitation with anti-FLAG M2 resin (Sigma) and immunoblotted with anti-FLAG M2 (Sigma) to detect FLAG-RalA and anti-Myc 9E10 (Invitrogen) to detect Myc-RalBP1 or Myc-Sec5. To detect phosphorylated RalA, cells expressing a vector control or HA-tagged Aurora-A were subjected to immunoprecipitation with anti-RalA antibody (BD Transduction Laboratories) and immunoblotted with antiphosphoserine (4A4; Millipore) to detect serine-phosphorylated RalA. For detection of endogenous GTP-bound small GTPases, GTP-RalA, GTP-Cdc42, and GTP-Rac1 were captured for pulldown analyses by incubating cell lysates with glutathione-agarose-bound recombinant glutathione S-transferase (GST)-RalBD for RalA or GST-PakBD for Cdc42 and Rac1 and detected with the anti-RalA, anti-Cdc42, and anti-Rac1 antibodies (BD Transduction Laboratories), respectively, as previously described (4, 65). The total amount of the corresponding small GTPase in the whole-cell lysate served as a loading control.
Soft agar assay.
Fifty thousand cells per 35-mm plate with a 2-mm grid were suspended in soft agar as described previously (17, 25). In the case of comparing RasG12V-transformed HEK-TtH cells in the absence or presence of HA-Aurora-AWT, HA-Aurora-AK162R, or HA-Aurora-AT288D, 104 cells were seeded for analysis. Colonies with >30 cells were scored after 5 weeks. Assays were done in triplicate and two to three times independently.
Isolation of internal membrane fractions.
Internal membrane fractions were prepared as described previously (8). Briefly, cells were resuspended in hypotonic lysis buffer (10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 250 mM sucrose, 1 mM dithiothreitol [DTT]) and homogenized with a Dounce homogenizer. Extract was centrifuged at 750 × g to pellet nuclei, and the supernatant was further centrifuged at 10,000 × g to pellet the crude membrane fraction. The crude membrane fraction was resuspended in MIB (250 mM sucrose, 40 mM KCl, 40 mM succinate, 40 mM HEPES, 20 mM EGTA, 800 mM mannitol) and layered over a discontinuous Percoll gradient composed of 42%, 37%, 30%, and 25% Percoll prepared in MIB. Percoll gradients were centrifuged at 25,000 rpm for 25 min in a Beckman TLS-55 rotor with the brake off. The endoplasmic reticulum (ER)-rich internal membrane fraction sedimented between the 25% and 30% bands.
Xenograft tumorigenesis assay.
Cells (107) mixed with Matrigel were injected subcutaneously into both flanks of SCID/beige mice (Charles River Laboratory), with four injection sites per cell line, and tumor volumes were determined twice per week and calculated as (length/2)2 × width. These experiments were approved by the Duke University Institutional Animal Care and Use Committee.
Immunofluorescence.
The described cells were plated on glass microslides the previous day, fixed, permeabilized, and incubated with anti-RalA or anti-RalB (BD Transduction Laboratories), anti-Myc 9E10 (Invitrogen), and/or EEA1 (BD Transduction Laboratories) primary antibodies, followed by incubation with the anti-mouse Alexa-488 secondary antibody (Molecular Probes) or Texas Red-phalloidin to detect endogenous RalA, RalB, actin, or ectopic Myc-tagged proteins as previously described (36). For visualization of transiently expressed GFP fusion proteins, HEK-TtH cells on glass microslides transfected with GFP-RalA fusion proteins with or without HA-tagged Aurora-A (WT, K162R, or T288D) and Myc-tagged RalBP1 or Sec5 were fixed, mounted, and visualized by confocal microscopy. A Zeiss LSM 410 confocal microscope, an Olympus Fluoview 300 laser scanning confocal imaging system configured with an IX70 fluorescence microscope fitted with a PlanApo 60× oil objective, or a Leica DMI6000CS scanning confocal imaging system fitted with a Leica Plan Apochromat 100×/1.4- to 0.70-numerical-aperture oil objective was used for imaging.
RESULTS AND DISCUSSION
Aurora-AT288D cooperates with the RalGEF pathway to promote Ras-mediated transformation.
Both oncogenic Ras mutations (7) and overexpression of Aurora-A occur in human cancers (22, 31), such as pancreatic cancer (27, 28, 35). In experimental systems, overexpression of Aurora-A enhanced Ras- and RalA-induced transformation of a murine cell line (59) and a canine cell line (66), respectively. Given this functional association between Ras oncogenic signaling and Aurora-A activation, we tested whether Aurora-A also enhances Ras-mediated transformation of human cells. For these experiments, we utilized HEK-TtH cells (normal HEK cells immortalized and rendered sensitive to Ras transformation by ectopic expression of the SV40 early region encoding large T and small t antigens and of the telomerase catalytic subunit hTERT) (24, 33). As these cells depend upon oncogenic Ras for tumorigenesis, and the other genetic changes required for tumorigenesis are known, Ras-transformed HEK-TtH cells provide a simplified, genetically defined, and malleable system for dissecting the relationship between Aurora-A and oncogenic Ras in transformation and tumorigenesis of human cells.
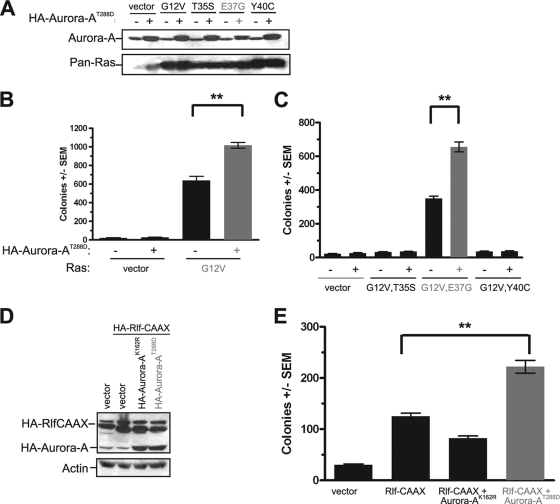
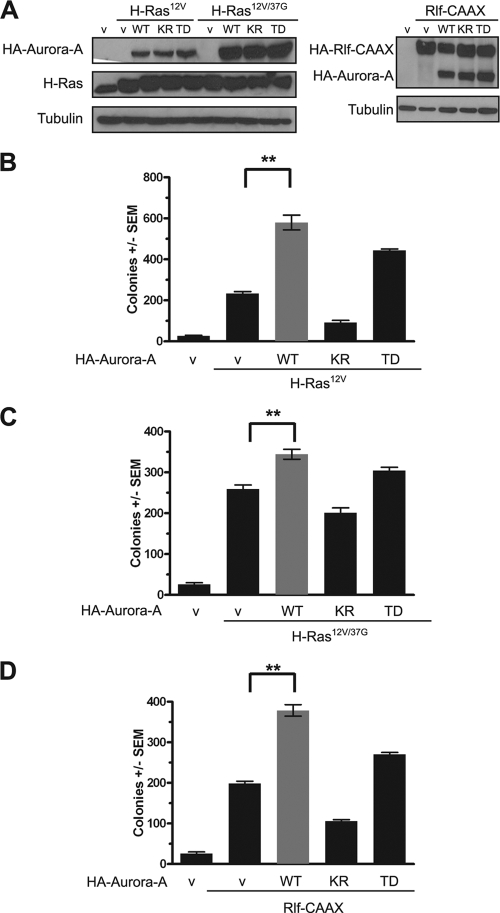
HEK-TtH cells were stably infected with a retrovirus either encoding a constitutively activated T288D mutant (5) of human Aurora-A (Aurora-AT288D) or carrying no transgene, in the absence or presence of oncogenic (G12V) H-Ras (RasG12V). Cell lines were verified to express the appropriate transgenes, as assessed by immunoblot analysis (Fig. 1A), and assayed for growth in soft agar as a measure of transformation. We found that activated Aurora-AT288D alone could not promote transformation of these cells in the absence of oncogenic Ras (Fig. 1B) but that Aurora-AT288D enhanced oncogenic Ras-mediated transformation twofold (Fig. 1B). These results indicated that the cooperative activity of Aurora-A and Ras signaling to promote growth transformation observed previously in murine fibroblast (59) and canine epithelial (66) cell lines can be extended to human cells.
FIG. 1.
Aurora-AT288D promotes Ras-induced transformation through the RalGEF pathway. (A and D) Appropriate expression, as detected by immunoblot analysis, of kinase-active HA-Aurora-AT288D, kinase-inactive HA-Aurora-AK162R, constitutively activated HA-Rlf-CAAX, and ectopic and endogenous Ras (Pan-Ras). Actin serves as a loading control. (B, C, and E) Anchorage-independent growth in soft agar of polyclonal HEK-TtH cells stably expressing the indicated transgenes, expressed as average numbers of colonies formed ± standard errors of the means (SEM) for six plates (two independent experiments conducted in triplicate). The same vector (with [+] or without [−] HA-Aurora-AT288D) was used as a control in all of these experiments. Significant P values (<0.001) are indicated by **. Tukey's multiple-comparison test was used to determine significance between cell lines.
To explore at what point Aurora-A may converge upon oncogenic Ras signaling, we tested whether Aurora-A cooperated with a specific Ras effector pathway to promote transformation. We focused our analyses on the three major Ras effectors involved in oncogenesis (Raf, PI3K, and RalGEF) by using oncogenic H-Ras effector domain mutants RasG12V,T35S, RasG12V,E37G, and RasG12V,Y40C, which retain preferential activation of the Raf, RalGEF, and PI3K pathways, respectively (34, 54, 62, 64). Kinase-active Aurora-AT288D or an empty vector as a negative control was therefore expressed in HEK-TtH cells in conjunction with no transgene or a transgene encoding each Ras effector domain mutant. Appropriate expression of Aurora-AT288D and the RasG12V effector mutants was verified by immunoblot analysis (Fig. 1A), and the resultant six cell lines in addition to vector controls were assayed for anchorage-independent growth. As previously reported (25), activation of the RalGEF pathway, but not the PI3K or mitogen-activated protein kinase (MAPK) pathway, promoted the growth of these cells in soft agar. Addition of kinase-active Aurora-AT288D did not endow anchorage-independent growth to either vector control cells or cells expressing Ras mutants activating the Raf or PI3K pathways but did enhance twofold the transformed growth of cells expressing RasG12V,E37G (Fig. 1C). Thus, Aurora-A cooperates synergistically with the RalGEF signaling arm of oncogenic Ras in cellular transformation.
To further validate these results, we tested whether Aurora-A similarly enhanced transformation of cells expressing an activated variant of the RalGEF protein Rlf/Rgl2 (Rlf-CAAX) (63) in place of RasG12V,E37G. Specifically, HEK-TtH cells were engineered to coexpress both Rlf-CAAX and Aurora-AT288D proteins, or, as a negative or positive control, an empty vector or Rlf-CAAX, respectively. Appropriate transgene expression was verified by immunoblot analysis (Fig. 1D). As expected, negative-control vector cells did not grow in soft agar, whereas the addition of Rlf-CAAX promoted transformed cell growth (Fig. 1E). In agreement with the ability of activated Aurora-AT288D to enhance transformation by RasG12V,E37G, cells expressing both the activated Rlf-CAAX and Aurora-AT288D proteins grew more robustly in soft agar (Fig. 1E). We next tested whether the kinase activity of Aurora-A was required to enhance RalGEF-mediated transformation by expressing a kinase-inactive mutant of Aurora-AK162R (5) in Rlf-CAAX-transformed cells (Fig. 1D). Whereas kinase-active Aurora-AT288D enhanced RalGEF-mediated transformation, the kinase-inactive Aurora-AK162R protein inhibited growth in soft agar (Fig. 1E). Thus, Aurora-A enhances Ras-RalGEF signaling to promote cellular transformation.
Aurora-AT288D-induced increase in RalGEF-induced transformation of human cells is lost upon mutation of the Aurora-A phosphorylation site S194 of RalA.
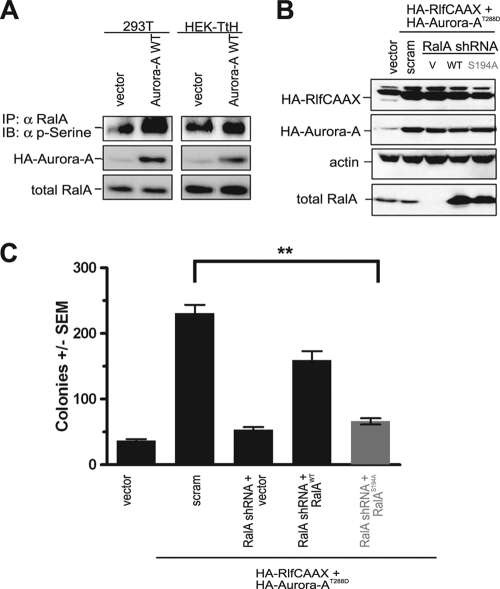
RalGEF proteins activate the RalA and RalB isoforms of Ral GTPases, which are related both structurally (82% sequence identity) and biochemically. Despite these similarities, however, the proteins have distinct roles in oncogenesis. Whereas RalA is required for Ras-mediated anchorage-independent growth (36, 38), RalB can instead be critical to cell viability, at least in some contexts (14, 15), particularly when cells are grown in the absence of substrate or during metastasis (38). The primary sequence differences between RalA and RalB are concentrated in their C-terminal-30-residue membrane-targeting sequences in the hypervariable domain (20), which have been shown to contribute to some of the functional differences described above (36). Upstream of the C-terminal CAAX tetrapeptide motif that signals for posttranslational modification by a geranylgeranyl isoprenoid required for membrane association, these divergent sequences target RalA to the plasma membrane and late endosomes but target RalB to the plasma membrane only (57). Aurora-A phosphorylates this C-terminal region at a serine residue, S194, found in RalA but not in RalB (66), and mutating this site inhibited the ability of an overexpressed and constitutively activated RalA mutant (G23V) to cooperate with ectopically expressed Aurora-A to promote the transformed growth of a canine cell line (66). Conversely, knockdown of PP2A Aβ in HEK cells expressing T-Ag, hTERT, and oncogenic Ras resulted in elevated phosphorylation of S183 and S194 of RalA, and when RalA was also knocked down in these cells, they were able to form tumors upon restoration with ectopic wild-type RalA but not with an S194A mutant version (56). Consistent with these observations, we found that endogenous RalA is phosphorylated at serines in both human 293T and HEK TtH cells (Fig. 2A), likely owing to expression of t-Ag in these cells (56), and that ectopically expressed wild-type Aurora-A increased the level of this phosphorylation (Fig. 2A). Similarly, we found that the Aurora-AT288D-mediated enhancement of RalGEF-mediated transformation of HEK-TtH cells requires phosphorylation of RalA at S194. First, in HEK-TtH cells stably expressing both activated Rlf-CAAX and Aurora-AT288D proteins, expression of endogenous RalA protein was stably knocked down by shRNA, as assessed by immunoblot analysis (Fig. 2B). Next, the loss of RalA expression was then complemented by a vector either carrying no transgene (negative control) or encoding an shRNA-resistant RalA protein in the wild-type (positive control) or S194A mutant (66) configuration, as assessed by immunoblot analysis (Fig. 2B). As expected (36), knockdown of RalA was found to reduce transformed cell growth in soft agar to the level of cells not transfected with activated RalGEF and Aurora-A. This loss of transformation was rescued, almost to the level of the positive control scramble control cells, by expression of shRNA-resistant wild-type RalA but not the S194A mutant of RalA (Fig. 2C). Lastly, the Aurora-AT288D-enhanced transformation of Rlf-CAAX-expressing cells was not reduced upon knockdown of RalB (see Fig. S1 in the supplemental material), which lacks the Aurora-A phosphorylation site (66). Thus, Aurora-AT288D enhancement of RalGEF-mediated transformation is lost specifically upon mutation of the Aurora-A substrate S194 of RalA.
FIG. 2.
Aurora-A potentiates RalGEF-transformation through phosphorylation of RalA S194. (A) Appropriate expression, as detected by immunoblot analysis, of wild-type HA-Aurora-A, endogenous RalA, and phosphorylated RalA (detected by immunoprecipitation [IP] of endogenous RalA followed by immunoblot analysis [IB] with an anti-phospho-serine [α p-serine] antibody) in 293T and HEK-TtH cells expressing the indicated transgenes. Total RalA serves as a loading control. (B) Appropriate expression, as detected by immunoblot analysis, of HA-Aurora-AT288D, HA-Rlf-CAAX, knockdown of endogenous RalA, and complementation by ectopic RalA resistant to RalA shRNA in HEK-TtH cells stably infected with retroviruses carrying no transgene (vector [V]) or the indicated transgenes in the presence of shRNA specific to RalA or a scrambled version (scram) of this sequence. Actin serves as a loading control. (C) Anchorage-independent growth in soft agar of the aforementioned polyclonal HEK-TtH cells infected with retroviruses encoding the indicated shRNAs and carrying transgenes, expressed as average numbers of colonies formed ± SEM for six plates (two independent experiments conducted in triplicate). Significant P values (<0.001) are indicated by **. Tukey's multiple-comparison test was used to determine significance between cell lines.
RalA S194 is required for transformed growth of human pancreatic cancer cell lines.
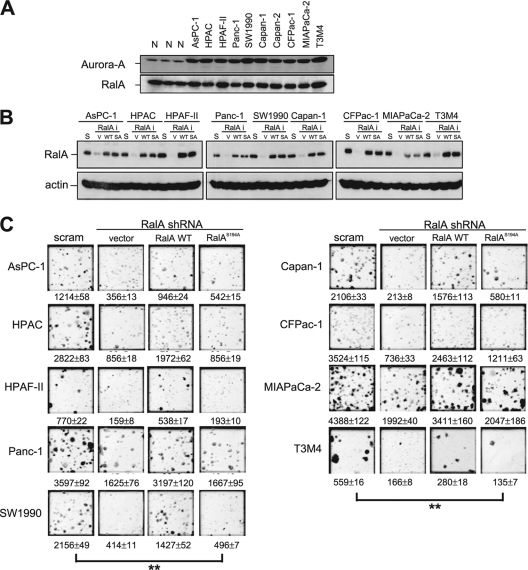
RalA is frequently activated in pancreatic cancers and is essential for transformed growth of pancreatic cancer cell lines in vitro and in vivo (38). Aurora-A protein is similarly overexpressed in this disease (35). Aurora-A and RalA mRNA overexpression is also associated with advanced human bladder cancer (58). Indeed, we detected elevated levels of this kinase in a panel of 10 KRAS mutation-positive pancreatic carcinoma cells, compared to the level for normal pancreatic cancer tissues, whereas the total protein levels of RalA did not differ between tumor cells and normal tissue (Fig. 3A). Thus, while RalA is upregulated at the level of protein activation (36, 38), Aurora-A is upregulated at the level of protein expression in pancreatic cancer cells. Given this correlation and the requirement of S194 of RalA for transformed or tumorigenic growth of canine (66), human HEK-TER (56), and HEK-TtH (Fig. 2C) cells, we tested whether S194 is also required for tumorigenic growth of human pancreatic cells. Specifically, a panel of pancreatic cancer cell lines, including AsPC-1, HPAC, HPAF-II, Panc-1, SW1990, Capan-1, CFPac-1, MIA PaCa-2, and T3M4 cells, were stably infected with a retrovirus encoding RalA shRNA or a scramble control, after which the RalA knocked-down cell lines were complemented by infection with retroviruses encoding shRNA-resistant forms of either wild-type RalA or the RalAS194A mutant. Immunoblot analysis verified appropriate knockdown of endogenous RalA and subsequent reexpression of the shRNA-resistant forms of RalA at levels roughly similar to that of endogenous RalA in all nine cell lines (Fig. 3B). All 36 cell lines were then assayed for anchorage-independent growth.
FIG. 3.
RalA S194 is broadly required for transformed growth of pancreatic cancer cell lines. (A) Detection of Aurora-A and RalA by immunoblot analysis of the indicated pancreatic cancer cell lines, compared with the results for three normal (N) pancreatic tissue specimens. (B) Immunoblot analysis of RalA in the indicated nine pancreatic cell lines, each retrovirally infected with a scramble sequence (S) or shRNA against RalA (RalAi) complemented by an empty vector (V), shRNA-resistant RalA in the wild-type (WT) or S194A mutant (SA) configuration. Actin serves as a loading control. (C) Photographs illustrating anchorage-independent growth in soft agar of the indicated polyclonal pancreatic cancer cells stably expressing either a RalA scramble sequence (scram) or RalA shRNA complemented with an empty vector (vector), the shRNA-resistant wild-type RalA protein (RalA WT), or the S194A mutant RalA protein (RalAS194A). Shown are average numbers of colonies formed ± SEM as calculated from triplicate plates. Data are from one representative experiment of two independent assays. Significant P values (<0.001) are indicated by **. Tukey's multiple-comparison test was used to determine significance between cell lines.
Consistent with the known role of RalA in transformation (36, 38), knockdown of RalA expression significantly impaired the anchorage-independent growth of all nine parental cell lines, ranging from 55% to 90% decrease in colony numbers. This decrease in transformed growth was, in large part, overcome by ectopic expression of the shRNA-resistant wild-type RalA protein. However, expression of a protein that was exactly the same except that it harbored the single point mutation at S194A that renders RalA resistant to Aurora-A phosphorylation extinguished the ability of RalA to rescue the loss of endogenous RalA and restore transformed cell growth in each of the nine cell lines (Fig. 3C). Based on these data, S194 appears to be broadly required for RalA promotion of anchorage-independent proliferation of human pancreatic cancer cells.
RalA S194 is required for tumorigenic growth of some human pancreatic cancer cell lines.
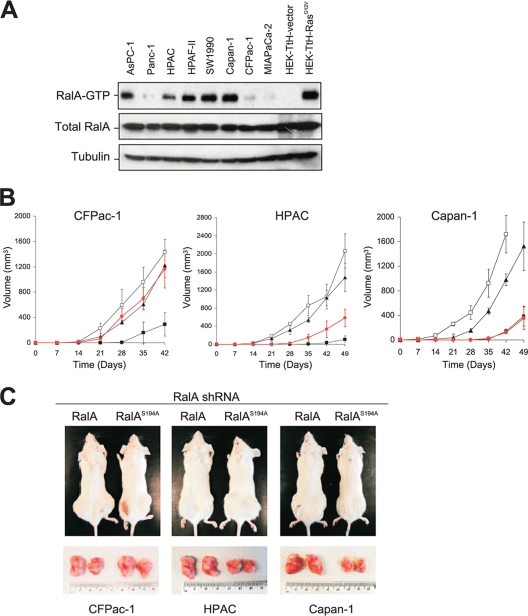
Since RalA is critical for tumorigenic growth of pancreatic cancer cells in vivo (38), we next determined whether S194 is also required for tumorigenesis of such cells. The CFPac-1, HPAC, and Capan-1 cell lines, which exhibit differing levels of RalA-GTP (Fig. 4A), were engineered to stably express scramble control shRNA, RalA shRNA, or a RalA protein resistant to RalA shRNA in either the wild-type or the S194A mutant configuration. These lines were injected subcutaneously into both flanks of immunocompromised mice. Compared to what was found for scramble control cells, loss of RalA prolonged the latency period of tumorigenesis at least twofold and impeded subsequent tumor growth in all three pancreatic cancer lines (Fig. 4B and C). In Capan-1 and HPAC cells, which have prominent RalA-GTP (Fig. 4A), RalAS194A was defective in restoring tumor growth to the same level as wild-type RalA (Fig. 4B and C), whereas in CFPac-1 cells, with weak RalA-GTP, wild-type and S194A RalA proteins were equally effective. The finding that RalAS194A did not rescue soft agar growth (Fig. 3C) but did rescue tumor growth of CFPac1 cells treated with RalA shRNA (Fig. 4B and C) suggests that RalA, but not Aurora-A, may be required for tumor growth. Whether there is an inverse correlation between the level of RalA-GTP (Fig. 4A) and the degree of tumor growth restoration by RalAS194A versus wild-type RalA (Fig. 4B and C) or this is simply a coincidence remains to be determined. If the former is the case, this occurrence may reflect the possibility that cells with higher basal levels of RalA-GTP are more dependent for tumorigenicity on RalA function and are therefore more sensitive to perturbations in RalA modulation. If the latter is the case, the variability observed may simply mean that not all oncogenic Ras-driven cancer cells equally require Aurora-A phosphorylation of RalA to promote tumorigenesis.
FIG. 4.
RalA S194 is required for tumorigenesis of pancreatic cancer cell lines. (A) RalA-GTP levels as detected in the indicated pancreatic cancer cell lines. HEK-TtH cells expressing empty vector or RasG12V serve as a negative or positive control, respectively. Total RalA and tubulin serve as loading controls. (B) Shown are tumor volumes (mm3) ± standard deviations versus times (days) observed for the indicated cell lines stably expressing a Ral scramble sequence (□), RalA-shRNA (▪), or RalA-shRNA complemented by expression of shRNA-resistant wild-type RalA (▴) or RalAS194A (•) injected into the flanks of immunocompromised mice. (C) Representative subcutaneous flank tumors observed in mice (top) and resected (bottom) from RalA shRNA-treated CFPac-1, HPAC, or Capan-1 cells transduced with shRNA-resistant wild-type RalA or the RalAS194A mutant at 42 days (CFPac-1) or 49 days (HPAC and Capan-1) after the cells were injected.
Aurora-AT288D promotes the translocation of endogenous RalA from the plasma membrane.
The C-terminal hypervariable domain of RalA, but not RalB, contains the S194 Aurora-A phosphorylation site, and the hypervariable regions of both proteins are known to account for the partially overlapping but distinct subcellular membrane localization patterns of the two proteins (36, 57). Further, phosphorylation by protein kinase C (PKC) of a similarly situated C-terminal serine in K-Ras4B (S181) can induce translocation of K-Ras4B from the plasma membrane to internal organelles (6). Given these observations, we speculated that in addition to its role in activating RalA (66), Aurora-A may alter the subcellular location of RalA.
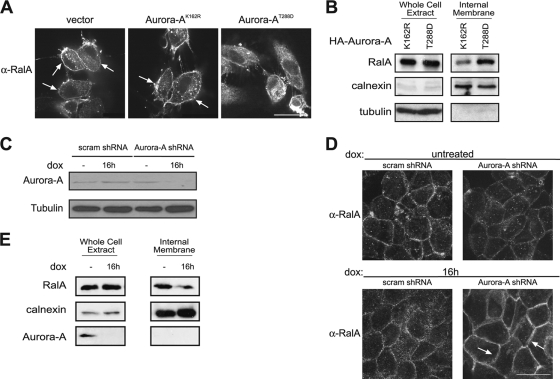
To address this possibility, we monitored the subcellular localization of endogenous RalA in both gain (ectopic Aurora-A)- and loss (Aurora-A shRNA)-of-function situations. In the gain-of-function approach, the subcellular distribution of endogenous RalA was first assessed by immunofluorescence in HEK-TtH cells. In cells that stably expressed either empty vector or kinase-inactive Aurora-AK162R, endogenous RalA was found abundantly at the plasma membrane as well as throughout the cytoplasm and at internal membranes. However, in cells expressing kinase-active Aurora-AT288D, there was a clear loss of RalA at the plasma membrane and a concomitant increase in the internal pool of protein (Fig. 5A), and some RalA colocalized with Rab11 and EEA1, markers of recycling endosomes and early endosomes, respectively (not shown). This effect of Aurora-A kinase activity on accumulation of RalA on internal membranes was borne out by biochemical fractionation. Specifically, in the presence of kinase-active Aurora-AT288D, RalA was greatly enriched in the isolated internal membrane fraction (defined by high levels of the ER-resident protein calnexin), compared to the amount seen in that fraction in the presence of kinase-inactive Aurora-AK162R (Fig. 5B). Furthermore, ectopic Aurora-A, whether in the kinase-active or -inactive mutant form, did not alter the levels of endogenous RalA (Fig. 5B).
FIG. 5.
Aurora-A regulates RalA subcellular localization. (A) Immunofluorescence demonstrating distribution of endogenous RalA by use of an anti-RalA antibody in HEK-TtH cells stably expressing an empty vector, kinase-inactive HA (epitope-tagged)-Aurora-AK162R, or kinase-active HA-Aurora-AT288D. Arrows, plasma membrane localization. Scale bar, 20 μm. (B) Immunoblot analysis of endogenous RalA, calnexin, and tubulin in a whole-cell extract and an internal membrane fraction isolated from HEK-TtH cells expressing HA-Aurora-AK162R or HA-Aurora-A-T288D. (C) Reduction in endogenous Aurora-A protein, as detected by immunoblot analysis, in HPAC cells expressing doxycycline (dox)-inducible Aurora-A shRNA treated with dox for 16 h, compared to the level for untreated cells or dox-treated cells expressing a dox-inducible scramble control shRNA. Tubulin serves as a loading control. (D) Relocalization of endogenous RalA from the cytoplasm to the plasma membrane, as detected by immunofluorescence, in HPAC cells expressing dox-inducible Aurora-A shRNA treated with dox for 16 h, compared to the level for untreated cells or dox-treated cells expressing a dox-inducible scramble control shRNA. Arrows, plasma membrane localization. Scale bar, 20 μm. (E) Immunoblot analysis of endogenous RalA, calnexin, and tubulin in a whole-cell extract and an internal membrane fraction isolated from HPAC cells expressing dox-inducible Aurora-A shRNA treated with dox for 16 h, compared to the level for untreated cells.
In the loss-of-function analysis, we first tested whether reducing the level of endogenous Aurora-A affected the localization of endogenous RalA in the human pancreatic cancer cell line HPAC (46), chosen as an example of cells that harbor a mutated KRAS allele (38), exhibit elevated levels of Aurora-A protein (Fig. 3A), and are sensitive to Aurora-A phosphorylation of RalAS194 for transformation (Fig. 3C) and tumorigenicity (Fig. 4B and C). Because constitutive knockdown of endogenous Aurora-A expression leads to rapid G2/M arrest and subsequently to apoptosis (26), HPAC cells were engineered to express a doxycycline-sensitive TET repressor and Aurora-A shRNA (or the scramble sequence) driven by a TET-ON operator (37), such that Aurora-A protein levels could be inducibly reduced in the presence of doxycycline (Fig. 5C). As expected, in scramble control cells with or without doxycycline in the culture medium, endogenous RalA was detected by immunofluorescence throughout the cytoplasm, at intracellular organelles, and at the plasma membrane along cell-cell borders. On the other hand, cells induced to express Aurora-A shRNA exhibited a noticeable increase in the concentration of RalA protein at the plasma membrane and a concomitant loss at internal membranes (Fig. 5D), which was even more obvious upon biochemical fractionation (Fig. 5E). Thus, by both gain- and loss-of-function analyses, we demonstrate that, akin to PKC promoting translocation of K-Ras4B from the plasma membrane (6), Aurora-A kinase activity promotes dissociation of endogenous RalA from the plasma membrane.
Phosphorylation of S194 by Aurora-AT288D is required for redistribution of RalA.
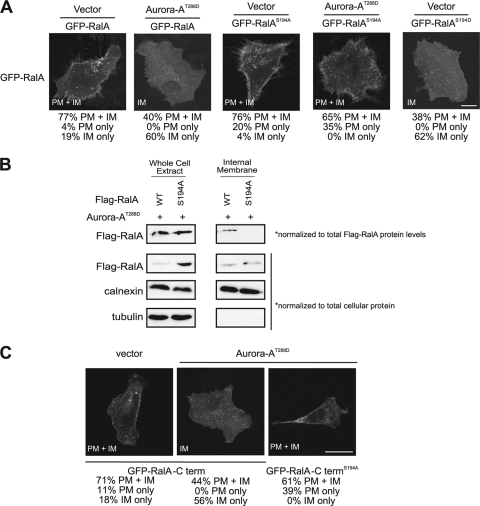
To address whether S194 of RalA is required for redistribution of RalA in the presence of Aurora-A, we expressed GFP-tagged RalAS194A in which the Aurora-A phosphorylation site was mutated in HEK-TtH cells ectopically expressing either kinase-active Aurora-AT288D or no transgene (vector). Cells expressing kinase-active Aurora-AT288D were smaller, possibly because constitutive activation of Aurora-A may promote more-rapid cell division (Fig. 6A). We found by immunofluorescence analysis that RalAS194A was retained in the plasma membrane, whether Aurora-A was activated or not. In contrast, GFP-RalAS194D, a putative phosphomimetic version of RalA, was observed primarily at internal membranes, consistent with the localization of GFP-RalA in the presence of kinase-active Aurora-AT288D (Fig. 6A). These observations were also supported by biochemical fractionation. In the presence of kinase-active Aurora-AT288D, a higher proportion of the total wild-type RalA was found in the internal membrane fraction than in the S194A mutant, after controlling for different levels of exogenous RalA expression (Fig. 6B). In support of a specific effect of Aurora-A on RalA, expression of kinase-active Aurora-AT288D did not alter the subcellular localization of GFP-RalB (see Fig. S3 in the supplemental material), which is highly similar to RalA but lacks the Aurora-A phosphorylation site (66), or of K-Ras4B (see Fig. S2 in the supplemental material), in which phosphorylation of a similar C-terminal serine residue by PKC displaces GFP-K-Ras from the plasma membrane (6). Lastly, we demonstrate that the last 20 amino acids of RalA are sufficient for Aurora-A-mediated relocalization of RalA. Specifically, a GFP fusion protein with the most-C-terminal 20 amino acids of RalA, comprising the hypervariable membrane targeting domain and containing the Aurora-A phosphorylation site, was relocalized from the plasma membrane upon expression of kinase-active Aurora-AT288D. Moreover, this relocalization was blocked when S194 was mutated to alanine (Fig. 6C). Thus, relocalization of RalA in the presence of Aurora-AT288D depends upon S194 of RalA.
FIG. 6.
Aurora-AT288D-mediated internalization of RalA depends upon S194. (A) Distribution of GFP-RalA constructs in HEK-TtH cells stably expressing vector or HA-Aurora-AT288D in combination with GFP-RalA or GFP-RalAS194A, compared to the level for HEK-TtH cells expressing the phosphomimetic mutant RalA protein GFP-RalAS194D with vector alone. GFP-RalA localization to both the plasma membrane and the internal membrane (PM+IM), the plasma membrane only (PM), or the internal membrane only (IM) in 50 cells was quantitated in two independent experiments for each condition. Representative images with the primary location are displayed. Scale bar, 20 μm. (B) Immunoblot analysis of endogenous RalA, calnexin, and tubulin in a whole-cell extract and an internal membrane fraction isolated from HEK-TtH cells expressing HA-Aurora-AT288D with either wild-type or S194A Flag-RalA. Eightfold more RalAS194A than wild-type RalA was expressed in whole-cell extract. Therefore, wild-type Flag-RalA levels in the whole-cell extract and the internal membrane fraction were normalized 8:1 to S194A Flag-RalA levels. (C) Distribution of the last 20 amino acids of the RalA C terminus fused to GFP (GFP-RalA-C term) or, as indicated, GFP-RalA-C termS194A in HEK-TtH cells stably expressing either an empty vector, HA-Aurora-AK162R, or HA-Aurora-AT288D. GFP-RalA localization to both the plasma membrane and the internal membrane (PM+IM), the plasma membrane only (PM), or the internal membrane only (IM) in 50 cells was quantitated in two independent experiments for each condition. Representative images with the primary location are displayed. Scale bar, 20 μm.
The RalA effector RalBP1 is translocated from the plasma membrane and activated in the presence of Aurora-AT288D.
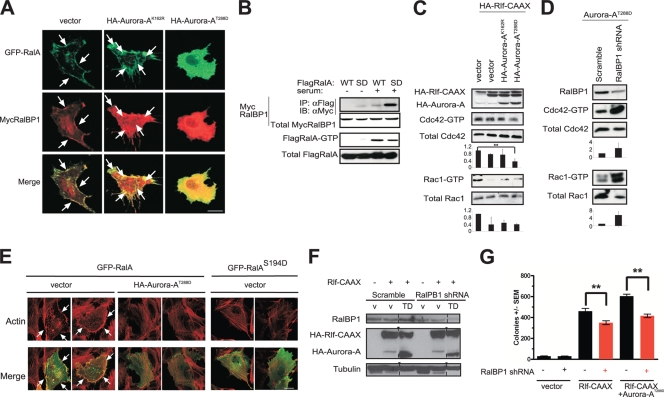
To explore the relationship of RalA with its effectors in the context of Aurora-A-mediated phosphorylation, we determined whether Aurora-A kinase activity, which alters RalA subcellular localization, also alters that of a key RalA effector, RalBP1/RLIP76 (10, 29, 50). We compared the localization of RalBP1 to that of ectopic RalA in HEK-TtH cells expressing kinase-inactive Aurora-AK162R versus kinase-active Aurora-AT288D. In the presence of kinase-inactive Aurora-AK162R, ectopic RalBP1 colocalized with RalA in the cytoplasm, internal structures, and protrusions of the plasma membrane. In the presence of kinase-active Aurora-AT288D, the plasma membrane pools of both RalA and RalBP1 repartitioned internally (Fig. 7A). These data indicate that Aurora-A coordinately regulates RalA and RalBP1. Expression of the effector domain mutant RalAD49N, which perturbs association of RalBP1 with RalA, did not result in changes in RalBP1 localization in the presence of kinase-inactive Aurora-AK162R versus kinase-active Aurora-AT288D (see Fig. S4 in the supplemental material), indicating that Aurora-A-mediated internalization of RalBP1 is dependent on the association between RalA and RalBP1. Furthermore, ectopic Aurora-A expression, either in the kinase-active or in the inactive mutant form, did not alter the levels of endogenous RalBP1 (see Fig. S5 in the supplemental material). To determine biochemically if this coordinate relocalization enhanced the association of RalBP1 with RalA, vectors encoding RalA (the wild type or the putative phosphomimetic S194D mutant) or RalBP1 were cotransfected into 293T cells, and the amount of RalBP1 coimmunoprecipitating with RalA was assessed by immunoblot analysis. In the absence of serum, when the majority of RalA is in the inactive GDP-bound state, RalBP1 did not readily coimmunoprecipitate with wild-type RalA (Fig. 7B) but did weakly associate with RalAS194D. Addition of serum to activate RalA promoted its association with RalBP1, and this interaction was enhanced nearly fivefold in the case of RalAS194D, compared to the level for wild-type RalA (Fig. 7B). However, Aurora-A activity did not affect all Ral effector interactions equally. Another validated Ral effector, Sec5, neither redistributed in the presence of kinase-active Aurora-AT288D nor preferentially bound RalAS194D (see Fig. S6 in the supplemental material). Compared to what was found for wild-type RalA, the association between RalAS194D and Sec5 was reduced by about one-third; however, the nature of this difference is unknown (see Fig. S6 in the supplemental material). Whether the selective influence of Aurora-A on effector association is due to the context of RalA activation, subcellular localization, or indirect effects, such as competition with RalB for the same effector, remains to be determined.
FIG. 7.
Aurora-A promotes cytoplasmic translocation and activation of RalBP1. (A) Distribution of GFP-RalA and Myc-tagged RalBP1 (MycRalBP1), visualized by immunofluorescence using an anti-Myc antibody in HEK-TtH cells stably expressing either a vector control, kinase-inactive HA-Aurora-AK162R, or kinase-active HA-Aurora-AT288D. Arrows, plasma membrane. Scale bar, 20 μm. (B) Aurora-A fosters the association of RalA with RalBP1. Immunoprecipitation (IP) of the Flag-tagged WT or the phosphomimetic S194D (SD) mutant version of RalA, followed by immunoblot analysis (IB) for detection of Flag-tagged, immunoprecipitated RalA protein or the presence or absence of coimmunoprecipitated Myc-tagged RalBP1 in the presence or absence of serum for activation of the RalA protein, as assessed by the level of Flag RalA-GTP. Total MycRalBP1 and Flag RalA serve as loading controls. (C) Aurora-A decreases Cdc42 and Rac1 activation. Shown are GTP-Cdc42 and GTP-Rac1 levels detected in HEK-TtH cells in which endogenous RalA was either not activated (vector) or activated by expressing HA-Rlf-CAAX in the presence, as assessed by immunoblot analysis, of either kinase-active HA-Aurora-AT288D or kinase-inactive HA-Aurora-AK162R. Total Cdc42 and Rac1 serve as loading controls. Cdc42-GTP and Rac1-GTP levels are normalized to levels for total Cdc42 and Rac1, expressed as fold changes ± standard deviations for three independent experiments (P = 0.029). (D) Knockdown of RalBP1 activates Cdc42 and Rac1. Shown are GTP-Cdc42 and GTP-Rac1 levels in HEK-TtH cells stably expressing shRNA against either the vector control or RalBP1. Total Cdc42 and Rac1 serve as loading controls. Cdc42-GTP and Rac1-GTP levels are normalized to the levels for total Cdc42 and Rac1, expressed as fold changes ± standard deviations for three independent experiments. (E) Distribution of GFP-RalA or GFP-RalAS194D and actin organization, visualized by immunofluorescence using Texas Red-phalloidin in HEK-TtH cells stably expressing either a vector control or kinase-active HA-Aurora-AT288D. Formation of filopodia and lamellipodia in 50 cells was quantitated in two independent experiments for each condition. Representative images are displayed. Arrows, filopodia or lamellipodia. Scale bar, 20 μm. (F) Appropriate expression, as detected by immunoblot analysis, of HA-Aurora-AT288D (TD), HA-Rlf-CAAX, or a knockdown of endogenous RalBP1 in HEK-TtH cells stably infected with retroviruses carrying no transgene (v) or the indicated transgenes in the presence of shRNA specific to RalBP1 or a scrambled version (scram) of this sequence. Tubulin serves as a loading control. (G) Anchorage-independent growth in soft agar of the aforementioned polyclonal HEK-TtH cells, infected with retroviruses encoding the indicated shRNAs and carrying the indicated transgenes, expressed as average numbers of colonies formed ± SEM for six plates (two independent experiments conducted in triplicate). Significant P values (<0.001) are indicated by **. Tukey's multiple-comparison test was used to determine significance between cell lines.
Given that compared to RalA, the RalAS194D mutant bound, if anything, more strongly to RalBP1 than to Sec5, we explored a possible relationship between RalBP1 and Aurora-A. Because RalBP1 has been shown to be a GTPase-activating protein (GAP) for two Rho family GTPases, Rac1 and Cdc42 (10, 29, 50), we investigated the signaling consequences of the observed increased association between RalA and RalBP1. Specifically, we examined the levels of endogenous activated GTP-bound Cdc42 and Rac1 as readouts of RalBP1 GAP activity and assessed whether the expression of kinase-inactive Aurora-AK162R or kinase-active Aurora-AT288D altered RalBP1 GAP activity. To do this, we first activated endogenous RalA in HEK-TtH cells by expression of Rlf-CAAX and measured the amounts of activated GTP-bound Cdc42 and Rac1 in the presence of either kinase-active Aurora-AT288D or kinase-inactive Aurora-AK162R. In cells expressing kinase-active Aurora-AT288D, activation of RalA signaling by Rlf-CAAX reduced the level of GTP-bound Cdc42 by one-half to one-third compared to the level for vector control cells (Fig. 7C). The nature of the selectivity toward decreased Cdc42-GTP levels compared to Rac1-GTP levels is unknown but, we speculate, could be due to differences in subcellular localization. As RalBP1 GAP activity is not altered upon binding activated RalA in vitro (50), recruitment of RalBP1 to specific subcellular sites may instead underlie the reduction in GTP-bound Cdc42. Furthermore, shRNA-mediated knockdown of RalBP1, as confirmed by immunoblot analysis, increased the levels of both activated GTP-bound Rac1 and Cdc42 both in the presence and in the absence of Aurora-AT288D (Fig. 7D; see also Fig. S7 in the supplemental material). Such a result is not surprising if indeed RalBP1 acts as a GAP for Cdc42 and Rac1. These results are consistent with a model in which Aurora-A-mediated phosphorylation of RalA leads to an enhanced interaction with RalBP1, perhaps to alter its subcellular localization, resulting in a decrease in GTP-bound Cdc42.
We next explored the cellular consequences of the increased association between RalA and RalBP1 and the decrease in GTP-bound Cdc42 promoted by Aurora-AT288D. Cdc42 activation causes formation of actin microspikes and filopodia, whereas Rac activation promotes concentration of actin at the leading edge of moving cells and the formation of lamellipodia. Therefore, we assayed for changes in cellular morphology in HEK-TtH cells transiently expressing GFP-RalA in the presence of either empty vector or kinase-active Aurora-AT288D. We first confirmed that stable expression of kinase-inactive Aurora-AK162R or kinase-active Aurora-AT288D did not alter actin cytoskeleton organization in the absence of exogenous RalA (see Fig. S8 in the supplemental material). Consistent with the biochemical decreases in Cdc42- and Rac1-GTP levels in the presence of kinase-active Aurora-AT288D (Fig. 7C), ∼76% of HEK-TtH cells expressing GFP-RalA and empty vector displayed filopodia and lamellipodia, whereas ∼30% of cells expressing both GFP-RalA and kinase-active Aurora-AT288D displayed filopodia and lamellipodia, indicative of decreases in both Cdc42 and Rac1 activations (Fig. 7E). In agreement, HEK-TtH cells transiently expressing a phosphomimetic mutant of RalA, GFP-RalAS194D, displayed an absence of filopodia and lamellipodia (Fig. 7E). Taken together, these data support the notion that Aurora-AT288D expression increases the association between RalA and its effector RalBP1, leading to enhanced RalBP1 function, as measured by decreased Cdc42- and Rac1-GTP levels and decreased Rho family GTPase-driven morphology.
These studies demonstrate that Aurora-A has a biological impact on RalA activation of RalBP1, but it remains to be determined whether such activation of RalBP1, or even suppression of Cdc42, fosters transformation. On one hand, RalBP1 contributes positively to transformation. Specifically, knockdown of RalBP1 in HEK-TtH cells transformed by Rlf-CAAX in the absence or presence of Aurora-AT288D, as confirmed by immunoblot analysis (Fig. 7F), exhibited reduced anchorage-independent growth, compared to the level for the scramble control counterparts (Fig. 7G). The basis of this decreased transformation is unknown, but given that RalBP1 knockdown cells can be cultured extensively with no overt impact on passaging (not shown) and that mice homozygous for a gene trap mutation of RALBP1 (RIP1 and RLIP76) are viable (3), the decrease in anchorage-independent growth may be related to a function RalBP1 plays in transformation, although this remains to be formally tested. Similarly, preventing RalA association with RalBP1 by introduction of the D49N effector domain mutation within a constitutively active RalAQ72L background slightly reduced transformation (33). On the other hand, RalA also contributes to transformation by pathways aside from RalBP1. Specifically, knockdown of RalBP1 (Fig. 7G) did not reduce transformation to the same extent as knockdown of RalA (Fig. 2C). Similarly, a D49E mutation, which inhibits binding of Sec5 and Exo84, reduced the transforming activity of RalAQ72L to a greater extent than the D49N mutation (33). Since RalBP1 could potentially serve as a GAP for other untested Rho GTPases (29) and has additional activities (2, 30, 67), it also remains to be tested if Cdc42 is the relevant target for RalBP1 in transformation. Moreover, activation of Cdc42 has been reported to both promote and suppress transformation. While RNA interference (RNAi) suppression of a Cdc42GEF or dominant-negative Cdc42 enhanced soft agar growth of human colon carcinoma cells (44), activation of Cdc42 promoted transformation of rodent fibroblasts (51). Thus, while activation of RalBP1 and suppression of Cdc42 constitute one plausible mechanism for promoting transformation, others are certainly possible.
Differential effects of Aurora-A versus Aurora-AT288D on RalA functions.
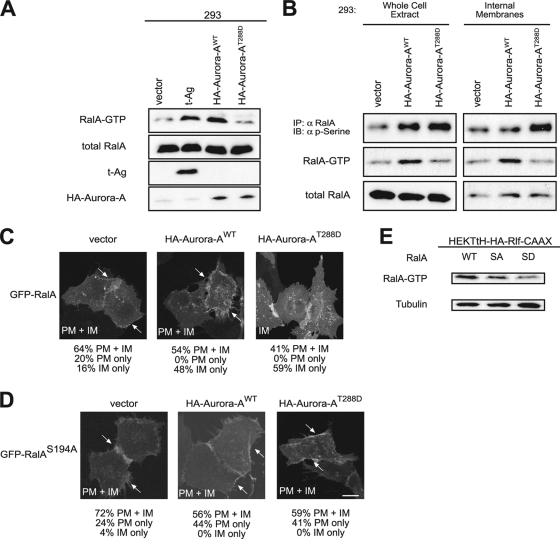
Aurora-A is upregulated, rather than mutationally activated, in human cancers (22, 31); hence, we also explored the effect on RalA function in cells expressing wild-type versus constitutively active Aurora-A. It has been demonstrated that expression of either version of Aurora-A resulted in elevated levels of GTP-bound and thus active RalA and that mutation of S194, but not S183, reduced this effect (66). Similarly, knockdown of the PP2A phosphatase subunit aβ has been found to lead to elevated levels of both RalA S183 and S194 phosphorylation and GTP-bound RalA (56). These data support the notion that phosphorylation of S194 activates wild-type RalA. We thus measured the level of RalA-GTP in human cells expressing Aurora-A versus Aurora-AT288D. Specifically, human 293 cells were transiently transfected with a vector carrying no transgene (as a negative control), encoding t-Ag (as a positive control, since t-Ag blocks the ability of PP2A to dephosphorylate S183 and S194 of RalA, resulting in elevated phosphorylation at these sites [56]), encoding Aurora-AT288D, or encoding wild-type Aurora-A. Expression of these transgenes and of endogenous RalA was confirmed by immunoblot analysis, and the levels of activated GTP-bound endogenous RalA were assessed by pulldown analyses (Fig. 8A). As previously reported (56), t-Ag promoted robust activation of RalA, compared to the level for vector control cells. Similarly, wild-type Aurora-A was highly effective in activating RalA. Unexpectedly, expression of Aurora-AT288D did not cause a significant increase in RalA-GTP, compared to the level for vector control cells (Fig. 8B). Since Aurora-AT288D potentiates Ral transforming activity (Fig. 2C; see also Fig. 9B, C, and D) yet does not robustly activate RalA (Fig. 8B), Aurora-A may foster Ral-mediated transformation by additional mechanisms independent of RalA-GTP formation.
FIG. 8.
Differential effects of Aurora-A versus Aurora-AT288D on RalA functions. (A) Appropriate expression, as detected by immunoblot analysis, of t-Ag, wild-type HA-Aurora-A, kinase-active HA-Aurora-AT288D, endogenous RalA, and GTP-bound RalA levels (detected by GST pulldown of endogenous RalA) in 293 cells expressing the indicated transgenes. Total RalA serves as a loading control. (B) Immunoblot analysis of phosphorylated RalA (detected by immunoprecipitation of endogenous RalA followed by immunoblot analysis with an antiphosphoserine [α p-serine] antibody) and RalA GTP-levels (detected by GST pulldown of endogenous RalA) in a whole-cell extract and an internal membrane fraction isolated from 293 cells expressing the indicated transgenes. (C, D) Distribution of GFP-RalA or GFP-RalAS194A in HEK-TtH cells stably expressing vector, HA-Aurora-AWT, or HA-Aurora-AT288D. Arrows, plasma membrane localization. GFP-RalA localization to both the plasma membrane and the internal membrane (PM+IM), the plasma membrane only (PM), or the internal membrane only (IM) in 50 cells was quantitated in two independent experiments for each condition. Representative images with the primary location are displayed. Arrows, plasma membrane localization. Scale bar, 20 μm. (E) Immunoblot analysis of RalA-GTP levels in HEK-TtH cells stably expressing HA-Rlf-CAAX and wild-type RalA (WT), RalAS194A (SA), or RalAS194D (SD). Tubulin serves as a loading control.
FIG. 9.
Wild-type versus kinase-active Aurora-A-mediated potentiation of Ras-induced transformation through the RalGEF pathway. (A) Appropriate expression, as detected by immunoblot analysis, of empty vector (v), wild-type HA-Aurora-A (WT), kinase-active HA-Aurora-AT288D (TD), kinase-inactive HA-Aurora-AK162R (KR), constitutively activated HA-Rlf-CAAX, or ectopic and endogenous Ras (Pan-Ras). Actin serves as a loading control. (B, C, D) Anchorage-independent growth in soft agar of polyclonal HEK-TtH cells stably expressing the indicated transgenes, expressed as average numbers of colonies formed ± SEM for six plates (two independent experiments conducted in triplicate). The same vector (v) (with [+] or without [−] HA-Aurora-A [WT], HA-Aurora-AT288D [TD], or HA-Aurora-AK162R [KR]) was used as a control in all of these experiments. Significant P values (<0.001) are indicated by **. Tukey's multiple-comparison test was used to determine significance between cell lines.
Given this result, we tested whether phosphorylation and subcellular localization of RalA were differentially affected by Aurora-A versus Aurora-AT288D. 293 cells were therefore stably infected with retroviruses carrying no transgene, encoding Aurora-A, or encoding Aurora-AT288D. RalA phosphorylation levels were measured by immunoprecipitation of endogenous RalA, followed by immunoblotting with a phospho-specific serine antibody, and endogenous RalA-GTP and total RalA levels were determined by pulldowns and immunoblot analyses total cellular lysates versus lysates derived from fractionated internal membranes. As expected (66), RalA serine phosphorylation was elevated upon expression of Aurora-A, and this was further increased in cells expressing constitutively active Aurora-AT288D. In contrast to their differential effects on RalA phosphorylation and GTP loading, both versions of this kinase resulted in higher levels of total RalA in the internal membrane fraction, although a greater proportion of the internal pool was phosphorylated in cells expressing Aurora-AT288D (Fig. 8B). This result was independently validated by immunofluorescence; GFP-RalA accumulation at internal membranes increased in cells expressing either version of Aurora-A (Fig. 8C), and in both cases, mutating S194A in RalA reduced this translocation (Fig. 8D). The reason for the increase in RalA phosphorylation with no corresponding increase in GTP loading in the presence of Aurora-AT288D is unclear. One possibility is that perhaps increased phosphorylation of RalA by Aurora-AT288D promotes translocation of phosphorylated RalA away from the plasma membrane, where RalGEF proteins associate with activated Ras to then reduce RalGEF-mediated RalA activation. In support of this model, RalAS194D, a putative phosphomimetic version of RalA, was enriched at internal membranes, compared to the level for wild-type RalA (Fig. 6A), but was less active (lower GTP levels), when coexpressed with an activated, plasma membrane-targeted RalGEF protein, than the wild-type or even the S194A mutant of RalA (Fig. 8E). Nevertheless, it still remains to be resolved why robust phosphorylation of RalA by Aurora-AT288D does not coincide with elevated RalA-GTP levels.
Wild-type versus activated-Aurora-A-mediated transformation.
Wild-type Aurora-A and Aurora-AT288D both increased RalA phosphorylation and promoted RalA translocation, whereas wild-type but not constitutively active Aurora-AT288D potently increased RalA-GTP levels. Capitalizing on the ability of these two forms of Aurora-A to differentially alter RalA functions, we tested whether elevated RalA-GTP or endomembrane translocation was associated with Aurora-A-mediated transformation in cells with activated Ral signaling. HEK-TtH cells in which the Ras-RalGEF pathway was activated at different levels by expression of oncogenic Ras (RasG12V), an oncogenic Ras mutant that preferentially activates RalGEF proteins (RasG12V,E37G) or an activated RalGEF protein (Rlf-CAAX), were stably infected with retroviruses either carrying no transgene or encoding kinase-inactive Aurora-AK162R (as a negative control), Aurora-A, or Aurora-AT288D. Expression was assessed by immunoblot analysis (Fig. 9A), and cells were assayed for anchorage-independent growth. As noted previously (Fig. 1), Aurora-AT288D, but not negative-control kinase-inactive Aurora-AK126R, cooperated to various degrees with all three activators of Ral to promote transformation. Wild-type Aurora-A enhanced this transformation only marginally better than Aurora-AT288D in RasG12V and RasG12V,E37G backgrounds (∼1.1-fold) (Fig. 9B and C) but approximately twofold in the Rlf-CAAX background (Fig. 9D). Given that only Aurora-A robustly activates RalA-GTP, whereas both Aurora-A and Aurora-AT288D promote translocation of RalA and growth transformation, their ability to enhance transformation was attributed more to endomembrane translocation than to formation of RalA-GTP in these experiments. Conversely, however, the ability of constitutively activated RalA (RalAQ72L) to promote anchorage-independent growth of HEK-TtH cells was not altered upon introduction of either the S194A mutation or the S194D mutation (see Fig. S9 in the supplemental material). Thus, in the context of constitutively active RalA, the loss of S194 phosphorylation has no effect on RalA transforming activity.
Summary.
Aurora-A normally functions as a mitotic kinase to ensure proper chromosome separation and cytokinesis (42). This kinase is frequently overexpressed in various human cancers and can become mislocalized to the cytoplasm, where it may phosphorylate inappropriate substrates, leading to both genomic instability and altered signaling, promoting cancerous development (22, 31, 32). We find that activated Aurora-A cooperates with the RalGEF-Ral effector signaling arm of oncogenic Ras to promote transformation in human model cells (Fig. 1 and 2) and, further, that transformation (Fig. 3) and in some cases also tumor growth (Fig. 4) of pancreatic cancer cells characterized by oncogenic Ras mutations depend upon S194 of RalA, the site phosphorylated by Aurora-A (66). In contrast to RalA, the nearly identical RalB protein cannot support anchorage-independent growth transformation or tumorigenicity of human cells (36, 38) and is neither phosphorylated (66) nor required for Aurora-A to promote RalGEF transformation of HEK-TtH cells (see Fig. S1 in the supplemental material). Thus, in addition to its effect on chromosome stability, aberrant overexpression or cell cycle-independent expression of Aurora-A in cancer may also foster transformation and tumorigenesis through phosphorylation of RalA, a key substrate of the oncogenic Ras-RalGEF effector pathway (66). Indeed, the finding (9) of Aurora-A overexpression in a cell cycle-independent fashion and its localization throughout tumor cells, as opposed to its restriction to the nuclei of normal cells, suggests a nonmitotic role for this kinase when it is aberrantly expressed. Overexpression of Aurora-A is not, however, the only mode of fostering the tumorigenic activity of RalA through phosphorylation. Phosphatase PP2A Aβ also affects both the phosphorylation status and the tumorigenicity of RalA in HEK cells (56). Thus, RalA activation through RalGEF stimulation by oncogenic Ras in cooperation with phosphorylation by kinase activation or phosphatase inactivation promotes tumorigenesis.
We also report that phosphorylation of RalA by Aurora-A leads to internalization of RalA and to elevated RalBP1 GAP activity (Fig. 5, 6, and 7). The significance of differential subcellular localization and effector utilization upon Aurora-A-mediated phosphorylation of RalA remains to be determined. We speculate that Aurora-A phosphorylation of RalA at S194 promotes internalization of RalA, and increases in the association of RalA with RalBP1 may spatially restrict the activation of Cdc42 and Rac1, thereby leading to changes in actin dynamics. It is not clear if these changes underlie the above-mentioned requirement for RalA phosphorylation in transformation and tumorigenesis. Increased RalA translocation upon expression of kinase-active Aurora-AT288D is transforming and depends upon S194 phosphorylation, even in the absence of robust additional activation of RalA-GTP. In contrast, mutating the S194 Aurora-A phosphorylation site did not alter the ability of constitutively activated RalA to transform cells (see Fig. S9 in the supplemental material). Whether this reflects pleiotropic effects of Aurora-AT288D or the ability of an oncogenic activated mutation in RalA to overcome the need for phosphorylation for transformation is unclear. Thus, while certainly the increased GTP loading of RalA in the presence of activated Aurora-A promotes transformation, it is unclear if the same holds true for internalization of RalA and, if so, whether this reflects suppression of Cdc42 activity or another effect.
Although our studies relate only to the situation in cancer cells, it is possible that regulation of RalA by Aurora-A in normal cells may also play a role in normal entry and exit from mitosis, as Aurora-A is required for chromosome separation and cytokinesis (1, 18, 41, 43, 68), RalA is also required for proper cytokinesis (11-13), and the RalBP1 homolog cytocentrin is involved in proper centrosome duplication and segregation (52). In normal cells, Aurora-A-mediated phosphorylation of RalA may promote the activation of the RalA-RalBP1 complex in a spatially restricted manner to promote the switching off of endocytosis during mitosis to ensure proper mitotic entry (13, 55). Even so, as both kinase-active and -inactive versions of Aurora-A can impair mitosis (43), yet only the kinase-active mutant transforms rodent cell lines (5) and, as we demonstrate here, human cells in cooperation with oncogenic Ras, the effect of Aurora-A on cell transformation may not be solely through chromosome instability. Phosphorylation of RalA either by a decrease in phosphatase PP2A aβ expression (56) or by ectopic expression of Aurora-A (66) has been found to increase the level of RalA-GTP.
In summary, first, we and others (56, 66) report that the concurrent activations of two seemingly disparate proteins, Ras and Aurora-A, converge through a common protein, RalA, to promote tumorigenesis. Aurora kinase inhibitors are currently under clinical evaluation for cancer treatment (32, 39). Thus, RalA may be a potential target for some of the antitumor activity of these inhibitors. In turn, RAS mutations may be a genetic determinant for patient response to these inhibitors and establish RalA phosphorylation at S194 as an important biomarker for their antitumor efficacy. Second, we found that Aurora-A promotes both internalization of RalA in an S194-dependent fashion and activation of RalBP1, as measured by reduced Cdc42-GTP levels. Whether this internalization of RalA plays a role in transformation or instead reflects another function of the protein remains to be determined.
Supplementary Material
Acknowledgments
We thank Mike White for plasmids pMT3-mycRalBP1 and pcDNA3-mycSec5.
This work is supported by NIH grants CA94184 and CA126903 (C.M.C.), CA42978 and CA67771 (C.J.D. and A.D.C.), and CA109550 (A.D.C.). C.M.C. is a Leukemia and Lymphoma Scholar, D.F.K. is a Leukemia and Lymphoma Fellow, and K.-H.L. and B.B.A. were Department of Defense Breast Cancer Research Predoctoral Scholars. D.C.B. is supported by an NIH T32 Training Fellowship.
Footnotes
Published ahead of print on 9 November 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Anand, S., S. Penrhyn-Lowe, and A. R. Venkitaraman. 2003. AURORA-A amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to Taxol. Cancer Cell 3:51-62. [DOI] [PubMed] [Google Scholar]
- 2.Awasthi, S., J. Cheng, S. S. Singhal, M. K. Saini, U. Pandya, S. Pikula, J. Bandorowicz-Pikula, S. V. Singh, P. Zimniak, and Y. C. Awasthi. 2000. Novel function of human RLIP76: ATP-dependent transport of glutathione conjugates and doxorubicin. Biochemistry 39:9327-9334. [DOI] [PubMed] [Google Scholar]
- 3.Awasthi, S., S. S. Singhal, S. Yadav, J. Singhal, K. Drake, A. Nadkar, E. Zajac, D. Wickramarachchi, N. Rowe, A. Yacoub, P. Boor, S. Dwivedi, P. Dent, W. E. Jarman, B. John, and Y. C. Awasthi. 2005. RLIP76 is a major determinant of radiation sensitivity. Cancer Res. 65:6022-6028. [DOI] [PubMed] [Google Scholar]
- 4.Bagrodia, S., S. J. Taylor, C. L. Creasy, J. Chernoff, and R. A. Cerione. 1995. Identification of a mouse p21Cdc42/Rac activated kinase. J. Biol. Chem. 270:22731-22737. [DOI] [PubMed] [Google Scholar]
- 5.Bischoff, J. R., L. Anderson, Y. Zhu, K. Mossie, L. Ng, B. Souza, B. Schryver, P. Flanagan, F. Clairvoyant, C. Ginther, C. S. Chan, M. Novotny, D. J. Slamon, and G. D. Plowman. 1998. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 17:3052-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bivona, T. G., S. E. Quatela, B. O. Bodemann, I. M. Ahearn, M. J. Soskis, A. Mor, J. Miura, H. H. Wiener, L. Wright, S. G. Saba, D. Yim, A. Fein, I. Perez de Castro, C. Li, C. B. Thompson, A. D. Cox, and M. R. Philips. 2006. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol. Cell 21:481-943. [DOI] [PubMed] [Google Scholar]
- 7.Bos, J. L. 1989. ras oncogenes in human cancer: a review. Cancer Res. 49:4682-4689. [PubMed] [Google Scholar]
- 8.Bozidis, P., C. D. Williamson, and A. M. Colberg-Poley. 2007. Isolation of endoplasmic reticulum, mitochondria, and mitochondria-associated membrane fractions from transfected cells and from human cytomegalovirus-infected primary fibroblasts. Curr. Protoc. Cell Biol. 37:3.27.1-3.27.23. [DOI] [PubMed] [Google Scholar]
- 9.Burum-Auensen, E., P. M. De Angelis, A. R. Schjolberg, K. L. Kravik, M. Aure, and O. P. Clausen. 2007. Subcellular localization of the spindle proteins Aurora A, Mad2, and BUBR1 assessed by immunohistochemistry. J. Histochem. Cytochem. 55:477-486. [DOI] [PubMed] [Google Scholar]
- 10.Cantor, S. B., T. Urano, and L. A. Feig. 1995. Identification and characterization of Ral-binding protein 1, a potential downstream target of Ral GTPases. Mol. Cell. Biol. 15:4578-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cascone, I., R. Selimoglu, C. Ozdemir, E. Del Nery, C. Yeaman, M. White, and J. Camonis. 2008. Distinct roles of RalA and RalB in the progression of cytokinesis are supported by distinct RalGEFs. EMBO J. 27:2375-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cawthon, R. M., P. O'Connell, A. M. Buchberg, D. Viskochil, R. B. Weiss, M. Culver, J. Stevens, N. A. Jenkins, N. G. Copeland, and R. White. 1990. Identification and characterization of transcripts from the neurofibromatosis 1 region: the sequence and genomic structure of EVI2 and mapping of other transcripts. Genomics 7:555-565. [DOI] [PubMed] [Google Scholar]
- 13.Chen, X. W., M. Inoue, S. C. Hsu, and A. R. Saltiel. 2006. RalA-exocyst-dependent recycling endosome trafficking is required for the completion of cytokinesis. J. Biol. Chem. 281:38609-38616. [DOI] [PubMed] [Google Scholar]
- 14.Chien, Y., S. Kim, R. Bumeister, Y. M. Loo, S. W. Kwon, C. L. Johnson, M. G. Balakireva, Y. Romeo, L. Kopelovich, M. Gale, Jr., C. Yeaman, J. H. Camonis, Y. Zhao, and M. A. White. 2006. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell 127:157-170. [DOI] [PubMed] [Google Scholar]
- 15.Chien, Y., and M. A. White. 2003. RAL GTPases are linchpin modulators of human tumour-cell proliferation and survival. EMBO Rep. 4:800-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choudhury, A., M. Dominguez, V. Puri, D. K. Sharma, K. Narita, C. L. Wheatley, D. L. Marks, and R. E. Pagano. 2002. Rab proteins mediate Golgi transport of caveola-internalized glycosphingolipids and correct lipid trafficking in Niemann-Pick C cells. J. Clin. Invest. 109:1541-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cifone, M. A., and I. J. Fidler. 1980. Correlation of patterns of anchorage-independent growth with in vivo behavior of cells from a murine fibrosarcoma. Proc. Natl. Acad. Sci. U. S. A. 77:1039-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cowley, D. O., J. A. Rivera-Perez, M. Schliekelman, Y. J. He, T. G. Oliver, L. Lu, R. O'Quinn, E. D. Salmon, T. Magnuson, and T. Van Dyke. 2009. Aurora-A kinase is essential for bipolar spindle formation and early development. Mol. Cell. Biol. 29:1059-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Downward, J. 2003. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 3:11-22. [DOI] [PubMed] [Google Scholar]
- 20.Feig, L. A. 2003. Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol. 13:419-425. [DOI] [PubMed] [Google Scholar]
- 21.Fukushige, S., F. M. Waldman, M. Kimura, T. Abe, T. Furukawa, M. Sunamura, M. Kobari, and A. Horii. 1997. Frequent gain of copy number on the long arm of chromosome 20 in human pancreatic adenocarcinoma. Genes Chromosomes Cancer 19:161-169. [DOI] [PubMed] [Google Scholar]
- 22.Giet, R., C. Petretti, and C. Prigent. 2005. Aurora kinases, aneuploidy and cancer, a coincidence or a real link? Trends Cell Biol. 15:241-250. [DOI] [PubMed] [Google Scholar]
- 23.Gigoux, V., S. L'Hoste, F. Raynaud, J. Camonis, and C. Garbay. 2002. Identification of Aurora kinases as RasGAP Src homology 3 domain-binding proteins. J. Biol. Chem. 277:23742-23746. [DOI] [PubMed] [Google Scholar]
- 24.Hahn, W. C., C. M. Counter, A. S. Lundberg, R. L. Beijersbergen, M. W. Brooks, and R. A. Weinberg. 1999. Creation of human tumour cells with defined genetic elements. Nature 400:464-468. [DOI] [PubMed] [Google Scholar]
- 25.Hamad, N. M., J. H. Elconin, A. E. Karnoub, W. Bai, J. N. Rich, R. T. Abraham, C. J. Der, and C. M. Counter. 2002. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev. 16:2045-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hata, T., T. Furukawa, M. Sunamura, S. Egawa, F. Motoi, N. Ohmura, T. Marumoto, H. Saya, and A. Horii. 2005. RNA interference targeting aurora kinase A suppresses tumor growth and enhances the taxane chemosensitivity in human pancreatic cancer cells. Cancer Res. 65:2899-2905. [DOI] [PubMed] [Google Scholar]
- 27.Hezel, A. F., A. C. Kimmelman, B. Z. Stanger, N. Bardeesy, and R. A. Depinho. 2006. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 20:1218-1249. [DOI] [PubMed] [Google Scholar]
- 28.Jones, S., X. Zhang, D. W. Parsons, J. C. Lin, R. J. Leary, P. Angenendt, P. Mankoo, H. Carter, H. Kamiyama, A. Jimeno, S. M. Hong, B. Fu, M. T. Lin, E. S. Calhoun, M. Kamiyama, K. Walter, T. Nikolskaya, Y. Nikolsky, J. Hartigan, D. R. Smith, M. Hidalgo, S. D. Leach, A. P. Klein, E. M. Jaffee, M. Goggins, A. Maitra, C. Iacobuzio-Donahue, J. R. Eshleman, S. E. Kern, R. H. Hruban, R. Karchin, N. Papadopoulos, G. Parmigiani, B. Vogelstein, V. E. Velculescu, and K. W. Kinzler. 2008. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321:1801-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jullien-Flores, V., O. Dorseuil, F. Romero, F. Letourneur, S. Saragosti, R. Berger, A. Tavitian, G. Gacon, and J. H. Camonis. 1995. Bridging Ral GTPase to Rho pathways. RLIP76, a Ral effector with CDC42/Rac GTPase-activating protein activity. J. Biol. Chem. 270:22473-22477. [DOI] [PubMed] [Google Scholar]
- 30.Jullien-Flores, V., Y. Mahe, G. Mirey, C. Leprince, B. Meunier-Bisceuil, A. Sorkin, and J. H. Camonis. 2000. RLIP76, an effector of the GTPase Ral, interacts with the AP2 complex: involvement of the Ral pathway in receptor endocytosis. J. Cell Sci. 113:2837-2844. [DOI] [PubMed] [Google Scholar]
- 31.Katayama, H., W. R. Brinkley, and S. Sen. 2003. The Aurora kinases: role in cell transformation and tumorigenesis. Cancer Metastasis Rev. 22:451-464. [DOI] [PubMed] [Google Scholar]
- 32.Keen, N., and S. Taylor. 2004. Aurora-kinase inhibitors as anticancer agents. Nat. Rev. Cancer 4:927-936. [DOI] [PubMed] [Google Scholar]
- 33.Kendall, S. D., S. J. Adam, and C. M. Counter. 2006. Genetically engineered human cancer models utilizing mammalian transgene expression. Cell Cycle 5:1074-1079. [DOI] [PubMed] [Google Scholar]
- 34.Khosravi-Far, R., M. A. White, J. K. Westwick, P. A. Solski, M. Chrzanowska-Wodnicka, L. Van Aelst, M. H. Wigler, and C. J. Der. 1996. Oncogenic Ras activation of Raf/mitogen-activated protein kinase-independent pathways is sufficient to cause tumorigenic transformation. Mol. Cell. Biol. 16:3923-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, D., J. Zhu, P. F. Firozi, J. L. Abbruzzese, D. B. Evans, K. Cleary, H. Friess, and S. Sen. 2003. Overexpression of oncogenic STK15/BTAK/Aurora A kinase in human pancreatic cancer. Clin. Cancer Res. 9:991-997. [PubMed] [Google Scholar]
- 36.Lim, K. H., A. T. Baines, J. J. Fiordalisi, M. Shipitsin, L. A. Feig, A. D. Cox, C. J. Der, and C. M. Counter. 2005. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell 7:533-745. [DOI] [PubMed] [Google Scholar]
- 37.Lim, K. H., and C. M. Counter. 2005. Reduction in the requirement of oncogenic Ras signaling to activation of PI3K/AKT pathway during tumor maintenance. Cancer Cell 8:381-392. [DOI] [PubMed] [Google Scholar]
- 38.Lim, K. H., K. O'Hayer, S. J. Adam, S. D. Kendall, P. M. Campbell, C. J. Der, and C. M. Counter. 2006. Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr. Biol. 16:2385-2394. [DOI] [PubMed] [Google Scholar]
- 39.Malumbres, M., and M. Barbacid. 2007. Cell cycle kinases in cancer. Curr. Opin. Genet. Dev. 17:60-65. [DOI] [PubMed] [Google Scholar]
- 40.Malumbres, M., and M. Barbacid. 2003. RAS oncogenes: the first 30 years. Nat. Rev. Cancer 3:459-465. [DOI] [PubMed] [Google Scholar]
- 41.Marumoto, T., S. Honda, T. Hara, M. Nitta, T. Hirota, E. Kohmura, and H. Saya. 2003. Aurora-A kinase maintains the fidelity of early and late mitotic events in HeLa cells. J. Biol. Chem. 278:51786-51795. [DOI] [PubMed] [Google Scholar]
- 42.Marumoto, T., D. Zhang, and H. Saya. 2005. Aurora-A—a guardian of poles. Nat. Rev. Cancer 5:42-50. [DOI] [PubMed] [Google Scholar]
- 43.Meraldi, P., R. Honda, and E. A. Nigg. 2002. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53-/- cells. EMBO J. 21:483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitin, N., L. Betts, M. E. Yohe, C. J. Der, J. Sondek, and K. L. Rossman. 2007. Release of autoinhibition of ASEF by APC leads to CDC42 activation and tumor suppression. Nat. Struct. Mol. Biol. 14:814-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moskalenko, S., D. O. Henry, C. Rosse, G. Mirey, J. H. Camonis, and M. A. White. 2002. The exocyst is a Ral effector complex. Nat. Cell Biol. 4:66-72. [DOI] [PubMed] [Google Scholar]
- 46.Norman, J., M. Franz, R. Schiro, S. Nicosia, J. Docs, P. J. Fabri, and W. R. Gower, Jr. 1994. Functional glucocorticoid receptor modulates pancreatic carcinoma growth through an autocrine loop. J. Surg. Res. 57:33-38. [DOI] [PubMed] [Google Scholar]
- 47.O'Hayer, K. M., and C. M. Counter. 2006. A genetically defined normal human somatic cell system to study ras oncogenesis in vivo and in vitro. Methods Enzymol. 407:637-647. [DOI] [PubMed] [Google Scholar]
- 48.Okada, T., T. Sawada, T. Osawa, M. Adachi, and K. Kubota. 2008. MK615 inhibits pancreatic cancer cell growth by dual inhibition of Aurora A and B kinases. World J. Gastroenterol. 14:1378-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pamonsinlapatham, P., R. Hadj-Slimane, F. Raynaud, M. Bickle, C. Corneloup, A. Barthelaix, Y. Lepelletier, P. Mercier, M. Schapira, J. Samson, A. L. Mathieu, N. Hugo, O. Moncorge, I. Mikaelian, S. Dufour, C. Garbay, and P. Colas. 2008. A RasGAP SH3 peptide aptamer inhibits RasGAP-Aurora interaction and induces caspase-independent tumor cell death. PLoS ONE 3:e2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park, S. H., and R. A. Weinberg. 1995. A putative effector of Ral has homology to Rho/Rac GTPase activating proteins. Oncogene 11:2349-2355. [PubMed] [Google Scholar]
- 51.Qiu, R. G., A. Abo, F. McCormick, and M. Symons. 1997. Cdc42 regulates anchorage-independent growth and is necessary for Ras transformation. Mol. Cell. Biol. 17:3449-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quaroni, A., and E. C. Paul. 1999. Cytocentrin is a Ral-binding protein involved in the assembly and function of the mitotic apparatus. J. Cell Sci. 112:707-718. [DOI] [PubMed] [Google Scholar]
- 53.Repasky, G. A., E. J. Chenette, and C. J. Der. 2004. Renewing the conspiracy theory debate: does Raf function alone to mediate Ras oncogenesis? Trends Cell Biol. 14:639-647. [DOI] [PubMed] [Google Scholar]
- 54.Rodriguez-Viciana, P., P. H. Warne, A. Khwaja, B. M. Marte, D. Pappin, P. Das, M. D. Waterfield, A. Ridley, and J. Downward. 1997. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell 89:457-467. [DOI] [PubMed] [Google Scholar]
- 55.Rosse, C., S. L'Hoste, N. Offner, A. Picard, and J. Camonis. 2003. RLIP, an effector of the Ral GTPases, is a platform for Cdk1 to phosphorylate epsin during the switch off of endocytosis in mitosis. J. Biol. Chem. 278:30597-30604. [DOI] [PubMed] [Google Scholar]
- 56.Sablina, A. A., W. Chen, J. D. Arroyo, L. Corral, M. Hector, S. E. Bulmer, J. A. DeCaprio, and W. C. Hahn. 2007. The tumor suppressor PP2A Abeta regulates the RalA GTPase. Cell 129:969-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shipitsin, M., and L. A. Feig. 2004. RalA but not RalB enhances polarized delivery of membrane proteins to the basolateral surface of epithelial cells. Mol. Cell. Biol. 24:5746-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith, S. C., G. Oxford, A. S. Baras, C. Owens, D. Havaleshko, D. L. Brautigan, M. K. Safo, and D. Theodorescu. 2007. Expression of ral GTPases, their effectors, and activators in human bladder cancer. Clin. Cancer Res. 13:3803-3813. [DOI] [PubMed] [Google Scholar]
- 59.Tatsuka, M., S. Sato, S. Kitajima, S. Suto, H. Kawai, M. Miyauchi, I. Ogawa, M. Maeda, T. Ota, and T. Takata. 2005. Overexpression of Aurora-A potentiates HRAS-mediated oncogenic transformation and is implicated in oral carcinogenesis. Oncogene 24:1122-1127. [DOI] [PubMed] [Google Scholar]
- 60.Wang, X., Y. X. Zhou, W. Qiao, Y. Tominaga, M. Ouchi, T. Ouchi, and C. X. Deng. 2006. Overexpression of aurora kinase A in mouse mammary epithelium induces genetic instability preceding mammary tumor formation. Oncogene 25:7148-7158. [DOI] [PubMed] [Google Scholar]
- 61.Warner, S. L., R. M. Munoz, D. J. Bearss, P. Grippo, H. Han, and D. D. Von Hoff. 2008. Pdx-1-driven overexpression of aurora A kinase induces mild ductal dysplasia of pancreatic ducts near islets in transgenic mice. Pancreas 37:e39-e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.White, M. A., C. Nicolette, A. Minden, A. Polverino, L. Van Aelst, M. Karin, and M. H. Wigler. 1995. Multiple Ras functions can contribute to mammalian cell transformation. Cell 80:533-541. [DOI] [PubMed] [Google Scholar]
- 63.Wolthuis, R. M., B. Bauer, L. J. van 't Veer, A. M. de Vries-Smits, R. H. Cool, M. Spaargaren, A. Wittinghofer, B. M. Burgering, and J. L. Bos. 1996. RalGDS-like factor (Rlf) is a novel Ras and Rap 1A-associating protein. Oncogene 13:353-362. [PubMed] [Google Scholar]
- 64.Wolthuis, R. M., N. D. de Ruiter, R. H. Cool, and J. L. Bos. 1997. Stimulation of gene induction and cell growth by the Ras effector Rlf. EMBO J. 16:6748-6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wolthuis, R. M., B. Franke, M. van Triest, B. Bauer, R. H. Cool, J. H. Camonis, J. W. Akkerman, and J. L. Bos. 1998. Activation of the small GTPase Ral in platelets. Mol. Cell. Biol. 18:2486-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu, J. C., T. Y. Chen, C. T. Yu, S. J. Tsai, J. M. Hsu, M. J. Tang, C. K. Chou, W. J. Lin, C. J. Yuan, and C. Y. Huang. 2005. Identification of V23RalA-Ser194 as a critical mediator for Aurora-A-induced cellular motility and transformation by small pool expression screening. J. Biol. Chem. 280:9013-9022. [DOI] [PubMed] [Google Scholar]
- 67.Yadav, S., S. S. Singhal, J. Singhal, D. Wickramarachchi, E. Knutson, T. B. Albrecht, Y. C. Awasthi, and S. Awasthi. 2004. Identification of membrane-anchoring domains of RLIP76 using deletion mutant analyses. Biochemistry 43:16243-16253. [DOI] [PubMed] [Google Scholar]
- 68.Zhang, D., T. Hirota, T. Marumoto, M. Shimizu, N. Kunitoku, T. Sasayama, Y. Arima, L. Feng, M. Suzuki, M. Takeya, and H. Saya. 2004. Cre-loxP-controlled periodic Aurora-A overexpression induces mitotic abnormalities and hyperplasia in mammary glands of mouse models. Oncogene 23:8720-8730. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.