Abstract
Upon DNA damage, eukaryotic cells activate a conserved signal transduction cascade known as the DNA damage checkpoint (DDC). We investigated the influence of DDC kinases on nucleotide excision repair (NER) in Saccharomyces cerevisiae and found that repair of both strands of an active gene is affected by Mec1 but not by the downstream checkpoint kinases, Rad53 and Chk1. Repair of the nontranscribed strand (by global genome repair) requires new protein synthesis, possibly reflecting the involvement of Mec1 in the activation of repair genes. In contrast, repair of the transcribed strand by transcription-coupled NER (TC-NER) occurs in the absence of new protein synthesis, and DNA damage results in Mec1-dependent but Rad53-, Chk1-, Tel1-, and Dun1-independent phosphorylation of the TC-NER factor Rad26, a member of the Swi/Snf group of ATP-dependent translocases and yeast homologue of Cockayne syndrome B. Mutation of the Rad26 phosphorylation site results in a decrease in the rate of TC-NER, pointing to direct activation of Rad26 by Mec1 kinase. These findings establish a direct role for Mec1 kinase in transcription-coupled repair, at least partly via phosphorylation of Rad26, the main transcription-repair coupling factor.
The genomes of living cells are constantly challenged by a variety of endogenous and exogenous agents capable of damaging DNA. In order to maintain genomic stability, elaborate pathways to repair DNA damage have evolved (22, 42). The most versatile of these is nucleotide excision repair (NER), which deals with a variety of helix-distorting lesions, including UV light-induced cyclobutane-pyrimidine dimers (CPDs) and those caused by the UV-mimetic 4-nitroquinoline-1-oxide (4NQO), as well as other bulky chemical adducts and various inter- and intrastrand cross-links (41, 60). The importance of NER is exemplified by the existence of various human syndromes that are linked to defects in this repair pathway, including xeroderma pigmentosum (XP), Cockayne syndrome (CS), and trichothiodystrophy (TTD) (10).
NER is a multistep process comprising lesion detection, helix opening, the formation of dual incisions around the site of damage, and, lastly, repair synthesis. The core reaction, involving more than 30 proteins, can be reconstituted in vitro using purified components from both humans (1, 35) and Saccharomyces cerevisiae (18). Two subpathways of NER exist: transcription-blocking lesions in the transcribed strand (TS) of active genes are repaired quickly by transcription-coupled NER (TC-NER), whereas the rest of the genome, including the nontranscribed strand (NTS) of active genes, is repaired more slowly by global genome NER (GG-NER) (8, 33, 34, 44, 54).
Damage detection in TC-NER is carried out by elongating RNA polymerase II (RNAPII) itself (reviewed in reference 50). When RNAPII stalls at a lesion in the TS of a gene, it somehow recruits a transcription-repair coupling factor that initiates NER. In humans, the best-understood TC-NER factor is the Cockayne syndrome B protein (CSB) (56). Mutations in CSB give rise to CS, a severe disease characterized by sensitivity to sunlight, neurological degeneration, growth defects, skeletal abnormalities, and mental retardation (10). The exact mechanism of eukaryotic TC-NER and the basis of most CS phenotypes remain unclear (36).
The Saccharomyces cerevisiae homologue of CSB, Rad26, was cloned based on sequence similarity and is required for normal TC-NER (58). However, in contrast to mammalian cells lacking CSB, yeast cells lacking RAD26 are not UV sensitive. In addition, budding yeast has a second TC-NER pathway, which is dependent on Rpb9, a nonessential subunit of RNAPII (29). The reason that rad26Δ cells are not UV sensitive is almost certainly that yeast cells can remove DNA damage also in the transcribed strand rather efficiently by GG-NER, a pathway requiring the RAD7 and RAD16 gene products. Indeed, cells lacking one of these GG-NER genes as well as RAD26 are much more UV sensitive than cells lacking only the GG-NER gene (59).
Another process utilized by eukaryotic cells to preserve genomic integrity in the presence of DNA damage is the complex signal transduction cascade known as the DNA damage checkpoint (DDC), which leads to temporary cell cycle arrest (reviewed in reference 37). This increases survival by allowing additional time for repair and by preventing replication and segregation of the damaged genome. Checkpoint mutants were first isolated because of their inability to delay cell cycle progression into mitosis after gamma irradiation (61, 62). Like many other signal transduction cascades, the DNA damage checkpoint proteins can be subdivided into groups of proteins acting at various steps: damage sensors, signal transducers, and effectors. Presently, about 20 proteins have been identified as components of the damage checkpoint (7). In Saccharomyces cerevisiae, the most important of these is the Mec1 protein kinase, the yeast homologue of human ATR (26, 39, 45). Mec1 controls the checkpoint by direct phosphorylation of target proteins but also via phosphorylation of downstream effector kinases, most notably the kinases Rad53 and Chk1 (11, 43, 49).
In addition to preventing cell cycle progression in the presence of DNA lesions, it is widely accepted, but rarely demonstrated, that the checkpoint may also enhance the repair capacity of cells. A primary mechanism by which this is achieved may be upregulation of genes encoding factors relevant for repair. Indeed, approximately 5% of the yeast genome is upregulated in response to various DNA-damaging agents (15, 23, 24). Many of these genes are regulated by Mec1, indicating an involvement of the DNA damage checkpoint in the DNA damage-related transcriptional response (15).
However, little is known about whether DNA repair factors are also directly regulated by checkpoint factors, for example via their phosphorylation. Examples of repair proteins that are phosphorylated by checkpoint kinases in budding yeast include Rad55 and Nej1 (3, 21), modification of which leads to enhanced repair of double-strand breaks. Less is known about direct phosphorylation of NER factors by the checkpoint kinases, although a central NER protein in human cells, XPA, is known to be phosphorylated by ATR after UV irradiation. Such phosphorylation is required for normal levels of survival after UV treatment. ATR also controls nuclear import of XPA after DNA damage, but this is possibly independent of phosphorylation (64, 65).
In this study, we show that NER in budding yeast is severely compromised in cells lacking the Mec1 kinase (but not in cells lacking the downstream checkpoint kinases Rad53 and Chk1), and we identify a novel Mec1 target, the TC-NER factor Rad26. Loss of Mec1-catalyzed Rad26 phosphorylation decreases the rate of TC-NER, indicating that this modification is required for optimal Rad26 function during DNA repair.
MATERIALS AND METHODS
Yeast strains.
All the Saccharomyces cerevisiae strains used were congenic with strain W303 (leu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-1) (55) and were grown and manipulated using standard techniques. A detailed list of the strains used in this study can be found in Table 1. Details about N-terminal tagging of RAD26 are available upon request.
TABLE 1.
- Saccharomyces cerevisiae strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| W303 1A | MATaleu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 | |
| W303 1B | MATα leu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 | |
| MGSC102 | MATα leu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 rad26Δ::HIS3 | 58 |
| MGSC126 | MATα leu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 rad16Δ::LEU2 | 59 |
| MGSC107 | MATα leu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 rad16Δ::LEU2 rad26Δ::HIS3 | 59 |
| JSY1105 | MATα leu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 URA3::MHRAD26 | This study |
| JSY1106 | MATα leu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 URA3::MHRAD26 rad16Δ::LEU2 | This study |
| JSY1107 | MATα leu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 rad16Δ::LEU2 rpb9::TRP1 | This study |
| JSY1108 | MATα leu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 rad16Δ::LEU2 rpb9::TRP1 rad26::HIS3 | This study |
| JSY1109 | MATα leu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 URA3::MHRAD26 mec1Δ::HIS3 sml1Δ::TRP1 | This study |
| JSY1110 | MATα leu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 URA3::MHRAD26 chk1Δ::HIS3 | This study |
| JSY1111 | MATα leu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 URA3::MHRAD26 rad53Δ::HIS3 sml1Δ::TRP1 | This study |
| JSY1112 | MATaleu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 mec1Δ::HIS3 sml1Δ::TRP1 | This study |
| JSY1113 | MATaleu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 chk1Δ::HIS3 | This study |
| JSY1114 | MATaleu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 rad53Δ::HIS3 sml1Δ::TRP1 | This study |
| JSY1115 | MATaleu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 mec1Δ::HIS3 sml1Δ::TRP1 rad26::KanMx | This study |
Plasmids.
The genomic MHRAD26 locus was amplified by PCR and cloned into pRS316 (47). For galactose-inducible overexpression of MHRAD26, the open reading frame was cloned into the pYC2 vector (Invitrogen). Site-directed mutagenesis was carried out using the QuikChange II XL kit (Stratagene) according to the manufacturer's instructions, and the presence of the point mutations was confirmed by sequencing. Plasmids pHAMec1wt and pHAMec1kd were a generous gift from George S. Brush.
Yeast growth and treatment.
Yeast strains were grown to early log phase (1 × 107 to 2 × 107 cells/ml) in appropriate medium. For UV treatment, the cells were collected by centrifugation and plated on agar plates and UV irradiation was performed using a UV Stratalinker 2400 (Stratagene). Afterwards, the cells were washed off the plate into the relevant prewarmed growth medium, and post-UV incubation was carried out for the indicated times. Treatment with methyl methanesulfonate (MMS; 99% stock solution; Sigma), bleomycin (5-mg/ml stock solution in water), H2O2 (1 M stock solution in water), and 4NQO (10-mg/ml stock solution in dimethyl sulfoxide [DMSO]) was performed by adding the compounds directly to the medium.
SDS-PAGE, Western blotting, and antibodies.
Fast preparation of yeast protein extracts was carried out as described previously (28). For detection of Rad26 phosphorylation, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using Criterion XT 3 to 8% gradient precast gels (Bio-Rad), run until the 100-kDa marker reached the bottom of the gel. Western blotting was carried out using standard techniques. Myc-tagged Rad26 was detected using monoclonal 9E10 antibody (12), and hemagglutinin (HA)-tagged Mec1 was detected using polyclonal anti-HA antibody (Abcam).
Galactose-induced protein overexpression.
Yeast cells were grown in medium containing raffinose to a cell density of 5 × 106 cells/ml. Galactose was added to a final concentration of 2%, and incubation continued for 3 h. 4NQO was added at a concentration of 5 μg/ml for purification of phosphorylated Rad26. For repression of HA-Mec1 expression, 2% glucose was added to the medium instead of galactose.
MS.
Polyacrylamide gel slices (1 to 2 mm) containing Rad26 were prepared for mass spectrometric (MS) analysis using the Janus liquid handling system (Perkin-Elmer). Briefly, the excised protein gel piece was placed in a well of a 96-well microtiter plate and destained with 50% (vol/vol) acetonitrile and 50 mM ammonium bicarbonate, reduced with 10 mM dithiothreitol (DTT), and alkylated with 55 mM iodoacetamide. After alkylation, Rad26 was digested with 6 ng/μl trypsin (Promega) overnight (o/n) at 37°C. The resulting peptides were extracted in 1% (vol/vol) formic acid, 2% (vol/vol) acetonitrile. The digest was analyzed by nanoscale capillary liquid chromatography-tandem MS (LC-MS/MS) using a nanoAcquity ultraperformance liquid chromatograph (UPLC; Waters) to deliver a flow of approximately 300 nl/min. A C18 Symmetry 5-μm, 180-μm by 20-mm μ-Precolumn (Waters) trapped the peptides prior to separation on a C18 BEH130 1.7-μm, 75-μm by 100-mm analytical UPLC column (Waters). Peptides were eluted with a gradient of acetonitrile. The analytical column outlet was directly interfaced via a modified nanoflow electrospray ionization source, with a two-dimensional (2-D) linear ion trap mass spectrometer (LTQ XL/ETD; ThermoScientific), equipped with a chemical ionization source to enable the generation and injection of fluoranthene radical anions for the electron transfer dissociation (ETD) reaction (9). The ETD process uses ion/ion chemistry to provide sequence information not available through conventional methods such as collision-induced dissociation (CID). Peptide fragmentation using an ETD approach augments the current methodologies available for the characterization of posttranslational modifications by more accurately identifying the specific amino acids that are modified. Data-dependent analysis was carried out in either CID or ETD mode, where automatic MS/MS spectra were acquired on multiply charged precursor ions in the m/z range 300 to 2,000. As predominantly doubly protonated ions are generated by tryptic digestion, a supplemental activation energy was used to improve fragmentation in the ETD experiments (51). All LC-MS/MS data were then searched against a protein database (UniProt 13.6) using the Mascot search engine program (Matrix Science, United Kingdom), with oxidized methionine, carbamidomethyl cysteine and phosphoserine, threonine, and tyrosine included as variable modifications (40).
Preparation of yeast extracts and immunoprecipitation of Rad26.
Cell pellets were resuspended in yeast lysis buffer (150 mM Tris-acetate, pH 7.8, 20% glycerol, 50 mM potassium acetate, 3 mM EDTA, 5 mM DTT, 0.1% Triton X-100, 1× protease inhibitors) (38). Silica beads were added, and cells were disrupted using a FastPrep-24 tissue and cell homogenizer (MP Biomedicals). Immunoprecipitation was carried out overnight at 4°C using protein A-coated beads coupled to 9E10 antibody. Beads were washed 3 times consecutively with lysis buffer containing 500 mM potassium acetate, followed by 2 washes with tobacco etch virus (TEV) cleavage buffer (10 mM Tris-HCl, pH 8, 150 mM NaCl, 1 mM β-mercaptoethanol). TEV elution was performed by resuspending the beads in 50 μl of TEV elution buffer and adding 3 μg of TEV protease for 6 h at 4°C. The supernatant containing the eluted protein was harvested after centrifugation.
Strand-specific nucleotide excision repair analysis.
NER assays were carried out as described previously (52, 53) with minor modifications. Cells were grown to a cell density of 4 × 107 cells/ml in 400 ml of appropriate medium, harvested by centrifugation, and resuspended in 800 ml ice-cold phosphate-buffered saline (PBS; cell density, 2 × 107 cells/ml). This cell suspension was irradiated with 100 J/m2 UV-C light using a germicidal lamp. After treatment, the cells were again collected by centrifugation, resuspended in fresh medium, and allowed to recover at 30°C in the dark. Aliquots were removed either before UV irradiation (U), immediately after UV irradiation (0), or at the indicated time points after UV treatment (usually up to 4 h). The cell pellets were resuspended in 5 ml spheroblasting buffer (0.9 M sorbitol, 100 mM Tris-HCl, pH 8.0, 100 mM EDTA, 28 mM β-mercaptoethanol, 5 mg Zymolyase 20 T) and incubated o/n at 4°C in the dark. The resulting spheroblasts were lysed by resuspension in lysis buffer (2 M urea, 100 mM NaCl, 50 mM Tris-HCl, pH 8.0, 5 mM CDTA, 0.25% [wt/vol] n-lauroyl sarcosine), and the genomic DNA was isolated and purified using phenol-chloroform extraction and ethanol precipitation. The DNA was resuspended in Tris-EDTA (TE) to a final concentration of about 0.5 mg/ml. Fifty micrograms of genomic DNA for each time point was then digested with HaeIII. After purification of digested DNA, the samples were incubated with protein extract from Micrococcus luteus in order to achieve incision at CPDs (details available upon request), and the DNA was again purified and resuspended in 100 μl TE. Analysis of a HaeIII restriction fragment of the RPB2 gene was achieved using biotinylated probes for either the TS (5′-biotin-GATAGCTTTTTTCCGTTTACCGATTATGTTAAGATCAAAGAA-3′) or the NTS (5′-biotin-GATAGCTTTTTTCCAATAATGGACCTGCCAAATCTAATCT-3′). Hybridization of the probes (1 μl of 2 mM stock) was carried out in the presence of 1 M NaCl. The mixture of DNA and probe was heated to 95°C for 5 min and then incubated at 55°C for 15 min. Streptavidin-coated magnetic beads were added for 10 min, and then the mixture was washed with BW buffer (1 M NaCl, 5 mM Tris-HCl, pH 8.0, 0.5 mM EDTA). The beads were finally resuspended in 4 μl of water, and labeling was carried out using the Sequenase v2.0 kit (USB) according to the manufacturer's instructions. After the beads were washed several times in water, the labeled fragments were eluted with formamide loading buffer and separated on a 6% denaturing gel.
Quantification of damage half-lives (time required to remove 50% of the damages [t50%]).
Damage signals (total signal without signal for the undamaged band at the top of the gel) were determined using a phosphorimager and then normalized for the total signal in the respective lanes to correct for differences in loading. The nonspecific signals detected in the unirradiated lane (background) were subtracted from lanes displaying samples with damaged DNA. The signal obtained for time zero was set to 1, and the signals for the later time points were set in relation to that. A graph with a trend line was made using Microsoft Excel software, and the time point at which the trend line crossed the 0.5 value was determined empirically.
RESULTS
NER is compromised in checkpoint-deficient cells.
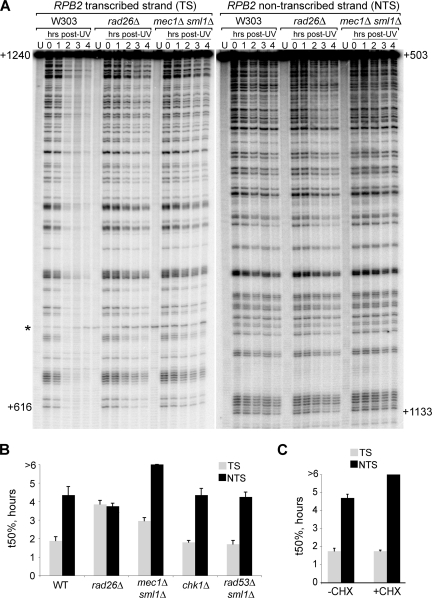
In order to investigate if, and to what extent, the central checkpoint kinase Mec1 affects NER in yeast, we measured the kinetics of CPD removal in a 740-bp fragment of the yeast RPB2 gene in a mec1Δ sml1Δ double mutant strain (deletion of SML1 is necessary to suppress the lethality of MEC1 deletion [66]), using a method which allows analysis of strand-specific repair at nucleotide resolution (52, 53). The repair rate in the checkpoint-deficient mutant was compared with that of the wild-type strain (W303; WT) and the TC-NER-deficient rad26Δ mutant. A representative gel is shown in Fig. 1A. The signals for the individual lanes were quantified using a phosphorimager, and the t50% was calculated for the individual strains (Fig. 1B). As expected, the difference in repair kinetics between the preferentially repaired TS and the NTS was evident in the WT strain, while no clear strand specificity was observed in TC-NER-deficient rad26Δ cells. Interestingly, cells lacking the checkpoint kinase Mec1 exhibited a dramatic impairment of NER, characterized by markedly slower repair of the TS and virtually no repair of the NTS at 4 h post-UV irradiation. A main role of Mec1 is to activate downstream checkpoint kinases, such as Rad53 and Chk1 (14). Interestingly, however, no NER defects were observed in the chk1Δ and rad53Δ sml1Δ strains (Fig. 1B and data not shown). Together, these results indicate that a normal DNA damage checkpoint, and more specifically the Mec1 kinase, is required for efficient repair by both NER subpathways.
FIG. 1.
Normal NER requires MEC1. (A) Representative sequencing gel showing a comparison of strand-specific repair in W303 (wild type), rad26Δ (TC-NER-deficient), and mec1Δ sml1Δ (checkpoint-deficient) cells. Numbers on the left and on the right of the gel indicate the nucleotide position relative to the RPB2 transcription start site on the TS and NTS, respectively. A nonspecific band appearing also in the unirradiated control sample (U) is marked with an asterisk. (B) Quantification of the signals from sequencing gels such as the one shown in panel A. Damages remaining at the different time points post-UV irradiation were calculated, and the time necessary for removal of 50% of the damages (t50%) was determined for both the TS and the NTS. Error bars show the standard error (2 independent experiments). (C) Quantification of NER in wild-type cells that were either left untreated (−CHX) or treated with 50 μg/ml cycloheximide (+CHX) for 1 h before UV treatment. Damage half-lives (t50%) are shown for the TS and the NTS, with error bars indicating the standard error (2 independent experiments).
Damage-induced de novo protein synthesis is necessary for GG-NER but not TC-NER.
The DNA damage checkpoint mediates transcriptional induction of many genes involved in a variety of cellular processes, including DNA repair (2, 15). Previous studies have shown that UV-induced de novo synthesis of proteins is required for repair of the NTS, but not the TS, of the active GAL10 and URA3 genes (5). In order to determine whether this is also the case for the RPB2 gene, we carried out NER assays on wild-type yeast treated with cycloheximide (CHX) (in order to abolish synthesis of new proteins) 1 h before UV treatment. Repair of the TS was not detectably affected by CHX treatment, while severely defective NTS repair was observed, similar to that observed in the mec1Δ sml1Δ strain (Fig. 1C). Together, these results are consistent with the idea that the defect in GG-NER observed in the checkpoint-deficient strain is due to defects in induced expression of factors required for dealing with DNA damage, while the defect in TC-NER must be caused by a preexisting factor(s).
Rad26 is phosphorylated after DNA damage.
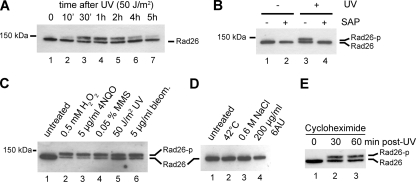
One possible explanation for the decrease in TC-NER in the mec1Δ sml1Δ strain is that a checkpoint kinase is phosphorylating a transcription-repair coupling factor to allow efficient damage removal in the TS. Since Rad26 is the best-characterized TC-NER protein in yeast, it was a primary candidate for investigation. We therefore generated a strain expressing Rad26, tagged N-terminally with 9 Myc epitopes and 8 histidine residues separated by 2 TEV cleavage sites (16), from its endogenous promoter within the genome (MHRAD26). Genetic characterization of the strain indicated that this protein is fully functional in TC-NER (data not shown). MHRAD26 cells were irradiated with UV-C light, and the SDS-PAGE mobility of tagged Rad26 was analyzed using an anti-Myc antibody. Interestingly, a slower-migrating form of the Rad26 protein became evident just 10 min after UV treatment, peaked between 1 and 2 h, and started disappearing thereafter (Fig. 2A). A longer exposure of the Western blots revealed that even in the absence of UV damage the mobility of a small amount of Rad26 was shifted (data not shown). To determine if phosphorylation of Rad26 was responsible for the observed shift in electrophoretic mobility, we immunoprecipitated Rad26 and treated the bead-bound material with shrimp alkaline phosphatase (SAP) to reverse phosphorylation. The slower-migrating protein was lost after incubation with phosphatase, indicating that it represents a phosphorylated form of Rad26 (Fig. 2B).
FIG. 2.
Damage-induced phosphorylation of Rad26. (A) Western blot analysis of Rad26 from wild-type cells harvested either before (0) or at various time points after UV irradiation. (B) Western blot analysis of Rad26 immunoprecipitated before and 2 h after UV treatment and treated either with buffer alone or with shrimp alkaline phosphatase. (C) Western blot analysis of Rad26 from undamaged cells and from cells treated with the indicated doses of various DNA-damaging agents for 1 h. (D) Western blot analysis of Rad26 from untreated wild-type cells or from cells treated with heat shock (shift from 30°C to 42°C), osmotic shock (addition of 600 mM NaCl to the medium), or 6AU (addition of 200 μg/ml to the medium) for 1 h. (E) Western blot analysis of Rad26 after UV treatment in the presence of the translation inhibitor cycloheximide.
Rad26 is phosphorylated by the Mec1 kinase.
To investigate whether Rad26 phosphorylation occurred specifically in response to UV irradiation or if it also occurs following other types of DNA damage, we now treated cells with various chemical agents to induce different types of DNA lesions. All of these DNA damage-inducing treatments caused Rad26 to be phosphorylated (Fig. 2C). Interestingly, other forms of cellular stress, including heat shock and osmotic shock, did not result in such modification (Fig. 2D). Because Rad26 is known to be recruited to stalled RNA polymerase II complexes and because UV-induced DNA lesions cause RNAPII stalling, we also tested whether 6-azauracil (6AU) could mediate Rad26 phosphorylation (6AU causes frequent RNAPII stalling by restricting nucleotide availability). However, treatment with 6AU did not cause a detectable increase in the phosphorylated form of Rad26 (Fig. 2D), suggesting that Rad26 modification is not a result of RNAPII stalling but rather represents a response to DNA damage. Finally, because we had observed that CHX addition affected GG-NER but not TC-NER, we also investigated the effect of CHX on Rad26 phosphorylation after DNA damage. CHX addition did not affect Rad26 phosphorylation (Fig. 2E).
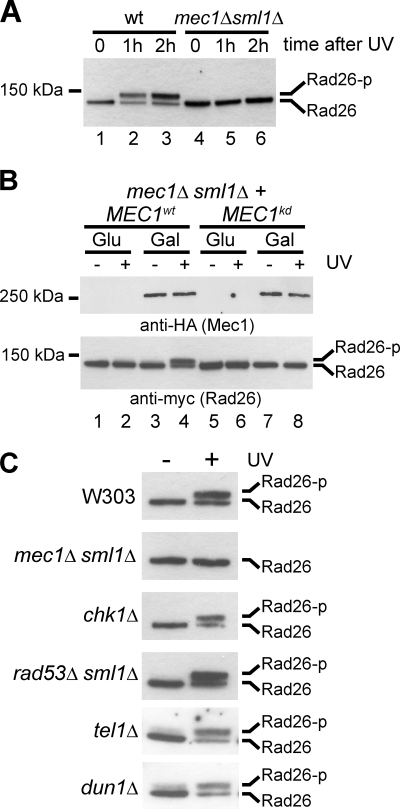
To determine whether a functional DNA damage checkpoint is required for Rad26 phosphorylation, we tested whether the modification occurs in the mec1Δ sml1Δ strain. Interestingly, it did not (Fig. 3A). To confirm that the lack of phosphorylation was due to the absence of Mec1 kinase activity and not an indirect result of concomitant MEC1 and SML1 deletion, we reintroduced galactose-inducible forms of HA-tagged wild-type MEC1 (MEC1wt) or kinase-dead MEC1 (MEC1kd) into these cells. Rad26 phosphorylation was recovered by growing cells carrying MEC1wt in galactose, but not glucose, while MEC1kd was unable to generate modified Rad26 despite being expressed at a level similar to the wild-type protein (Fig. 3B, compare lanes 3 and 4 with lanes 7 and 8). This indicates that the kinase activity of Mec1 is required for Rad26 phosphorylation.
FIG. 3.
The DNA damage checkpoint kinase Mec1 is required for Rad26 phosphorylation. (A) Western blot analysis of Rad26 phosphorylation before and after UV irradiation in wild-type and checkpoint-deficient mec1Δ sml1Δ cells. (B) Western blot analysis of Rad26 phosphorylation in mec1Δ sml1Δ cells carrying expression plasmids for HA-tagged wild-type or kinase-dead Mec1. Extracts were prepared from cells in which expression of Mec1 was either induced with galactose (Gal) or repressed with glucose (Glu), before and 1 h after UV irradiation. Mec1 and Rad26 proteins were detected using antibodies against HA and Myc epitopes, respectively. (C) Western blot analysis of Rad26 phosphorylation before and after UV irradiation in cells of the indicated genotype.
As shown above, Mec1, but not its downstream signal transducers, Chk1 and Rad53, is required for normal TC-NER. To determine if Chk1 or Rad53 (either directly or indirectly) is required for phosphorylation of Rad26, we deleted CHK1 or RAD53 and examined the phosphorylation state of Rad26 in these cells after UV irradiation. Moreover, we also tested the effect of deletion of TEL1 (encoding the yeast homologue of the ATM kinase) and DUN1 (encoding a kinase acting downstream of Rad53 in the DNA damage checkpoint pathway) on Rad26 phosphorylation. Damage-induced phosphorylation was abrogated only in cells lacking the Mec1 kinase, supporting the idea that Mec1 directly phosphorylates Rad26 without the involvement of downstream kinases (Fig. 3C). This result is also in agreement with the observation that MEC1 deletion, but not CHK1 or RAD53 deletion, leads to a defect in TC-NER (Fig. 1B).
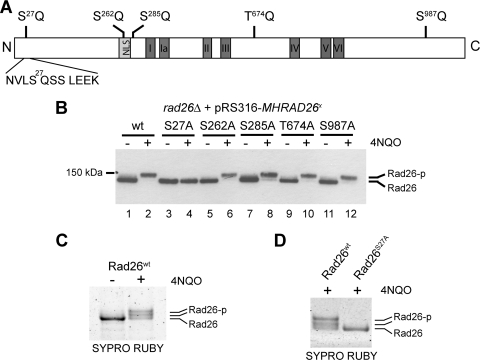
Rad26 is phosphorylated primarily on serine 27.
Mec1 phosphorylates serine and threonine residues that are immediately adjacent to a glutamine (SQ/TQ motifs) (14). Rad26 possesses 1 TQ and 4 SQ consensus motifs (Fig. 4A). We cloned the MHRAD26 locus into the pRS316 vector (47), mutated these sites individually to alanine, and then analyzed the phosphorylation status of Rad26 after treatment with the UV-mimetic compound 4NQO (Fig. 4B). 4NQO was used in place of UV treatment because this drug can be used in a more easily controlled manner and because it also leads to posttranslational modification of Rad26 (Fig. 2C). The different versions of the Rad26 protein were expressed at similar levels (compare lanes 1, 3, 5, 7, 9, and 11). Mutation of Rad26 serine 27 abolished the 4NQO-induced shift, whereas all the other single point mutants behaved like the wild-type protein (Fig. 4B, compare lane 4 with lanes 2, 6, 8, 10, and 12, respectively), suggesting that S27 is the only phosphorylated residue in Rad26. However, we could not rule out the possibility that a residual modification escapes detection by SDS-PAGE mobility shift. We therefore also utilized MS to investigate sites of Rad26 phosphorylation. Due to the low abundance of Rad26, we were unable to purify enough modified protein for MS analysis. To try to circumvent this problem, the tagged gene was placed behind a galactose-inducible promoter on a centromeric plasmid (pYC2-MHRAD26) and introduced into a rad26Δ mutant. Expression was induced by growth in galactose-containing medium, after which cellular DNA was damaged by treatment with 4NQO. Rad26 was purified using Myc affinity resin and eluted using TEV protease. Interestingly, the availability of more protein made it possible to visualize two slower-migrating forms of Rad26 after extended SDS-PAGE and staining of the gel (Fig. 4C), suggesting that more than one phosphate group can be added to Rad26 after DNA damage. Indeed, MS analysis indicated that both S27 and S30 were phosphorylated in this sample. These sites of phosphorylation were identified following peptide fragmentation using both CID and ETD. Moreover, while this work was in progress, a proteome-wide study of protein phosphorylation reported that S27 and S29 of Rad26 are phosphorylated in a Mec1-dependent manner (4). Surprisingly, neither S29 nor S30 exists in the context of a proper Mec1 consensus motif (Fig. 4A). We therefore speculated that S27 is the primary Rad26 phosphorylation site but that serine residues nearby might become targeted as “innocent bystanders” in connection with S27 phosphorylation. In order to investigate this idea, we mutated S27 in the pYC2-MHRAD26 plasmid (creating plasmid pYC2-MHRAD26S27A) and analyzed both the wild-type and the mutant protein after 4NQO treatment. Indeed, whereas the wild-type protein showed 2 distinct slower-migrating bands, no shift was observed with the S27A mutant (Fig. 4D). The absence of phosphorylated residues in this mutant was also confirmed by mass spectrometry. Together, these data indicate that Mec1 phosphorylates S27 of Rad26 after DNA damage and that this event may also occasionally result in modification of S29 and/or S30 as well.
FIG. 4.
Identification of serine 27 as the Rad26 phosphorylation site. (A) Schematic representation of the SQ/TQ motifs in the Rad26 protein. Gray boxes show the positions of the nuclear localization signal (NLS) and the 7 conserved SWI2/SNF2 translocase domains. (B) Western blot analysis of Rad26 phosphorylation before and after 4NQO treatment (5 μg/ml) in rad26Δ cells carrying a plasmid for expression of either wild-type Rad26 or mutant versions lacking one of the five SQ/TQ phosphorylation sites. (C) Sypro Ruby-stained gel showing wild-type Rad26 purified after overexpression in rad26Δ cells either without (−) or with (+) treatment with 4NQO (5 μg/ml for 10 min). (D) Sypro Ruby-stained gel showing wild-type and S27A Rad26 purified after overexpression in rad26Δ cells after 4NQO treatment (5 μg/ml for 10 min).
Rad26 phosphorylation enhances TC-NER efficiency.
Given that MEC1 deletion affects NER and that Rad26 appears to be a direct target of the Mec1 kinase, we asked whether Rad26 phosphorylation plays a role in TC-NER. We first tried to test this genetically, taking advantage of the fact that the rad16Δ rad26Δ double mutant is much more UV sensitive than either single mutant (59). Point mutants in which Rad26 serine 27 was mutated either to alanine to prevent phosphorylation (rad26SA) or to glutamic acid to mimic phosphorylation (rad26SE) were introduced into rad16Δ rad26Δ cells, and the UV sensitivity of these strains relative to control strains was tested by spotting assays. However, like wild-type Rad26, both mutated forms of Rad26 rescued the rad16Δ rad26Δ double mutant so that its UV sensitivity was similar to that of the rad16Δ single mutant (data not shown). Simultaneous mutation of all five Mec1 consensus target motifs (SQ/TQ), or concomitant mutation of serines 27, 29 and 30, also did not lead to increased UV sensitivity (data not shown). In order to exclude the possibility that the effect of the point mutations in Rad26 is masked by the second TC-NER pathway in yeast, which is mediated by the Rpb9 protein (29), we also carried out the same assay using a rad16Δ rad26Δ rpb9Δ triple mutant. Again, we failed to detect a defect in the phosphorylation mutants that could be uncovered by this assay (data not shown).
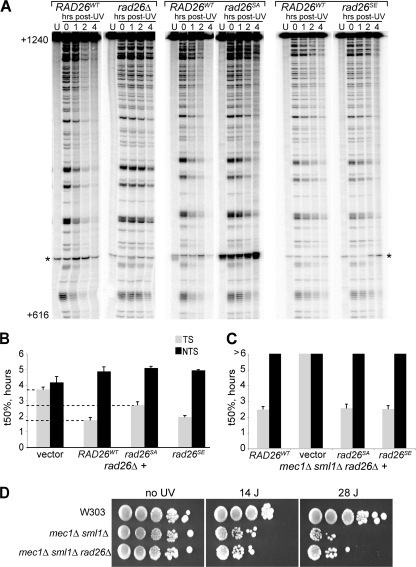
These results indicate that phosphorylation of Rad26 is not absolutely required for the function of the protein in repair but did not rule out the possibility that the modification has an effect on TC-NER kinetics that is not severe enough to cause altered sensitivity to UV light in phenotypic assays. In order to investigate TC-NER and GG-NER directly, we again utilized the strand-specific NER assay. rad26Δ mutants carrying wild-type RAD26 (RAD26WT) were compared to those carrying no RAD26, rad26SA, or rad26SE. Figure 5A and B shows that, as expected, severely delayed TC-NER was observed in cells lacking RAD26 compared to the wild-type control. Importantly, although the effect was less pronounced than that observed in rad26Δ cells, cells expressing the Rad26 alanine mutant exhibited a clear and highly reproducible delay in TC-NER (Fig. 5A, rad26SA), so that damage half-life was extended from ∼1.7 h in the presence of wild-type RAD26 to ∼2.6 h in the rad26SA mutant, while damage half-life in rad26Δ cells was >3.5 h (Fig. 5B). TC-NER in the rad26SE mutant (where glutamate mimics persistent phosphorylation) was largely unaffected (Fig. 5A and B). Repair of the NTS was not affected by any of the mutations. Together, these results indicate that damage-induced phosphorylation of Rad26 by the Mec1 kinase enhances the rate of TC-NER.
FIG. 5.
S27A mutation decreases the rate of TC-NER. (A) Representative gels showing side-by-side comparisons of cells expressing wild-type Rad26 (RAD26WT) and cells carrying either empty vector (rad26Δ) or cells expressing Rad26 in which serine 27 was replaced with an alanine (rad26SA) or a glutamic acid (rad26SE) residue, respectively. Only gels for the TS are shown (NTS repair was not affected). Numbers on the left indicate the nucleotide position relative to the RPB2 transcription start site. A nonspecific band appearing also in the unirradiated control sample (U) is marked with an asterisk. (B) Comparison of the times required for repair of 50% of the damages (t50%), obtained from quantifications of gels such as the one in panel A. Error bars indicate the standard error (3 independent experiments). (C) Quantification of NER kinetics in the mec1Δ sml1Δ rad26Δ triple mutant expressing either no Rad26 (vector), wild-type Rad26 (RAD26WT), or Rad26 in which serine 27 was replaced with an alanine (rad26SA) or a glutamic acid (rad26SE) residue. (D) Spotting assay comparing the UV sensitivities of the wild-type strain (W303), the mec1Δ sml1Δ double mutant, and the mec1Δ sml1Δ rad26Δ triple mutant.
Expression of the rad26SE mutant is not sufficient to rescue the TC-NER defects of a mec1Δ sml1Δ mutant.
The finding that the Rad26 phosphomimic mutation does not significantly affect TC-NER kinetics in the wild-type background led us to investigate whether introducing this mutant into the mec1Δ sml1Δ background could bypass the requirement for the Mec1 kinase for efficient TC-NER, i.e., whether Rad26 is the only important Mec1 target in this process. We therefore deleted RAD26 in the mec1Δ sml1Δ strain and reintroduced RAD26WT, rad26SA, rad26SE, or the empty vector as control. Analysis of TC-NER kinetics in these strains showed that the remaining TC-NER in the mec1Δ sml1Δ cells is highly dependent on the presence of Rad26 (Fig. 5C). Reintroduction of either RAD26WT or rad26SA brought the rate of TC-NER back to the level of the mec1Δ sml1Δ strain, showing that, as expected, mutating the phosphorylation site in Rad26 to alanine does not further diminish TC-NER if the strain already lacks the responsible Mec1 kinase. Mutation of Rad26 S27 to a glutamic acid residue did not lead to faster TC-NER in the checkpoint-deficient strain, showing that expression of a phosphorylation-mimicking mutant of Rad26 is not sufficient to bypass the requirement for Mec1 (Fig. 5C). Because of the severe inhibition of TC-NER in the mec1Δ sml1Δ rad26Δ strain (compared to the mec1Δ sml1Δ strain), it might be expected that this mutant would be considerably more UV sensitive than the mec1Δ sml1Δ strain. Surprisingly, however, this was not the case (Fig. 5D). As a matter of fact, RAD26 deletion had little effect or appeared to slightly suppress the UV sensitivity of the mec1Δ sml1Δ strain at high UV doses. This result indicates that cellular characteristics other than the rate of TC-NER repair are important for cell survival after UV irradiation and reinforces the idea that slower kinetics of TC-NER do not necessarily result in increased sensitivity to UV irradiation, as observed previously with the rad26SA mutant.
DISCUSSION
In this study we investigated the interplay between the DNA damage checkpoint and NER by examining the efficiency of DNA repair in cells lacking checkpoint kinases. Our data indicate that repair of both strands of an active gene is defective in cells lacking Mec1 but not in cells lacking Rad53 and Chk1, indicating that the DDC—and specifically Mec1—affects both TC-NER and GG-NER.
Although we did not investigate it in detail, the role of the DNA damage checkpoint in GG-NER appears to rely primarily on induction of repair factors, which is in agreement with previously published work (5), though we cannot rule out that impairment of checkpoint activation by cycloheximide treatment is partly responsible for the observed effect. The targets of the checkpoint that are responsible for this effect are not yet known, but potential candidates include the NER factors Rad2, Rad7, Rad16, and Rad23 (6, 25, 31, 46). In contrast, the checkpoint appears to function in TC-NER primarily via modification of repair factors, since inhibition of new protein synthesis via cycloheximide treatment had little or no effect on this repair pathway. Supporting this idea, we identified Rad26 as a target for phosphorylation upon DNA damage. Rad26 phosphorylation required Mec1, but not Rad53, Chk1, Tel1, or Dun1. Several of these checkpoint kinases affect cell cycle progression after DNA damage (14), but only Mec1 affects the rate of TC-NER, indicating that the effect of mutating this kinase is via decreased Rad26 phosphorylation (and possibly decreased phosphorylation of other repair factors) rather than of defects in cell cycle progression. Rad26 is primarily modified on serine 27 (S27), part of an SQ motif, which is also the general consensus recognition motif for Mec1 and Tel1 kinases. Most importantly, mutation of this phosphorylation site to an alanine (preventing phosphorylation), but not to a negatively charged glutamic acid residue (mimicking persistent phosphorylation), leads to a clear delay in TC-NER. This delay is most evident shortly after DNA damage induction, which correlates well with the rapid phosphorylation of Rad26 observed in response to UV irradiation. In further support of the conclusion that S27 is the only functionally important Mec1 phosphorylation site, the rates of TC-NER in a rad26 mutant in which all 5 SQ/TQ sites were mutated to alanine and one in which S27/S29/S30 were mutated together were similar to that of the rad26S27A mutant (data not shown).
How might Mec1-catalyzed phosphorylation modify the function of Rad26 to enhance TC-NER? Unfortunately, the mechanism of eukaryotic TC-NER is still not completely understood, and no reconstituted biochemical assay exists in yeast with which the activities of unmodified versus modified Rad26 can be compared. As the Rad26 protein is a DNA-dependent ATPase (17), we speculated that this activity might be influenced by phosphorylation. However, our initial studies suggest that the DNA-stimulated ATPase activity of phosphorylated Rad26 is not significantly different from that of the unmodified protein in vitro (data not shown). Rad26 interacts with Def1, a factor controlling ubiquitylation and degradation of the largest subunit of RNAPII, Rpb1 (63). In theory, the TC-NER defect observed for the rad26S27A mutant might thus potentially be explained if Rad26 phosphorylation interfered with Def1 interaction and thereby the function of this protein. However, we failed to detect significant changes in Rpb1 ubiquitylation and degradation in cells expressing Rad26 phosphorylation mutants (data not shown). Another possibility is that phosphorylation of Rad26 regulates its interaction with other TC-NER factors. We have sought to identify proteins that might interact differently with the modified form of Rad26. However, in our hands, few proteins associate stably with Rad26 so that they can be identified by MS analysis. More work is thus necessary to determine the exact function of Mec1-catalyzed Rad26 phosphorylation in TC-NER.
In contrast to rad26SA, the rad26SE mutation resulted in little or no defect in TC-NER, indicating that mutation of serine 27 to glutamate indeed acts as a phosphomimic. Nevertheless, introduction of this mutant into the mec1Δ sml1Δ strain did not alleviate the TC-NER defect in these cells. We were somewhat surprised by this finding, as the decrease observed in TC-NER in mec1Δ sml1Δ cells was not dramatically larger than that observed in the rad26SA mutant. This result might indicate a requirement for other factors regulated by Mec1. Thus, additional phosphorylation events on one or more other factors, carried out by one or more kinases in the DDC, appear to also be required for efficient TC-NER. This is not surprising, as proteomic screens have identified several substrates for checkpoint kinases (4, 32, 48), many of which are involved in DNA repair. The identification of the functionally relevant target(s) is an important future goal.
Somewhat surprisingly, although rad26SA—but not rad26SE—significantly increased the half-life of UV-induced DNA damage in the transcribed strand, we failed to uncover phenotypic consequences of it. We also failed to uncover a phenotypic effect of rad26 or Rad26 phosphorylation on MMS, bleomycin, or hydrogen peroxide sensitivity (data not shown). This indicates that the increase in repair efficiency observed when S27 is phosphorylated is not enough to affect viability after DNA damage. Strikingly, even though TC-NER is virtually undetectable at the RPB2 gene after deletion of RAD26 in the mec1Δ sml1Δ strain, no significant increase in UV sensitivity was observed in the mec1Δ sml1Δ rad26Δ strain, showing that a decrease in TC-NER kinetics does not necessarily lead to a an increase in UV sensitivity. This helps explain the lack of a clear phenotypic consequence of rad26S27A mutation: other characteristics than rate of DNA repair must be of greater importance once a certain level of repair impairment has been reached. Identifying other cellular processes and mechanisms that determine UV sensitivity represents an important future challenge.
Interestingly, TC-NER seems to play a more central role in the response of higher cells to UV irradiation. For example, mammalian cells lacking CSB are UV sensitive (56), while yeast cells lacking RAD26 are not (58). The involvement of the checkpoint in NER described here might well be conserved in evolution and could potentially be more important for the overall human DNA damage response. Indeed, we note that CSB has numerous potential SQ/TQ motifs and that one of these was recently identified as a target for ATM/ATR (32). We found that mutation of the CSB site identified by mass spectrometric analysis does not result in UV sensitivity in human cell lines (data not shown), but other preliminary data suggest that more sites in CSB may also be phosphorylated, so a more comprehensive analysis of checkpoint kinase-mediated CSB phosphorylation will be required before firm conclusions can be made.
Interestingly, some of the clinical features of patients suffering from Seckel syndrome (which involves deficiencies in the DNA damage checkpoint [27]) and Cockayne syndrome are overlapping, with both diseases resulting in microencephaly, “birdlike” facial features, and severe growth defects (13, 20). Although other possibilities cannot be ruled out, these similarities are at least consistent with a link between the DNA damage checkpoint and TC-NER also in humans. Indeed, the reason why Seckel syndrome patients display these abnormalities is presently unclear. Similarly, why CS patients display them is also poorly understood, but it is almost certainly not due to persistence of NER lesions in the transcribed strand of active genes, as these lesions also persist in several classes of XP patients (XPA, XPD, XPB, XPF, and XPG) who do not display these severe symptoms (10). An attractive possibility is that some of the phenotypes are caused by a defect of CS cells in the transcription-coupled removal of oxidative lesions, which also appear to be substrates for the base excision repair (BER) pathway (57). An involvement of CSB in the repair of such lesions has been proposed (reviewed in references 19 and 36) and might potentially explain the severe brain phenotypes exhibited by CS patients, since oxygen consumption in the brain is much higher than that of any other tissue, which in turn results in high levels of harmful metabolic by-products, including reactive oxygen species (ROS). Prolonged stalling of RNAPII at sites of DNA damage is a strong signal for apoptosis (30), which would be particularly detrimental in the brain, where cells cannot be replaced. An important future line of research will therefore be to investigate the potential effect of checkpoint-dependent CSB phosphorylation on the repair of DNA damage in higher cells.
Acknowledgments
This work was supported by a grant from the European Community (Integrated Project DNA repair, grant no. LSHG-CT-2005-512113) (to J.Q.S.), by in-house grants from Cancer Research UK (to J.Q.S. and J.M.S.), by the Medical Research Council (to R.W.), and by Cancer Research Wales (to Y.T. and H.G.).
We thank George S. Brush for providing Mec1 plasmids.
Footnotes
Published ahead of print on 9 November 2009.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Aboussekhra, A., M. Biggerstaff, M. K. Shivji, J. A. Vilpo, V. Moncollin, V. N. Podust, M. Protic, U. Hubscher, J. M. Egly, and R. D. Wood. 1995. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell 80:859-868. [DOI] [PubMed] [Google Scholar]
- 2.Aboussekhra, A., J. E. Vialard, D. E. Morrison, M. A. de la Torre-Ruiz, L. Cernakova, F. Fabre, and N. F. Lowndes. 1996. A novel role for the budding yeast RAD9 checkpoint gene in DNA damage-dependent transcription. EMBO J. 15:3912-3922. [PMC free article] [PubMed] [Google Scholar]
- 3.Ahnesorg, P., and S. P. Jackson. 2007. The non-homologous end-joining protein Nej1p is a target of the DNA damage checkpoint. DNA Repair (Amst.) 6:190-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albuquerque, C. P., M. B. Smolka, S. H. Payne, V. Bafna, J. Eng, and H. Zhou. 2008. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell. Proteomics 7:1389-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Moghrabi, N. M., I. S. Al-Sharif, and A. Aboussekhra. 2003. UV-induced de novo protein synthesis enhances nucleotide excision repair efficiency in a transcription-dependent manner in S. cerevisiae. DNA Repair (Amst.) 2:1185-1197. [DOI] [PubMed] [Google Scholar]
- 6.Bang, D. D., V. Timmermans, R. Verhage, A. M. Zeeman, P. van de Putte, and J. Brouwer. 1995. Regulation of the Saccharomyces cerevisiae DNA repair gene RAD16. Nucleic Acids Res. 23:1679-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartek, J., and J. Lukas. 2007. DNA damage checkpoints: from initiation to recovery or adaptation. Curr. Opin. Cell Biol. 19:238-245. [DOI] [PubMed] [Google Scholar]
- 8.Bohr, V. A., C. A. Smith, D. S. Okumoto, and P. C. Hanawalt. 1985. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell 40:359-369. [DOI] [PubMed] [Google Scholar]
- 9.Coon, J. J., J. E. P. Syka, J. C. Schwartz, J. Shabanowitz, and D. F. Hunt. 2004. Anion dependence in the partitioning between proton and electron transfer in ion/ion reactions. Int. J. Mass Spectrom. 236:33-42. [Google Scholar]
- 10.de Boer, J., and J. H. Hoeijmakers. 2000. Nucleotide excision repair and human syndromes. Carcinogenesis 21:453-460. [DOI] [PubMed] [Google Scholar]
- 11.de la Torre-Ruiz, M. A., C. M. Green, and N. F. Lowndes. 1998. RAD9 and RAD24 define two additive, interacting branches of the DNA damage checkpoint pathway in budding yeast normally required for Rad53 modification and activation. EMBO J. 17:2687-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evan, G. I., G. K. Lewis, G. Ramsay, and J. M. Bishop. 1985. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 5:3610-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falik-Zaccai, T. C., M. Laskar, N. Kfir, W. Nasser, H. Slor, and M. Khayat. 2008. Cockayne syndrome type II in a Druze isolate in Northern Israel in association with an insertion mutation in ERCC6. Am. J. Med. Genet. A 146A:1423-1429. [DOI] [PubMed] [Google Scholar]
- 14.Friedel, A. M., B. L. Pike, and S. M. Gasser. 2009. ATR/Mec1: coordinating fork stability and repair. Curr. Opin. Cell Biol. 21:237-244. [DOI] [PubMed] [Google Scholar]
- 15.Gasch, A. P., M. Huang, S. Metzner, D. Botstein, S. J. Elledge, and P. O. Brown. 2001. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol. Biol. Cell 12:2987-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenwood, C., L. A. Selth, A. B. Dirac-Svejstrup, and J. Q. Svejstrup. 2009. An iron-sulfur cluster domain in Elp3 important for the structural integrity of elongator. J. Biol. Chem. 284:141-149. [DOI] [PubMed] [Google Scholar]
- 17.Guzder, S. N., Y. Habraken, P. Sung, L. Prakash, and S. Prakash. 1996. RAD26, the yeast homolog of human Cockayne's syndrome group B gene, encodes a DNA-dependent ATPase. J. Biol. Chem. 271:18314-18317. [DOI] [PubMed] [Google Scholar]
- 18.Guzder, S. N., Y. Habraken, P. Sung, L. Prakash, and S. Prakash. 1995. Reconstitution of yeast nucleotide excision repair with purified Rad proteins, replication protein A, and transcription factor TFIIH. J. Biol. Chem. 270:12973-12976. [DOI] [PubMed] [Google Scholar]
- 19.Hanawalt, P. C. 2000. DNA repair. The bases for Cockayne syndrome. Nature 405:415-416. [DOI] [PubMed] [Google Scholar]
- 20.Harper, R. G., E. Orti, and R. K. Baker. 1967. Bird-beaded dwarfs (Seckel's syndrome). A familial pattern of developmental, dental, skeletal, genital, and central nervous system anomalies. J. Pediatr. 70:799-804. [DOI] [PubMed] [Google Scholar]
- 21.Herzberg, K., V. I. Bashkirov, M. Rolfsmeier, E. Haghnazari, W. H. McDonald, S. Anderson, E. V. Bashkirova, J. R. Yates III, and W. D. Heyer. 2006. Phosphorylation of Rad55 on serines 2, 8, and 14 is required for efficient homologous recombination in the recovery of stalled replication forks. Mol. Cell. Biol. 26:8396-8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoeijmakers, J. H. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411:366-374. [DOI] [PubMed] [Google Scholar]
- 23.Jelinsky, S. A., P. Estep, G. M. Church, and L. D. Samson. 2000. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol. Cell. Biol. 20:8157-8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jelinsky, S. A., and L. D. Samson. 1999. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc. Natl. Acad. Sci. U. S. A. 96:1486-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, J. S., L. Prakash, and S. Prakash. 1990. Regulated expression of the Saccharomyces cerevisiae DNA repair gene RAD7 in response to DNA damage and during sporulation. Nucleic Acids Res. 18:3281-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato, R., and H. Ogawa. 1994. An essential gene, ESR1, is required for mitotic cell growth, DNA repair and meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 22:3104-3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerzendorfer, C., and M. O'Driscoll. 2009. Human DNA damage response and repair deficiency syndromes: linking genomic instability and cell cycle checkpoint proficiency. DNA Repair (Amst.) 8:1139-1152. [DOI] [PubMed] [Google Scholar]
- 28.Kushnirov, V. V. 2000. Rapid and reliable protein extraction from yeast. Yeast 16:857-860. [DOI] [PubMed] [Google Scholar]
- 29.Li, S., and M. J. Smerdon. 2002. Rpb4 and Rpb9 mediate subpathways of transcription-coupled DNA repair in Saccharomyces cerevisiae. EMBO J. 21:5921-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ljungman, M., and F. Zhang. 1996. Blockage of RNA polymerase as a possible trigger for u.v. light-induced apoptosis. Oncogene 13:823-831. [PubMed] [Google Scholar]
- 31.Madura, K., and S. Prakash. 1990. Transcript levels of the Saccharomyces cerevisiae DNA repair gene RAD23 increase in response to UV light and in meiosis but remain constant in the mitotic cell cycle. Nucleic Acids Res. 18:4737-4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuoka, S., B. A. Ballif, A. Smogorzewska, E. R. McDonald III, K. E. Hurov, J. Luo, C. E. Bakalarski, Z. Zhao, N. Solimini, Y. Lerenthal, Y. Shiloh, S. P. Gygi, and S. J. Elledge. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316:1160-1166. [DOI] [PubMed] [Google Scholar]
- 33.Mellon, I., V. A. Bohr, C. A. Smith, and P. C. Hanawalt. 1986. Preferential DNA repair of an active gene in human cells. Proc. Natl. Acad. Sci. U. S. A. 83:8878-8882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mellon, I., G. Spivak, and P. C. Hanawalt. 1987. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 51:241-249. [DOI] [PubMed] [Google Scholar]
- 35.Mu, D., C. H. Park, T. Matsunaga, D. S. Hsu, J. T. Reardon, and A. Sancar. 1995. Reconstitution of human DNA repair excision nuclease in a highly defined system. J. Biol. Chem. 270:2415-2418. [DOI] [PubMed] [Google Scholar]
- 36.Nouspikel, T. 2008. Nucleotide excision repair and neurological diseases. DNA Repair (Amst.) 7:1155-1167. [DOI] [PubMed] [Google Scholar]
- 37.Nyberg, K. A., R. J. Michelson, C. W. Putnam, and T. A. Weinert. 2002. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36:617-656. [DOI] [PubMed] [Google Scholar]
- 38.Otero, G., J. Fellows, Y. Li, T. de Bizemont, A. M. Dirac, C. M. Gustafsson, H. Erdjument-Bromage, P. Tempst, and J. Q. Svejstrup. 1999. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 3:109-118. [DOI] [PubMed] [Google Scholar]
- 39.Paulovich, A. G., and L. H. Hartwell. 1995. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell 82:841-847. [DOI] [PubMed] [Google Scholar]
- 40.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 41.Prakash, S., and L. Prakash. 2000. Nucleotide excision repair in yeast. Mutat. Res. 451:13-24. [DOI] [PubMed] [Google Scholar]
- 42.Sancar, A., L. A. Lindsey-Boltz, K. Unsal-Kacmaz, and S. Linn. 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73:39-85. [DOI] [PubMed] [Google Scholar]
- 43.Sanchez, Y., B. A. Desany, W. J. Jones, Q. Liu, B. Wang, and S. J. Elledge. 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271:357-360. [DOI] [PubMed] [Google Scholar]
- 44.Selby, C. P., E. M. Witkin, and A. Sancar. 1991. Escherichia coli mfd mutant deficient in “mutation frequency decline” lacks strand-specific repair: in vitro complementation with purified coupling factor. Proc. Natl. Acad. Sci. U. S. A. 88:11574-11578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siede, W., J. B. Allen, S. J. Elledge, and E. C. Friedberg. 1996. The Saccharomyces cerevisiae MEC1 gene, which encodes a homolog of the human ATM gene product, is required for G1 arrest following radiation treatment. J. Bacteriol. 178:5841-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siede, W., G. W. Robinson, D. Kalainov, T. Malley, and E. C. Friedberg. 1989. Regulation of the RAD2 gene of Saccharomyces cerevisiae. Mol. Microbiol. 3:1697-1707. [DOI] [PubMed] [Google Scholar]
- 47.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smolka, M. B., C. P. Albuquerque, S. H. Chen, and H. Zhou. 2007. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. U. S. A. 104:10364-10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun, Z., D. S. Fay, F. Marini, M. Foiani, and D. F. Stern. 1996. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 10:395-406. [DOI] [PubMed] [Google Scholar]
- 50.Svejstrup, J. Q. 2002. Mechanisms of transcription-coupled DNA repair. Nat. Rev. Mol. Cell Biol. 3:21-29. [DOI] [PubMed] [Google Scholar]
- 51.Swaney, D. L., G. C. McAlister, M. Wirtala, J. C. Schwartz, J. E. Syka, and J. J. Coon. 2007. Supplemental activation method for high-efficiency electron-transfer dissociation of doubly protonated peptide precursors. Anal. Chem. 79:477-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teng, Y., S. Yu, S. H. Reed, and R. Waters. 2009. Lux ex tenebris: nucleotide resolution DNA repair and nucleosome mapping. Methods 48:23-34. [DOI] [PubMed] [Google Scholar]
- 53.Teng, Y., Y. Yu, J. A. Ferreiro, and R. Waters. 2005. Histone acetylation, chromatin remodelling, transcription and nucleotide excision repair in S. cerevisiae: studies with two model genes. DNA Repair (Amst.) 4:870-883. [DOI] [PubMed] [Google Scholar]
- 54.Terleth, C., C. A. van Sluis, and P. van de Putte. 1989. Differential repair of UV damage in Saccharomyces cerevisiae. Nucleic Acids Res. 17:4433-4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619-630. [DOI] [PubMed] [Google Scholar]
- 56.Troelstra, C., A. van Gool, J. de Wit, W. Vermeulen, D. Bootsma, and J. H. Hoeijmakers. 1992. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne's syndrome and preferential repair of active genes. Cell 71:939-953. [DOI] [PubMed] [Google Scholar]
- 57.Tsutakawa, S. E., and P. K. Cooper. 2000. Transcription-coupled repair of oxidative DNA damage in human cells: mechanisms and consequences. Cold Spring Harbor Symp. Quant. Biol. 65:201-215. [DOI] [PubMed] [Google Scholar]
- 58.van Gool, A. J., R. Verhage, S. M. Swagemakers, P. van de Putte, J. Brouwer, C. Troelstra, D. Bootsma, and J. H. Hoeijmakers. 1994. RAD26, the functional S. cerevisiae homolog of the Cockayne syndrome B gene ERCC6. EMBO J. 13:5361-5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Verhage, R. A., A. J. van Gool, N. de Groot, J. H. Hoeijmakers, P. van de Putte, and J. Brouwer. 1996. Double mutants of Saccharomyces cerevisiae with alterations in global genome and transcription-coupled repair. Mol. Cell. Biol. 16:496-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waters, R., Y. Teng, Y. Yu, S. Yu, and S. H. Reed. 2009. Tilting at windmills? The nucleotide excision repair of chromosomal DNA. DNA Repair (Amst.) 8:146-152. [DOI] [PubMed] [Google Scholar]
- 61.Weinert, T. A., and L. H. Hartwell. 1988. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241:317-322. [DOI] [PubMed] [Google Scholar]
- 62.Weinert, T. A., G. L. Kiser, and L. H. Hartwell. 1994. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 8:652-665. [DOI] [PubMed] [Google Scholar]
- 63.Woudstra, E. C., C. Gilbert, J. Fellows, L. Jansen, J. Brouwer, H. Erdjument-Bromage, P. Tempst, and J. Q. Svejstrup. 2002. A Rad26-Def1 complex coordinates repair and RNA pol II proteolysis in response to DNA damage. Nature 415:929-933. [DOI] [PubMed] [Google Scholar]
- 64.Wu, X., S. M. Shell, Y. Liu, and Y. Zou. 2007. ATR-dependent checkpoint modulates XPA nuclear import in response to UV irradiation. Oncogene 26:757-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu, X., S. M. Shell, Z. Yang, and Y. Zou. 2006. Phosphorylation of nucleotide excision repair factor xeroderma pigmentosum group A by ataxia telangiectasia mutated and Rad3-related-dependent checkpoint pathway promotes cell survival in response to UV irradiation. Cancer Res. 66:2997-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao, X., E. G. Muller, and R. Rothstein. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2:329-340. [DOI] [PubMed] [Google Scholar]