Abstract
Dyskerin is a component of small nucleolar ribonucleoprotein complexes and acts as a pseudouridine synthase to modify newly synthesized ribosomal, spliceosomal, and possibly other RNAs. It is encoded by the DKC1 gene, the gene mutated in X-linked dyskeratosis congenita, and is also part of the telomerase complex. The yeast ortholog, Cbf5, is an essential protein, but in mammals the effect of dyskerin ablation at the cellular level is not known. Here we show that mouse hepatocytes can survive after induction of a Dkc1 deletion. In the absence of dyskerin, rRNA processing is inhibited with the accumulation of large precursors, and fibrillarin does not accumulate in nucleoli. A low rate of apoptosis is induced in the hepatocytes, which show an induction of the p53-dependent cell cycle checkpoint pathway. Signs of liver damage including an increase in serum alanine aminotransferase activity and a disordered structure at the histological and macroscopic levels are observed. In response to carbon tetrachloride administration, when wild-type hepatocytes mount a rapid proliferative response, those without dyskerin do not divide. We conclude that hepatocytes can survive without dyskerin but that the role of dyskerin in RNA modification is essential for cellular proliferation.
Modification of specific bases of newly synthesized ribosomal and spliceosomal RNAs is an early posttranscriptional event and is carried out by specialized nucleolar complexes, the snoRNPs. There are 2 classes of snoRNPs, H/ACA RNPs, which pseudouridylate specific uridines, and C/D RNPs, which catalyze the methylation of specific residues. In both cases the complexes consist of a guide RNA and 4 proteins (1, 12, 55). In the case of H/ACA RNPs the guide RNA is an H/ACA snoRNA which uses base pairing to guide the complex to specific uridines in the nascent target RNA (14). The 4 proteins are dyskerin (20, 35), NOP10 (21), NHP2 (21, 52), and GAR1 (16); dyskerin is the pseudouridine synthase (29). RNA modification is an ancient process, and the snoRNP proteins are highly conserved. Despite this, the role of RNA modifications is not clearly understood. Pseudouridines are frequently found in regions of rRNAs that are functionally important, and they are thought to increase the stability of folded domains. Though depletion of individual snoRNAs does not seem to affect growth in yeast, the yeast ortholog of dyskerin, Cbf5, is essential (26). Interestingly, mutations that render Cbf5 unable to form pseudouridine permit cell survival but lead to a severe reduction in ribosome synthesis (56).
H/ACA snoRNPs have other cellular functions as well as pseudouridylation of rRNA (34). Some are involved in cleavage of rRNA precursors, and a class of H/ACA RNPs is localized in the Cajal body rather than the nucleolus (9). These “scaRNPs” are characterized by the presence in the scaRNA of a conserved Sca motif (43) and function in the modification of snRNAs (9), the small RNAs essential for pre-mRNA splicing. In vertebrates the 3′ domain of telomerase RNA, TERC, is an H/ACA RNA (36), and this RNA, which provides the template for synthesis of telomere repeats, is found in an RNP complex with the same 4 proteins found in other snoRNPs as well as telomerase reverse transcriptase and other proteins (13, 37, 48, 49). TERC RNA contains a Sca motif and is found in Cajal bodies (25). Finally, some H/ACA RNPs have no known function, their integral RNA having no known target. In one case of such an orphan RNA its expression appears to be restricted to the brain (8), opening up the possibility that H/ACA RNAs can effect tissue-specific gene expression.
In Drosophila the dyskerin ortholog, minifly, is essential for viability (15). Minifly mutations that retain some function lead to small size, developmental delay, and reduced fertility through defects in pseudouridylation and rRNA processing. In mice complete knockout of the gene encoding dyskerin, Dkc1, is embryonic lethal (19). In humans hypomorphic mutations in the DKC1 gene on the X chromosome cause X-linked dyskeratosis congenita (DC) (20, 28), an inherited bone marrow failure syndrome characterized by the presence of hypo- and hyperpigmentation, nail dystrophy, and mucosal leukoplakia (50). Interestingly most of the pathological effects of the dyskerin mutations in DC patients seem to be due to problems with telomere maintenance, since other forms of DC are caused by mutations in other components of telomerase (51) and biochemical investigations show a decrease in levels of telomerase RNA, TERC, and telomerase activity but no detectable change in rRNA processing or snoRNA metabolism (53, 54). Mice, or mouse embryonic stem (ES) cells, containing dyskerin amino acid changes that cause DC in humans so far show variable effects, some having decreased Terc RNA and telomerase and mild defects in ribosome biogenesis (18, 38). So far none of these mice has shown the pathological features of DC, probably due to the fact that laboratory mice have very long telomeres. In the complete absence of telomerase, mice show no striking defects in the first generation but they pass shortened telomeres to their offspring and generation 3 and later generations show some features similar to those seen in DC (3, 32). Recently we described a mouse line produced by gene targeting which contained a copy of a human mutation consisting of a 3′ truncation of the Dkc1 gene, encoding a protein with the last 21 amino acids (aa) missing (17). These mice showed the slower growth of proliferative tissues characteristic of DC. The slow growth was dependent on telomerase, independent of telomere length, and associated with the accumulation of DNA damage.
In mammalian cells, though dyskerin is essential for embryonic development, it is not known if it is essential for cell survival or for cell division. In this paper we used an inducible knockout strategy to delete the Dkc1 gene in adult mice and found that while most tissues were reconstituted from cells that had escaped the deletion the liver survived for weeks with the majority of its cells containing the deleted gene. This gave us the opportunity of studying mammalian cells that did not contain dyskerin. These cells, despite their long-term survival, were defective in rRNA synthesis, showed signs of a cell cycle checkpoint response and the induction of apoptosis, and were defective in cell division following liver injury. We conclude that dyskerin is not required for cell survival but is essential for cell division.
MATERIALS AND METHODS
Generation of Dkc1 conditional KO mice.
The generation of the Dkc1lox allele in C57BL/6 mice has been previously described (19). Mice carrying the Alb-Cre transgene (42) and mice carrying the Mx1-Cre transgene were obtained from the Jackson Laboratory (Bar Harbor, ME). Homozygous floxed dyskerin mice were bred to Alb-Cre mice, and male mice (9 weeks old unless specified) which carry a floxed dyskerin allele and Alb-Cre were selected for study (Dkc1loxTg:AlbCre, referred to as knockout [KO] mice). For all experiments and analysis, mice with the genotype Dkc1lox or Dkc1+Tg:AlbCre were used as controls and are referred to as wild type (WT) throughout. Similarly, male Dkc1loxTg:Mx1Cre mice were selected for study (KO) along with their Dkc1lox or Dkc1+Tg:Mx1Cre littermates (WT). For induction of the Mx1-Cre transgene, mice (5 weeks old) were injected with poly(I:C) (250 μg intraperitoneally [i.p.]) every other day for a total of 3 injections. Mice were sacrificed 4 weeks after the last injection.
All experiments on mice were performed under the strict guidelines of the National Institutes of Health and the Institutional Animal Use and Care Committee at Washington University. Age- and sex-matched conditional knockout mice and control littermates were weighed and sacrificed (n = 3 to 5). Livers were isolated, and the wet weights were recorded to calculate the liver weight-to-body weight ratio. Livers were also used for DNA and RNA isolation, protein extraction, histology, and immunohistochemistry.
CCl4 treatment.
Nine-week-old male Dkc1loxTg:AlbCre mice and controls were subjected to carbon tetrachloride (CCl4) (Sigma-Aldrich, St. Louis, MO) treatment (2 μg/g of body weight; i.p.) to induce liver damage. Equal numbers of KO and WT mice were sacrificed by CO2 asphyxiation at different times after CCl4 treatment. Mice were injected i.p. with 2.5 mg/mouse bromodeoxyuridine (BrdU; Sigma-Aldrich, St. Louis, MO) 1 h before sacrifice. The regenerating livers were harvested and used for DNA isolation, total RNA isolation, protein extraction, and paraffin embedding.
DNA, RNA, and protein analysis.
Liver tissues from the WT and KO mice were used for isolating and purifying DNA and RNA. DNA was isolated by the standard sodium dodecyl sulfate (SDS)-proteinase K method. Southern blotting to distinguish between the Dkc1lox and the Dkc1ΔDC1 alleles was performed as described previously (19). RNA was isolated using Trizol (Invitrogen, Carlsbad, CA).
Nuclear protein extraction was performed using NE-PER nuclear and cytoplasmic extraction reagents (Pierce, Rockford, IL). Twenty micrograms of protein was resolved by SDS-polyacrylamide gel electrophoresis (PAGE) analysis and transferred to a Hybond-ECL nitrocellulose membrane (GE Healthcare, Piscataway, NJ). Antibodies were detected by ECL plus Western blotting detection reagents (GE Healthcare) and visualized by autoradiography. Primary antibodies used in this study were polyclonal antiserum to 2 dyskerin peptides, DCSNPLKREIGDYIR (aa 74 to 88) and GLLDKHGKPTDNTPAC (aa 382 to 396 + C) (38); a polyclonal antiserum against the dyskerin peptide DRKPLQEDDVAEIQHA (aa 18 to 33); anti-TATA box binding protein (1:2,000; Abcam, Cambridge, MA); anti-p53 (1:1,000; Abcam); anti-p21 (1:200; Abcam); and antifibrillarin (1:1,000; Abcam). The secondary antibody was either horseradish peroxidase-conjugated goat anti-mouse antibody (1:10,000; Abcam) or horseradish peroxidase-conjugated goat anti-rabbit antibody (1:10,000; Abcam).
Pre-rRNA analysis.
Pre-rRNA analysis by Northern blotting was as described previously (44) using synthetic oligonucleotide DNA probes. RNA (10 μg) was electrophoresed in 1% (wt/vol) agarose-formaldehyde gels and blotted to Hybond-N+ membranes (GE Healthcare) overnight. Oligonucleotides (10 pmol) were end labeled with [γ-32P]ATP and T4 polynucleotide kinase (Fisher, Pittsburgh, PA). The temperature of hybridization and washing was determined by the melting temperature (Tm) of each probe. The probes used were ITS1, 5′-CTCTCACCTCACTCCAGACACCTCGCTCCA-3′; ITS2, 5′-ACCCACCGCAGCGGGTGACGCGATTGATCG-3′; and 28S, 5′-CACCTTTTCTGGGGTCTGAT-3′.
Sucrose gradient centrifugation.
Sucrose gradient centrifugation for polysome analysis was performed as previously described (24). One gram of liver tissue, 200 μl hypotonic buffer (1.5 mM KCl, 2.5 mM MgCl2, and 5.0 mM Tris-Cl, pH 7.4), and 200 μl hypotonic lysis buffer (1.5 mM KCl, 2.5 mM MgCl2, and 5.0 mM Tris-Cl, pH 7.4; 2% sodium deoxycholate, 2% Triton X-100, and 2.5 mM dithiothreitol [DTT]) were gently homogenized using a Dounce homogenizer. The lysates were centrifuged at 8,000 × g for 10 min at 4°C. The supernatant (∼800 μl) was supplemented with 4 μl RNase inhibitor (Invitrogen) and 80 μl heparin and stored at −80°C. Linear 10% to 45% sucrose gradients (80 mM NaCl, 5 mM MgCl2, 20 mM Tris-Cl, pH 7.4, and 1 mM DTT) were formed using a Gradient Master (BioComp). Gradients were centrifuged at 38,000 rpm for 3 h at 4°C and analyzed with an ISCO fractionator (ISCO).
Histology and immunohistochemistry.
Liver sections from the age-matched knockout mice and control littermates were analyzed by hematoxylin and eosin (HE) staining to count mitotic bodies characteristic of cells in M phase of the cell cycle. Indirect immunohistochemistry for BrdU was used to identify proliferating cells in S phase. Terminal deoxynucleotidyltransferase-mediated dUTP nick-end labeling (TUNEL) was used to detect apoptotic nuclei.
Double staining of dyskerin and fibrillarin or dyskerin and BrdU was used to determine their expression and/or localization. Briefly, 4-μm-thick paraffin sections were passed through xylene and graded alcohol and rinsed in phosphate-buffered saline (PBS). Slides were then treated with Diva Decloaking solution (BioCare Medical, Walnut Creek, CA) in a pressure cooker for 3 min. Slides were rinsed with PBS three times. Slides were then incubated with primary rabbit anti-dyskerin antibody (1:200) and mouse antifibrillarin (1:1,000) or with antidyskerin and rat anti-BrdU (1:400; Accurate Chemical & Scientific Corp, Westbury, NY) overnight at 4°C. After washes, the sections were incubated in the secondary anti-rabbit Cy3-conjugated antibody (1:500; Jackson ImmunoResearch, West Grove, PA) and anti-mouse fluorescein isothiocyanate (FITC)-conjugated antibody (Jackson ImmunoResearch) or with anti-rabbit Cy3 and anti-rat FITC-conjugated antibody (1:200; Jackson ImmunoResearch) for 1 h at room temperature. Sudan black autofluorescence inhibitor (Chemicon International, Temecula, CA) was used to quench the background fluorescence. Slides were rinsed with distilled water (dH2O) and counterstained with DAPI (4′,6-diamidino-2-phenylindole). Slides were viewed under an Axioskop 40 (Zeiss) upright research microscope, and digital images were obtained with a Nikon Coolpix camera.
The positive cells were counted in 5 low-power fields (200×) in 3 sections for HE, BrdU, and TUNEL staining or a higher-power field (1,000×; up to 300 cells) from 3 different knockout or control livers for double staining.
mRNA processing and quantitative reverse transcription-PCR (qRT-PCR).
The levels of pre-mRNA and mRNA of glyceraldehyde-3-phosphate dehydrogenase (GADPH) and nucleolin were measured by RT-PCR. 28S was used as a control. The SYBR green master mix (Applied Biosystems, Foster City, CA) was used to follow RNA amplification in the real-time reaction. Reverse transcription and PCRs were performed according to the manufacturer's recommendations for the SuperScript III first-strand synthesis system for RT-PCR (Invitrogen). The primers were specifically designed with Primer Express 3.0 software (Applied Biosystems). For nucleolin, the forward primer for spliced mRNA was 5′-CATGTGACTAAGTCCTTCATGACTGA-3′, the forward primer specific for unspliced pre-mRNA was 5′-GGATGGGAAAAGTAAAGGGATTG-3′, and the reverse primer was 5′-GCCTTTGACCTTTCTCTCCAGTAT-3′. For GAPDH, the forward primer for spliced mRNA was 5′-TTGTGGAAGGGCTCATGACCACAGTCCA-3′, the forward primer for unspliced pre-mRNA was 5′-CTCACTCATTGCCCCCGTGTTTTCTA-3′, and the reverse primer was 5′-TCAGATCCACGACGGACACATTGGGGG-3′. For U2, the forward primer was 5′-TTTGGCTAAGATCAAGTGTAGTATCTGTT-3′ and the reverse primer was 5′-CCAACTCCTACTTCCAAAAATCCA-3′. For 28S, the forward primer was 5′-GGTAGCCAAATGCCTCGTCAT-3′ and the reverse primer was 5′-CCCTTGGCTGTGGTTTCG-3′. Fluorescence was measured using an Applied Biosystems 7500 Fast Real-Time PCR system (Applied Biosystems).
Statistical analysis.
An unpaired two-tailed Student t test was used to compare hepatocellular BrdU incorporation, TUNEL staining, mitotic body index, liver weight-to-body weight ratio, alanine aminotransferase (ALT) activity, and DNA recombination efficiency with significance set at 0.05.
RESULTS
Dkc1 knockout mouse models.
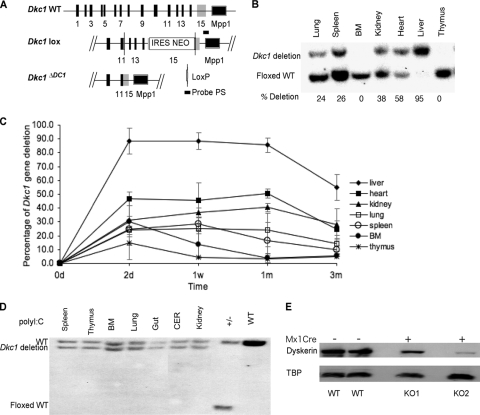
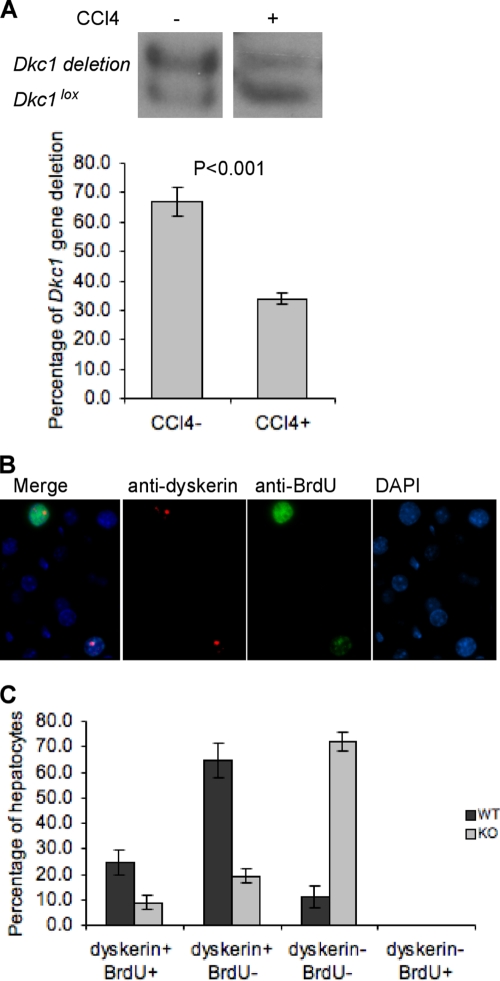
We previously described the construction of the Dkc1lox allele by gene targeting (19). The Dkc1lox allele has a lox element in intron 11 and an internal ribosome entry site (IRES)-Neo selection cassette followed by a second lox element in the 3′ untranslated region (UTR) (Fig. 1A). In the presence of the Cre recombinase, the lox elements recombine, removing exons 12 to 15, producing the deleted allele Dkc1ΔDC1. In experiments with mouse embryonic fibroblast (MEF) cells (not shown), we do not detect any truncated protein corresponding to the expected product from the deleted allele, and so the Dkc1ΔDC1 allele is essentially a null allele. Having found that the Dkc1ΔDC1 allele is embryonic lethal (19), we were interested in whether adult tissues could survive in the absence of dyskerin. We thus bred male Dkc1loxTg:Mx1Cre mice, taking advantage of the Mx1 interferon-inducible promoter, which can be conveniently induced by double-stranded RNA in the form of poly(I:C). After Cre induction mice were sacrificed at 2 days, 1 week, 1 month, and 3 months, and DNA was extracted from different tissues and examined by Southern blotting for the proportions of Dkc1lox and Dkc1ΔDC1 alleles. The results (Fig. 1B and C) show that in most tissues the proportion of surviving cells containing the deletion is about 40% (heart and kidney) or close to zero (bone marrow, thymus, spleen, and lung). In the liver, however, the proportion of surviving cells is much higher, greater than 80% in some animals; it reaches a maximum after 2 days, remains high for 4 weeks, and then slowly falls afterwards. These results almost certainly arise from competitive outgrowth of the few cells that escape the Dkc1 deletion populating most of the adult organs and tissues, as has been seen in experiments involving the deletion of essential genes in adult mice (46). In experiments in which female Dkc1lox/+ animals were injected, most tissues consisted of 50% deleted and 50% WT alleles, indicating that the efficiency of deletion was close to 100% and the surviving cells were presumably the ones with the WT allele on the active X chromosome (Fig. 1D). Thus, we conclude that hematopoietic tissues with high rates of cellular proliferation cannot survive without dyskerin, while tissues with a lower proliferative rate, such as the uninjured adult liver, can tolerate high levels of Dkc1 deletion.
FIG. 1.
Deletion of the Dkc1 gene in various tissues of Dkc1loxTg:Mx1Cre transgenic mice at different times. Groups of 4 mice (6 weeks old) were injected with poly(I:C) (i.p.; 250 μg) 3 times every other day. Genomic DNA was prepared from the indicated organs, and Cre-mediated deletion of the Dkc1 gene was detected by Southern blotting. (A) Diagram of Dkc1 gene and the targeted alleles. The Dkc1lox allele has a lox element in intron 11 and an IRES Neo selection cassette followed by a second lox element in the 3′ UTR. In the presence of the Cre recombinase, the lox elements recombine, removing exons 12 to 15, producing the deleted allele Dkc1ΔDC1. (B) Southern blotting for the proportions of Dkc1lox and Dkc1ΔDC1 in various tissues collected 4 weeks after poly(I:C) injection. BM, bone marrow. (C) Quantification of the Dkc1 recombination efficiency in various tissues of Dkc1loxTg:Mx1Cre mice over time. The surviving cells in liver are more than 80% recombined at the Dkc1 locus, while those in thymus and bone marrow are only 0 to ∼20% recombined. (D) Cells in various tissues of female Dkc1lox/+ mice consist of 50% deleted and 50% WT alleles after poly(I:C) treatment, showing that the induction of the deletion is efficient. In this experiment the mice were Dkc1DC7 with differently placed lox sites (19). CER, cerebellum. (E) Western blot showing the decrease in the amount of dyskerin in the liver of 2 mice, one with a 60% reduction and one with a 90% reduction in the amount of dyskerin. TBP, TATA box binding protein.
Deletion of Dkc1 in mouse liver.
We wanted to investigate in more detail the induction of the Dkc1 deletion in the liver in the Dkc1loxTg:Mx1Cre mice after poly(I:C) injection. The proportion of cells with the deletion reached 90% after 2 days and remained at 90% until 1 month after injection before gradually decreasing (Fig. 1C). We therefore chose to use livers 1 month after injection for our studies. The maximum extent of deletion was variable as measured either by Southern blotting or by Western blotting. Figure 1E shows the decrease in the amount of dyskerin in the livers of 2 mice, one with a 60% reduction and one with a 90% reduction in the amount of dyskerin. The antibody used in this experiment (38) was raised against a mixture of 2 peptides, one of which is deleted as part of the Dkc1ΔDC1 deletion. It is thus possible that a truncated protein is present in the livers after deletion that is not detected by this antibody. With a second antibody made against an N-terminal peptide, however, no truncated peptide is detected (see Fig. S1 in the supplemental material), indicating that if this peptide is made, it is very unstable.
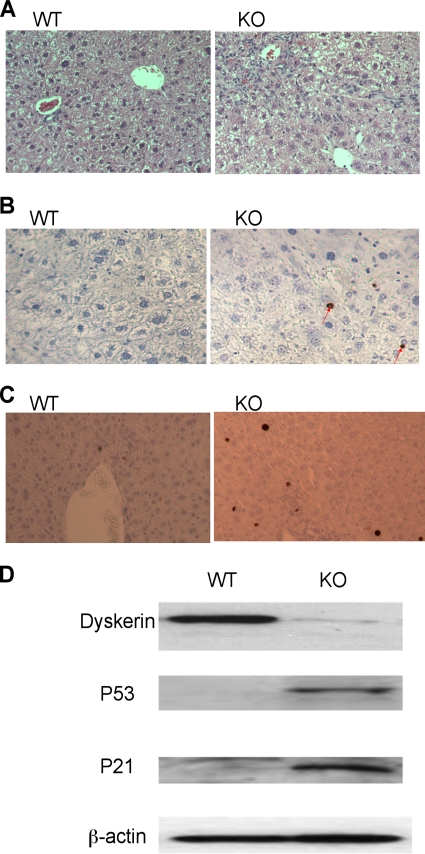
The loss of dyskerin from the liver in these mice was confirmed by immunohistochemistry. In WT hepatocytes dyskerin colocalizes with fibrillarin as dense spots at the nucleoli, but in livers from the knockout mice dyskerin is not detectable in many of the hepatocytes (Fig. 2A). Interestingly, in many hepatocytes lacking dyskerin, fibrillarin is not localized in dense spots as before. Fibrillarin is a component of C/D box snoRNAs which participate in methylation of nascent rRNA molecules during the same stages of rRNA biogenesis in which H/ACA snoRNAs act (7). To ask if the lack of detectable fibrillarin is due to destabilization of fibrillarin caused in some way by lack of dyskerin, we looked at fibrillarin by Western blotting (Fig. 2B). There was no difference between WT and knockout livers, suggesting that in the absence of dyskerin C/D snoRNPs are not maintained in nucleoli.
FIG. 2.
Loss of dyskerin and fibrillarin in hepatocyte nucleoli in Dkc1loxTg:Mx1Cre transgenic mice by immunohistochemistry. (A) Dyskerin localizes to the nucleolus and colocalizes with the C/D snoRNP protein fibrillarin. The liver tissue was paraffin embedded and incubated with rabbit antidyskerin and mouse antifibrillarin antibody. The nucleoli were counterstained with DAPI. (B) Western blotting showed that the amount of nuclear fibrillarin protein in the KO mice was not changed. TATA box binding protein (TBP) was used as a loading control.
To confirm our result that mice can tolerate lack of dyskerin in liver and to use a system independent of interferon induction, we next decided to induce the Dkc1 deletion by using the Cre recombinase under a liver-specific promoter, i.e., the mouse albumin promoter, AlbCre (41). We studied the appearance of the Dkc1 deletion in male Dkc1loxTg:AlbCre mice. Southern blot analysis showed a peak of 70% of liver cells containing the deletion at 6 weeks with this level being maintained until 12 weeks and then declining (Fig. 3A). Because AlbCre mice express Cre only in hepatocytes and possibly in bile duct and hepatocytes make up only about 70% of liver cells, these calculations underestimate the amount of deletion. A similar decrease in dyskerin was detected at the protein level (Fig. 3B). Immunofluorescence with dyskerin and fibrillarin antibodies was similar to that observed with Dkc1loxTg:Mx1Cre mice.
FIG. 3.
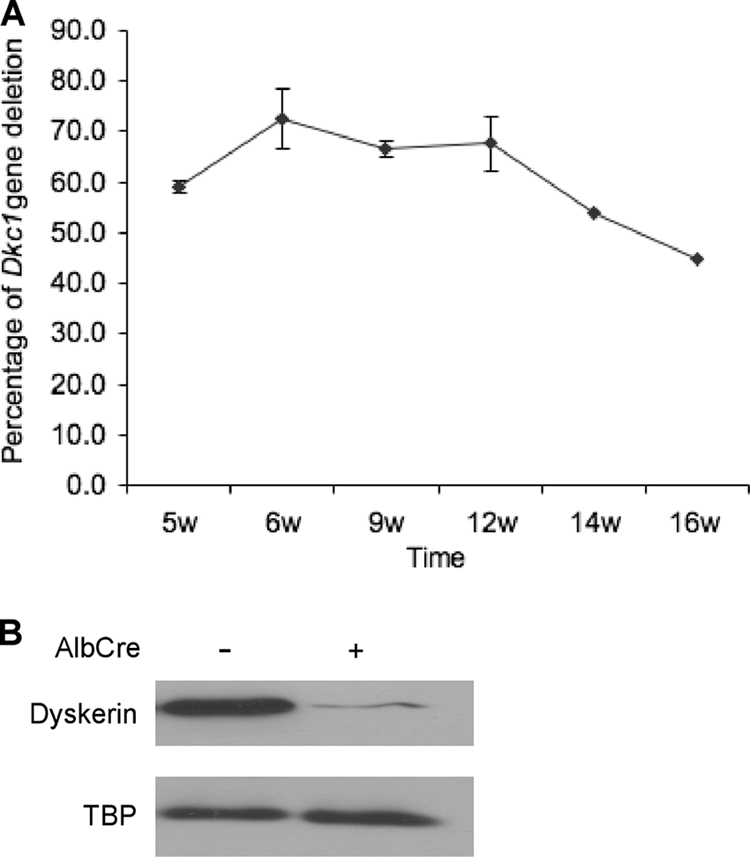
Deletion of Dkc1 gene in liver of Dkc1loxTg:AlbCre transgenic mice at different times. (A) Dkc1 recombination efficiency in mice of different ages determined by Southern blot analysis. Liver DNA was isolated from 5- to ∼16-week-old mice and subjected to Southern blot analysis as shown in Fig. 1; the percentage of Dkc1ΔDC1 alleles reached a peak of ∼70% at 6 to 12 weeks. (B) Western blotting showed that the dyskerin protein in liver is significantly decreased in the KO mice. TBP is TATA box binding protein used as a loading control.
Dkc1loxTg:AlbCre mice show signs of chronic liver damage and impaired rRNA processing.
The histology of the livers depleted of dyskerin by Mx1-Cre induction appeared normal (data not shown). However, in the livers of Dkc1loxTg:AlbCre mice at 9 weeks of age marked abnormalities were observed. These were not present at 6 weeks (data not shown), indicating that they may develop several weeks after Dkc1 deletion. For example, these livers exhibited fibrotic regions and small proliferating cells in periportal areas (Fig. 4A). In addition the liver weight and body weight of the KO mice were significantly reduced compared with WT mice (Table 1). To further assess liver damage in these mice, we measured serum ALT activity and bilirubin. Comparable and normal levels of bilirubin were observed in the WT and KO mice. However, the KO mice showed a 3.5-fold increase in serum ALT, indicating the presence of liver damage. We also used TUNEL staining to measure the extent of apoptosis in the liver tissue and found that 0.2 to 0.3% of hepatocytes in the knockout livers stained positive while positive-staining hepatocytes in WT age-matched animals were virtually undetectable (Fig. 4B). Finally, we performed analysis of hepatocellular BrdU incorporation, which revealed that 1.3% of hepatocytes in KO mice but less than 0.5% in WT mice incorporated BrdU, reflecting increased cellular turnover in KO livers (Fig. 4C).
FIG. 4.
Dyskerin deletion activates a p53-dependent cell cycle arrest pathway in Dkc1loxTg:AlbCre mice. (A) HE staining of liver section showing the fibrotic regions and the appearance of small proliferating cells in the portal areas in KO mice (200×). (B) Detection of apoptotic hepatocytes in KO mice by TUNEL staining (200×; P < 0.05). The arrows show positive nuclei. (C) Increased hepatocyte proliferation in KO mice by BrdU staining (200×; P < 0.05). (D) Western blot showing the elevation of p53 and p21 protein in the livers of KO mice. β-Actin was used as a loading control.
TABLE 1.
Body weight and liver weight in 9-week-old WT (Dkc1lox) and KO (Dkc1loxTg:AlbCre) mice
| Variable | WT | KO |
|---|---|---|
| Liver wt (g) | 1.310 ± 0.045 | 1.178 ± 0.051a |
| Body wt (g) | 28.503 ± 0.991 | 24.620 ± 0.629a |
| Liver/body wt ratio | 0.046 | 0.049 |
P < 0.05.
Because impaired ribosome biogenesis, a possible effect of dyskerin ablation, is known to promote p53-dependent cell cycle arrest and apoptosis (33, 40), we next quantified the expression of p53 and the p53 target p21 in KO and WT livers. Figure 4D shows increased levels of both proteins in KO liver, indicating that the p53 cell cycle arrest pathway is activated in hepatocytes lacking dyskerin.
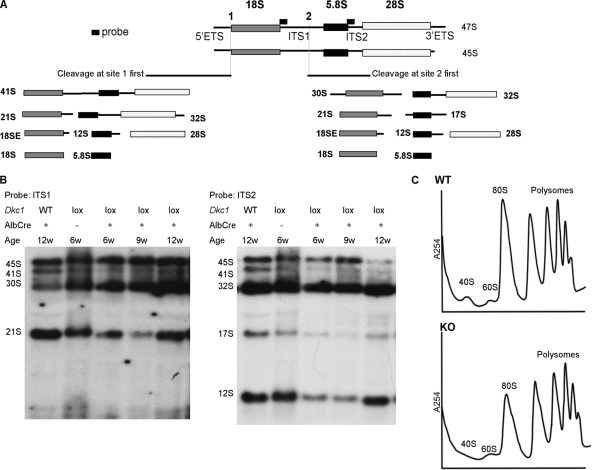
Together, our observations that lack of dyskerin leads to mislocalization of fibrillarin (Fig. 2A) and induces a p53-dependent cell cycle arrest (Fig. 4D) suggested that ribosome biogenesis may be impaired in the hepatocytes lacking dyskerin. Although the effect of dyskerin ablation on ribosome biogenesis in mammalian cells has not been investigated, Cbf5-deficient yeasts exhibit a complete block in rRNA synthesis (6). Furthermore, yeast Cbf5 mutants that fail to pseudouridylate RNA display a growth defect and impaired ribosome production but do not show abnormal accumulation of rRNA processing intermediates (56). Based on these observations, we were interested in the effects of dyskerin ablation on rRNA processing in mouse liver. To investigate this, hepatic rRNA from Dkc1loxTg:AlbCre mice and their wild-type littermates of different ages was analyzed by Northern blotting. The results show that dyskerin deficiency results in changes in the accumulation of rRNA precursors including decreased 21S, 41S, 17S, and 12S RNA. Of the products of the ITS1 cleavage, the 30S species increases while the 32S species is not altered (Fig. 5A and B; see also Fig. S2 in the supplemental material). Finally we analyzed the polysome profile of WT and KO livers, which showed a decrease in the relative amount of 80S ribosomes in the KO livers (Fig. 5C).
FIG. 5.
Dyskerin ablation disrupts rRNA processing in the Dkc1loxTg:AlbCre mice. (A) Scheme of pre-rRNA processing in mouse liver (5). The probes at ITS1 and ITS2 were used to detect the precursors of 18S and 5.8S, respectively. (B) Northern blot of RNA from livers of male mice of different genotypes. With the ITS1 probe a decrease in the amount of the 21S and accumulation of the 30S, which are the precursors of the mature 18S RNA, are observed in the KO mice. With the ITS2 probe a decrease in the precursors of 41S, 17S, and 12S was observed. (C) Effect of dyskerin deletion on polysomes. Polysomes from liver extracts were sedimented through 10 to 45% sucrose gradients, showing a decrease in the amount of 80S ribosomes but no significant change in the polysome profile. The 40S and 60S subunits were present in wild-type and KO livers in very small amounts.
Dkc1loxTg:AlbCre mice exhibit an impaired hepatocellular proliferative response to acute liver damage.
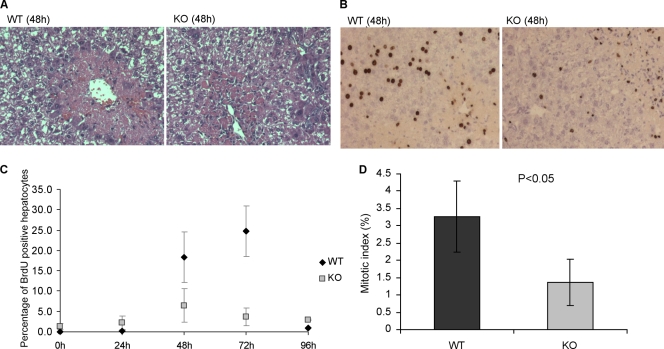
The data described above, showing that hepatocytes can survive for months after Dkc1 deletion, suggest that dyskerin is not essential for cell survival (Fig. 1B and D). However, the modestly increased liver damage seen in these mice raises the question of whether the slow loss of Dkc1-deleted liver cells over time is the result of a relative proliferative advantage of wild-type hepatocytes over Dkc1-null cells during the hepatic regenerative response to such injury. To answer this question, we examined the degree of injury and hepatic regenerative response to CCl4 in Dkc1loxTg:AlbCre and Dkc1lox mice. Both KO and WT mice showed comparable perivenous damage (Fig. 6A), indicating that dyskerin deletion did not affect the susceptibility of liver cells to CCl4 damage. In contrast, analysis of hepatocellular BrdU incorporation showed significantly (P < 0.05) increased hepatocellular proliferation at 48 and 72 h after CCl4 injection in WT compared with KO mice (Fig. 6B and C). The reduced proliferative index in KO cells was further demonstrated by the significantly diminished mitotic index 72 h after CCl4 treatment in these animals (Fig. 6D). Interestingly, the proportion of cells that are BrdU positive at 48 h to those in mitosis at 72 h is the same in WT and KO livers, implying that there is no block at the G2-M transition in KO mouse hepatocytes. Figure 6B also illustrates the presence of a population of nonparenchymal periportal liver cells that are labeled with BrdU. These cells are much more abundant in KO than WT livers. Although we have not yet identified them, we suspect that they are infiltrating immunological cells. These data show that hepatocellular proliferation is impaired in Dkc1loxTg:AlbCre mice.
FIG. 6.
Hepatocyte proliferation before and after CCl4 treatment in Dkc1loxTg:AlbCre mice. (A) Similar perivenous damage in the livers of WT and KO mice after CCl4 treatment by HE staining (200×). (B) Impaired hepatocyte proliferation in the liver of KO mice 72 h after CCl4 treatment by BrdU staining (200×). (C) Quantification of BrdU-positive hepatocytes after CCl4 treatment at different time points. At time zero, BrdU incorporation was seen in 1.3% of hepatocytes in KO mice but in less than 0.5% of hepatocytes in WT mice. The peak of the response in WT liver was seen at 72 h with 24.7% hepatocytes positive, but only 6.5% hepatocytes were positive in KO livers (P < 0.05 at time zero, 48 h, and 72 h). (D) Decreased mitotic index in the KO hepatocytes due to the poor proliferative response.
Dyskerin is necessary for cell division.
The observation that hepatocellular proliferation is reduced in Dkc1loxTg:AlbCre versus control mice and that some hepatocytes in Dkc1loxTg:AlbCre mice escape the Dkc1 deletion raises the possibility that the small number of proliferating hepatocytes in the KO livers were those that escaped deletion. Southern blotting of liver DNA from the KO livers before and 48 h after the CCl4 treatment shows that the proportion of the Dkc1lox allele expressing WT dyskerin goes from 30% before to 70% after (Fig. 7A), which is consistent with this possibility. Immunohistochemical analysis showed that 48 h after CCl4 treatment all the BrdU-positive hepatocytes in Dkc1loxTg:AlbCre mice were positive for dyskerin expression (Fig. 7B and C). These data provide convincing evidence that dyskerin is essential for hepatocellular proliferation.
FIG. 7.
Dyskerin is necessary for cell division. (A) Southern blot of liver DNA from the Dkc1loxTg:AlbCre mice before and 48 h after the CCl4 treatment. The proportion of the WT Dkc1lox allele in the KO liver goes from 30% before to 70% after (P < 0.001). (B) Immunofluorescent detection of the presence of BrdU and dyskerin in the hepatocytes of Dkc1loxTg:AlbCre mice 48 h after treatment with CCl4. Paraffin-embedded liver sections were double stained with antidyskerin and anti-BrdU antibodies, and the nucleoli were counterstained with DAPI. Hepatocellular BrdU incorporation was correlated with dyskerin in the dyskerin-null mice. (C) Quantification of dyskerin- and BrdU-positive hepatocytes. Three hundred hepatocytes were counted, and all those that had incorporated BrdU were dyskerin positive.
mRNA processing in the absence of dyskerin.
We have observed that dyskerin can be ablated from a large proportion of mouse hepatocytes without any overt effect on the health of the animal. As well as being involved in ribosome biogenesis and telomere maintenance, dyskerin is involved indirectly in mRNA splicing, since snRNAs are modified by pseudouridylation guided by H/ACA scaRNAs. Inhibition of mRNA splicing may be expected to cause a more severe phenotype than we observed. For this reason we wanted to examine mRNA splicing in WT and KO livers (31). We thus chose 2 genes expressed in liver, GAPDH and nucleolin, and measured the steady-state levels of unspliced and spliced transcripts in WT and KO animals, reasoning that the ratio between them would reflect the efficiency of splicing. The calculated ratio between spliced and unspliced transcripts did not show any significant change for either gene, implying that dyskerin ablation did not affect splicing efficiency (Fig. 8A). This result was not expected since spliceosomal RNAs, including U2, contain a high proportion of modified nucleotides and in the case of U2 it has been shown in vitro that pseudouridines are required for efficient splicing (10). We thus examined the steady-state level of U2 RNA in WT and KO livers by qRT-PCR and found no significant difference (Fig. 8B). These data indicate that there is no gross defect in mRNA processing in the absence of dyskerin.
FIG. 8.

Dkc1 deletion does not affect mRNA processing. (A) Levels of nucleolin and GAPDH pre-mRNA and mRNA in KO and WT liver. Dkc1 deletion has no effect on the RNA levels or the pre-mRNA/mRNA ratio. 28S RNA was used as an internal control. (B) Measurement of U2 RNA levels in KO and WT liver by qRT-PCR.
DISCUSSION
Most of our knowledge about dyskerin and snoRNPs and their role in modification of rRNA and snRNA comes from genetic studies of yeast and structural studies of archaebacteria. Modification using guide RNAs is a highly conserved process, and the proteins associated with H/ACA RNA and C/D RNAs are also highly conserved (22). In humans mutations in DKC1 cause X-linked dyskeratosis congenita (20), and there is good evidence that the major effect of these pathogenic dyskerin mutations is to decrease telomerase activity, causing increased telomere shortening and exhaustion of stem cells in highly proliferating tissues (50). It is possible, however, that defects in snoRNP function also contribute to X-linked DC (38, 45). In mice a Dkc1 knockout is embryonic lethal (19), while other mutations cause variable effects on telomerase activity and rRNA biogenesis (17, 18, 38, 45). Here we have used an inducible deletion strategy to knock out Dkc1 in adult mice. We found that while most tissues could not survive without dyskerin and became populated by cells that had escaped the deletion, the liver could survive with a large proportion of its Dkc1 genes deleted for several months. In fact the rate of turnover of hepatocytes in the livers of knockout animals was higher than that in wild-type animals. This was most likely due to the induction of apoptosis in a small percentage of cells and their replacement by the division of hepatocytes that had, we presume, escaped the deletion. Deletion in solid tissues using the Mx1-Cre system is never complete, but since we achieved the maximum possible efficiency of deletion of about 50% in Dkc1lox/+ females, we assume that the efficiency of deletion in our mice was high.
Ribosome synthesis is inhibited in the absence of dyskerin.
One of our primary objectives was to determine to what extent lack of dyskerin affects rRNA processing and ribosome biogenesis. Our experiments suggest that in the absence of dyskerin ribosome biogenesis cannot proceed. Firstly, in total liver RNA probed for rRNA precursors by Northern blotting we see a marked reduction in more mature processed rRNA precursors and also an accumulation of the 30S precursor. In Dkc1loxTg:AlbCre mice the dyskerin gene is deleted in hepatocytes, which comprise about 70% of the liver cells, and as seen in the immunofluorescence experiments, some hepatocytes escape the deletion. Therefore, a decrease in the amount of an RNA precursor likely means that it is absent in cells with the deletion. This assumption is supported by the differences in the rRNA processing pattern between Alb-Cre and Mx1-Cre experiments. With the Dkc1loxTg:Mx1Cre mice after poly(I:C) treatment, the dyskerin deletion extends to nonhepatocytes, and in these experiments, we see a more extreme change in the pattern of rRNA processing (data not shown). The precise nature and sequence of processing events that take place during the maturation of the primary 47S transcript to the mature 18S, 5.8S, and 28S rRNAs in mouse are not completely established (27, 47). However, two alternative early events are cleavage in the 5′ external transcribed spacer (ETS) to give a 41S precursor or cleavage within ITS1 to give a 30S and a 32S RNA. Very little 41S RNA is detected in KO livers, suggesting that dyskerin is required for the early cleavage event in the 5′ ETS (27). The 30S and 32S precursors in normal mouse cells are further processed, 30S being processed via a 21S intermediate to mature 18S RNA and 32S being processed via 17S and 12S intermediates to mature 5.8S RNA. The probes that we used (Fig. 5) hybridize to the 30S, 32S, 17S, 12S, and 21S species and show that while the early products of ITS1 cleavage accumulate, the amount of 12S, 17S, and 21S intermediates is reduced in KO compared with WT livers. Together these data suggest that in the absence of dyskerin the initial ITS1 cleavage event takes place but the 30S and 32S precursors are not processed further, indicating that further processing requires pseudouridylation of the nascent transcripts and/or an H/ACA RNA-dependent cleavage event. Pseudouridylation of rRNA is thought to take place on the early transcripts, prior to cleavage in ITS1 (4, 11). Our results are compatible with recent studies showing that while most cleavage events in rRNA processing, including those in the 5′ ETS, require the participation of snoRNPs, a cleavage site within ITS1 requires, at least in yeast, only the action of the catalytic RNA RNase MRP (23, 30).
More compelling evidence that ribosome biogenesis is halted in cells without dyskerin is our observation that in these cells immunofluorescence with a fibrillarin antibody does not show a brightly staining nucleolus as seen in wild-type cells. Fibrillarin is the active methylase present in C/D snoRNPs, and in ribosome biogenesis it is active on the same molecules as is dyskerin. These molecules will not become pseudouridylated in the absence of dyskerin, and our results suggest that lack of pseudouridylation affects the action of C/D snoRNAs. The mechanism underlying the apparent linkage between these 2 processes will be the subject of further experiments—it will be interesting to know if lack of methylation inhibits pseudouridylation. Moreover, the factors that maintain snoRNPs in the nucleolus may be amenable to analysis in this system.
Dyskerin is essential for cell division.
The second major finding from our experiments is the absence of any cell division in cells with no dyskerin. The dyskerin-depleted livers that we studied showed a much-reduced proliferative response to liver damage induced by CCl4. Moreover, regeneration was accompanied by an increase in the relative amount of wild-type DNA compared with DNA containing the Dkc1 deletion, indicating that much of the regeneration had involved division of cells that did not contain a deleted gene. Compelling evidence that absence of dyskerin is incompatible with cell division is our observation that when Alb-Cre mice were treated with CCl4 and allowed to recover from the resulting liver damage very few cells in the knockout livers were replicating their DNA as assessed by BrdU incorporation. Remarkably the few cells that did incorporate BrdU were all among the minority of cells in the livers that stained positive with the dyskerin antibody. Presumably these were cells that had escaped the deletion. Whether the block in the cell cycle is caused by nucleolar disruption activating the p53 checkpoint response (39, 41) or is due to an unknown mechanism that couples cell division and ribosome biogenesis (2, 41) remains to be determined.
Conclusions.
Using conditional deletion of the mouse Dkc1 gene in hepatocytes, we have produced a model in which we can study mammalian cells without any dyskerin. This model should prove useful in delineating the role of dyskerin and H/ACA snoRNAs in ribosome biogenesis, telomere maintenance, and mRNA expression. Furthermore, the influence of ribosome biogenesis on cell cycle progression may be amenable to experimentation using this model. The experiments presented here show that lack of dyskerin blocks rRNA processing after cleavage of the primary transcript in ITS1 and leads to disruption of ribosome biogenesis. In the absence of dyskerin, hepatocytes are not able to divide.
Supplementary Material
Acknowledgments
We are grateful to Dennis Dietzen for determination of ALT and bilirubin levels. We thank Sandra Navarette for skilled technical assistance, Debbie Laflamme for care of mice, and Rachel Idol for advice and help with polysome analysis. We acknowledge the help of the DDRCC Morphology Core Facility.
We thank the NCI and NIH for financial support through grants to P.J.M. (CA106995), M.B. (HL079556 and CA105312), and D.A.R. (DK068219). D.A.R. is also supported by a grant from CDHNF/TAP.
Footnotes
Published ahead of print on 16 November 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Balakin, A. G., L. Smith, and M. J. Fournier. 1996. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell 86:823-834. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein, K. A., F. Bleichert, J. M. Bean, F. R. Cross, and S. J. Baserga. 2007. Ribosome biogenesis is sensed at the Start cell cycle checkpoint. Mol. Biol. Cell 18:953-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blasco, M. A., H. W. Lee, M. P. Hande, E. Samper, P. M. Lansdorp, R. A. DePinho, and C. W. Greider. 1997. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91:25-34. [DOI] [PubMed] [Google Scholar]
- 4.Boisvert, F. M., S. van Koningsbruggen, J. Navascues, and A. I. Lamond. 2007. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8:574-585. [DOI] [PubMed] [Google Scholar]
- 5.Bowman, L. H., B. Rabin, and D. Schlessinger. 1981. Multiple ribosomal RNA cleavage pathways in mammalian cells. Nucleic Acids Res. 9:4951-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cadwell, C., H. J. Yoon, Y. Zebarjadian, and J. Carbon. 1997. The yeast nucleolar protein Cbf5p is involved in rRNA biosynthesis and interacts genetically with the RNA polymerase I transcription factor RRN3. Mol. Cell. Biol. 17:6175-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caffarelli, E., M. Losito, C. Giorgi, A. Fatica, and I. Bozzoni. 1998. In vivo identification of nuclear factors interacting with the conserved elements of box C/D small nucleolar RNAs. Mol. Cell. Biol. 18:1023-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavaille, J., K. Buiting, M. Kiefmann, M. Lalande, C. I. Brannan, B. Horsthemke, J. P. Bachellerie, J. Brosius, and A. Huttenhofer. 2000. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. U. S. A. 97:14311-14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darzacq, X., B. E. Jady, C. Verheggen, A. M. Kiss, E. Bertrand, and T. Kiss. 2002. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 21:2746-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donmez, G., K. Hartmuth, and R. Luhrmann. 2004. Modified nucleotides at the 5′ end of human U2 snRNA are required for spliceosomal E-complex formation. RNA 10:1925-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313-318. [DOI] [PubMed] [Google Scholar]
- 12.Filipowicz, W., P. Pelczar, V. Pogacic, and F. Dragon. 1999. Structure and biogenesis of small nucleolar RNAs acting as guides for ribosomal RNA modification. Acta Biochim. Pol. 46:377-389. [PubMed] [Google Scholar]
- 13.Fu, D., and K. Collins. 2007. Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol. Cell 28:773-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganot, P., M. Caizergues-Ferrer, and T. Kiss. 1997. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 11:941-956. [DOI] [PubMed] [Google Scholar]
- 15.Giordano, E., I. Peluso, S. Senger, and M. Furia. 1999. minifly, a Drosophila gene required for ribosome biogenesis. J. Cell Biol. 144:1123-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Girard, J. P., H. Lehtonen, M. Caizergues-Ferrer, F. Amalric, D. Tollervey, and B. Lapeyre. 1992. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J. 11:673-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu, B. W., M. Bessler, and P. J. Mason. 2008. A pathogenic dyskerin mutation impairs proliferation and activates a DNA damage response independent of telomere length in mice. Proc. Natl. Acad. Sci. U. S. A. 105:10173-10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He, J., B. W. Gu, J. Ge, Y. Mochizuki, M. Bessler, and P. J. Mason. 2009. Variable expression of Dkc1 mutations in mice. Genesis 47:366-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, J., S. Navarrete, M. Jasinski, T. Vulliamy, I. Dokal, M. Bessler, and P. J. Mason. 2002. Targeted disruption of Dkc1, the gene mutated in X-linked dyskeratosis congenita, causes embryonic lethality in mice. Oncogene 21:7740-7744. [DOI] [PubMed] [Google Scholar]
- 20.Heiss, N. S., S. W. Knight, T. J. Vulliamy, S. M. Klauck, S. Wiemann, P. J. Mason, A. Poustka, and I. Dokal. 1998. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat. Genet. 19:32-38. [DOI] [PubMed] [Google Scholar]
- 21.Henras, A., Y. Henry, C. Bousquet-Antonelli, J. Noaillac-Depeyre, J. P. Gelugne, and M. Caizergues-Ferrer. 1998. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 17:7078-7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henras, A. K., C. Dez, and Y. Henry. 2004. RNA structure and function in C/D and H/ACA s(no)RNPs. Curr. Opin. Struct. Biol. 14:335-343. [DOI] [PubMed] [Google Scholar]
- 23.Henras, A. K., J. Soudet, M. Gerus, S. Lebaron, M. Caizergues-Ferrer, A. Mougin, and Y. Henry. 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 65:2334-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Idol, R. A., S. Robledo, H. Y. Du, D. L. Crimmins, D. B. Wilson, J. H. Ladenson, M. Bessler, and P. J. Mason. 2007. Cells depleted for RPS19, a protein associated with Diamond Blackfan Anemia, show defects in 18S ribosomal RNA synthesis and small ribosomal subunit production. Blood Cells Mol. Dis. 39:35-43. [DOI] [PubMed] [Google Scholar]
- 25.Jady, B. E., E. Bertrand, and T. Kiss. 2004. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J. Cell Biol. 164:647-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang, W., K. Middleton, H. J. Yoon, C. Fouquet, and J. Carbon. 1993. An essential yeast protein, CBF5p, binds in vitro to centromeres and microtubules. Mol. Cell. Biol. 13:4884-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kent, T., Y. R. Lapik, and D. G. Pestov. 2009. The 5′ external transcribed spacer in mouse ribosomal RNA contains two cleavage sites. RNA 15:14-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knight, S. W., N. S. Heiss, T. J. Vulliamy, S. Greschner, G. Stavrides, G. S. Pai, G. Lestringant, N. Varma, P. J. Mason, I. Dokal, and A. Poustka. 1999. X-linked dyskeratosis congenita is predominantly caused by missense mutations in the DKC1 gene. Am. J. Hum. Genet. 65:50-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lafontaine, D. L., C. Bousquet-Antonelli, Y. Henry, M. Caizergues-Ferrer, and D. Tollervey. 1998. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 12:527-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lygerou, Z., C. Allmang, D. Tollervey, and B. Seraphin. 1996. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science 272:268-270. [DOI] [PubMed] [Google Scholar]
- 31.Ma, X., C. Yang, A. Alexandrov, E. J. Grayhack, I. Behm-Ansmant, and Y. T. Yu. 2005. Pseudouridylation of yeast U2 snRNA is catalyzed by either an RNA-guided or RNA-independent mechanism. EMBO J. 24:2403-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marciniak, R., and L. Guarente. 2001. Human genetics. Testing telomerase. Nature 413:370-371. [DOI] [PubMed] [Google Scholar]
- 33.McGowan, K. A., J. Z. Li, C. Y. Park, V. Beaudry, H. K. Tabor, A. J. Sabnis, W. Zhang, H. Fuchs, M. H. de Angelis, R. M. Myers, L. D. Attardi, and G. S. Barsh. 2008. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat. Genet. 40:963-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meier, U. T. 2005. The many facets of H/ACA ribonucleoproteins. Chromosoma 114:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meier, U. T., and G. Blobel. 1994. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J. Cell Biol. 127:1505-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitchell, J. R., J. Cheng, and K. Collins. 1999. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell. Biol. 19:567-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitchell, J. R., E. Wood, and K. Collins. 1999. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402:551-555. [DOI] [PubMed] [Google Scholar]
- 38.Mochizuki, Y., J. He, S. Kulkarni, M. Bessler, and P. J. Mason. 2004. Mouse dyskerin mutations affect accumulation of telomerase RNA and small nucleolar RNA, telomerase activity, and ribosomal RNA processing. Proc. Natl. Acad. Sci. U. S. A. 101:10756-10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Opferman, J. T., and G. P. Zambetti. 2006. Translational research? Ribosome integrity and a new p53 tumor suppressor checkpoint. Cell Death Differ. 13:898-901. [DOI] [PubMed] [Google Scholar]
- 40.Panic, L., J. Montagne, M. Cokaric, and S. Volarevic. 2007. S6-haploinsufficiency activates the p53 tumor suppressor. Cell Cycle 6:20-24. [DOI] [PubMed] [Google Scholar]
- 41.Pestov, D. G., Z. Strezoska, and L. F. Lau. 2001. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol. Cell. Biol. 21:4246-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Postic, C., and M. A. Magnuson. 2000. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis 26:149-150. [DOI] [PubMed] [Google Scholar]
- 43.Richard, P., X. Darzacq, E. Bertrand, B. E. Jady, C. Verheggen, and T. Kiss. 2003. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. EMBO J. 22:4283-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robledo, S., R. A. Idol, D. L. Crimmins, J. H. Ladenson, P. J. Mason, and M. Bessler. 2008. The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA 14:1918-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruggero, D., S. Grisendi, F. Piazza, E. Rego, F. Mari, P. H. Rao, C. Cordon-Cardo, and P. P. Pandolfi. 2003. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science 299:259-262. [DOI] [PubMed] [Google Scholar]
- 46.Ruzankina, Y., C. Pinzon-Guzman, A. Asare, T. Ong, L. Pontano, G. Cotsarelis, V. P. Zediak, M. Velez, A. Bhandoola, and E. J. Brown. 2007. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell 1:113-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strezoska, Z., D. G. Pestov, and L. F. Lau. 2002. Functional inactivation of the mouse nucleolar protein Bop1 inhibits multiple steps in pre-rRNA processing and blocks cell cycle progression. J. Biol. Chem. 277:29617-29625. [DOI] [PubMed] [Google Scholar]
- 48.Venteicher, A. S., E. B. Abreu, Z. Meng, K. E. McCann, R. M. Terns, T. D. Veenstra, M. P. Terns, and S. E. Artandi. 2009. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 323:644-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Venteicher, A. S., Z. Meng, P. J. Mason, T. D. Veenstra, and S. E. Artandi. 2008. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell 132:945-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vulliamy, T., and I. Dokal. 2006. Dyskeratosis congenita. Semin. Hematol. 43:157-166. [DOI] [PubMed] [Google Scholar]
- 51.Vulliamy, T. J., and I. Dokal. 2008. Dyskeratosis congenita: the diverse clinical presentation of mutations in the telomerase complex. Biochimie 90:122-130. [DOI] [PubMed] [Google Scholar]
- 52.Watkins, N. J., A. Gottschalk, G. Neubauer, B. Kastner, P. Fabrizio, M. Mann, and R. Luhrmann. 1998. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA 4:1549-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong, J. M., and K. Collins. 2006. Telomerase RNA level limits telomere maintenance in X-linked dyskeratosis congenita. Genes Dev. 20:2848-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong, J. M., M. J. Kyasa, L. Hutchins, and K. Collins. 2004. Telomerase RNA deficiency in peripheral blood mononuclear cells in X-linked dyskeratosis congenita. Hum. Genet. 115:448-455. [DOI] [PubMed] [Google Scholar]
- 55.Yang, Y., C. Isaac, C. Wang, F. Dragon, V. Pogacic, and U. T. Meier. 2000. Conserved composition of mammalian box H/ACA and box C/D small nucleolar ribonucleoprotein particles and their interaction with the common factor Nopp140. Mol. Biol. Cell 11:567-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zebarjadian, Y., T. King, M. J. Fournier, L. Clarke, and J. Carbon. 1999. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol. Cell. Biol. 19:7461-7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.