Abstract
Wnt signaling is crucial in the organization and maintenance of the human intestinal epithelium, and somatic mutations that result in deregulated Wnt signaling are an early event in the development of colorectal cancer. The Wnt ligand ultimately results in the stabilization of cytoplasmic β-catenin, which is then free to enter the nucleus and activate transcription through its interaction with the transcription factor TCF4. Our laboratory recently found that KLF4, a transcription factor highly expressed in the adult intestine and critical for intestinal differentiation, interacts with β-catenin and inhibits Wnt signaling. In this study, we characterize the molecular mechanisms of KLF4-mediated inhibition of Wnt/β-catenin signaling. We find that the KLF4 directly interacts with the C-terminal transactivation domain of β-catenin and inhibits p300/CBP recruitment by β-catenin. KLF4 inhibits p300/CBP-mediated β-catenin acetylation as well as histone acetylation on Wnt target genes. In addition, we observe that KLF4 directly interacts with TCF4 independently of β-catenin and that KLF4 and TCF4 are expressed in similar patterns within the large intestine, with greatest staining near the epithelial surface. These results provide a deeper understanding of the regulation of β-catenin in the intestine and will have important implications in cancer and stem cell research.
The Wnt/β-catenin signaling pathway plays important roles in early development (19, 31), stem cell renewal (22, 34), and tumorigenesis (20, 35). In addition, Wnt signaling is crucial in the organization and maintenance of the human intestinal epithelium (46, 51), and somatic mutations that result in deregulated Wnt signaling are an early event in the development of colorectal cancer (37, 39, 47). In this pathway, many different components work together to transduce an external signal into changes in gene expression within the target cell. Upon binding its receptor, the Wnt ligand ultimately results in the stabilization of cytoplasmic β-catenin (44), which is then free to enter the nucleus and activate transcription through its interaction with the TCF/LEF family of transcription factors (1, 18, 36).
In the adult intestine, TCF4 is the primary TCF/LEF family member involved in mediating active Wnt/β-catenin signaling (25, 26). In contrast, TCF1 and TCF3 are primarily transcriptional repressors (14, 49), whereas LEF1 is not expressed in the adult intestine (59). In the absence of β-catenin, TCF4 recruits corepressors such as CtBP (4), HDAC1 (3, 24), and Groucho/TLE (5, 28, 50), in order to silence expression of target genes. However, binding of β-catenin results in displacement of these corepressors (7). β-Catenin then recruits coactivators through its N-terminal and C-terminal transactivation domains. Its N-terminal transactivation domain interacts with BCL9/Legless, which in turn recruits Pygopus (2, 27, 45, 55), whereas its C-terminal domain recruits p300/CBP. p300 and CBP are closely related proteins that promote transcriptional activation through several mechanisms, including recruitment of the basal transcriptional machinery and acetylation of nearby histones (12, 41). Notably, recruitment of p300/CBP by β-catenin is required for Wnt signaling in vivo (17, 54).
Krüppel-like factor 4 (KLF4) is a transcription factor highly expressed in the adult intestine (52) and is critical in the process of differentiation (21). Our laboratory recently found that KLF4 interacts with β-catenin and inhibits Wnt signaling (61). Given the critical role of β-catenin in mediating Wnt signaling and in the development of colorectal cancer, a better understanding of the mechanism of KLF4-mediated inhibition will lead to novel therapies for colorectal cancer. However, the precise molecular mechanisms of how KLF4 inhibits β-catenin are not entirely clear. In this study, we aim to more fully characterize the interactions between KLF4 and β-catenin. Earlier work demonstrated that KLF4 interacts with the C-terminal transactivation domain of β-catenin, the same domain known to interact with p300/CBP. Thus, we hypothesize that KLF4 inhibits Wnt/β-catenin signaling by blocking the recruitment of the transcriptional coactivator p300/CBP.
MATERIALS AND METHODS
Plasmid DNA constructs.
pCS2-KLF4 (N-terminal Flag tag), pCS2-β-catenin (N-terminal Flag tag), pcDNA3-Myc-β-catenin-C, pRC-CMV-mCBP-HA, SuperTOPFlash, Myc-TCF4, and glutathione S-transferase (GST)-p300 (CH3) have been described previously (9, 32, 61). Truncation mutants KLF4Δ155-399(ΔM), KLF4Δ402-483(ΔC), KLF4-N(1-157), KLF4-M(157-401), KLF4ΔM1(1-157;351-483), KLF4(300-483), KLF4(350-483), KLF4(350-459), KLF4(350-429), and KLF4(350-401) were generated by PCR and cloned into the pCS2 vector. HA-TCF4 was generated by subcloning the Myc-TCF4 plasmid by PCR. pcDNA4-Flag-ICAT was generated by cloning the ICAT open reading frame from an SW480 cDNA library using PCR and then inserting the resultant DNA sequence into the pcDNA4-TO-2xFLAG vector. KLF4, β-catenin, TCF4(1-65), and TCF4(265-496) were subcloned into the pMBP-C5x vector (purchased from NEB) by PCR. GST-β-catenin, GST-β-catenin-N(14-150), and GST-β-catenin-C(594-781) were generated by PCR and cloned into the pGEX-6P3 vector. All constructs were verified by DNA sequencing. Primers for these constructs are available upon request.
Cell culture and transient transfection.
HEK293T and HCT116 cells were grown in Dulbecco modified Eagle medium (DMEM) (Mediatech) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. LS174T cells were grown in RPMI medium (Mediatech) supplemented with 5% fetal bovine serum and 1% penicillin/streptomycin. Stable cell line LS174T-tet/on-KLF4 has been described previously (61). HEK293T cells were transiently transfected using the calcium phosphate method as described previously (61).
Western blotting and immunoprecipitation (IP).
Western blotting and immunoprecipitation were performed as described previously (61). For acetylation experiments, 5 mM sodium butyrate and 5 mM nicotinamide (Acros Organics) were added to cells 6 h prior to harvest and to lysis buffer to inhibit deacetylases. KLF4 and its mutants were immunoprecipitated, eluted with 0.2 mg/ml Flag peptide, and analyzed by Western blotting with an antibody that specifically recognizes acetylated lysine (catalog no. 9441; Cell Signaling) or acetyl-K49-β-catenin (catalog no. 9534S; Cell Signaling). Other antibodies used include mouse anti-Flag (catalog no. F1804; Sigma), rabbit anti-Flag (catalog no. F7425; Sigma), rat anti-HA (catalog no. 3F10; Roche), β-catenin (catalog no. C2206; Sigma), and p300 (catalog no. sc-584; Santa Cruz).
GST pull-down.
Cells were lysed in the appropriate volume of lysis buffer (50 mM HEPES, 150 mM NaCl, 100 μM ZnSO4, 10% glycerol, 1 mM dithiothreitol [DTT], 1% Triton X-100, with protease inhibitors, pH 7.4). GST-tagged expression vectors were expressed in Escherichia coli, and the resultant protein was purified with GST beads (glutathione Sepharose; Sigma). GST beads containing purified GST-tagged protein were then incubated with cell lysate at 4°C for 1 h; then, beads were washed three times with lysis buffer and boiled in 1× sodium dodecyl sulfate (SDS) sample buffer, followed by analysis via Western blotting.
RT-PCR.
LS174T-tet/on-KLF4 cells were plated at approximately 1 × 106 cells per plate in a 10-cm2 dish. The following day, doxycycline (1 μg/ml) was added to the culture medium. After 48 h of incubation, RNA was isolated using the RNeasy kit (Qiagen). Reverse transcriptase PCR (RT-PCR) was performed as described previously (61). The following primers were used: β-actin, 5′-CAACCGCGAGAAGATGAC-3′ and 5′-AGGAAGGCTGGAAGAGTG-3′; survivin, 5′-CATTCGTCCGGTTGCGCTTTCC-3′ and 5′-GCGCACTTTCTCCGCAGTTTCC-3′; and c-myc, 5′-TGGGCTGTGAGGAGGTTTG-3′ and 5′-TATGTGGAGCGGCTTCTCG-3′.
Luciferase assays.
HEK293T cells were transiently transfected in a 12-well plate with 0.2 μg of the Super8xTOPFlash reporter, 0.05 μg of Renilla luciferase, and 0.5 μg of each plasmid DNA. Total DNA transfected in each well was normalized using pCS2 DNA as needed. Two days later, cells were harvested and luciferase activity was measured. All conditions were done in triplicate, and each experiment was carried out at least two times.
Adenovirus construction.
Ad-GFP and Ad-KLF4 adenoviruses were prepared according to the protocol described in reference 16. Briefly, pCS2-Flag-KLF4 was cut by HindIII and XbaI and inserted into the pAdTrack vector. The GFP-KLF4 cassette was then transferred into the pAdEasy vector via homologous recombination in the E. coli BJ5183 cell line after electroporation and selection on kanamycin agar plates. Resistant colonies were then grown in 200 ml LB/Kan+ plates, and pAdEasy-GFP-KLF4 DNA was harvested using a MidiPrep (Qiagen). DNA was then transfected into 293-Ad cells using the calcium phosphate method. Several days later, adenovirus-containing medium was harvested and used to reinfect fresh 293-Ad cells. After several rounds of enrichment, high-titer adenovirus was collected and stored at −80°C. Viral titers were determined based on the percentage of infected cells expressing green fluorescent protein (GFP) and estimated to be 1 × 108 PFU/ml. HCT116 cells were infected at a multiplicity of infection of 10.
Interference RNA and immunohistochemistry.
Interference RNA and immunohistochemistry were tested as described previously (61). For staining, the following antibodies were used: KLF4 (61) and TCF4 (catalog no. C9B9; Cell Signaling).
Chromatin immunoprecipitation (ChIP).
ChIP assays were performed according to the protocol developed by Nowak et al. (40) with some modifications. HEK293T cells (1 × 106) were plated on 10-cm2 dishes. The following day, doxycycline was added to a final concentration of 1 μg/ml, and cells were grown for an additional 36 h. The cells were cross-linked with disuccimidyl glutarate (catalog no. 20593; Pierce) and formaldehyde at room temperature. Cells were pelleted at 3,000 rpm for 1 min and resuspended in 900 μl L1 buffer (50 mM Tris, 2 mM EDTA, 0.1% IGEPAL, 10% glycerol, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, protease inhibitor cocktail, pH 8.0) and allowed to sit on ice for 15 min. After centrifugation at 4,000 rpm for 5 min, the supernatant was removed and the nuclear pellet was resuspended in 500 μl of ChIP lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris, pH 8.0, protease inhibitor cocktail). Cell lysate was sonicated four times for 10 s at setting 5 on a Branson Sonifier 150 on ice, with a 30-s break between sonications. After centrifugation at 13,200 rpm for 10 min at 4°C, the supernatant was transferred to a fresh tube. The A260 of a 1:50 dilution of each sample was measured using a spectrophotometer to estimate DNA content. For each assay, 100 μl of the most dilute sample was used, and the more concentrated samples were diluted in lysis buffer so that each condition received the same amount of total DNA. Lysate was diluted and incubated with 4 μg of the appropriate antibody overnight, followed by incubation with 100 μl of protein A-agarose-salmon sperm DNA 50% slurry (catalog no. 16-157; Upstate) for 3 h at 4°C. Beads were then washed with a series of washes, and bound DNA-protein complexes were eluted and de-cross-linked. DNA was then purified by phenol-chloroform extraction and ethanol precipitation. Pelleted DNA was resuspended in 20 μl TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0), and 1 μl was used for PCR. Optimal cycling parameters to ensure operation in the linear range were primer specific, but typically 32 to 40 cycles were done. The following primers were used: SuperTOPFlash, 5′-CAACGCGTGTACGGGAGGTACTTGGAG-3′ and 5′-CAGGATCCGTGGCTTTACCAACAGTAC-3′; survivin, 5′-GGGGCGCTAGGTGTGGG-3′ and 5′-TTCAAATCTGGCGGTTAATGGC-3′; and c-myc, 5′-TATGTGGAGCGGCTTCTCG-3′ and 5′-TGGGCTGTGAGGAGGTTTG-3′. For ChIP assays, antibodies used include preimmune rabbit immunoglobulin G (IgG) and acetylated histone H3 (catalog no. 06-599; Millipore).
RESULTS
KLF4 levels are decreased in intestinal adenomas from ApcMin/+ mice.
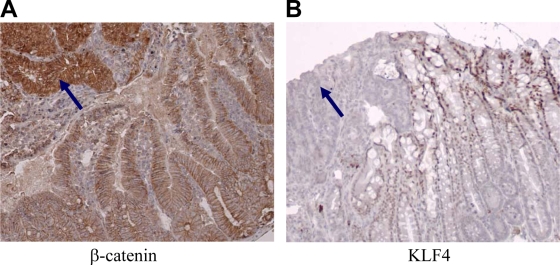
We and others have shown that KLF4 levels are reduced in a subset of human cancers (6, 42, 60, 61). However, KLF4 levels are increased in other cancers, such as breast cancer (10, 43). To further examine KLF4 expression in a genetically defined tumor model, we performed immunohistochemistry on intestinal tissue from ApcMin/+ mice (38). In the normal intestine, KLF4 is expressed in the nuclei of differentiated epithelial cells. In adenomas, KLF4 levels were significantly decreased (Fig. 1B). In contrast, β-catenin levels were significantly increased in the nuclei of adenomas (Fig. 1A). These results demonstrate an inverse relationship between the expression levels of β-catenin and KLF4 in both normal and adenomatous epithelia. We previously demonstrated that KLF4 inhibits Wnt/β-catenin signaling (61), and others have shown that hemizygous deletion of the Klf4 gene resulted in an increased size and number of adenomas in ApcMin/+ mice (11). Collectively, these data suggest that KLF4 represses the development of β-catenin-induced colorectal tumors. However, the mechanism delineating precisely how KLF4 inhibits β-catenin is unclear. This is the primary aim of our current study.
FIG. 1.
KLF4 expression in the intestine of the ApcMin/+ mouse. (A) β-Catenin levels are increased in the nuclei of adenomas (arrow) compared with their levels in adjacent normal intestine. (B) KLF4 levels are decreased in adenomas (arrow) compared with their levels in adjacent normal intestine.
KLF4 directly interacts with the C terminus of β-catenin.
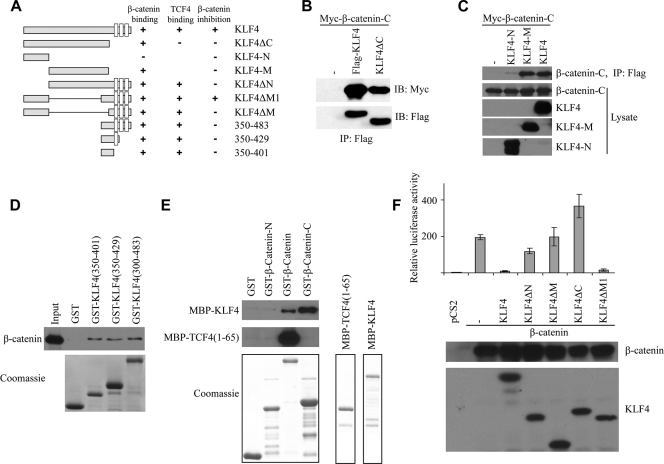
Previous work in our lab demonstrated that KLF4 interacts with the C-terminal transactivation domain of β-catenin (61). To further characterize this interaction, we used various domains of KLF4 and individually tested their ability to immunoprecipitate the β-catenin C-terminal transactivation domain (see Fig. 2A for all KLF4 mutants used in this article). As shown in Fig. 2B, both full-length KLF4 and KLF4ΔC interact with β-catenin, suggesting that KLF4 without its C-terminal zinc fingers is sufficient for β-catenin binding. To more finely map the interacting region of KLF4, we designed several additional mutants. The N-terminal transactivation domain of KLF4 (amino acids 1 to 157) does not interact with β-catenin, whereas the middle region of KLF4 (amino acids 157 to 401) is sufficient for this interaction (Fig. 2C). To confirm these results, we performed an in vitro binding assay using GST-tagged KLF4 mutants and incubated each mutant with colon cancer cell lysate containing endogenous β-catenin (SW480 cells). We found that KLF4(350-401) was sufficient for interacting with β-catenin (Fig. 2D). To test if KLF4 directly or indirectly binds β-catenin, we performed an in vitro binding assay using purified β-catenin and KLF4 (Fig. 2E). We found that both full-length β-catenin and the C terminus of β-catenin bind directly KLF4. As a control, the N terminus of TCF4 binds only the full-length β-catenin but not the N terminus or C terminus of β-catenin (Fig. 2E).
FIG. 2.
The N-terminal and C-terminal domains of KLF4 are required for inhibition of the TOPFlash reporter. (A) Diagram of all KLF4 constructs used in this paper. All constructs contain an N-terminal Flag tag. Full-length KLF4 contains 483 amino acids. The open boxes shown in the C termini represent zinc finger motifs. The binding affinity and activity of each construct are summarized in adjacent columns. (B) HEK293T cells were transfected with Myc-tagged β-catenin C-terminal transactivation domain and full-length KLF4 or KLF4ΔC (deletion of C-terminal zinc fingers). Anti-Flag immunoprecipitation was performed, followed by anti-Myc Western blotting. (C) HEK293T cells were transfected with a Myc-tagged β-catenin C terminus and either the N-terminal (amino acids 1 to 157) or middle (amino acids 157 to 401) domain of the full-length KLF4 protein. After anti-Flag immunoprecipitation, a Myc Western blot assay was performed (top row). Western blotting was performed on cell lysate as a control. (D) SW480 cell lysate was incubated with various GST-tagged KLF4 constructs as shown. After several washings, bound protein was eluted and an anti-β-catenin Western blot assay was performed. Coomassie staining demonstrates the expression level of each GST-tagged protein (lower panel). (E) KLF4 directly interacts with β-catenin. The GST-tagged β-catenin N terminus, β-catenin, and β-catenin C terminus were incubated with MBP-tagged KLF4 or TCF4(1-65). β-Catenin proteins were pulled down with GST beads, and KLF4 and TCF4 were analyzed by Western blotting with an anti-MBP antibody. Coomassie staining demonstrates expression level of each purified protein (lower panel). (F) HEK293T cells were plated in 12-well plates, and each well was transfected with 0.2 μg Super8xTOPFlash, 0.05 μg Renilla luciferase, 0.5 μg of β-catenin DNA, and 0.5 μg of one of the various KLF4 constructs shown. Cells were harvested 48 h later and analyzed for luciferase activity. Renilla was used as a control for transfection efficiency and used to normalize raw luciferase data. Data points were performed in triplicate, and values are expressed as fold increases relative to controls (empty vector only). Western blotting demonstrates similar levels of expression of β-catenin between conditions and of expression levels of various KLF4 constructs (lower panel).
The N-terminal and C-terminal domains of KLF4 are required for inhibition of the TOPFlash reporter.
We next addressed the functional significance of these interactions using the TOPFlash reporter assay, which provides an index of the overall level of β-catenin/TCF activity within a cell (61). As previously demonstrated, full-length KLF4 represses TOPFlash activity (Fig. 2F) (61). Deletion of the N-terminal region, the middle region, or the three C-terminal zinc fingers effectively abolished the ability of KLF4 to repress the TOPFlash reporter. Moreover, the N terminus of KLF4 or a fragment containing amino acids 350 to 483 alone was unable to repress the TOPFlash reporter (not shown). However, a fusion of these two domains (amino acids 1 to 157 and 350 to 483, labeled as ΔM1) effectively repressed the TOPFlash reporter. KLF4ΔC(1-401) is able to interact with β-catenin (Fig. 2B) but cannot repress the TOPFlash reporter. These data collectively suggest that the N-terminal transactivation domain, the C-terminal portion of the middle domain, and the C-terminal DNA-binding domain are all required for KLF4-mediated inhibition of the TOPFlash reporter, whereas region 158 to 349 appears to be dispensable.
Expression patterns of KLF4 and TCF4 in the mouse intestine overlap.
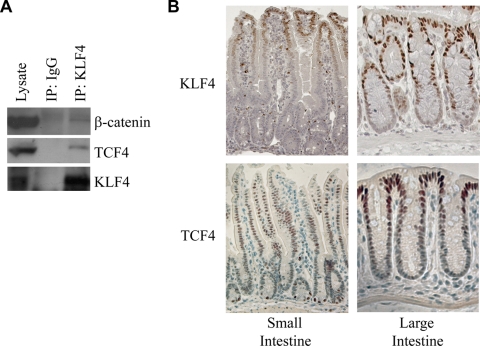
Since KLF4 and β-catenin clearly interact, and it is well known that β-catenin also interacts with TCF4 in order to activate target genes (26), we decided to test whether all three proteins could be found in the same complex. We immunoprecipitated endogenous KLF4 from HCT116 cells and then performed Western blotting for β-catenin and TCF4. We detected both β-catenin and TCF4 in the immunoprecipitate, suggesting that KLF4, β-catenin, and TCF4 coimmunoprecipitate (Fig. 3A). Next, we decided to investigate whether these proteins are expressed in similar compartments in the mouse small and large intestines. In the small intestine, KLF4 was most strongly expressed at the tips of villi, with decreasing expression along the crypt-villus axis toward the crypts (Fig. 3B). Expression in the large intestine followed a similar pattern, with the most intense staining at the epithelial surface and the weakest at the base of the crypts. Expression of TCF4 followed a similar pattern to that of KLF4 in the colon. However, we observed a few exceptions to these general patterns in the small intestine. First, we noted that in the small intestine, in a seemingly random pattern, a cell would stain intensely for KLF4. Presumably these represent goblet cells, given the role of KLF4 in goblet cell differentiation (21). Second, cells at the base of small intestinal crypts stained intensely for TCF4, possibly representing stem cells that typically reside there.
FIG. 3.
KLF4 and TCF4 interact. (A) Endogenous KLF4 was immunoprecipitated from HCT116 cells, and Western blotting was performed on the eluate. Both β-catenin and TCF4 were detectable in the eluate. IgG was used as a negative control. (B) Immunohistochemistry of the mouse small and large intestine. Expression patterns of KLF4 and TCF4 in the mouse intestine overlap, with the strongest expression near the epithelial surface. Tissue was harvested from the small and large intestines of an adult mouse, embedded in paraffin, and stained using either a KLF4 or TCF4 antibody. The base of the crypts is at the bottom of each photo, whereas the tip of the villus (small intestine) or luminal surface (large intestine) is at the top of the photo.
KLF4 directly binds TCF4.
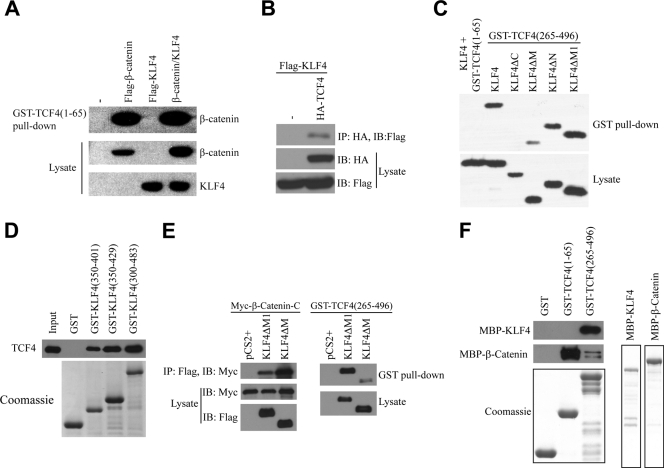
To delineate the mechanisms of KLF4-mediated inhibition of β-catenin, we tested whether KLF4 could interfere with binding between β-catenin and TCF4. The N terminus of TCF4 interacts with β-catenin (48). Thus, we performed a GST pulldown assay using GST-tagged TCF4(1-65), incubated with cell lysate containing endogenous β-catenin with or without overexpressed KLF4 (Fig. 4A). We found that β-catenin interacted strongly with the N terminus of TCF4, as expected, whereas KLF4 did not interact, nor did it modulate interactions between TCF4(1-65) and β-catenin. These data suggest that KLF4 does not interfere with TCF4/β-catenin binding. Given that KLF4, β-catenin, and TCF4 coimmunoprecipitate (Fig. 3A), we decided to test whether KLF4 and TCF4 could directly interact as well. To test this, we performed immunoprecipitation using HA-tagged TCF4 and Flag-tagged KLF4. As shown in Fig. 4B, after immunoprecipitation of HA-tagged full-length TCF4, we could detect KLF4 protein in the eluate, suggesting that KLF4 and TCF4 indeed interact. To elucidate the necessary domains for this interaction, we performed a GST pulldown assay using either the β-catenin-interacting domain of TCF4 (amino acids 1 to 65) or the DNA-binding HMG domain of TCF4 (amino acids 265 to 496), and incubated it with either full-length KLF4 or various deletion mutants. Full-length KLF4 interacts with the DNA-binding domain of TCF4 but not the N-terminal β-catenin binding domain (Fig. 4C). Moreover, deletion of the three C-terminal zinc fingers of KLF4, but not other domains, effectively abolished interactions between KLF4 and TCF4. Next, we decided to further qualify these results using full-length TCF4 and various GST-tagged mutants of KLF4. The C-terminal fragment of the middle domain of KLF4 (amino acids 350 to 401), which lies just N-terminal to the first zinc finger, is sufficient for interactions with full-length TCF4 (Fig. 4D). When all three zinc fingers are added to this GST-tagged peptide (amino acids 350 to 483), KLF4 appears to interact with TCF4 more strongly. Collectively, these data suggest that the DNA-binding domain of TCF4 interacts with the C-terminal region of KLF4. Since KLF4ΔM1 but not KLF4ΔM inhibits TOPFlash activity, we compared their relative binding affinities with β-catenin. Surprisingly, we found that both KLF4ΔM1 and KLF4ΔM bind the C terminus of β-catenin (Fig. 4E), raising the question of why they have different activities in Wnt inhibition. We then compared the bindings of KLF4ΔM and KLFΔM1 to TCF4 and found that TCF4(265-496) binds KLF4ΔM1 more strongly than KLF4ΔM (Fig. 4E), similar to the result in Fig. 4C, suggesting that KLF4 needs to interact with both β-catenin and TCF4 to inhibit Wnt signaling. β-Catenin and TCF4 may bind both amino acids 350 to 401 and zinc finger domains of KLF4 or bind KLF4 indirectly in this experiment. To test if TCF4 directly or indirectly binds KLF4, GST-TCF4(1-65) and GST-TCF4(265-496) were incubated with purified KLF4 (Fig. 4F). We found that KLF4 directly binds TCF4(265-496). As a control, β-catenin binds TCF4(1-65). These experiments confirmed that KLF4 directly binds both β-catenin and TCF4.
FIG. 4.
The C-terminal domain of KLF4 and the DNA-binding domain of TCF4 interact. (A) HEK293T cells were transfected with Flag-β-catenin, Flag-KLF4, or both, and after 48 h, lysate was collected and incubated with GST-tagged TCF4(1-65). After several washings, bound proteins were eluted and an anti-Flag Western blot assay was performed. (B) HEK293T cells were transfected with Flag-tagged KLF4, with or without HA-tagged TCF4, lysate was harvested, and anti-HA immunoprecipitation was performed. An anti-Flag Western blot assay was then performed on the bound proteins. (C) GST-TCF4(1-65) or GST-TCF4(265-496) was incubated with lysate from HEK293T cells overexpressing various Flag-tagged KLF4 constructs as shown. After several washings, bound proteins were eluted and an anti-Flag Western blot assay was performed. (D) HEK293T cells were transfected with a Myc-tagged TCF4 construct. Forty-eight hours later, cell lysate was collected and incubated with various GST-tagged KLF4 constructs. After several washings, bound proteins were eluted and an anti-Myc Western blot assay was performed. Coomassie staining was performed to demonstrate expression levels of the various GST-tagged proteins used. (E) KLF4ΔM1 binds both β-catenin and TCF4. Left: the Myc-tagged β-catenin C terminus was cotransfected with Flag-tagged KLF4ΔM and KLF4ΔM1 into HEK293T cells. β-Catenin was immunoprecipitated with an anti-Myc antibody, and KLF4ΔM and KLF4ΔM1 were analyzed by Western blotting with an anti-Flag antibody. Right: Flag-tagged KLF4ΔM and KLF4ΔM1 were transfected into HEK293T cells and then incubated with GST-TCF4(265-496). TCF4 was pulled down with GST beads, and KLF4ΔM and KLF4ΔM1 were analyzed by Western blotting. (F) KLF4 directly interacts with TCF4. GST-tagged TCF4(1-65) and TCF4(265-496) were incubated with MBP-tagged KLF4 or β-catenin. TCF4 proteins were pulled down with GST beads, and KLF4 and β-catenin were analyzed by Western blotting with an anti-MBP antibody. Coomassie staining demonstrates the expression level of each purified protein (lower panel).
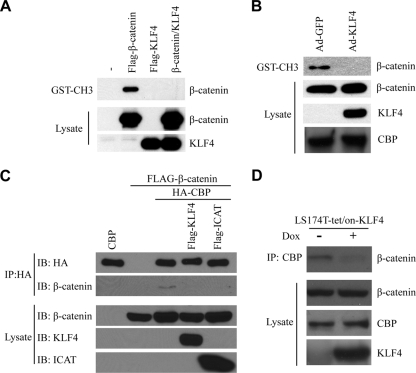
KLF4 inhibits binding between β-catenin and p300/CBP.
After binding the promoter of a Wnt target gene, TCF4 recruits β-catenin. β-Catenin then recruits transcriptional coactivators such as p300/CBP (17, 54). Since KLF4 does not affect binding between β-catenin and TCF4 (Fig. 4), we hypothesized that KLF4 might interfere with the recruitment of coactivators instead. To test this, we incubated Flag-tagged β-catenin or KLF4 with the CH3 domain of p300 and performed a GST pulldown assay. As shown in Fig. 5A, β-catenin interacts with the CH3 domain of p300, as expected. However, overexpression of KLF4 effectively blocked the ability of β-catenin to interact with the CH3 domain. We performed a similar experiment using endogenous β-catenin from HCT116 cells and found that overexpression of KLF4 using an adenoviral construct blocked binding between β-catenin and GST-tagged p300-CH3 (Fig. 5B). To test whether KLF4 could inhibit binding between β-catenin and full-length CBP, we overexpressed HA-tagged CBP and Flag-tagged β-catenin in 293T cells (Fig. 5C). After immunoprecipitation using an anti-HA antibody, we were able to detect β-catenin in the eluate (Fig. 5C, second row, third column). However, overexpression of KLF4 blocked this interaction without affecting expression of CBP or β-catenin. ICAT, a protein known to interact with β-catenin and inhibit its interaction with p300 (8), also blocked β-catenin/CBP interactions in this assay. Finally, we decided to demonstrate similar results using endogenous β-catenin and CBP. In this experiment, we immunoprecipitated CBP from LS174T cells and were able to detect β-catenin in the eluate (Fig. 5D). Overexpression of KLF4 in these cells effectively blocked binding between CBP and β-catenin without affecting expression of either protein. Collectively, these data suggest that KLF4 interferes with binding between β-catenin and p300/CBP.
FIG. 5.
KLF4 inhibits binding between β-catenin and p300/CBP. (A) HEK293T cells were transfected with Flag-β-catenin, Flag-KLF4, or both, and after 48 h, lysate was collected and incubated with the GST-tagged CH3 domain of p300. After several washings, bound proteins were eluted and an anti-Flag Western blot assay was performed. (B) HCT116 cells were treated with either control adenovirus (Ad-GFP) or a KLF4-expressing adenovirus (Ad-KLF4). After 48 h, cell lysate was collected and incubated with the GST-tagged CH3 domain of p300. After several washings, bound proteins were eluted and an anti-β-catenin Western blot assay was performed. Western blotting against CBP demonstrates that overexpression of KLF4 did not affect the expression of CBP. (C) HEK293T cells were transfected with HA-tagged CBP and Flag-tagged β-catenin, with or without Flag-KLF4 or ICAT. Anti-HA immunoprecipitation was performed on the cell lysate, and bound proteins were analyzed via anti-Flag Western blotting. (D) Expression of KLF4 was induced in LS174T-tet/on-KLF4 cells via treatment with doxycycline for 48 h. Anti-CBP immunoprecipitation was performed on cell lysate, and bound proteins were analyzed via an anti-β-catenin Western blot assay.
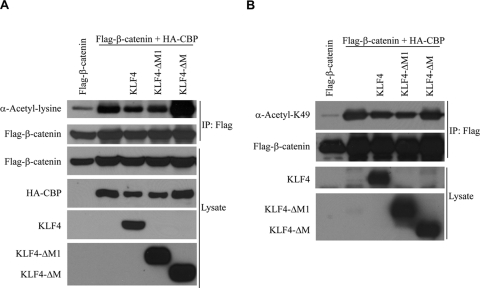
KLF4 inhibits acetylation of β-catenin by p300/CBP.
β-Catenin is acetylated by p300/CBP (29, 58). Thus, if KLF4 interferes with interactions between β-catenin and p300/CBP, it follows that KLF4 should inhibit acetylation of β-catenin as well. To test this hypothesis, we overexpressed CBP and Flag-tagged β-catenin, immunoprecipitated β-catenin, and performed a Western blot assay using a general anti-acetyl-lysine antibody. Under these conditions, β-catenin was indeed acetylated (Fig. 6A), and coexpression of KLF4 effectively decreased the total level of acetylated β-catenin. KLF4ΔM1, which inhibits β-catenin-mediated activation of the TOPFlash reporter (see Fig. 2F), also blocks acetylation of β-catenin. In contrast, KLF4ΔM, which cannot inhibit the TOPFlash reporter, does not block acetylation of β-catenin.
FIG. 6.
KLF4 inhibits acetylation of β-catenin. (A) HEK293T cells were transfected with Flag-β-catenin, HA-CBP, and various Flag-tagged KLF4 constructs. After 48 h, anti-Flag immunoprecipitation was performed on cell lysate. After several washings, bound proteins were analyzed via an anti-acetylated lysine Western blot assay. (B) HEK293T cells were transfected with Flag-β-catenin, HA-CBP, and various Flag-tagged KLF4 constructs. After 48 h, cell lysate was collected and directly analyzed via an anti-acetylated lysine 49-β-catenin Western blot assay.
β-Catenin is preferentially acetylated at lysine 49 (57, 58), and antibodies to β-catenin acetylated at this location are commercially available. Using this antibody, we found that KLF4 effectively blocked CBP-mediated acetylation of β-catenin at lysine 49 as well (Fig. 6B) and found similar results for KLF4ΔM1 and KLF4ΔM, as was seen in Fig. 6A. These data further confirm that KLF4 interferes with interactions between β-catenin and p300/CBP by demonstrating that overexpression of KLF4 results in decreased β-catenin acetylation as well.
KLF4 inhibits histone acetylation on β-catenin/TCF4-regulated promoters.
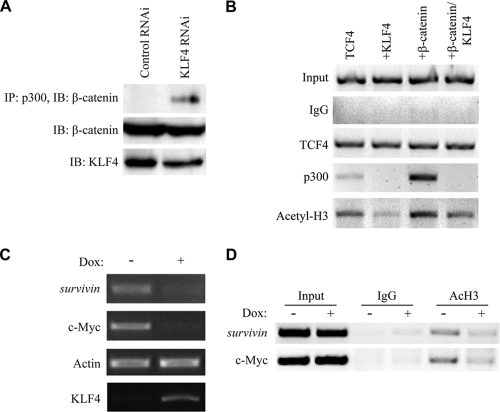
To test the physiologic significance of these interactions using endogenous proteins, we immunoprecipitated p300 from HCT116 cells and performed a Western blot assay for β-catenin. Interestingly, this interaction was more easily detectable when expression of endogenous KLF4 was decreased using RNA interference (RNAi) (Fig. 7A). If KLF4 inhibits β-catenin by interfering with binding between β-catenin and p300/CBP, it follows that overexpression of KLF4 should decrease the relative occupancy of p300/CBP on β-catenin/TCF4-regulated promoters as well. To test this hypothesis, we first performed chromatin immunoprecipitation (ChIP) on the TOPFlash promoter. As expected, overexpression of β-catenin resulted in increased occupancy of p300, as well as increased localized histone acetylation (Fig. 7B). However, coexpression of KLF4 effectively blocked p300 occupancy on the TOPFlash promoter and resulted in decreased localized histone acetylation. To test whether these results are applicable to endogenous gene targets as well, we first tested the ability of KLF4 to inhibit expression of c-myc and survivin, two known targets of β-catenin/TCF signaling (33, 56). As expected, overexpression of KLF4 decreased expression of both c-myc and survivin (Fig. 7C). Next, we performed ChIP assays using primers targeting the proximal promoters of these two genes, and we found that overexpression of KLF4 decreased localized histone acetylation on these endogenous promoters as well (Fig. 7D). Collectively, these data suggest that KLF4 inhibits p300/CBP-mediated histone acetylation on β-catenin/TCF target genes.
FIG. 7.
KLF4 inhibits β-catenin-mediated recruitment of p300/CBP. (A) HCT116 cells were transfected with either control RNAi or KLF4 RNAi, and after 48 h, anti-p300 immunoprecipitation was performed on the cell lysate. Bound proteins were washed and analyzed via anti-β-catenin Western blotting. (B) HEK293T cells were transfected with the Super8xTOPFlash plasmid as well as the expression plasmids shown. After 48 h, cells were cross-linked and sonicated, and immunoprecipitation was performed using the antibodies shown at left as row labels. Bound DNA-protein complexes were eluted and de-cross-linked, and free DNA was amplified using primers specific to the promoter region of the Super8xTOPFlash reporter. (C) Expression of KLF4 was induced in LS174T-tet/on-KLF4 cells via treatment with doxycycline for 48 h. Total RNA was then harvested, and cDNA was produced using reverse transcriptase and amplified via PCR using primers specific to the genes shown. Actin was included as a negative control. (D) Expression of KLF4 was induced in LS174T-tet/on-KLF4 cells via treatment with doxycycline for 48 h. After 48 h, cells were cross-linked and sonicated, and immunoprecipitation was performed using antibodies shown at left as row labels. Bound DNA-protein complexes were eluted and de-cross-linked, and free DNA was amplified using primers specific to the promoter region of the genes shown.
DISCUSSION
Our previous study demonstrated that KLF4 interacts with β-catenin and represses Wnt signaling. In this study, we found that KLF4 directly interacts with both β-catenin and TCF4. Via GST pulldown assays and immunoprecipitation, we demonstrate that KLF4 inhibits binding between β-catenin and p300/CBP (Fig. 5). Using ChIP assays, we find that KLF4 inhibits the recruitment of p300/CBP by β-catenin to the TOPFlash promoter, resulting in decreased localized histone acetylation and transcriptional activation of the TOPFlash reporter gene (Fig. 7B). In addition, we demonstrate that KLF4 blocks p300/CBP-mediated acetylation of β-catenin (Fig. 6) and inhibits histone acetylation on c-myc and survivin genes. Previous studies have implicated the role of β-catenin acetylation in stability (57), transcription (58), and protein-protein interactions (29). Thus, we would expect KLF4 to antagonize at least one of these roles for β-catenin acetylation.
We demonstrated that KLF4 interacts with the C-terminal transactivation domain of β-catenin, whereas conversely the middle region of KLF4, extending from amino acid 350 to amino acid 401, and the zinc finger domains interact with β-catenin (Fig. 2). These data are physiologically relevant, as after immunoprecipitation of endogenous KLF4 from HCT116 cells, we could detect endogenous β-catenin (Fig. 3A). In addition, we found that endogenous KLF4 and TCF4 interact via immunoprecipitation assays (Fig. 3). Further investigation demonstrates that the C-terminal region of KLF4, extending from amino acid 350 to amino acid 483, interacts with TCF4(265-496), which contains the HMG domain. Note that the HMG domain of TCF4 is distinct from the domain that interacts with β-catenin (the N terminus, amino acids 1 to 65). Since the N terminus of TCF4 is not required for KLF4 binding, these data suggest that TCF4 binds to KLF4 independently of β-catenin. Given that KLF4 interacts with the HMG domain of TCF4, a plausible mechanism for KLF4-mediated inhibition of Wnt signaling might be that KLF4 prevents TCF4 from binding its consensus sequence. However, our ChIP data found that overexpression does not affect TCF4 occupancy on the TOPFlash promoter (Fig. 7B), arguing against this possibility.
The C terminus of β-catenin binds KLF4 independently of TCF4. We note that the β-catenin and TCF4-binding domain of KLF4(350-483) is required for repressing Wnt/β-catenin signaling. However, this domain must be combined with the N-terminal transactivation domain of KLF4(1-157) in order to effectively repress β-catenin (Fig. 2E). These results suggest that multiple domains within KLF4 perform discrete functions in repressing transcription mediated by β-catenin.
How KLF4 inhibits recruitment of p300/CBP by β-catenin is not known. Since both KLF4 and p300/CBP bind the C terminus region of β-catenin, KLF4 may compete with p300/CBP for β-catenin binding. ICAT, a protein previously characterized to inhibit Wnt/β-catenin (8), similarly interferes with binding between β-catenin and p300/CBP. Interestingly, ICAT uses a three-helix bundle to interact with the C terminus of β-catenin (13). Since the crystal structure of the KLF4 protein has not yet been determined, it is not clear if KLF4 interfaces with β-catenin in a similar manner. Recently, it was reported that WT1, another C2H2 zinc finger protein, competes with TCF4 for CBP binding and thus inhibits Wnt signaling (23). Since KLF4 also binds p300/CBP (9), KLF4 may compete with β-catenin/TCF4 for p300/CBP binding as well. However, the roles of p300 and CBP in Wnt signaling are very complicated in that they are bimodal regulators of Wnt signaling. It has been reported that p300 and CBP have differential roles in the expression of the survivin gene (33). They can also inhibit Wnt signaling by interacting with TCF (30). In addition, KLF4 also interacts with both p300 and CBP (9). Structural biology approaches to studying these questions will be required to better understand the functional interactions between KLF4 and the β-catenin/TCF4/CBP complex.
In addition, we make several interesting observations regarding the expression of KLF4 in the intestine. First, we analyzed the expression of KLF4 and β-catenin in the normal large intestine and adenomas from ApcMin/+ mice (Fig. 1). We found that expression of KLF4 and that of β-catenin are inversely related, consistent with our model that KLF4 represses Wnt signaling by inhibiting β-catenin (61). We observe that KLF4 is downregulated in adenomas from ApcMin/+ mice. Since these tumors are caused by an Apc truncation that results in activation of β-catenin, β-catenin may repress KLF4 expression through unknown mechanisms. Further investigation into the cross talk between β-catenin and KLF4 will provide more insights into the mechanisms of colon tumorigenesis. Second, we observe that KLF4 and TCF4 are expressed in similar patterns within the large intestine (Fig. 3). The pattern of immunohistochemical staining that we found for the TCF4 protein in mouse intestine is consistent with in situ hybridization data looking at expression patterns of TCF4 mRNA that have been previously published (15). The function of TCF4 in intestinal stem cells has been well studied (25). However, its role in differentiated intestinal epithelial cells is not entirely clear. Given that KLF4 generally functions as a differentiation-associated factor within the intestine (21), we speculate that in the absence of active Wnt signaling, KLF4 interacts with TCF4 in order to maintain the differentiated state. Recently, KLF4 has been found to be one of the four factors that regulate stem cell reprogramming (53). It will be interesting to examine whether KLF4 inhibits Wnt signaling in inducible pluripotent stem cells (iPS cells) or embryonic stem cells.
Acknowledgments
We thank Tianxin Yu, Jun Yang, and Mark Evers for helpful discussions and suggestions.
P.M.E. was supported in part by a Multidisciplinary Training in Cancer Research Predoctoral Training Grant from the Sealy Center for Cancer Cell Biology and the National Institutes of Health (T32CA117834). C.L. was supported by R01 DK071976 from the NIH.
Footnotes
Published ahead of print on 9 November 2009.
REFERENCES
- 1.Behrens, J., J. P. von Kries, M. Kuhl, L. Bruhn, D. Wedlich, R. Grosschedl, and W. Birchmeier. 1996. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature 382:638-642. [DOI] [PubMed] [Google Scholar]
- 2.Belenkaya, T. Y., C. Han, H. J. Standley, X. Lin, D. W. Houston, J. Heasman, and X. Lin. 2002. Pygopus encodes a nuclear protein essential for Wingless/Wnt signaling. Development 129:4089-4101. [DOI] [PubMed] [Google Scholar]
- 3.Billin, A. N., H. Thirlwell, and D. E. Ayer. 2000. Beta-catenin-histone deacetylase interactions regulate the transition of LEF1 from a transcriptional repressor to an activator. Mol. Cell. Biol. 20:6882-6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brannon, M., J. D. Brown, R. Bates, D. Kimelman, and R. T. Moon. 1999. XCtBP is a XTcf-3 co-repressor with roles throughout Xenopus development. Development 126:3159-3170. [DOI] [PubMed] [Google Scholar]
- 5.Cavallo, R. A., R. T. Cox, M. M. Moline, J. Roose, G. A. Polevoy, H. Clevers, M. Peifer, and A. Bejsovec. 1998. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature 395:604-608. [DOI] [PubMed] [Google Scholar]
- 6.Dang, D. T., K. E. Bachman, C. S. Mahatan, L. H. Dang, F. M. Giardiello, and V. W. Yang. 2000. Decreased expression of the gut-enriched Kruppel-like factor gene in intestinal adenomas of multiple intestinal neoplasia mice and in colonic adenomas of familial adenomatous polyposis patients. FEBS Lett. 476:203-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daniels, D. L., and W. I. Weis. 2005. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat. Struct. Mol. Biol. 12:364-371. [DOI] [PubMed] [Google Scholar]
- 8.Daniels, D. L., and W. I. Weis. 2002. ICAT inhibits beta-catenin binding to Tcf/Lef-family transcription factors and the general coactivator p300 using independent structural modules. Mol. Cell 10:573-584. [DOI] [PubMed] [Google Scholar]
- 9.Evans, P. M., W. Zhang, X. Chen, J. Yang, K. K. Bhakat, and C. Liu. 2007. Kruppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation. J. Biol. Chem. 282:33994-34002. [DOI] [PubMed] [Google Scholar]
- 10.Foster, K. W., A. R. Frost, P. McKie-Bell, C. Y. Lin, J. A. Engler, W. E. Grizzle, and J. M. Ruppert. 2000. Increase of GKLF messenger RNA and protein expression during progression of breast cancer. Cancer Res. 60:6488-6495. [PubMed] [Google Scholar]
- 11.Ghaleb, A. M., B. B. McConnell, M. O. Nandan, J. P. Katz, K. H. Kaestner, and V. W. Yang. 2007. Haploinsufficiency of Kruppel-like factor 4 promotes adenomatous polyposis coli dependent intestinal tumorigenesis. Cancer Res. 67:7147-7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldman, P. S., V. K. Tran, and R. H. Goodman. 1997. The multifunctional role of the co-activator CBP in transcriptional regulation. Recent Prog. Horm. Res. 52:103-119; discussion 119-120. [PubMed] [Google Scholar]
- 13.Graham, T. A., W. K. Clements, D. Kimelman, and W. Xu. 2002. The crystal structure of the beta-catenin/ICAT complex reveals the inhibitory mechanism of ICAT. Mol. Cell 10:563-571. [DOI] [PubMed] [Google Scholar]
- 14.Gregorieff, A., and H. Clevers. 2005. Wnt signaling in the intestinal epithelium: from endoderm to cancer. Genes Dev. 19:877-890. [DOI] [PubMed] [Google Scholar]
- 15.Gregorieff, A., D. Pinto, H. Begthel, O. Destree, M. Kielman, and H. Clevers. 2005. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology 129:626-638. [DOI] [PubMed] [Google Scholar]
- 16.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. U. S. A. 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hecht, A., K. Vleminckx, M. P. Stemmler, F. van Roy, and R. Kemler. 2000. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 19:1839-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huber, O., R. Korn, J. McLaughlin, M. Ohsugi, B. G. Herrmann, and R. Kemler. 1996. Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mech. Dev. 59:3-10. [DOI] [PubMed] [Google Scholar]
- 19.Huelsken, J., R. Vogel, V. Brinkmann, B. Erdmann, C. Birchmeier, and W. Birchmeier. 2000. Requirement for beta-catenin in anterior-posterior axis formation in mice. J. Cell Biol. 148:567-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jen, J., S. M. Powell, N. Papadopoulos, K. J. Smith, S. R. Hamilton, B. Vogelstein, and K. W. Kinzler. 1994. Molecular determinants of dysplasia in colorectal lesions. Cancer Res. 54:5523-5526. [PubMed] [Google Scholar]
- 21.Katz, J. P., N. Perreault, B. G. Goldstein, C. S. Lee, P. A. Labosky, V. W. Yang, and K. H. Kaestner. 2002. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development 129:2619-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kielman, M. F., M. Rindapaa, C. Gaspar, N. van Poppel, C. Breukel, S. van Leeuwen, M. M. Taketo, S. Roberts, R. Smits, and R. Fodde. 2002. Apc modulates embryonic stem-cell differentiation by controlling the dosage of beta-catenin signaling. Nat. Genet. 32:594-605. [DOI] [PubMed] [Google Scholar]
- 23.Kim, M. K., T. J. McGarry, P. O'Broin, J. M. Flatow, A. A. Golden, and J. D. Licht. 2009. An integrated genome screen identifies the Wnt signaling pathway as a major target of WT1. Proc. Natl. Acad. Sci. U. S. A. 106:11154-11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kioussi, C., P. Briata, S. H. Baek, D. W. Rose, N. S. Hamblet, T. Herman, K. A. Ohgi, C. Lin, A. Gleiberman, J. Wang, V. Brault, P. Ruiz-Lozano, H. D. Nguyen, R. Kemler, C. K. Glass, A. Wynshaw-Boris, and M. G. Rosenfeld. 2002. Identification of a Wnt/Dvl/beta-Catenin →Pitx2 pathway mediating cell-type-specific proliferation during development. Cell 111:673-685. [DOI] [PubMed] [Google Scholar]
- 25.Korinek, V., N. Barker, P. Moerer, E. van Donselaar, G. Huls, P. J. Peters, and H. Clevers. 1998. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet. 19:379-383. [DOI] [PubMed] [Google Scholar]
- 26.Korinek, V., N. Barker, P. J. Morin, D. van Wichen, R. de Weger, K. W. Kinzler, B. Vogelstein, and H. Clevers. 1997. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science 275:1784-1787. [DOI] [PubMed] [Google Scholar]
- 27.Kramps, T., O. Peter, E. Brunner, D. Nellen, B. Froesch, S. Chatterjee, M. Murone, S. Zullig, and K. Basler. 2002. Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear beta-catenin-TCF complex. Cell 109:47-60. [DOI] [PubMed] [Google Scholar]
- 28.Levanon, D., R. E. Goldstein, Y. Bernstein, H. Tang, D. Goldenberg, S. Stifani, Z. Paroush, and Y. Groner. 1998. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc. Natl. Acad. Sci. U. S. A. 95:11590-11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levy, L., Y. Wei, C. Labalette, Y. Wu, C. A. Renard, M. A. Buendia, and C. Neuveut. 2004. Acetylation of beta-catenin by p300 regulates beta-catenin-Tcf4 interaction. Mol. Cell. Biol. 24:3404-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, J., C. Sutter, D. S. Parker, T. Blauwkamp, M. Fang, and K. M. Cadigan. 2007. CBP/p300 are bimodal regulators of Wnt signaling. EMBO J. 26:2284-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, P., M. Wakamiya, M. J. Shea, U. Albrecht, R. R. Behringer, and A. Bradley. 1999. Requirement for Wnt3 in vertebrate axis formation. Nat. Genet. 22:361-365. [DOI] [PubMed] [Google Scholar]
- 32.Luo, J., M. Li, Y. Tang, M. Laszkowska, R. G. Roeder, and W. Gu. 2004. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 101:2259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma, H., C. Nguyen, K. S. Lee, and M. Kahn. 2005. Differential roles for the coactivators CBP and p300 on TCF/beta-catenin-mediated survivin gene expression. Oncogene 24:3619-3631. [DOI] [PubMed] [Google Scholar]
- 34.Miyabayashi, T., J. L. Teo, M. Yamamoto, M. McMillan, C. Nguyen, and M. Kahn. 2007. Wnt/beta-catenin/CBP signaling maintains long-term murine embryonic stem cell pluripotency. Proc. Natl. Acad. Sci. U. S. A. 104:5668-5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyoshi, Y., H. Nagase, H. Ando, A. Horii, S. Ichii, S. Nakatsuru, T. Aoki, Y. Miki, T. Mori, and Y. Nakamura. 1992. Somatic mutations of the APC gene in colorectal tumors: mutation cluster region in the APC gene. Hum. Mol. Genet. 1:229-233. [DOI] [PubMed] [Google Scholar]
- 36.Molenaar, M., M. van de Wetering, M. Oosterwegel, J. Peterson-Maduro, S. Godsave, V. Korinek, J. Roose, O. Destree, and H. Clevers. 1996. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell 86:391-399. [DOI] [PubMed] [Google Scholar]
- 37.Morin, P. J., A. B. Sparks, V. Korinek, N. Barker, H. Clevers, B. Vogelstein, and K. W. Kinzler. 1997. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science 275:1787-1790. [DOI] [PubMed] [Google Scholar]
- 38.Moser, A. R., H. C. Pitot, and W. F. Dove. 1990. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science 247:322-324. [DOI] [PubMed] [Google Scholar]
- 39.Munemitsu, S., I. Albert, B. Rubinfeld, and P. Polakis. 1996. Deletion of an amino-terminal sequence beta-catenin in vivo and promotes hyperphosporylation of the adenomatous polyposis coli tumor suppressor protein. Mol. Cell. Biol. 16:4088-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nowak, D. E., B. Tian, and A. R. Brasier. 2005. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. Biotechniques 39:715-725. [DOI] [PubMed] [Google Scholar]
- 41.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953-959. [DOI] [PubMed] [Google Scholar]
- 42.Ohnishi, S., S. Ohnami, F. Laub, K. Aoki, K. Suzuki, Y. Kanai, K. Haga, M. Asaka, F. Ramirez, and T. Yoshida. 2003. Downregulation and growth inhibitory effect of epithelial-type Kruppel-like transcription factor KLF4, but not KLF5, in bladder cancer. Biochem. Biophys. Res. Commun. 308:251-256. [DOI] [PubMed] [Google Scholar]
- 43.Pandya, A. Y., L. I. Talley, A. R. Frost, T. J. Fitzgerald, V. Trivedi, M. Chakravarthy, D. C. Chhieng, W. E. Grizzle, J. A. Engler, H. Krontiras, K. I. Bland, A. F. LoBuglio, S. M. Lobo-Ruppert, and J. M. Ruppert. 2004. Nuclear localization of KLF4 is associated with an aggressive phenotype in early-stage breast cancer. Clin. Cancer Res. 10:2709-2719. [DOI] [PubMed] [Google Scholar]
- 44.Papkoff, J., B. Rubinfeld, B. Schryver, and P. Polakis. 1996. Wnt-1 regulates free pools of catenins and stabilizes APC-catenin complexes. Mol. Cell. Biol. 16:2128-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parker, D. S., J. Jemison, and K. M. Cadigan. 2002. Pygopus, a nuclear PHD-finger protein required for Wingless signaling in Drosophila. Development 129:2565-2576. [DOI] [PubMed] [Google Scholar]
- 46.Pinto, D., A. Gregorieff, H. Begthel, and H. Clevers. 2003. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 17:1709-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Powell, S. M., N. Zilz, Y. Beazer-Barclay, T. M. Bryan, S. R. Hamilton, S. N. Thibodeau, B. Vogelstein, and K. W. Kinzler. 1992. APC mutations occur early during colorectal tumorigenesis. Nature 359:235-237. [DOI] [PubMed] [Google Scholar]
- 48.Poy, F., M. Lepourcelet, R. A. Shivdasani, and M. J. Eck. 2001. Structure of a human Tcf4-beta-catenin complex. Nat. Struct. Biol. 8:1053-1057. [DOI] [PubMed] [Google Scholar]
- 49.Roose, J., G. Huls, M. van Beest, P. Moerer, K. van der Horn, R. Goldschmeding, T. Logtenberg, and H. Clevers. 1999. Synergy between tumor suppressor APC and the beta-catenin-Tcf4 target Tcf1. Science 285:1923-1926. [DOI] [PubMed] [Google Scholar]
- 50.Roose, J., M. Molenaar, J. Peterson, J. Hurenkamp, H. Brantjes, P. Moerer, M. van de Wetering, O. Destree, and H. Clevers. 1998. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature 395:608-612. [DOI] [PubMed] [Google Scholar]
- 51.Sansom, O. J., K. R. Reed, A. J. Hayes, H. Ireland, H. Brinkmann, I. P. Newton, E. Batlle, P. Simon-Assmann, H. Clevers, I. S. Nathke, A. R. Clarke, and D. J. Winton. 2004. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 18:1385-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shields, J. M., R. J. Christy, and V. W. Yang. 1996. Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J. Biol. Chem. 271:20009-20017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takahashi, K., and S. Yamanaka. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663-676. [DOI] [PubMed] [Google Scholar]
- 54.Takemaru, K. I., and R. T. Moon. 2000. The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J. Cell Biol. 149:249-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thompson, B., F. Townsley, R. Rosin-Arbesfeld, H. Musisi, and M. Bienz. 2002. A new nuclear component of the Wnt signalling pathway. Nat. Cell Biol. 4:367-373. [DOI] [PubMed] [Google Scholar]
- 56.van de Wetering, M., E. Sancho, C. Verweij, W. de Lau, I. Oving, A. Hurlstone, K. van der Horn, E. Batlle, D. Coudreuse, A. P. Haramis, M. Tjon-Pon-Fong, P. Moerer, M. van den Born, G. Soete, S. Pals, M. Eilers, R. Medema, and H. Clevers. 2002. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111:241-250. [DOI] [PubMed] [Google Scholar]
- 57.Winer, I. S., G. T. Bommer, N. Gonik, and E. R. Fearon. 2006. Lysine residues Lys-19 and Lys-49 of beta-catenin regulate its levels and function in T cell factor transcriptional activation and neoplastic transformation. J. Biol. Chem. 281:26181-26187. [DOI] [PubMed] [Google Scholar]
- 58.Wolf, D., M. Rodova, E. A. Miska, J. P. Calvet, and T. Kouzarides. 2002. Acetylation of beta-catenin by CREB-binding protein (CBP). J. Biol. Chem. 277:25562-25567. [DOI] [PubMed] [Google Scholar]
- 59.Wong, M. H., J. Huelsken, W. Birchmeier, and J. I. Gordon. 2002. Selection of multipotent stem cells during morphogenesis of small intestinal crypts of Lieberkuhn is perturbed by stimulation of Lef-1/beta-catenin signaling. J. Biol. Chem. 277:15843-15850. [DOI] [PubMed] [Google Scholar]
- 60.Yang, Y., B. G. Goldstein, H. H. Chao, and J. P. Katz. 2005. KLF4 and KLF5 regulate proliferation, apoptosis and invasion in esophageal cancer cells. Cancer Biol. Ther. 4:1216-1221. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, W., X. Chen, Y. Kato, P. M. Evans, S. Yuan, J. Yang, P. G. Rychahou, V. W. Yang, X. He, B. M. Evers, and C. Liu. 2006. Novel cross talk of Kruppel-like factor 4 and beta-catenin regulates normal intestinal homeostasis and tumor repression. Mol. Cell. Biol. 26:2055-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]