Abstract
The central hallmark of telomerases is repetitive copying of a short, defined sequence within its integral RNA subunit. We sought to identify structural determinants of this unique activity in the catalytic protein subunit telomerase reverse transcriptase (TERT) of telomerase. Residues within the highly conserved telomerase-specific T motif of human TERT were mutationally probed, leading to variant telomerases with increased repeat extension rates and wild-type processivity. The extension rate increases were independent of template sequence composition and only moderately correlated to telomerase RNA (TR) binding. Importantly, analysis of substrate primer elongation showed that the extension rate increases primarily resulted from increases in the repeat (type II) translocation rate. Our findings indicate a participatory role for the T motif in repeat translocation, an obligatory event for repetitive telomeric DNA synthesis. Thus, the T motif serves as a restrictive determinant of repetitive reverse transcription.
In eukaryotes, chromosomal telomeres serve to maintain genomic integrity, functioning both as buffers against genetic erosion and as suppressors of chromosome fusion and deleterious recombinational events (7). A distinguishing feature of the DNA component of these nucleoprotein structures is the presence of multiple short, tandem repeats. De novo synthesis of telomeric DNA is performed by the cellular polymerase telomerase. This specialized ribonucleoprotein reverse transcriptase (RT) accomplishes this via repetitive copying of a short, defined sequence within its integral RNA subunit (TR) by its catalytic protein subunit, telomerase reverse transcriptase (TERT).
Investigations into TERT sequence/function relationships have led to the elucidation of a basic structural framework for the catalytic subunit. The TERT subunit is comprised of a central RT domain with conserved sequence motifs shared with retroviral and other RTs, which is flanked by N- and C-terminal extensions specific to telomerases (Fig. 1). The N-terminal extension (NTE) is further subdivided into two domains, the N-terminal-most TEN (TERT essential N-terminal)/GQ/RID1 (RNA interaction domain-1) domain and the TRBD (TERT RNA binding domain)/RID2 (RNA interaction domain-2) domain, separated by a nonconserved linker. The catalysis of nucleotide addition (polymerization) is performed by the RT domain, while other functional roles have been ascribed to the remaining three telomerase-specific domains. The TEN/GQ/RID1 domain displays binding affinity for single-stranded telomeric DNA as well as low affinity for TR (25, 34, 39, 42). These interactions provide the underlying basis for the implicated important role of the TEN/GQ/RID1 domain in DNA substrate recognition and repeat addition processivity. The TRBD displays a relatively high affinity for TR, and this TERT-TR interaction is thought to promote stable assembly of the ribonucleoprotein complex (4, 14, 30, 38). Among the enzymatic functions linked to the C-terminal extension (CTE) are participation in nucleotide addition processivity (23, 24) and, in the case of human TERT (hTERT), dimerization (1).
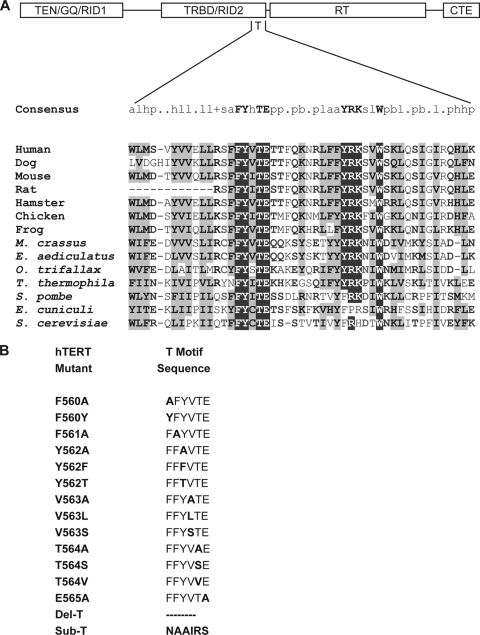
FIG. 1.
Sequence alignment of TERT protein T motifs and hTERT T-motif mutants. (A) Alignment of TERT protein T-motif sequences from human (37, 40), dog (41), mouse (16), rat (GenBank accession no. AAF62177.1), hamster (18), chicken (11), frog (28), Moneuplotes crassus (26), Euplotes aediculatus (32), Oxytricha trifallax (6), Tetrahymena thermophila (6, 9), Schizosaccharomyces pombe (40), Encephalitozoon cuniculi (27), and Saccharomyces cerevisiae (32). The structural domain organization of TERT protein is shown above the sequence alignment. (B) Sequences of hTERT T-motif mutants studied in this work.
The unique ability of telomerase to repeatedly copy the short templating segment within the TR is the result of several well-coordinated events. The templating residues must initially be properly placed within the active site. Once completely copied, the template region must then be efficiently translocated and repositioned (referred to as type II translocation [33]) for reuse. Integral to this process is the maintenance of stable associations between DNA substrate, enzyme, and product. Processive elongation of single-stranded DNA (ssDNA) termini requires that the DNA substrate remain bound to the enzyme while being extended with telomeric repeats. This interaction is thought to be mediated by contacts with one or more primer/product anchor/alignment sites (or PAS) within the TERT protein (2, 8, 10, 21, 31). Biochemical and structural analyses suggest that at least one PAS resides within the TEN/GQ/RID1 domain (25, 34, 47). Along with proper primer/product anchoring, successful repetitive template copying requires that TR be stably anchored to TERT. Most experimental evidence implicates the TRBD as the main TR anchoring domain in TERT. Mutation or deletion of this region results in severe reduction or abrogation of TR binding (3-5, 29, 38). Furthermore, this domain in isolation can bind TR (30).
While elements involved in repeat synthesis related to substrate/product interaction and TR binding have been identified, it is unclear precisely which region(s) of TERT plays a role in translocation during the repeat addition cycle. Obvious candidates would be highly conserved motifs within the telomerase-specific domains of TERT. Of the three telomerase-specific domains, the TRBD appears to contain the greatest number of phylogenetically conserved sequence motifs, including the T motif, the hallmark of telomerases (1). The T motif was the first conserved element unique to TERTs to be described (40), identified via the nearly universal conservation of the FYXTE telomerase signature sequence within it (Fig. 1). Initial structure/function studies of hTERT revealed that mutations in this signature sequence could reduce or abrogate enzymatic activity (45). This observation, in conjunction with the telomerase-specific nature of the motif, led to early speculation that the T motif may play an important role in TERT-TR binding interactions and/or repetitive template copying (45). The involvement of the T motif in TR (RNA) binding has subsequently been demonstrated by several studies of yeast, ciliate, and human telomerases (4, 5, 29, 38). However, a role for the T motif in repetitive template copying has yet to be established. In the current study, we sought to determine whether the T motif plays a role in telomerase's unique repetitive reverse transcription. We report here that residues within the FYXTE telomerase signature sequence exert considerable influence on repeat extension rate. Targeted mutagenesis of this sequence led to mutants with significant increases in extension rate, increases which could be achieved only via an increase in the repeat translocation rate. These results provide the first evidence that the telomerase T motif directly participates in the process of repetitive template copying.
MATERIALS AND METHODS
Site-directed mutagenesis of the T motif of hTERT.
A cassette strategy (12) was used to generate hTERT T-motif mutants (Fig. 1). Briefly, an intermediate vector was generated from pNFLAGhTERT (12) by PCR in which the region encoding amino acids 558 to 565 (containing the T motif) was replaced with a pair of SapI endonuclease sites. Following SapI digestion, pairs of complementary oligonucleotides encoding specific amino acid substitutions (for example, CTCAGGTCTTTCTTTTATGTCGCCGAG [T564A Top] and GGTCTCGGCGACATAAAAGAAAGACCT [T564A Bottom] were used to make the T564A mutant) were annealed and ligated into the plasmid to generate specific T-motif variant expression constructs.
In vitro reconstitution of human telomerase.
Telomerase activity was reconstituted in vitro by synthesizing hTERT protein in the presence of gel-purified human telomerase RNA (hTR) as previously described (12). Typical reaction mixtures contained 0.77 pmol (0.04 μg)/μl wild-type or mutant pNFLAGhTERT and 0.41 pmol (0.6 μg)/μl purified hTR. Human telomerase was reconstituted in vitro at 30°C for 3.5 h, then flash frozen in liquid nitrogen, and stored at −80°C until needed. Aliquots of the reconstituted telomerase were also analyzed via SDS-PAGE to quantify the in vitro-synthesized hTERT protein.
Telomerase activity assay.
Telomerase activity was detected via a direct primer extension assay. Typical primer extension reaction mixtures contained 2 μl in vitro-reconstituted (IVR) telomerase, 1 pmol 5′-32P end-labeled substrate primer d(TTAGGG)3, 1 mM dATP, 1 mM dGTP, and 1 mM TTP in 1× PE reaction buffer (50 mM Tris-Cl [pH 8.3], 50 mM KCl, 2 mM dithiothreitol [DTT], 3 mM MgCl2, and 1 mM spermidine) in a total volume of 10 μl. Reaction mixtures were incubated at 30°C for 30 min, the reactions were then terminated, and the reaction mixtures were processed as previously described (12). The recovered reaction products were dissolved in urea PAGE sample buffer (95% formamide-20 mM EDTA) and resolved on 8 M urea-10% polyacrylamide gels. The gels were then dried, and the reaction products were detected and analyzed via a phosphorimager (Molecular Dynamics) using ImageQuant software (Molecular Dynamics).
Competitive primer challenge assay.
A competitive primer challenge (“bind and chase”) variation of the direct primer extension assay was used to assess activity, repeat extension rate, and processivity (12). IVR telomerase was prebound to 5′-32P-labeled substrate primer d(TTAGGG)3 in 1× PE reaction buffer for 5 min at 30°C. Primer extension synthesis was then initiated by adding 160-fold molar excess of unlabeled competitor (chase) primer plus dATP, dGTP, and TTP in 1× PE reaction buffer (final concentration of deoxynucleoside triphosphate [dNTP] of 1 mM). The reaction mixture was incubated at 30°C, aliquots (10 μl) were removed at 0.25 to 30 min, and synthesis was terminated with an equal volume of stop solution. The products of the primer extension reactions were then processed and analyzed as described above for the standard telomerase activity assay.
Relative enzyme activities (total repeats synthesized/time), repeat extension rates (repeats added/time), and repeat processivities (probability that a substrate primer will be extended with an additional repeat rather than being released by the enzyme following template copying) were calculated as previously described (13). Briefly, relative enzyme activity was calculated from the intensity (corrected for background and normalized against unextended primer) and length of the most abundant (modal) product band generated by an enzyme at a given time. Because the modal product is readily quantified, it provides a practical and definitive measure of product synthesis. In contrast, accurately quantifying signal intensity and length of each of the individual end-labeled products is quite difficult (or impossible) when there are hundreds of bands per reaction to measure, particularly the closely spaced longer product bands. Repeat extension rate was determined by dividing the length of the modal band (in number of repeats) by time. Repeat processivity was determined by calculating the fraction of total products that have been extended beyond a given repeat as a function of repeat number.
The relative contributions of the template copying rate and repeat translocation rate to the extension rate were calculated from the observed intensities of the set of 6 bands associated with synthesis (template copying and translocation) of a single repeat (containing actively polymerizing complexes [12]). To obtain the intensity value for actively translocating species, the observed band intensity of the fully copied repeat (6 nucleotides added to the primer [+6-nt]) band was adjusted by subtracting the signal contributions of stalled/dissociated species within the band and the contribution of copying the terminal template nucleotide (C46). The percentage of band signal contributed by stalled/dissociated species was calculated from the repeat processivity (P). Specifically, the band intensity of the stalled/dissociated species within the +6-nt band that originated from the population of complexes which continued to be elongated into larger (>+6-nt) products, was calculated using the equation IS/D = [(100 − P)/100][(100/P)IE], where IS/D is the band intensity of the stalled/dissociated species and IE is the summed band intensity of all products beyond the +6-nt band. The signal corresponding to copying of C46 (IC46) was derived from the averaged signal of bands representing copying of the other 5 template nucleotides, i.e., IC46 = (sum +1-nt to +5-nt band signals)/5. Thus, the band intensity of translocating species, ITR (corrected for repeat processivity) was calculated using the equation ITR = [I+6 − (IS/D + IC46)]/(100 − P), where I+6 is the observed +6-nt band intensity. The band intensity corresponding to template copying events, ITC, was calculated as (6/5)(sum +1-nt to +5-nt band signals).
hTR binding assay.
An immunoprecipitation-based assay similar to the method first described by Bryan et al. (5) and later modified by Moriarity et al. (39) was used to measure hTR binding to TERT protein. Radiolabeled full-length hTR RNA was prepared by in vitro transcription in the presence of 800 ci/mmol CTP, using gel-purified FspI-digested phTRF (12) as a template. The radiolabeled RNA was gel purified, and the specific activity was adjusted to 2 × 105 to 5 × 105 cpm/pmol with unlabeled hTR RNA prior to use.
Human telomerase was reconstituted with radiolabeled hTR, and then the TERT/TR complexes were immunoprecipitated with M2 anti-FLAG agarose (Sigma). M2 anti-FLAG agarose suspension (12.5 μl) was preequilibrated in binding buffer (50 mM Tris-Cl [pH 7.4], 150 mM NaCl, 1 mM EDTA) (final volume of 90 μl) and combined with 10-μl IVR reaction mixture, and the mixture was slowly rotated for 1.5 h at 4°C. The resin was then gently pelleted and washed three times with 100 μl binding buffer. The washed resin pellet was then resuspended in Laemmli buffer, and the immunopurified products were separated on SDS-polyacrylamide gels. The resolved products (32P-labeled TR and 35S-labeled TERT) in the fixed, dried gels were detected and quantified via phosphorimaging (with 35S-labeled TERT background signal removed for TR quantification) as described previously (5). Relative hTR binding was measured as the ratio of the amount of hTR in a given lane divided by the amount of hTERT in the same lane, compared to the same ratio for wild-type TERT and TR on the same gel. A control immunoprecipitation with an IVR reaction mixture lacking pNFLAGhTERT was used to determine nonspecific hTR binding, and this background value was subtracted from hTR values obtained from TERT-containing immunoprecipitations.
RESULTS
Enzymatic activity of T-motif mutants of human TERT.
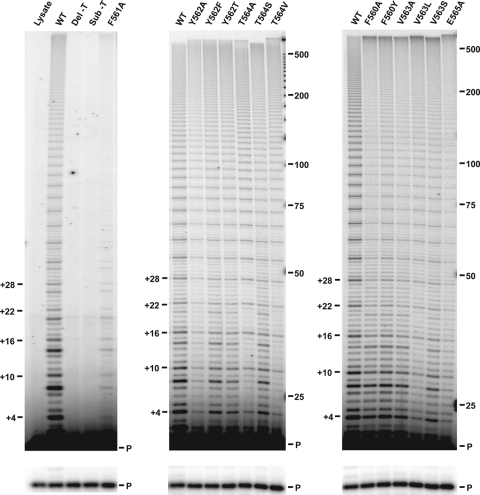
To assess the contribution of hTERT T-motif residues to telomerase enzymatic activity and function, a panel of hTERT protein mutants was generated (Fig. 1). Specifically, the residues of the FYXTE signature sequence (hTERT amino acid residues 561 to 565) as well the N-terminally flanking phenylalanine (F560), were targeted. In addition to several point mutants, variants were generated where residues 560 to 565 were deleted (Del-T) or replaced with the inherently flexible NAAIRS sequence (46) (Sub-T), a sequence that is minimally disruptive to native protein structure (36). Reconstituted telomerase complexes were assembled in rabbit reticulocyte lysates, with hTERT protein synthesized in vitro in the presence of purified hTR, and the reconstituted enzymes were assayed for activity in a direct primer extension assay. Most of the mutants were active to some extent (Fig. 2 and Table 1), and in general, the overall activities of the mutants were lower than the activity with wild-type hTERT. While some substitutions resulted in modest reductions in activity (e.g., Y562T), others had severe effects (e.g., F561A), while complete substitution or deletion of the FYXTE sequence motif (Sub-T and Del-T) resulted in inactive enzymes (Table 1). The Y562F, T564A, and T564V mutants were the only exceptions, displaying activities near or slightly greater than the wild-type enzyme activity. It should be noted that because these activity assays were carried out using an end-labeled substrate primer, all of the products are equivalently labeled, regardless of length. Thus, the overall signal intensity of the products in each lane is not necessarily reflective of enzyme activity level, since the length of products also needs to be considered.
FIG. 2.
Primer extension by hTERT T-motif mutants. In vitro-reconstituted telomerase mutants were assayed for telomerase activity via direct primer extension as described in Materials and Methods. The numbers to the left of the gels (+4, +10, etc.) indicate the positions of products corresponding to the end of each round of template copying (expressed as the number of nucleotides added to the primer). Marker sizes (in nucleotides) are indicated to the right of the gels. The lighter exposure under each gel shows the unextended d(TTAGGG)3 substrate primer (P) within each reaction, which served as a loading control (as it represented >95% of the recovered material). WT, wild type.
TABLE 1.
Telomerase activities, repeat extension rates, and processivities of hTERT T-motif mutantsa
| hTERT | Relativeb telomerase activity (avg ± SD) | Extension ratec |
Processivityc |
||
|---|---|---|---|---|---|
| No. of repeats/min (avg ± SD) | Relativeb | Value (%) (avg ± SD) | Relativeb | ||
| WT | 1.00 | 6.79 ± 0.52 | 1.00 | 97.05 ± 0.79 | 1.00 |
| F560A | 0.41 ± 0.09 | 7.42 ± 0.12 | 1.09 | 98.26 ± 1.30 | 1.01 |
| F560Y | 0.40 ± 0.06 | 7.42 ± 0.14 | 1.09 | 98.29 ± 0.87 | 1.01 |
| F561A | 0.09 ± 0.01 | 6.83 ± 0.71 | 1.01 | 98.03 ± 1.19 | 1.01 |
| Y562A | 0.49 ± 0.01 | 12.78 ± 1.27 | 1.88 | 98.06 ± 1.61 | 1.01 |
| Y562F | 1.09 ± 0.18 | 12.50 ± 1.41 | 1.84 | 97.97 ± 0.55 | 1.01 |
| Y562T | 0.76 ± 0.05 | 11.08 ± 1.61 | 1.63 | 98.14 ± 0.32 | 1.01 |
| V563A | 0.31 ± 0.05 | 6.42 ± 0.59 | 0.94 | 98.40 ± 0.44 | 1.01 |
| V563L | 0.27 ± 0.04 | 9.92 ± 0.35 | 1.46 | 98.28 ± 0.97 | 1.01 |
| V563S | 0.33 ± 0.07 | 6.58 ± 0.35 | 0.97 | 98.34 ± 1.22 | 1.01 |
| T564A | 1.38 ± 0.18 | 28.83 ± 1.92 | 4.25 | 97.47 ± 0.74 | 1.00 |
| T564S | 0.50 ± 0.03 | 8.42 ± 0.35 | 1.24 | 97.79 ± 0.59 | 1.01 |
| T564V | 0.79 ± 0.20 | 22.44 ± 0.86 | 3.30 | 97.57 ± 0.42 | 1.01 |
| E565A | 0.34 ± 0.05 | 15.92 ± 0.82 | 2.34 | 98.84 ± 1.09 | 1.02 |
| Del-T | 0.00 | ND | ND | ||
| Sub-T | 0.00 | ND | ND | ||
Telomerase activities, extension rates, and processivities were determined from competitive primer challenge assays. Telomerase activities and extension rates represent averages of three independent experiments, while processivity values represent averages of two independent experiments.
Relative to that of the wild-type hTERT.
ND, not determined.
T-motif modifications affect repeat extension rate but not processivity.
While mutation of T-motif residues of hTERT generally led to decreased overall enzymatic activity, template copying generally appeared to be similar to wild-type hTERT (Fig. 2). However, the maximum size of the products generated by several of the mutants (e.g., Y562A and E565A mutants) appeared larger than those from the wild-type enzyme (Fig. 2), suggesting altered repeat extension rate and/or processivity. In order to establish whether such changes had occurred, the active mutants were tested in primer extension assays performed under competitive primer challenge (“bind and chase”) conditions (Fig. 3). When assayed under these conditions, both the repeat extension rate and processivity (the extent to which a substrate is elongated with repeats before being released by the enzyme) can be readily determined (12).
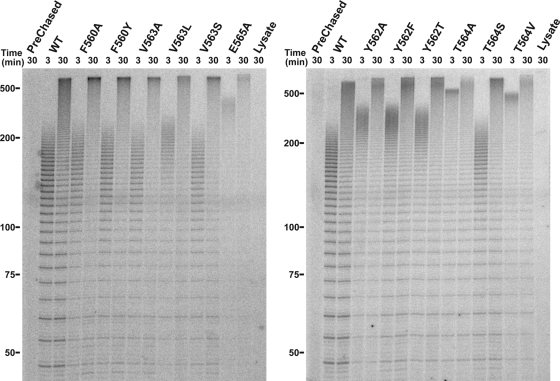
FIG. 3.
Competitive primer challenge assay with hTERT T-motif mutants. Primer extension reactions were carried out under competitor challenge conditions. Following 5-min binding of the radiolabeled substrate primer, extension reactions were initiated and chased with excess cold competitor primer. Postchase aliquots were taken at 3 and 30 min and analyzed via PAGE. The prechased lane contains 30-min extension reaction mixture in which excess competitor primer was added before IVR wild-type telomerase was added. Marker sizes (in nucleotides) are indicated to the left of the gels.
The results of this testing revealed that only small (<2%) increases in processivity resulted from the T-motif substitutions (Table 1). In contrast, the repeat extension rate (Table 1) was substantially influenced by several of the T-motif mutations. Specifically, all Tyr562 substitutions and the V563L substitution led to increases (1.46- to 1.88-fold) in the repeat extension rate compared to the wild-type enzyme. Even larger increases were seen with the T564V and E565A enzymes (3.30- and 2.34-fold over the wild-type enzyme, respectively). The greatest increase in extension rate was exhibited by the T564A variant, whose rate was 4.25-fold that of the wild type. All of the other T-motif mutants tested (F560A, F560Y, F561A, V563A, V563S, and T564S) displayed wild-type enzyme-like repeat extension rates (Table 1). Based on these findings, residues Y562, T564, E565, and to a lesser extent, V563 appear to be the determinants of the telomerase extension rate. Importantly, a direct correlation was seen between the repeat extension rate (Table 1) and largest products synthesized (Fig. 2 and 3). Taken together, these data indicate that the observed difference between the length of the longest products of the wild-type enzyme and the T-motif variants reflects increased repeat extension rate and not higher processivity.
T-motif mutants can assemble into stable hTERT/hTR complexes.
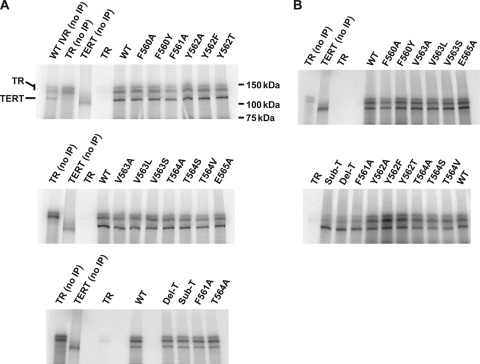
In order to better understand the basis for the effects on the extension rates seen for the T-motif mutants, we examined TR binding by the mutants. We used an immunoprecipitation-based assay to measure TR binding by hTERT. Standard in vitro reconstitutions of telomerase were carried out using radiolabeled hTR, and the TERT/TR complexes were then immunoprecipitated with anti-FLAG-agarose. These assays revealed that mutations in the FYXTE sequence of hTERT had minimal effect on hTR binding, as even the enzymatically inactive Del-T and Sub-T mutants bound TR at nearly wild-type enzyme levels (Fig. 4A and Table 2).
FIG. 4.
hTR binding by hTERT T-motif mutants. In vitro reconstitution of telomerase activity in the presence of radiolabeled hTR were carried out, followed by anti-FLAG immunoprecipitation (IP) of the TERT/TR complexes. The immunopurified products were analyzed via SDS-PAGE followed by phosphorimaging. The positions of hTR and hTERT are indicated. (A) Reconstitutions with radiolabeled hTR at a final concentration of hTR of 400 nM. (B) Reconstitutions with radiolabeled hTR at a final concentration of hTR of 40 nM.
TABLE 2.
hTR binding by hTERT T-motif mutants
| hTERT | Relative bindinga (avg ± SD) with: |
|
|---|---|---|
| 400 nM hTR | 40 nM hTR | |
| WT | 1.00 | 1.00 |
| F560A | 1.09 ± 0.25 | 0.72 ± 0.06 |
| F560Y | 1.21 ± 0.09 | 0.94 ± 0.11 |
| F561A | 0.83 ± 0.06 | 0.35 ± 0.07 |
| Y562A | 1.06 ± 0.05 | 1.26 ± 0.09 |
| Y562F | 1.06 ± 0.20 | 1.39 ± 0.03 |
| Y562T | 0.93 ± 0.25 | 1.51 ± 0.04 |
| V563A | 1.16 ± 0.13 | 0.89 ± 0.08 |
| V563L | 1.25 ± 0.25 | 0.71 ± 0.17 |
| V563S | 1.35 ± 0.10 | 0.63 ± 0.02 |
| T564A | 1.11 ± 0.18 | 1.56 ± 0.06 |
| T564S | 1.08 ± 0.20 | 1.72 ± 0.25 |
| T564V | 0.94 ± 0.16 | 1.93 ± 0.56 |
| E565A | 1.03 ± 0.22 | 0.69 ± 0.07 |
| Del-T | 0.83 ± 0.08 | 0.36 ± 0.09 |
| Sub-T | 0.74 ± 0.13 | 0.35 ± 0.11 |
Relative to the wild type. Values represent averages of two independent experiments.
Our standard reconstitution conditions have been optimized for robust enzymatic activity, including the use of relatively high TR concentration (400 nM). Although the hTR concentration used here (400 nM) was within the linear range of the binding assay with wild-type hTERT (data not shown), we repeated the assay at a lower hTR (40 nM) concentration to detect possible differences in high-affinity binding. When assayed at 40 nM hTR, differences were evident (Fig. 4B), with relative binding ranging from 0.35 to 1.93 compared to the wild-type hTERT (Table 2). Notably, the mutants displaying the lowest enzymatic activity—the inactive Del-T and Sub-T mutants and the minimally active F561A mutant—were also the least competent for TR binding. For the remaining mutants, little correlation could be made between binding and enzyme activity (compare Table 1 and Table 2). However, hTR binding did correlate moderately with the extension rate, with the enzymes that displayed faster extension rates being better binders (compare Table 1 and Table 2). Thus, these findings imply that the influence of T-motif residues on the repeat extension rate may be related to hTR binding interactions. Importantly, these experiments reveal that at higher hTR concentrations, all of the T-motif mutant TERTs can stably associate with TR.
The influence of T-motif residues on the extension rate is independent of template/product sequence composition.
We have previously shown that the TR template sequence is a determinant of the telomerase extension rate; therefore, we sought to determine whether the T motif's influence on extension rate may be mediated via interactions with the template sequence. Several of the T-motif hTERT mutant proteins were reconstituted with the hTR variant MH3 (12). MH3 differs from wild-type hTR only in its template sequence, containing two base substitutions (A48U and A54U) (Fig. 5A). We had earlier demonstrated that reconstitution of telomerase with MH3 hTR and wild-type hTERT yields an enzyme with an extension rate nearly 2-fold faster than telomerase reconstituted with wild-type hTR (12). When the T-motif mutant TERTs were tested, primer extension assays (Fig. 5B) revealed that the MH3 hTR-reconstituted T-motif variants also displayed increased extension rates compared to enzymes reconstituted with wild-type hTR (Table 3). Furthermore, the relative extension rates of the enzymes mirrored the rates seen when TERT proteins were reconstituted with wild-type hTR (Tables 1 and 3). Thus, the intrinsic extension rates of the mutant and wild-type proteins were increased by the same degree by the MH3 template sequence, as evidenced by the nearly identical MH3 hTR extension rate/wild-type hTR extension rate ratios (MH3hTRER/WThTRER) (Table 3). Importantly, the lack of influence by the MH3 sequence on the relative extension rates of the mutants implies that the T motif is not interacting with the bases of the template sequence/product residues. From these data, we conclude that the T-motif residues and the template sequence residues/composition modulate telomerase extension rate by independent mechanisms.
FIG. 5.
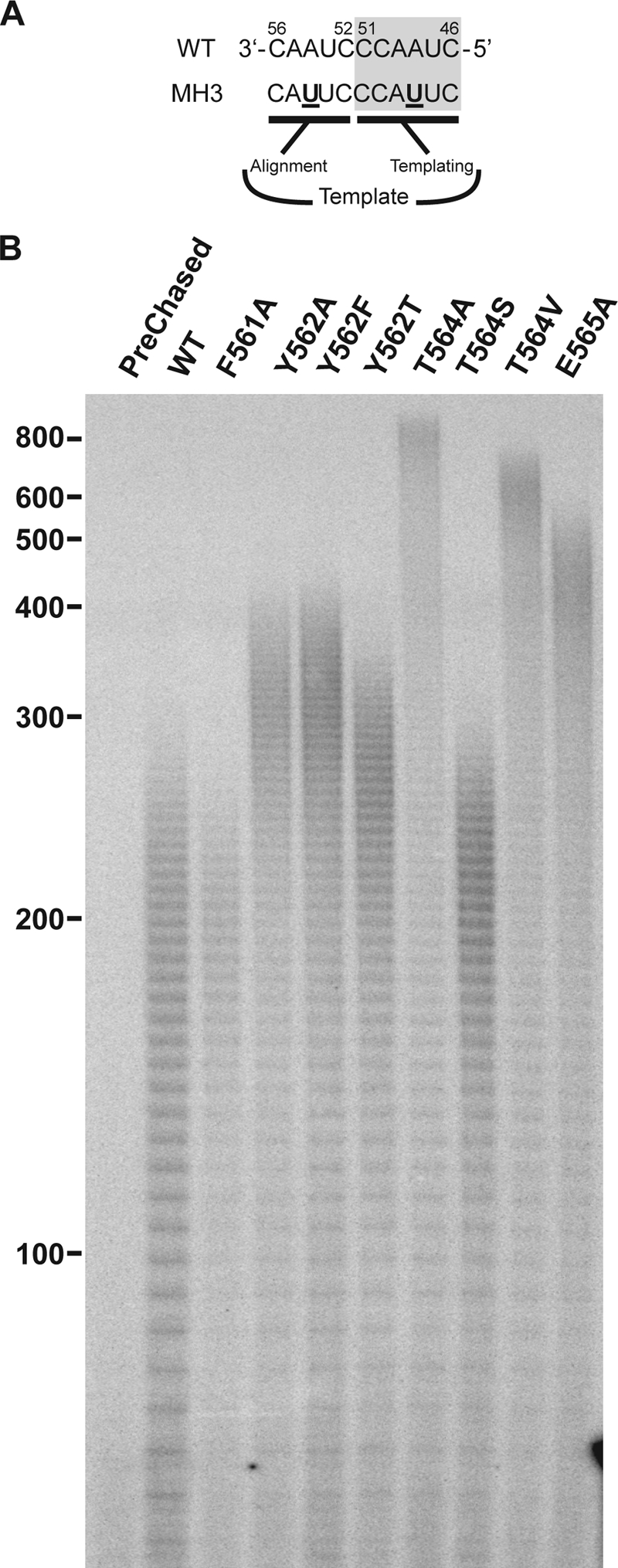
Effect of the hTR template sequence on primer extension by hTERT T-motif mutants. (A) Wild-type and T-motif mutant hTERT proteins were reconstituted in the presence of wild-type and template mutant MH3 hTR RNA (12) shown. (B) Primer extension reactions were carried out under competitor challenge conditions as detailed in Materials and Methods. Postchase aliquots were taken at 3 min and analyzed via PAGE (4% gel). The prechased lane shows a 3-min extension reaction where excess competitor primer was added before IVR wild-type telomerase was added. Marker sizes (in nucleotides) are indicated to the left of the gel.
TABLE 3.
Extension rate of hTERT T-motif mutants reconstituted in the presence of MH3 template mutant hTR
| hTERT | Extension rate |
MH3hTRER/WThTRERb | |
|---|---|---|---|
| No. of repeats/min (avg ± SD) | Relativea | ||
| WT | 12.23 ± 1.28 | 1.00 | 1.80 |
| F561A | 12.99 ± 0.76 | 1.06 | 1.90 |
| Y562A | 23.43 ± 1.94 | 1.92 | 1.83 |
| Y562F | 21.90 ± 0.74 | 1.79 | 1.75 |
| Y562T | 19.05 ± 1.23 | 1.56 | 1.72 |
| T564A | 55.42 ± 6.12 | 4.53 | 1.92 |
| T564S | 14.77 ± 0.62 | 1.21 | 1.76 |
| T564V | 42.79 ± 1.17 | 3.50 | 1.91 |
| E565A | 30.36 ± 3.67 | 2.48 | 1.91 |
Relative extension rates are expressed relative to the rate for the wild-type hTERT.
Ratio of the extension rate with MH3 hTR to the extension rate with WT hTR (see Table 1). Rates represent averages of two independent experiments.
Extension rate increases due to increase in translocation rate.
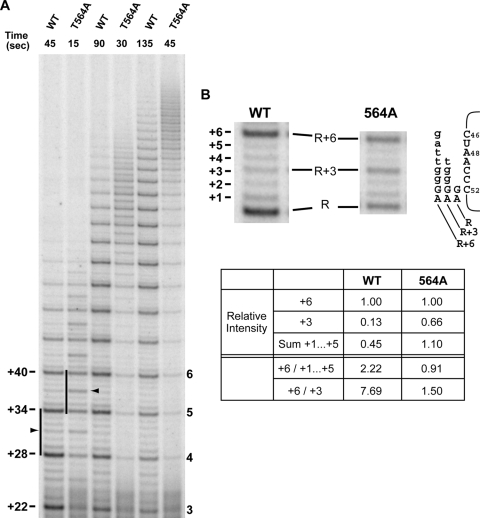
The repeat extension rate is determined by the combined rates of template copying and repeat translocation (type II translocation). To measure the relative contribution of each to the extension rate, we performed a quantitative comparison of the band intensities of the set of six accumulated products of a single repeat (Fig. 6), associated with actively polymerizing complexes. As these bands represent a snapshot of primarily actively polymerizing enzyme complexes, their relative abundance (accumulation) provides a relative measure of duration of that step in the repeat cycle. Primer extension assays were carried out under competitive primer challenge conditions, which allow for repeats consisting of products associated with active polymerization to be readily distinguished (12). These assays revealed that for the wild-type enzyme, the fully copied repeat (+6-nt) product was approximately two times more abundant than the summed total of the 5 partial repeat (i.e., +1-nt, +2-nt,… ,+5-nt) products (Fig. 6B). However, the observed +6-nt band is comprised not only of translocating molecules but also of stalled/dissociated species no longer being actively extended. Based on the quantification of products beyond the 5th repeat (i.e., >+34-nt band [Fig. 6]), the +6-nt band intensity attributable to stalled/dissociated species, IS/D, formed during synthesis of products beyond the 5th repeat is equivalent to approximately 5% of the observed +6-nt signal. Furthermore, molecules corresponding to copying of the terminal template nucleotide (C46) also contribute to the +6-nt band signal. We estimate the +6-nt band intensity attributable to C46 copying, IC46, based on the averaged intensity of the +1-nt to +5-nt bands (Fig. 6B) to be equivalent to ∼9% of the observed +6-nt band intensity. Consequently, we calculate the relative band intensity of actively translocating species, ITR (at 97% processivity [Table 1]) to be 0.83 and the relative intensity of products representing template copying (ITC) to be 0.54. Thus, the relative proportions of the elongating species indicate that for wild-type human telomerase, translocation takes approximately 1.55 times as long as copying the entire template.
FIG. 6.
Competitive primer challenge assay with wild-type and T564A mutant hTERT. (A) Primer extension reactions were carried out under competitor challenge conditions. Following 5-min binding of radiolabeled substrate primer, extension reactions were initiated and chased with excess cold competitor primer. Postchase aliquots were taken at the time points (in seconds) indicated and analyzed via PAGE. The black arrowheads indicate product accumulation prior to copying template residue A48. Regions of active polymerization (12) for wild-type hTERT (containing the 5th full repeat) and 564A hTERT (containing the 6th full repeat) are indicated by the black bars. The numbers to the left of the gel (+22, +28, etc.) indicate positions of products corresponding to the end of each round of template copying (expressed as number of nucleotides added to the primer), with the number of full repeats shown to the right of the gel. (B) Enlarged view of the active polymerization regions marked in panel A. Schematic diagram indicates alignments of repeat + 3 (R + 3) and R + 6 major synthesis products with the template RNA. Nucleotides added during each round of primer elongation are shown in lowercase type. hTR nucleotide positions are indicated next to the template sequence. Relative band intensities of the +3 (R + 3), +6 (R + 6), and sum of +1 to +5 products in the repeats shown were quantified via phosphorimaging.
We then examined the product synthesis pattern obtained with T564A hTERT, the T-motif variant with the greatest increase in the extension rate. In contrast to the wild-type hTERT, the repeat synthesis pattern (Fig. 3) showed that the relative band intensity of the +6-nt product was less than the summed total of the +1- to +5-nt partial repeat products (Fig. 6B). This implied that for the A564 mutant, template copying takes longer than translocation. In determining the relative band intensities for actively translocating species versus products of template copying, we calculated two sets of values for ITR and ITC, based on different IC46 values. In addition to estimating IC46 from the averaged intensity of the summed +1-nt to +5-nt bands (Fig. 6B), IC46 was also determined from the averaged intensity of the summed +1-, +2-, +4-, and +5-nt bands. This was done to address the possibility that the +3-nt (repeat [R] + 3) band (which was much more abundant than any of the other partial products) may disproportionately contribute to the calculation of IC46. When the signal intensities were calculated (for the 6th repeat) using the two different IC46 values, we found that the relative band intensity of the actively translocating species ranged from 0.7 to 0.82, while the relative intensity of the products of template copying ranged from 1.20 to 1.32. Thus, we determine that for A564 telomerase, copying the entire template takes approximately 1.46 to 1.89 times as long as translocation.
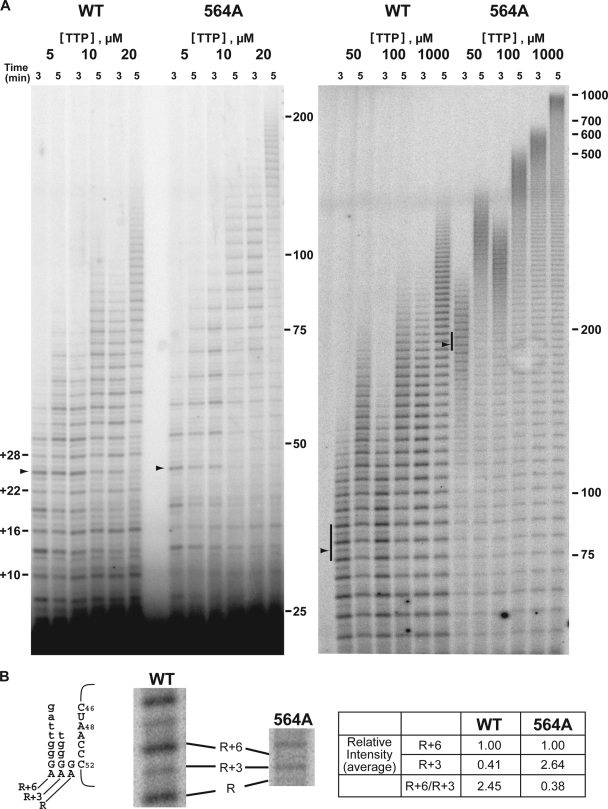
To further demonstrate the effect of the T564A mutation on the translocation rate, we performed primer extension reactions at reduced nucleotide concentrations. Under limiting nucleotide concentrations, template copying is slowed, which would have a more profound effect on the extension rate of an enzyme whose template copying is slower than translocation (e.g., T564A enzyme) compared to one where translocation is slower than template copying (e.g., wild-type enzyme). To limit the effects of lowered nucleotide concentration exclusively to template copying, only TTP was reduced. We have previously shown that reducing the TTP concentration primarily affects copying at a single mid-template residue, nucleotide A48 (13). In addition, studies have shown that dGTP can play a role in ciliate telomerase processivity and possibly translocation, independent of its function as an incorporating nucleotide (19, 20, 22). Therefore, primer extension assays were carried out in the presence of different TTP concentrations, and the results are shown in Fig. 7. As expected, the rates of repeat synthesis by both the wild-type and A564 mutant telomerases were sensitive to TTP concentration. A general reduction in the overall extension rate was observed as the TTP concentration was reduced (Table 4). Michaelis-Menten fitting of the data in Table 4 indicated apparent Kms for extension of 62 μM and 145 μM for wild-type and A564 telomerases, respectively. This reflected the more profound effect of TTP reduction on the A564 mutant, as the percent decrease in extension rate seen for the A564 mutant was greater than the percent changes observed for the wild type (at each TTP concentration tested) (Table 4). Furthermore, as the TTP concentration was reduced, extension rates became almost identical (Table 4), suggesting that the rate of template copying across A48 had become the major limiting determinant of the extension rate. This was clearly evident in the product patterns, which shifted from the characteristic R + 6-dominated pattern to one where the R + 3 product was the major species as TTP concentration decreased. Notably, comparison of the R + 6/R + 3 band intensity ratios of products associated with active polymerizing complexes (12) of the wild-type and A564 enzymes at 50 μM TTP (Fig. 7) reflected the previous finding at nonlimited TTP concentration (Fig. 6). Specifically, the R + 3 product was proportionally more abundant for the mutant. For the wild-type enzyme, the R + 6 product band was on average 2.45-fold more intense than the R + 3 band (measured at the 9th and 10th repeats of the 3-min time point) (Fig. 7). For the A564 mutant, the R + 6 band was on average 2.64-fold less intense than the R + 3 band (measured at the 26th and 27th repeats of the 3-min time point) (Fig. 7), representing an R + 6/R + 3 ratio 6.48-fold lower than wild type. These data indicated that, for the mutant enzyme, translocation was less of an impediment to repeat synthesis than copying across template residue A48. Importantly, even with a higher proportion of R + 3 accumulation, the extension rate of the A564 mutant was still over 3-fold faster than that of the wild type.
FIG. 7.
Competitive primer challenge assay with hTERT T-motif mutants and different TTP concentrations. (A) Primer extension synthesis by wild-type and T564A telomerases was measured in primer extension reaction mixtures containing 1 mM dATP and 1 mM dGTP in the presence of increasing concentrations of TTP. Postchase aliquots were taken at 3 and 5 min and analyzed via PAGE (left gel, primer extension reaction mixtures with 5 to 20 μM TTP on 8% gel; right gel, primer extension reaction mixtures with 50 to 1,000 μM TTP on 4% gel). The black arrowheads indicate product accumulation prior to copying template residue A48. Regions of active polymerization (12) for the wild-type telomerase (containing the 9th and 10th repeats) and 564A telomerase (containing the 26th and 27th repeats) are indicated by a black bar. The numbers to the left of the gels (+4, +10, etc.) indicate positions of products corresponding to the end of each round of template copying. Marker sizes (in nucleotides) are indicated to the right of the gels. (B) Enlarged view of active polymerization regions marked in panel A. Schematic diagram indicates alignments of R + 3 and R + 6 major products with the template RNA. Nucleotides added during each round of primer elongation are shown in lowercase type. hTR nucleotide positions are indicated next to the template sequence. The average relative band intensities of the R + 3 and R + 6 products in the two repeats shown were quantified via phosphorimaging.
TABLE 4.
Extension rates of wild-type and T564A hTERT at different TTP concentrations
| TTP concn (μM) | Extension rate of the WT hTERT |
Extension rate of the T564A hTERT |
564AER/564TERb | ||
|---|---|---|---|---|---|
| No. of repeats/min (avg ± SD) | Relativea | No. of repeats/min (avg ± SD) | Relative | ||
| 5 | 0.82 ± 0.18 | 0.13 | 1.00 ± 0.15 | 0.04 | 1.22 |
| 10 | 1.27 ± 0.03 | 0.20 | 1.87 ± 0.22 | 0.07 | 1.47 |
| 20 | 1.73 ± 0.42 | 0.27 | 3.70 ± 0.32 | 0.13 | 2.13 |
| 50 | 2.90 ± 0.46 | 0.46 | 9.03 ± 0.41 | 0.32 | 3.11 |
| 100 | 3.47 ± 0.27 | 0.56 | 12.67 ± 1.90 | 0.45 | 3.65 |
| 1,000 | 6.23 ± 0.59 | 1.00 | 28.23 ± 1.61 | 1.00 | 4.53 |
Extension rates are expressed relative to the extension rate for 1,000 μM TTP for the given hTERT.
Ratio of the extension rate of 564A hTERT to the extension rate of 564T hTERT. The values are averages of two independent experiments.
Thus, from our results of primer extension assays performed under saturating and limiting nucleotide conditions, we conclude that the translocation rate of the T-motif mutant is faster than the wild-type enzyme and that the mutant's increased extension rate is primarily due to an increased translocation rate.
DISCUSSION
Iterative template copying, the defining property of telomerase, is fundamental to carrying out its role in telomere biosynthesis. To explore the participation of the T motif in this process (telomeric repeat synthesis), residues within the T-motif signature sequence were mutationally probed, leading to variant telomerases whose repeat extension rates were faster than the rate of the wild-type enzyme. To our knowledge, these T-motif variants are the first TERT mutants to be identified with increased rates of repeat extension synthesis. Similar to wild-type telomerase, repeat synthesis by the mutants was highly processive, indicating that extension rate increases reflected more rapid repetitive copying of the template sequence.
Increases in the repeat extension rate require that one or both of the key contributing components to the extension rate, template copying and translocation, be increased. Mechanistically, faster template copying is achievable by increasing substrate affinity and/or catalysis rate (turnover rate [kcat]). As the extension rate determinations (Table 1) were carried out at a dNTP concentration (1 mM) which was saturating, it is doubtful that the extension rate increases observed here reflect increased substrate affinity. In fact, for the 564A mutant, the apparent Km for extension was increased. It also seems improbable that mutating non-RT domain residues would affect the chemistry of polymerization in a manner that would lead to an increase in catalytic rate sufficient to produce a 4-fold increase (as in the case of the 564A enzyme) in the extension rate. Moreover, given that for the wild-type enzyme, template copying comprises roughly one third of the duration of the synthesis of a single repeat (see above), even if template copying time were reduced to zero, the resulting effect on extension rate would be an increase of only 1.5-fold. Consequently, increases in the extension rate of greater than 1.5-fold must involve a change in the translocation rate. Similarly, extension rate increases of over 3-fold, the maximum increase possible if translocation occurred instantaneously, would require changes in template copying rate as well. Thus, the functional implications of the increases in the extension rate of the T-motif mutants, particularly the 564A variant, point to a role for this motif in template/repeat translocation and to some extent, template copying. Furthermore, in participating in translocation, the T motif serves as a determinant of repetitive reverse transcription.
Interestingly, we observed that T-motif modifications affect repeat translocation rate but not processivity. Processivity is the result of an equilibrium between translocation efficiency (i.e., how successfully primer is translocated per round of template copying) and the rate of primer dissociation. Because these modifications increase translocation rate but not efficiency, it would not be expected a priori that they would affect processivity. Moreover, changes in type II translocation rate would not necessarily be accompanied by altered primer/product association with anchoring regions in the enzyme, particularly those outside of the active site. These anchoring interactions, which act as the primary forces that determine the primer dissociation rates, are the main determinants of processivity. Consequently, it would not be anticipated that an increased repeat translocation rate would necessarily result in increased processivity.
How does the T motif contribute to repeat translocation? As both repeat translocation and template copying are influenced by T-motif interactions, there is reasonable likelihood that this motif is involved in template placement and/or template movement through the active site. This could be accomplished either by direct contact with TR or by mediating proposed conformational changes/rearrangements that move/position the template during repeat synthesis. Data from RNA binding studies (in this work and by others [4, 5, 29]) indicate a participation of the T motif in TR binding, suggesting direct contact. Additional insight may be gleaned from the high-resolution crystal structures that have recently been solved for the TRBD in isolation (from Tetrahymena) and as a part of the full-length TERT protein (from the beetle Tribolium castaneum) (15, 43). Both structures possess a common, shared architecture, suggesting that TRBD structure is highly conserved. In both crystal structures, residues of the FYXTE signature sequence are located within a deep cleft comprising the putative RNA binding pocket of the TRBD. Specifically, they are positioned at the base of a hairpin which extends from the pocket and is predicted to be placed in proximity of the active site. With the exception of the phenylalanine residue, the side chains of these residues are solvent exposed. When a DNA-RNA heteroduplex was modeled into the predicted active site of the T. castaneum TERT structure, the TRBD hairpin containing the FYXTE sequence in the resultant complex was within close proximity of the 5′ terminus of the RNA strand (15). This suggests that these residues could contact the template region of the TR at its 5′ end while being copied. Thus, while we do not rule out the possibility of the T motif influencing translocation/template copying by modulating telomerase conformational changes, both TR binding studies and available TRBD crystal structural information favor a mechanism involving direct interaction with TR.
A key observation lending support to a direct template interaction role is that stable immunoprecipitable ribonucleoprotein complexes can be assembled (in the presence of a sufficiently high concentration of hTR) with TERTs containing complete deletion or substitution of the FYXTE signature sequence (Table 2) but they are inactive. Although we cannot discount the possibility that the TR/TERT complexes of the inactive Del-T and Sub-T mutants are in some way misassembled, these findings imply that while residues of the signature sequence are not required for maintenance of stable hTR/hTERT association, they are essential for enzymatic activity. Previous studies of T-motif function, mainly on Tetrahymena TERT (tTERT), attributed the reduction or loss of activity in FYXTE substitution and deletion mutants to impaired TR binding leading to the inability to form TR/TERT complexes (4, 5, 29). Our binding studies at reduced concentration of hTR (concentration similar to that in tTERT studies) support an involvement of FYXTE residues in the interactions required for TR assembly into the complex. However, the ability to assemble inactive complexes at a high concentration of hTR seems to indicate that a primary function of the signature sequence is to aid in repeat synthesis, via TR interaction.
On the basis of our data and TERT structural modeling (see above), we propose that in addition to securing hTR to hTERT as previously proposed (5), the T motif is actively participating in telomeric DNA synthesis by assisting in the presentation of the template to the catalytic site. During initial TR binding, the T motif places/positions the template into the active site. As we find that mutant hTR templates can be efficiently presented for copying by T-motif mutant enzymes, the recognition/placement of the template sequence by the T motif appears to be independent of the template sequence composition. This is supported by studies of wild-type telomerase demonstrating that completely nontelomeric sequences can serve as templates (44). Thus, it is likely that the FYXTE residues are not interacting with the template bases, but rather with the phosphodiester backbone of the template (or immediately adjacent regions), in close proximity to the active site. The T-motif hairpin maintains contact with the TR near the template region as it is reverse transcribed, until the end of the template is reached, at which point translocation occurs and the template is repositioned. During translocation, the T motif guides the template back to the active site as the template is reset into place.
In summary, we have shown that residues in the FYXTE signature sequence of the telomerase-specific T motif can substantially influence the repeat extension rate. The results of our biochemical analysis indicate that these residues modulate the repeat translocation process, providing insight toward understanding how telomerase carries out repeat synthesis. The precise mechanistic nature of their contribution awaits further investigation, including determination of the high-resolution structure of the hTERT/hTR ribonucleoprotein complex.
Acknowledgments
We thank the AECOM DNA Sequencing Facility.
This work was supported by Public Health Service grants K01 CA87542 (Howard Temin Award) to W.C.D. and R37 AI030861 to V.R.P.
Footnotes
Published ahead of print on 16 November 2009.
REFERENCES
- 1.Autexier, C., and N. F. Lue. 2006. The structure and function of telomerase reverse transcriptase. Annu. Rev. Biochem. 75:493-517. [DOI] [PubMed] [Google Scholar]
- 2.Baran, N., Y. Haviv, B. Paul, and H. Manor. 2002. Studies on the minimal lengths required for DNA primers to be extended by the Tetrahymena telomerase: implications for primer positioning by the enzyme. Nucleic Acids Res. 30:5570-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beattie, T. L., W. Zhou, M. O. Robinson, and L. Harrington. 2000. Polymerization defects within human telomerase are distinct from telomerase RNA and TEP1 binding. Mol. Biol. Cell 11:3329-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosoy, D., Y. Peng, I. S. Mian, and N. F. Lue. 2003. Conserved N-terminal motifs of telomerase reverse transcriptase required for ribonucleoprotein assembly in vivo. J. Biol. Chem. 278:3882-3890. [DOI] [PubMed] [Google Scholar]
- 5.Bryan, T. M., K. J. Goodrich, and T. R. Cech. 2000. Telomerase RNA bound by protein motifs specific to telomerase reverse transcriptase. Mol. Cell 6:493-499. [DOI] [PubMed] [Google Scholar]
- 6.Bryan, T. M., J. M. Sperger, K. B. Chapman, and T. R. Cech. 1998. Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytricha trifallax. Proc. Natl. Acad. Sci. U. S. A. 95:8479-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, S. R., and E. H. Blackburn. 2004. Telomeres and telomerase. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359:109-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins, K. 2006. The biogenesis and regulation of telomerase holoenzymes. Nat. Rev. Mol. Cell Biol. 7:484-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins, K., and L. Gandhi. 1998. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc. Natl. Acad. Sci. U. S. A. 95:8485-8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins, K., and C. W. Greider. 1993. Tetrahymena telomerase catalyzes nucleolytic cleavage and nonprocessive elongation. Genes Dev. 7:1364-1376. [DOI] [PubMed] [Google Scholar]
- 11.Delany, M. E., and L. M. Daniels. 2004. The chicken telomerase reverse transcriptase (chTERT): molecular and cytogenetic characterization with a comparative analysis. Gene 339:61-69. [DOI] [PubMed] [Google Scholar]
- 12.Drosopoulos, W. C., R. Direnzo, and V. R. Prasad. 2005. Human telomerase RNA template sequence is a determinant of telomere repeat extension rate. J. Biol. Chem. 280:32801-32810. [DOI] [PubMed] [Google Scholar]
- 13.Drosopoulos, W. C., and V. R. Prasad. 2007. The active site residue valine 867 in human telomerase reverse transcriptase influences nucleotide incorporation and fidelity. Nucleic Acids Res. 35:1155-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman, K. L., and T. R. Cech. 1999. Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes Dev. 13:2863-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillis, A. J., A. P. Schuller, and E. Skordalakes. 2008. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature 455:633-637. [DOI] [PubMed] [Google Scholar]
- 16.Greenberg, R. A., R. C. Allsopp, L. Chin, G. B. Morin, and R. A. DePinho. 1998. Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene 16:1723-1730. [DOI] [PubMed] [Google Scholar]
- 17.Reference deleted.
- 18.Guo, W., M. Okamoto, Y. M. Lee, M. A. Baluda, and N. H. Park. 2001. Enhanced activity of cloned hamster TERT gene promoter in transformed cells. Biochim. Biophys. Acta 1517:398-409. [DOI] [PubMed] [Google Scholar]
- 19.Hammond, P. W., and T. R. Cech. 1997. dGTP-dependent processivity and possible template switching of euplotes telomerase. Nucleic Acids Res. 25:3698-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammond, P. W., and T. R. Cech. 1998. Euplotes telomerase: evidence for limited base-pairing during primer elongation and dGTP as an effector of translocation. Biochemistry 37:5162-5172. [DOI] [PubMed] [Google Scholar]
- 21.Hammond, P. W., T. N. Lively, and T. R. Cech. 1997. The anchor site of telomerase from Euplotes aediculatus revealed by photo-cross-linking to single- and double-stranded DNA primers. Mol. Cell. Biol. 17:296-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hardy, C. D., C. S. Schultz, and K. Collins. 2001. Requirements for the dGTP-dependent repeat addition processivity of recombinant Tetrahymena telomerase. J. Biol. Chem. 276:4863-4871. [DOI] [PubMed] [Google Scholar]
- 23.Hossain, S., S. Singh, and N. F. Lue. 2002. Functional analysis of the C-terminal extension of telomerase reverse transcriptase. A putative “thumb” domain. J. Biol. Chem. 277:36174-36180. [DOI] [PubMed] [Google Scholar]
- 24.Huard, S., T. J. Moriarty, and C. Autexier. 2003. The C terminus of the human telomerase reverse transcriptase is a determinant of enzyme processivity. Nucleic Acids Res. 31:4059-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobs, S. A., E. R. Podell, and T. R. Cech. 2006. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat. Struct. Mol. Biol. 13:218-225. [DOI] [PubMed] [Google Scholar]
- 26.Karamysheva, Z., L. Wang, T. Shrode, J. Bednenko, L. A. Hurley, and D. E. Shippen. 2003. Developmentally programmed gene elimination in Euplotes crassus facilitates a switch in the telomerase catalytic subunit. Cell 113:565-576. [DOI] [PubMed] [Google Scholar]
- 27.Katinka, M. D., S. Duprat, E. Cornillot, G. Metenier, F. Thomarat, G. Prensier, V. Barbe, E. Peyretaillade, P. Brottier, P. Wincker, F. Delbac, H. El Alaoui, P. Peyret, W. Saurin, M. Gouy, J. Weissenbach, and C. P. Vivares. 2001. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414:450-453. [DOI] [PubMed] [Google Scholar]
- 28.Kuramoto, M., K. Ohsumi, T. Kishimoto, and F. Ishikawa. 2001. Identification and analyses of the Xenopus TERT gene that encodes the catalytic subunit of telomerase. Gene 277:101-110. [DOI] [PubMed] [Google Scholar]
- 29.Lai, C. K., M. C. Miller, and K. Collins. 2002. Template boundary definition in Tetrahymena telomerase. Genes Dev. 16:415-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai, C. K., J. R. Mitchell, and K. Collins. 2001. RNA binding domain of telomerase reverse transcriptase. Mol. Cell. Biol. 21:990-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, M. S., and E. H. Blackburn. 1993. Sequence-specific DNA primer effects on telomerase polymerization activity. Mol. Cell. Biol. 13:6586-6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lingner, J., T. R. Hughes, A. Shevchenko, M. Mann, V. Lundblad, and T. R. Cech. 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276:561-567. [DOI] [PubMed] [Google Scholar]
- 33.Lue, N. F. 2004. Adding to the ends: what makes telomerase processive and how important is it? Bioessays 26:955-962. [DOI] [PubMed] [Google Scholar]
- 34.Lue, N. F. 2005. A physical and functional constituent of telomerase anchor site. J. Biol. Chem. 280:26586-26591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reference deleted.
- 36.Marsilio, E., S. H. Cheng, B. Schaffhausen, E. Paucha, and D. M. Livingston. 1991. The T/t common region of simian virus 40 large T antigen contains a distinct transformation-governing sequence. J. Virol. 65:5647-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyerson, M., C. M. Counter, E. N. Eaton, L. W. Ellisen, P. Steiner, S. D. Caddle, L. Ziaugra, R. L. Beijersbergen, M. J. Davidoff, Q. Liu, S. Bacchetti, D. A. Haber, and R. A. Weinberg. 1997. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell 90:785-795. [DOI] [PubMed] [Google Scholar]
- 38.Moriarty, T. J., S. Huard, S. Dupuis, and C. Autexier. 2002. Functional multimerization of human telomerase requires an RNA interaction domain in the N terminus of the catalytic subunit. Mol. Cell. Biol. 22:1253-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moriarty, T. J., D. T. Marie-Egyptienne, and C. Autexier. 2004. Functional organization of repeat addition processivity and DNA synthesis determinants in the human telomerase multimer. Mol. Cell. Biol. 24:3720-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakamura, T. M., G. B. Morin, K. B. Chapman, S. L. Weinrich, W. H. Andrews, J. Lingner, C. B. Harley, and T. R. Cech. 1997. Telomerase catalytic subunit homologs from fission yeast and human. Science 277:955-959. [DOI] [PubMed] [Google Scholar]
- 41.Nasir, L., E. Gault, S. Campbell, M. Veeramalai, D. Gilbert, R. McFarlane, A. Munro, and D. J. Argyle. 2004. Isolation and expression of the reverse transcriptase component of the Canis familiaris telomerase ribonucleoprotein (dogTERT). Gene 336:105-113. [DOI] [PubMed] [Google Scholar]
- 42.O'Connor, C. M., C. K. Lai, and K. Collins. 2005. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J. Biol. Chem. 280:17533-17539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rouda, S., and E. Skordalakes. 2007. Structure of the RNA-binding domain of telomerase: implications for RNA recognition and binding. Structure 15:1403-1412. [DOI] [PubMed] [Google Scholar]
- 44.Ware, T. L., H. Wang, and E. H. Blackburn. 2000. Three telomerases with completely non-telomeric template replacements are catalytically active. EMBO J. 19:3119-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weinrich, S. L., R. Pruzan, L. Ma, M. Ouellette, V. M. Tesmer, S. E. Holt, A. G. Bodnar, S. Lichtsteiner, N. W. Kim, J. B. Trager, R. D. Taylor, R. Carlos, W. H. Andrews, W. E. Wright, J. W. Shay, C. B. Harley, and G. B. Morin. 1997. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 17:498-502. [DOI] [PubMed] [Google Scholar]
- 46.Wilson, I. A., D. H. Haft, E. D. Getzoff, J. A. Tainer, R. A. Lerner, and S. Brenner. 1985. Identical short peptide sequences in unrelated proteins can have different conformations: a testing ground for theories of immune recognition. Proc. Natl. Acad. Sci. U. S. A. 82:5255-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wyatt, H. D., D. A. Lobb, and T. L. Beattie. 2007. Characterization of physical and functional anchor site interactions in human telomerase. Mol. Cell. Biol. 27:3226-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]