Abstract
The administration of high-dose clindamycin (CLI) along with penicillin is recommended for the treatment of streptococcal toxic shock syndrome. However, the prevalence of CLI-resistant Streptococcus pyogenes strains is increasing worldwide, and the effect of CLI on CLI-resistant S. pyogenes strains remains unknown. We aimed to evaluate the effect of CLI on the in vitro production of three major virulent exoproteins, namely, streptolysin O (Slo), NAD glycohydrolase (Nga), and streptokinase (Ska), by CLI-resistant S. pyogenes strains. After the incubation of M1 serotype CLI-resistant S. pyogenes D2TY in the presence of 1 μg/ml CLI, the amounts of Slo, Nga, and Ska and the levels of slo, nga, and ska mRNA in the supernatant were analyzed by Northern blotting and Western blotting, respectively. The results of both assays showed that the production of Slo, Nga, and Ska was higher with CLI treatment than without CLI treatment. We evaluated the role of the sensor kinase CovS, which is involved in the two-component system of S. pyogenes, in the CLI-induced production of these three exoproteins. Northern blotting analysis revealed that CLI induced the expression of covS mRNA in wild-type strain D2TY. Furthermore, both Northern blotting and Western blotting analyses showed that CLI decreased the levels of expression of Slo, Nga, and Ska in isogenic covS mutant D2TYcovS. These results suggest that CLI increases the production of three virulent exoproteins in CLI-resistant S. pyogenes strains via the action of CovS.
Streptococcus pyogenes is a gram-positive bacterium that is responsible for pharyngitis and postinfection diseases such as rheumatic fever and glomerulonephritis. In addition, S. pyogenes causes streptococcal toxic shock syndrome (STSS) (7).
The management of STSS requires intensive antibiotic treatment, and the present consensus regarding the antibiotic treatment of STSS is the administration of a high dose of clindamycin (CLI) together with penicillin (16). CLI is a lincosamide that binds to the 50S subunit of the bacterial ribosome and inhibits protein synthesis (25). With regard to the inhibition of protein synthesis, it has been shown that the synthesis of several streptococcal exoproteins, including virulence factors (e.g., SpeA, SpeB, and M protein), is inhibited by CLI at sub-MICs (6, 18, 21). Another reason for the use of CLI is that it transfers to tissues well (24).
In recent years, macrolide- and lincosamide-resistant S. pyogenes strains have gradually spread worldwide (15, 20, 23). Although the prevalence of macrolide- and lincosamide-resistant S. pyogenes strains has been well investigated, the effects of these drugs on macrolide- and lincosamide-resistant S. pyogenes strains has not been studied. We have previously shown by using two-dimensional gel electrophoresis that the production of streptolysin O (Slo), NAD glycohydrolase (Nga), and streptokinase (Ska) in cultures of S. pyogenes is increased by treatment with CLI at sub-MICs (24). In addition, we examined the role of the two-component sensor kinase, CovS, in the effect of CLI at sub-MICs. The increase in the amounts of exoproteins detected in the wild-type strain was considerably inhibited in an isogenic covS mutant strain (21).
Therefore, we attempted to clarify the relationship between CLI and the production of three major exoproteins by S. pyogenes, namely, Slo, Nga, and Ska, and that between CLI and the two-component sensor kinase CovS in CLI-resistant S. pyogenes strains.
MATERIALS AND METHODS
Bacterial strains.
The S. pyogenes strain of the M1 serotype used in this study, strain D2TY, was isolated from a patient in Japan with STSS. The bacteria (strain D2TY and its derived knockout mutants) were cultured at 37°C without agitation in brain heart infusion broth (Eiken Chemical Co., Tokyo, Japan) containing 0.3% yeast extract (Difco Laboratories, Detroit, MI) (BHI-YE). The antibiotic used in this study was CLI hydrochloride (Sigma Chemical Co., St. Louis, MO). Escherichia coli JM109 (Promega, Madison, WI) was cultured in Luria-Bertani (LB) agar (Difco Laboratories) or LB broth with aeration at 37°C. The following antibiotics were used at the indicated concentrations, as appropriate: ampicillin (Sigma Chemical Co.) at 100 μg/ml (E. coli) and spectinomycin (Sigma Chemical Co.) at 100 μg/ml (E. coli) and 100 μg/ml (S. pyogenes).
Preparation of covS-knockout mutants.
Mutants with nonpolar inactivated covS genes were constructed by introducing double-crossover allelic replacements in the chromosome of S. pyogenes D2TY. We used a suicide vector (pcsrS::aad9/pFW12), which was constructed in a previous study (21), and successfully performed double-crossover replacement, as described elsewhere (21). We confirmed the successful crossover replacement by using direct DNA sequencing (ABI Prism 3100 genetic analyzer; Life Technologies Corp., Carlsbad, CA). We designated this mutant D2TYcovS.
Susceptibility test.
The MICs of CLI for strains D2TY and D2TYcovS were assayed by the broth microdilution method and by the Etest method, in accordance with the guidelines of the Clinical and Laboratory Standards Institute (CLSI; formerly the National Committee for Clinical Laboratory Standards) (5). According to the recommendations of the CLSI, the MIC of CLI for S. pyogenes D2TY is 256 μg/ml. Since strains for which the MIC of CLI exceeds 1 μg/ml are defined as resistant, we used a CLI concentration of 1 μg/ml in our experiments. We also performed the double-disk diffusion test (the D-zone test) with D2TY by using a 15-μg erythromycin disk and a 2-μg CLI disk placed 16 mm apart, as described previously (20).
Detection of lincosamide resistance genes.
Bacterial chromosomes were extracted by using a DNA extraction kit (Promega), according to the manufacturer's instructions. Two lincosamide resistance genes, namely, ermB and ermTR, were amplified by PCR as described before (20). The sequences of the PCR primers used for the amplification of these genes are shown in Table 1.
TABLE 1.
Oligonucleotide primers used in this study
| Gene target | Primer designation | Primer sequence | Product size (bp) | Reference or source |
|---|---|---|---|---|
| ermB | ermB-F | CGAGTGAAAAAGTACACAACC | 616 | 20 |
| ermB-R | GGCGTGTTTCATTGCTTGATG | 20 | ||
| ermTR | ermTR-F | GCATGACATAAACCTTCA | 206 | 20 |
| ermTR-R | AGGTTATAATGAAACAGA | 20 | ||
| slo | Slo-EcoRI | GGAATTCCTACTTATAAGTAATCGAAC | 1,622 | 24 |
| Slo-BamHI | CGGATCCGAATCGAACAAACAAAACAC | 24 | ||
| nga | Nga-BamHI | CGGATCCGTTAGTGGCAAAGAAAATAA | 1,355 | 24 |
| Nga-XhoI | CCTCGAGTTACTTGGTATCTTGCATTT | 24 | ||
| ska | Ska-F | ATTTGGAACAGTCAAGCCTGTCC | 1,263 | 11 |
| Ska-R | TTAGGGTTATCAGGTATAGGTG | 11 | ||
| covS | csrS-n4 | ACAAGGCTCATTTGAGGGAC | 500 | This study |
| csrS-c1 | GCACTGCAGCTAACTCTCTTTAGACTGGG | This study | ||
| covR | csrR-n4 | GGCTAGCTTAATCGTGGCGACGATGAG | 240 | This study |
| csrR-c1 | GCACTGCAGTTATTTCTCACGAATAACGT | This study |
RNA isolation and Northern blotting analysis.
Total RNA was extracted and purified as described previously (24). In brief, bacterial cells were cultured in 5 ml BHI-YE to which 1 μg/ml CLI was or was not added at the beginning of culture. The cells were harvested when the optical density at 660 nm was approximately 0.8 (late log phase). Total RNA was extracted and purified with an RNA-protected bacterial reagent (Qiagen, Hilden, Germany) and was then treated with RNase-free DNase (Qiagen). Approximately 1 μg of each total RNA preparation was electrophoresed on 1% agarose gels containing 1.1 M formaldehyde. The RNA was transferred to a Hybond-N+ membrane (GE Healthcare); and mRNA was detected with 32P-labeled nga, slo, ska, covS, and covR DNAs (random primer DNA labeling kit, version 2; Takara, Ohtsu, Japan). The membranes were then autoradiographed and analyzed at room temperature with a bioimaging analyzer (BAS-1800II; Fujifilm, Tokyo, Japan). Triplicate assays were performed with RNA from at least two independent cultures.
Western blotting analysis.
An aliquot of S. pyogenes stock solution frozen at −80°C was cultured in 5 ml BHI-YE at 37°C without agitation. The protease inhibitor E-64 (Roche Diagnosis, Indianapolis, IN) was added to the BHI-YE, as described by Aziz et al. (1). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described before (24). After SDS-PAGE, bacterial proteins were electrophoretically transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA). Anti-Slo antibody (dilution, 1:2,000; BioAcademia, Osaka, Japan), anti-Nga antibody (which was kindly provided by Tohru Akiyama; dilution, 1:2,000), and anti-Ska antibody (dilution, 1:2,000; BioAcademia) were then incubated for 16 h at room temperature with the antigen-containing membranes. The antigen-antibody complexes were detected by using horseradish peroxidase goat anti-rabbit immunoglobulin G (Acris Antibodies GmbH, Hiddenhausen, Germany), which was used as the secondary antibody at a dilution of 1:2,000. Detection was performed with an immunostaining kit (Konica-Minoruta Inc., Tokyo, Japan), according to the manufacturer's instructions. Triplicate assays were performed with proteins from at least two independent cultures.
RESULTS
Single lincosamide resistance gene in CLI-resistant S. pyogenes D2TY.
We performed PCR analysis to assess the genetic factors for CLI resistance. S. pyogenes D2TY possessed only the ermB gene; another gene, ermTR, was not detected by PCR (data not shown). The double-disk diffusion test indicated that D2TY had the constitutive macrolide-lincosamide-streptogramin B (MLSB) resistance phenotype (Fig. 1).
FIG. 1.
Double-disk diffusion test with S. pyogenes D2TY. D2TY, which is resistant to erythromycin (EM) and CLI, had the constitutive MLSB resistance phenotype.
Effect of CLI on production of exoproteins Slo, Nga, and Ska.
We used Northern blot analysis to determine whether the levels of transcription of slo, nga, and ska increased after CLI treatment. The results of Northern blotting analysis showed that the levels of slo, nga, and ska mRNA after CLI treatment had increased compared with the levels before CLI treatment (Fig. 2A). We also used Western blotting to analyze the posttranscription levels of the products encoded by the genes mentioned above. Figure 2B shows that CLI treatment remarkably increased the levels of Slo, Nga, and Ska.
FIG. 2.
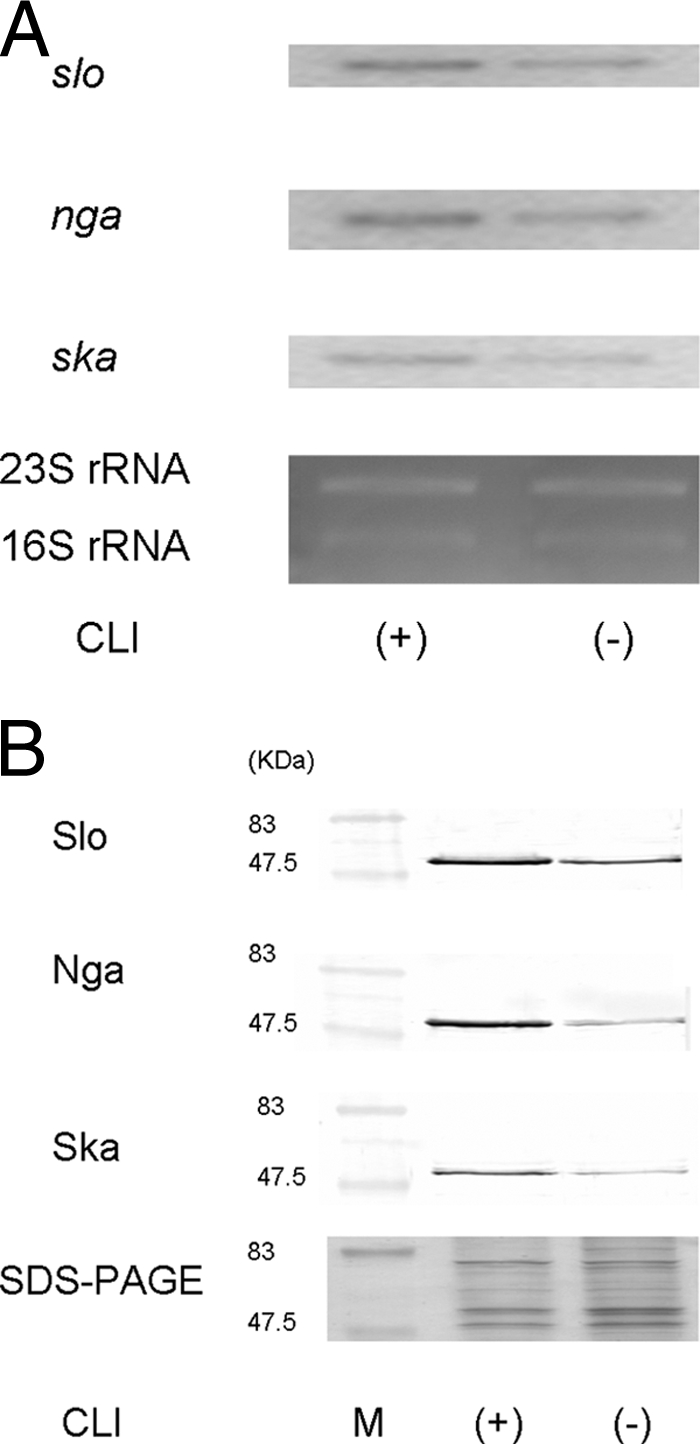
Expression of Slo, Nga, and Ska in CLI-resistant S. pyogenes D2TY after CLI treatment, as assessed by Northern blotting analysis (A) and Western blotting analysis (B). D2TY was cultured in BHI-YE supplemented with 1 μg/ml CLI (+) or without CLI (−). Total RNA was extracted from S. pyogenes D2TY, electrophoresed, and blotted. 32P-labeled DNA fragments of slo, nga, and ska were used as probes. The levels of expression of 23S and 16S rRNAs were used as internal controls to assess the results of the Northern blotting analysis. The Slo, Nga, and Ska proteins in the supernatant were analyzed by Western blotting with a polyclonal antibody. The supernatant was used as a control to asses the result of the Western blotting analysis by SDS-PAGE. These experiments were performed more than three times, and the same expression patterns were shown in all three experiments.
The results of those two assays suggest that CLI affects the expression of the three exoproteins in CLI-resistant strains.
Effect of CLI on CovS transcriptional levels.
In our previous study, we suggested that CovS contributes to the effects of CLI treatment (22). Hence, using Northern blotting, we first analyzed whether CLI induced the expression of covS. As expected, after CLI treatment, the level of covS mRNA was increasing in comparison with the level without CLI treatment (Fig. 3).
FIG. 3.
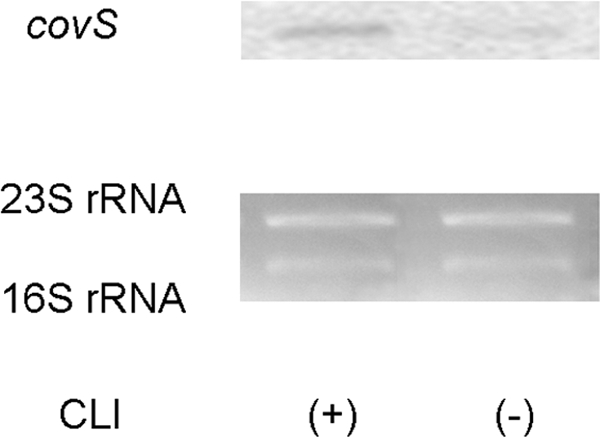
Expression of covS in CLI-resistant S. pyogenes D2TY after CLI treatment, as assessed by Northern blotting analysis. D2TY was cultured in BHI-YE supplemented with 1 μg/ml CLI (+) or without CLI (−). Total RNA was extracted from S. pyogenes D2TY, electrophoresed, and blotted. A 32P-labeled fragment of covS DNA was used as the probe. The levels of expression of 23S and 16S rRNAs were used as internal controls to assess the results of the Northern blotting analysis. These experiments were performed more than three times, and the same expression patterns were shown in all three experiments.
Effect of CovS on CLI-induced production of exoproteins.
We prepared an isogenic ΔcovS mutant strain (D2TYcovS) from D2TY. Inactivation of the covS gene in this mutant was confirmed by Northern blotting analysis. covS mRNA was not expressed in D2TYcovS, and the level of expression of the mRNA of covR, a two-component regulator, was not altered (data not shown).
Finally, we evaluated the differences in the levels of slo, nga, and ska mRNA in the isogenic ΔcovS mutant by Northern blotting analysis. Figure 4A shows that CLI treatment decreased the levels of expression of slo, nga, and ska mRNA in the ΔcovS mutant. We also confirmed that CLI treatment decreased the levels of expression of Nga, Slo, and Ska by Western blotting analysis (Fig. 4B).
FIG. 4.
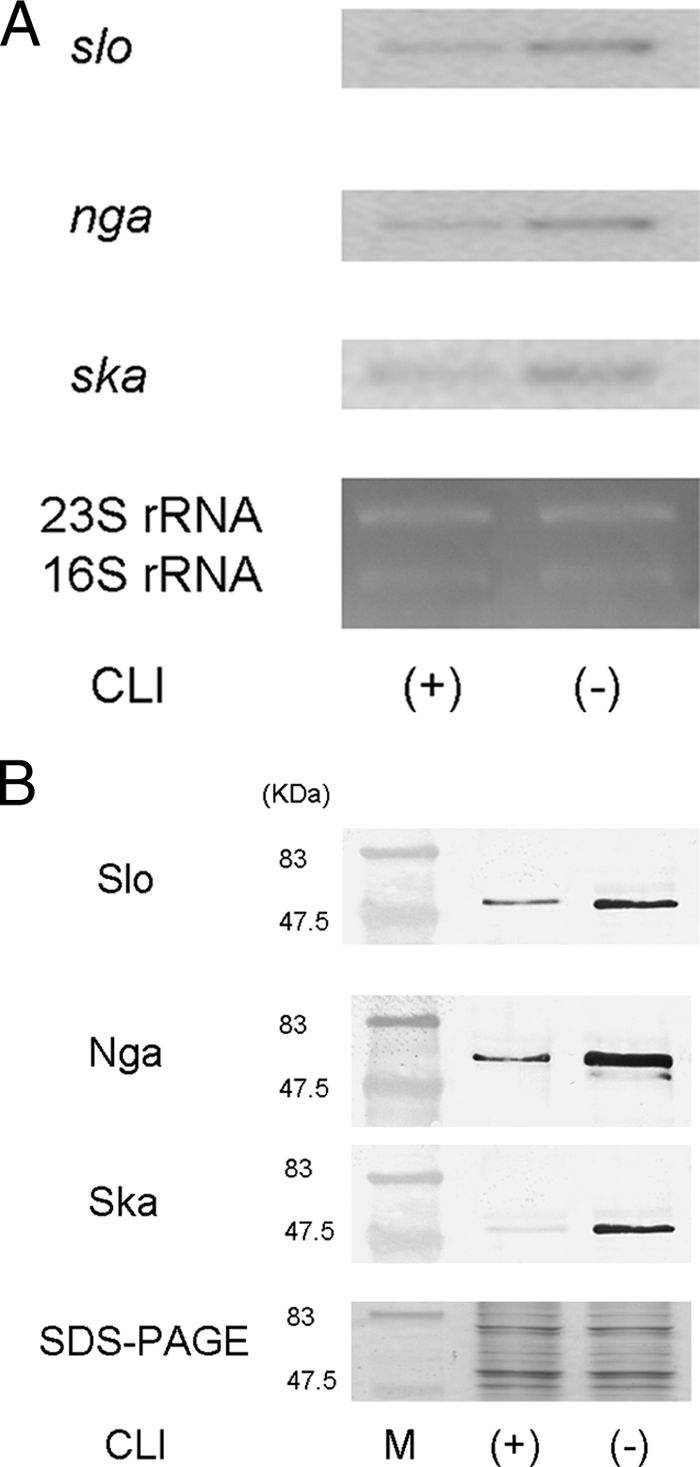
Expression of Slo, Nga, and Ska in CLI-resistant S. pyogenes D2TYcovS after CLI treatment, as assessed by Northern blotting analysis (A) and Western blotting analysis (B). D2TYcovS was cultured in BHI-YE supplemented with 1 μg/ml CLI (+) or without CLI (−).Total RNA was extracted from S. pyogenes D2TYcovS, electrophoresed, and blotted. 32P-labeled DNA fragments of slo, nga, and ska were used as probes. The levels of expression of 23S and 16S rRNAs were used as internal controls to assess the results of the Northern blotting analysis. The Slo, Nga, and Ska proteins in the supernatant were analyzed by Western blotting with a polyclonal antibody. The supernatant was used as a control to assess the result of the Western blotting analysis by SDS-PAGE. These experiments were performed more than three times, and the same expression patterns were shown in all three experiments.
DISCUSSION
This is the first study to characterize virulence gene regulation by the two-component regulatory systems (TCSs) of S. pyogenes in response to the antibiotic CLI. It has been reported that Mg2+ (11, 12) and the cationic antimicrobial peptide LL-37 (13) may regulate the genes required for growth via the action of CovS in S. pyogenes. We demonstrated that CLI would induce new signal stimulations through the CovS system. Our result is supported by a previous report that sub-MICs of antibiotics trigger the general stress response in some bacteria (2).
The CovS/CovR system is one of the TCSs in S. pyogenes, and the activity of this major regulatory system influences 15% of all chromosomal genes in S. pyogenes and also the system itself (10). The sensor kinase (CovS) is a surface protein that responds to environmental signals by undergoing autophosphorylation and is cotranscribed with CovR (9). CovS is required for the growth of S. pyogenes under different conditions of general stress (39°C, pH 6, or high salt concentrations) (8). Dalton and Scott explained the role of CovS as follows (8): stress signals activate the phosphatase activity of CovS, resulting in the dephosphorylation of CovR. The dephosphorylated CovR dissociates from the promoters to which it is bound, leading to the inhibition of the promoters. CovR-regulated promoters may exhibit various degrees of sensitivity to CovR phosphorylation. CovR-regulated genes may exhibit differential expression under normal and stressful conditions. Our finding of CLI-induced Ska expression is consistent with the theory of Dalton and Scott (8). Ska is more abundant in isogenic covR mutant strains than in wild-type strains (10). CLI may suppress the activity of CovR and may thus have increased the amount of Ska. With regard to Staphylococcus aureus, it was suggested that CLI blocks the translation of several other negative-regulation factors, thereby increasing the levels of transcription of exoproteins (14). The exoprotein levels may increase because of inhibition of the synthesis of suppressive regulators; this may nullify the suppressive effects and enable the effect of decreased translation to overcome the direct effects of antibiotic treatment.
Our findings regarding Slo and Nga expression are not consistent with the results of Graham et al. The expression of these two exoproteins in CovR mutants is lower than that in wild-type strains (10). We speculated that there were two possible explanations for the CLI-induced regulation of Slo and Nga by CovS. One possibility is that other regulatory pathways control translation and/or transcription by CovS. CovS may function as a direct positive signal in the presence of CLI; it is located upstream of the positive pathway that increases the levels of production of exoproteins by CLI. These proteins may be necessary for the bacteria to survive in stressful surroundings, and the levels of these proteins may be increased because of increased levels of transcription. The TCS may interact with many regulators, leading to cross talk. Positive regulators of CovS-mediated phosphorylation have not yet been found. CovS may regulate a gene required for growth in the presence of CLI independently of CovR, i.e., by cross talk with another two-component response regulator. The other possibility is that the increase in exoprotein levels might be caused by the inhibition of the streptococcal cysteine protease SpeB. SpeB protease degrades Slo, Nga, and Ska; the decrease in SpeB levels in the presence of CLI therefore increases the levels of these three exoproteins (1, 24). However, the increase in the levels of these exoproteins was greater in the presence of both CLI and the specific cysteine protease inhibitor E-64 than in the presence of only E-64. Since the possibility of the involvement of other proteases cannot be ruled out, this mechanism warrants further investigation.
In our previous study of sub-MICs of CLI, CovS inactivation decreased the levels of many exoproteins in CLI-susceptible S. pyogenes strains (21). In this study, the production of three major exoproteins, namely, Slo, Nga, and Ska, was increased in a CLI-resistant wild-type S. pyogenes strain after treatment with 1 μg/ml CLI. However, the levels of production of these proteins in the CLI-resistant isogenic S. pyogenes ΔcovS strain decreased after treatment with CLI. We also found that this change in the levels of expression of slo and nga occurred at the transcriptional level. Our result regarding the CLI-resistant strain is consistent with that of a previous study with a CLI-susceptible strain (21). Moreover, we demonstrated that CLI induced the expression of covS by increasing the level of gene transcription. We also analyzed the levels of transcription of slo, nga, and ska and found that their levels of transcription were also increased. In antibiotic-susceptible strains, inhibitors of protein synthesis directly interact with the ribosomes and terminate peptide translation from mRNA. However, CLI is not effective at the suppression of the transcription of slo, nga, and ska in CLI-resistant strains. This result is consistent with the ineffective suppression of some virulent proteins in CLI-susceptible strains by sub-MICs of CLI (24).
Slo and Nga appear to be functionally linked. Slo exerts its cytolytic function by forming large homopolymeric pores in membranes (3). Nga, which is transferred through the Slo-induced pores, contributes to bacterial pathogenesis by modulating host-cell signaling pathways to inhibit internalization, augment Slo-mediated cytotoxicity, and induce target cell apoptosis (4). Ska is a potent enzyme that activates the blood clot-dissolving protein plasminogen in the host. Thus, because Ska aids the movement of bacteria through host tissue, it is considered to contribute to the spread of S. pyogenes (23). Since the virulence of S. pyogenes shows interindividual differences, CLI administration might adversely affect STSS treatment, as judged solely on the basis of the exoprotein production. Further studies are required to assess the choice of antibiotics for the treatment of STSS.
From the clinical perspective, our findings show that the inappropriate use of antibiotics for the treatment of infections caused by antibiotic-resistant strains may worsen clinical outcomes by increasing the levels of production of virulent exoproteins. Although susceptibility tests should be performed before antibiotic administration for the treatment of infections with antibiotic-resistant bacteria, it usually takes several days to confirm the result of the antibiotic susceptibility assay with S. pyogenes. STSS is especially a dramatically rapidly progressive invasive infection and leads to death within several hours (7). The present consensus regarding the use of antibiotic treatment for STSS recommended that antibiotic therapy be started as soon as an S. pyogenes infection is diagnosed. Thus, it is impossible from a clinical perspective to start antibiotic therapy after confirmation of the results of the susceptibility assay. A new therapy for STSS caused by CLI-resistant S. pyogenes strains is urgently needed, as indicated by the findings of our investigation. We found that CLI suppresses the production of three major virulent proteins, namely, Slo, Nga, and Ska, via the inactivation of covS. Thus, CovS may be a new target for drugs for the treatment of severe streptococcal infections. CLI itself is known to suppress exoprotein production, especially SpeB protease production. Owing to the suppression of virulent exoproteins, combination therapy with CLI and a chemical or antibody targeting CovS may be more effective than ordinary antistreptococcal therapy, even in the case of infection with CLI-resistant S. pyogenes strains. Further analysis of the interaction of CLI with CovS is desired.
In summary, we suggest that CLI increases the levels of production of Slo, Nga, and Ska in CLI-resistant S. pyogenes strains via the action of CovS.
Acknowledgments
We thank Hideyuki Matsui for technical support. The anti-Nga antibody was kindly provided by Tohru Akiyama.
This study was supported by a Grant-in-Aid for Research from the Nagoya City University, Nagoya, Japan, and by a grant (grant 21590485) from the Ministry of Education, Science, and Culture of Japan.
Footnotes
Published ahead of print on 5 October 2009.
REFERENCES
- 1.Aziz, R. K., M. J. Pabst, A. Jeng, R. Kansal, D. E. Low, V. Nizet, and M. Kotb. 2004. Invasive M1T1 group A Streptococcus undergoes a phase-shift in vivo to prevent proteolytic degradation of multiple virulence factors by SpeB. Mol. Microbiol. 51:123-134. [DOI] [PubMed] [Google Scholar]
- 2.Bandow, J. E., H. Brötz, and M. Hecker. 2002. Bacillus subtilis tolerance of moderate concentrations of rifampin involves the σB-dependent general and multiple stress response. J. Bacteriol. 184:459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billington, S. J., B. H. Jost, and J. G. Songer. 2000. Thiol-activated cytolysins: structure, function and role in pathogenesis. FEMS Microbiol. Lett. 182:197-205. [DOI] [PubMed] [Google Scholar]
- 4.Bricker, A. L., C. Cywes, C. D. Ashbaugh, and M. R. Wessels. 2002. NAD+-glycohydrolase acts as an intracellular toxin to enhance the extracellular survival of group A streptococci. Mol. Microbiol. 44:257-269. [DOI] [PubMed] [Google Scholar]
- 5.CLSI/NCCLS. 2003. Method for dilution antimicrobial susceptibility test for bacteria that grow aerobically. Approved standard M7-A6, 6th ed. CLSI/NCCLS, Wayne, PA.
- 6.Coyle, E. A., R. Cha, and M. J. Rybak. 2003. Influences of linezolid, penicillin, and clindamycin, alone and in combination, on streptococcal pyrogenic exotoxin A release. Antimicrob. Agents Chemother. 47:1752-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalton, T. L., and J. R. Scott. 2004. CovS inactivates CovR and is required for growth under conditions of general stress in Streptococcus pyogenes. J. Bacteriol. 186:3928-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalton, T. L., R. I. Hobb, and J. R. Scott. 2006. Analysis of the role of CvR and CovS in the dissemination of Streptococcus pyogenes in invasive skin disease. Microb. Pathog. 40:221-227. [DOI] [PubMed] [Google Scholar]
- 10.Graham, M. R., L. M. Smoot, C. A. Migliaccio, K. Virtaneva, D. E. Sturdevant, S. F. Porcella, M. J. Federle, G. J. Adams, J. R. Scott, and J. M. Musser. 2002. Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc. Natl. Acad. Sci. U. S. A. 99:13855-13860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gryllos, I., J. C. Levin, and M. R. Wessels. 2003. The CsrR/CsrS two-component system of group A Streptococcus responds to environmental Mg2+. Proc. Natl. Acad. Sci. U. S. A. 100:4227-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gryllos, I., R. Grifantini, A. Colaprico, S. Jiang, E. DeForce, A. Hakansson, J. L. Telford, G. Grandi, and M. R. Wessels. 2007. Mg2+ signaling defines the group A streptococcal CsrRS (CovRS) regulon. Mol. Microbiol. 65:671-683. [DOI] [PubMed] [Google Scholar]
- 13.Gryllos, I., H. J. Trans-Winkler, M. F. Cheng, H. Chung, R. Bolcome III, W. Lu, R. I. Lehrer, and M. R. Wessels. 2008. Induction of group A Streptococcus virulence by a human antimicrobial peptide. Proc. Natl. Acad. Sci. U. S. A. 105:16755-16760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herbert, S., P. Barry, and R. P. Novick. 2001. Subinhibitory clindamycin differentially inhibits transcription of exoprotein genes in Staphylococcus aureus. Infect. Immun. 69:2996-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikebe, T., K. Hirasawa, R. Suzuki, J. Isobe, D. Tanaka, C. Katsukawa, R. Kawahara, M. Tomita, K. Ogata, M. Endoh, R. Okuno, H. Watanabe, and the Working Group for Group A Streptococci in Japan. 2005. Antimicrobial susceptibility survey of Streptococcus pyogenes isolated in Japan from patients with severe invasive group A streptococcal infection. Antimicrob. Agents Chemother. 49:788-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lappin, E., and A. J. Ferguson. 2009. Gram-positive toxic shock syndrome. Lancet Infect. Dis. 9:281-290. [DOI] [PubMed] [Google Scholar]
- 17.Reference deleted.
- 18.Mascini, E. M., M. Jansze, L. M. Schouls, J. Verhoef, and H. Van Dijk. 2001. Penicillin and clindamycin differentially inhibit the production of pyrogenic exotoxins A and B by group A streptococci. Int. J. Antimicrob. Agents 18:395-398. [DOI] [PubMed] [Google Scholar]
- 19.Reference deleted.
- 20.Richter, S. S., K. P. Heilmann, S. E. Beekmann, N. J. Miller, A. L. Miller, C. L. Rice, C. D. Doern, S. D. Reid, and G. V. Doern. 2005. Macrolide-resistant Streptococcus pyogenes in the United States, 2002-2003. Clin. Infect. Dis. 41:599-608. [DOI] [PubMed] [Google Scholar]
- 21.Sawai, J., T. Hasegawa, T. Kamimura, A. Okamoto, D. Ohmori, and M. Ohta. 2007. Growth phase-dependent effect of clindamycin on production of exoproteins by Streptococcus pyogenes. Antimicrob. Agents Chemother. 51:461-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun, H., U. Ringdahl, J. W. Homeister, W. P. Fay, N. C. Engleberg, and A. Y. Yang. 2004. Plasminogen is a critical host pathogenicity factor for group A streptococcal infection. Science 305:1283-1286. [DOI] [PubMed] [Google Scholar]
- 23.Tamayo, J., E. Perez-Trallero, J. L. Gomez-Garces, and J. I. Alos on behalf of the Spanish Group for the Study of Infection in the Primary Health Care Setting (IAP-SEIMC). 2005. Resistance to macrolides, clindamycin and telithromycin in Streptococcus pyogenes isolated in Spain during 2004. J. Antimicrob. Chemother. 56:780-782. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka, M., T. Hasegawa, A. Okamoto, K. Torii, and M. Ohta. 2005. Effect of antibiotics on group A Streptococcus exoprotein production analyzed by two-dimensional gel electrophoresis. Antimicrob. Agents Chemother. 49:88-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tension, T., M. Lovmar, and M. Ehrenberg. 2003. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J. Mol. Biol. 330:1005-1014. [DOI] [PubMed] [Google Scholar]



