Abstract
Simocyclinone D8, a coumarin derivative isolated from Streptomyces antibioticus Tü 6040, represents an interesting new antiproliferative agent. It was originally suggested that this drug recognizes the GyrA subunit and interferes with the gyrase catalytic cycle by preventing its binding to DNA. To further characterize the mode of action of this antibiotic, we investigated its binding to the reconstituted DNA gyrase (A2B2) as well as to its GyrA and GyrB subunits and the individual domains of these proteins, by performing protein melting and proteolytic digestion studies as well as inhibition assays. Two binding sites were identified, one (anticipated) in the N-terminal domain of GyrA (GyrA59) and the other (unexpected) at the C-terminal domain of GyrB (GyrB47). Stabilization of the A subunit appears to be considerably more effective than stabilization of the B subunit. Our data suggest that these two distinct sites could cooperate in the reconstituted enzyme.
Resistance to antibiotics is a constantly increasing clinical concern, particularly in hospitals and other health care settings (16, 22). In fact, antibiotic-resistant pathogens cause serious infections for which there are limited therapeutic interventions and that are often life threatening. Hence, the discovery and development of new, effective antibacterial drugs that possibly exhibit new mechanisms of action represent challenging and urgent goals. Possible successful approaches include (i) the exploitation of new drug targets in the pathogen and (ii) the identification of drugs that act at a novel site(s) of known targets (35). In either case, detailed information on the molecular mechanism(s) of drug action is required to rationally develop new effective agents.
In this connection, a major target for antimicrobial intervention is DNA gyrase (19). This enzyme is a prokaryotic type II topoisomerase that modulates the topological state of DNA through cleaving and resealing steps (24, 31). Besides performing reactions such as decatenation, unknotting, and relaxation common to the type II family members, DNA gyrase is also able to introduce negative supercoils, a reaction coupled to ATP hydrolysis (9, 10). DNA gyrase works as a tetramer (A2B2) formed by two A subunits (GyrA) and two B subunits (GyrB) (28). The catalytic tyrosine covalently linked to DNA in the cleavage complex is located in the N-terminal domain of GyrA (GyrA59). The C-terminal domain of GyrA (GyrA33) facilitates the wrapping of DNA around the enzyme. GyrB contains the ATP-binding and hydrolysis site in its N-terminal domain (GyrB43), whereas its C-terminal portion (GyrB47) is involved in DNA and GyrA binding.
A number of drugs are effective in impairing the activity of DNA gyrase (Fig. 1), among which the fluoroquinolones are the therapeutically most relevant (3, 5). As suggested by biological and chemico-physical studies, they act at the cleavage site by trapping the cleavage complex and interacting principally with GyrA but also with GyrB and DNA (12, 27, 34, 37). Recently, the structures of a bacterial topoisomerase II with DNA and quinolones have been determined, confirming such a model (15).
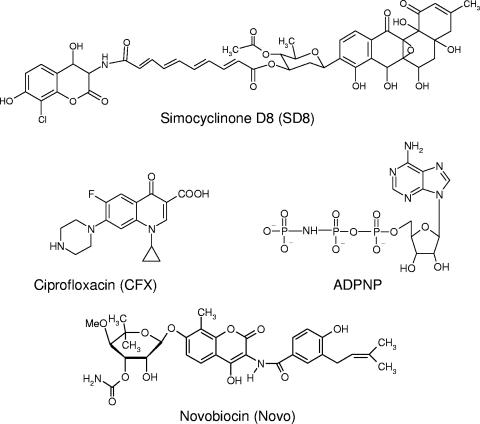
FIG. 1.
Chemical structures of the DNA gyrase ligands tested.
Another well-known class of gyrase inhibitors is represented by the coumarin derivatives (20, 25, 36). Unfortunately, these compounds exhibit an inadequate pharmacological profile that prevents their widespread clinical application. This notwithstanding, they have proved very useful in providing detailed information on the mechanism of enzyme action and the molecular details of the drug-protein interaction (1, 11, 21). Compared to the quinolones, the coumarins act in a completely different way and at a distinct site located on GyrB, which partially overlaps the ATP-binding site (17). Thus, they prevent the ATP hydrolysis required for the enzymatic cycle.
More recently, novel angucyclinone-type DNA gyrase inhibitors, simocyclinone D4 and simocyclinone D8 (SD8), were isolated from the mycelium extract of Streptomyces antibioticus Tü 6040 (8, 13, 30, 38). They were shown to exhibit antiproliferative activity on gram-positive bacteria as well as on cancer cell lines (26, 29). In particular, SD8 has been shown to be even more effective than novobiocin at inhibiting gyrase-catalyzed supercoiling (7). Its mechanism of action is thought to involve the prevention of DNA binding by gyrase, and the structure of the N-terminal domain of GyrA (GyrA59) with SD8 bound has recently been solved (6). This novel mode of action suggests that SD8 is a potential new lead molecule for drug design.
In previous work, we utilized protein melting studies to characterize metal ion structural effects on both DNA gyrase subunits as well as quinolone binding to GyrA (32-34). In the study described here, we used the same experimental approach, along with activity assays and limited proteolysis experiments, as an effective means of monitoring the interaction of SD8 with DNA gyrase with the principal aim of locating the drug binding site(s) on the protein. As reference compounds, we used the established gyrase inhibitors ciprofloxacin (CFX), a fluoroquinolone, and novobiocin (Novo), an aminocoumarin. Additionally, we also utilized ADPNP, a nonhydrolyzable analog of ATP, which efficiently binds to the enzyme (Fig. 1). Our results indicate that there are at least two sites for SD8 binding to DNA gyrase: one is located in the A subunit and the other is located in the B subunit.
MATERIALS AND METHODS
Proteins.
All proteins were produced and purified according to previously reported protocols (23, 34). Protein concentrations were assessed from the UV absorbance and by the Bradford method (Bio-Rad).
Drugs.
Ciprofloxacin was provided by Glaxo Wellcome (Verona, Italy). Stock solutions were made in double-distilled water and diluted to the working concentration in the desired buffer. SD8 was kindly provided by L. Heide, University of Tübingen. Stock solutions (≈1.5 mg/ml) were prepared in methanol. The solutions were further diluted with water, and their final concentrations were determined by measurement of the UV absorbance (ɛ at 345 nm = 11,000 M−1 cm−1). ADPNP was purchased from Sigma Chemical Co. (St. Louis, MO).
CD measurements.
Circular dichroism (CD) measurements were performed in 10 mM Tris-HCl (pH 7.4)-20 mM KCl in the presence or the absence MgCl2 ions up to 4 mM. CD spectra were recorded by using 1- to 10-mm-path-length cells on a Jasco J810 spectropolarimeter. For each measurement, three scans were run and recorded with a 1-nm step resolution. The observed ellipticity was converted to the mean residue ellipticity, [θ], in degrees × cm2 × dmol−1. For the thermal denaturation experiments, protein solutions (≈0.01 mg/ml) were equilibrated at 25°C, and then the signal at 220 nm was recorded while the temperature was increased at 0.8°C/min and the protein solution was stirred to allow equilibration. The melting temperature (Tm) was determined by locating the maxima/minima of the first derivative of the curve describing the melting profile (CD versus temperature). The melting temperature experiments were performed in triplicate, and the corresponding curves were practically superimposable.
Proteolysis measurements.
Test proteins (3 μg) were mixed with 0.04 mg of trypsin in 50 mM Tris-HCl (pH 7.5)-50 mM KCl-4 mM MgCl2-4 mM dithiothreitol (DTT)-6.5% glycerol. After various times at 37°C, the reaction was stopped with 1 volume of stop mixture (62 mM Tris-HCl, pH 6.8, 2% sodium dodecyl sulfate [SDS], 10% glycerol, 0.1% bromophenol blue). Alternatively, proteolysis was performed by incubating the test proteins (1 to 3 μg) with thermolysin (0.16 mg/ml) for 30 s in 20 mM Tris-HCl-50 mM NaCl-10 mM CaCl2 in a total volume of 10 μl. At the end of the incubation, samples were heated at 95°C and loaded onto a 12% SDS-polyacrylamide gel. The gels were stained with Coomassie brilliant blue, photographed, and quantified by use of a Bio-Rad Gel Doc 1000 apparatus.
DNA gyrase assays.
Supercoiled plasmid pBR322 (0.125 μg; Fermentas) was incubated with DNA gyrase (60 nM) in a total volume of 20 μl in 35 mM Tris-HCl (pH 7.5)-24 mM KCl-1.8 mM spermidine-2 mM DTT-0.1 mg/ml bovine serum albumin-6.5% (wt/vol) glycerol for 30 min at 37°C in the presence and the absence of increasing drug concentrations. The reaction products were resolved on 1% agarose gels in 0.5×TBE (45 mM Tris, 45 mM boric acid, 1 mM disodium EDTA), and the bands were visualized by ethidium bromide staining and photographed. The relative amounts of the different DNA topoisomers were quantified on the Bio-Rad Gel Doc 1000 apparatus.
Gel shift assay.
A 5′-end-labeled DNA fragment corresponding to the sequence of pBR322 from positions 906 to 1064 was prepared by PCR. Before amplification, one synthetic primer was labeled with [γ-32P]ATP and T4 polynucleotide kinase in the appropriate buffer. The labeled DNA (2,000 cpm) was incubated with different amounts of protein at 25°C in the presence or the absence of SD8 in 50 mM Tris-HCl (pH 7.5)-4 mM MgCl2-55 mM KCl-5 mM DTT containing 5% glycerol. The mixture was incubated for 15 min at 25°C, loaded onto a 5% native polyacrylamide gel (75:1), and run at low voltage (35 V/cm) at 20°C in 90 mM Tris-90 mM boric acid-10 mM MgCl2. The bands were visualized by autoradiography and quantified with a Storm 840 apparatus (GE Healthcare).
RESULTS
SD8 efficiently binds to reconstituted DNA gyrase.
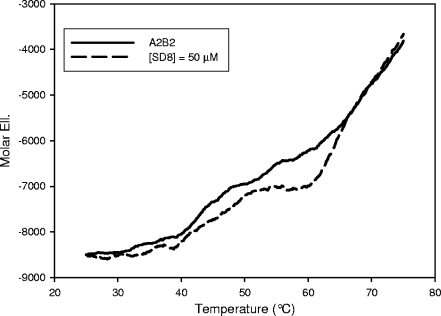
The available data indicate that DNA gyrase is the biological target of SD8 (6, 7, 26). To monitor this interaction, we performed CD measurements using the reconstituted protein. Addition of SD8 did not induce significant changes in the CD spectra in the 200- to 250-nm range. Moreover, no apparent role was played by the presence of Mg2+ up to 4 mM. This points to the absence of extensive protein structural changes as a result of protein complexation. In our previous work, we were able to show gyrase-ligand interactions by monitoring the changes in protein thermal stability. Indeed, when SD8 was added to the reconstituted enzyme, we observed a dramatic shift to higher temperatures in the protein melting transitions (Fig. 2). This shift was a function of the SD8 concentration, thus suggesting the formation of a protein-drug complex. The identification of the protein subdomains involved in the interaction with SD8 could be attempted from the analysis of the melting profiles. However, as we previously pointed out (32), it is not safe to perform such an analysis due to several overlapping transitions. Thus, we preferred to localize drug binding from the changes in the protein structure or thermal stability using each isolated protein subunit/domain.
FIG. 2.
Melting profiles of DNA gyrase acquired by reading the protein CD signal at 220 nm in 10 mM Tris-HCl (pH 7.5)-20 mM KCl-4 mM MgCl2 in the absence (solid line) and in the presence (dashed line) of 50 μM SD8. The protein concentration was 0.05 μM. Mol. Ell., molar ellipticity.
SD8 affects the thermal transitions of both GyrA and GyrB.
The chiroptical properties of GyrA, GyrA59, GyrB, GyrB43, and GyrB47 have recently been investigated (33-34). Additionally, well-characterized ligands are available for GyrA (quinolones) and GyrB (Novo, ADPNP), and we utilized those ligands in our experiments to fully validate our results. In agreement with the behavior of the reconstituted enzyme discussed above, no significant changes in the protein CD spectra occurs upon addition of SD8 or the other ligands tested (data not shown). Again, here we detected drug binding from the changes in protein thermal stability upon addition of putative binders.
(i) GyrA.
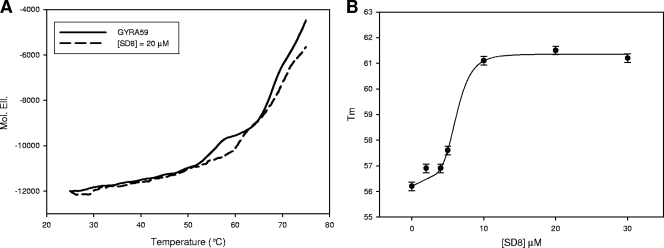
We previously reported that when the GyrA thermal melting profile was recorded in the presence or absence of CFX, a drug-mediated shift to a higher Tm was observed (34). However, neither of the other ligands tested was able to induce an appreciable modification in the thermal stability of GyrA. By using GyrA59 as a substrate, the same was true for ADPNP and Novo (see Fig. S1 in the supplemental material), whereas addition of SD8 produces a concentration-dependent increment in the protein Tm (Fig. 3A). This effect is not very remarkable (maximal change in Tm [ΔTm] ≈ 5.5°C), but the maximal variation occurred at a low drug concentration (10 μM). Additionally, by plotting Tm versus the SD8 concentrations, a sigmoidal curve was obtained, which suggests that a cooperative conformational change was produced upon drug binding (Fig. 3B).
FIG. 3.
SD8 modulation of the thermal stability of GyrA59 in 10 mM Tris-HCl (pH 7.5)-20 mM KCl-4 mM MgCl2. (A) Melting profiles of GyrA59 acquired by reading the protein CD signal at 220 nm in the absence (solid line) and in the presence (dashed line) of SD8. The protein concentration was 0.2 μM. (B) Variation of the Tm for the thermal transition of GyrA59 as a function of SD8 concentration.
(ii) GyrB.
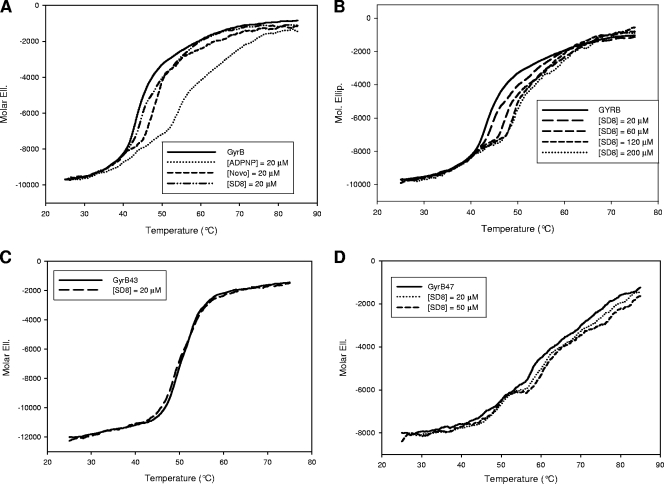
A clear modulation of the GyrB melting profile was observed in the presence of all ligands tested except CFX. In particular, ADPNP, Novo, and SD8 produced a concentration-dependent increase in the Tm of GyrB, indicating a stabilization of the protein structure (Fig. 4A and B). The ligand-GyrB complexes exhibited distinct features in terms of the extent of thermal shift and the metal ion requirement (Fig. 5).
FIG. 4.
Modulation of the melting profiles of GyrB and related subdomains acquired by recording the protein CD signal at 220 nm in 10 mM Tris-HCl (pH 7.5)-20 mM KCl-4 mM MgCl2. (A) Effects of 20 μM ligands on the melting profile of GyrB; (B) effects of increasing SD8 concentrations on the melting profile of GyrB; (C) effects of 20 μM SD8 on the melting profile of GyrB43; (D) melting profile of GyrB47 in the presence or absence of SD8. The protein concentration was 0.2 μM.
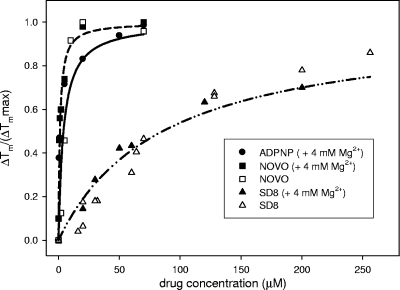
FIG. 5.
Incremental variation of the melting temperature (ΔT/ΔTmax, where ΔT is the change in temperature and ΔTmax is the change in the maximum temperature) for the higher-temperature thermal transition of GyrB as a function of the ligand concentrations. The data refer to the melting profiles acquired by reading the CD signal at 220 nm in 10 mM Tris-HCl (pH 7.5)-20 mM KCl in the presence (full symbols) or the absence (empty symbols) of 4 mM MgCl2. In the presence of ADPNP, it was not possible to calculate this value when Mg2+ was not included in the buffer (see Fig. S1 in the supplemental material).
Interestingly, the temperature required to unfold the protein in the presence of ADPNP was strictly dependent upon Mg2+ (see Fig. S2 in the supplemental material). Indeed, under our experimental conditions, a maximal shift in the Tm was not observed in the absence of the divalent metal ion, thus supporting Mg2+-mediated ligand-protein interactions. In contrast, the interaction of SD8 and Novo with the protein was not sensitive to the presence or absence of Mg2+ in solution. This suggests a lack of any direct interaction between these drugs and divalent ions (data not shown). The difference in ΔTm values recorded under the two conditions depends on the divalent metal ion effect on the protein melting profile (33). The ranking order for GyrB stabilization was Novo ≥ ADPNP > SD8 (Fig. 5).
To identify the protein domain(s) actually involved in the drug binding process, similar experiments were performed with the GyrB N- and C-terminal domains, GyrB43 and GyrB47, respectively. Interestingly, SD8 did not significantly affect the GyrB43 thermal transitions (Fig. 4C), whereas it effectively stabilized the native conformation of GyrB47 (Fig. 4D).
SD8 does not share its binding site(s) with coumarins.
The data presented above show that there are distinct patterns of drug-mediated stabilization of each gyrase subunit. To further map possible overlaps of binding sites, we examined the protein melting profile obtained in the simultaneous presence of two of the ligands tested. Typical examples are reported in Fig. 6, in which the effects of the presence of two competitors (ADPNP and Novo; Fig. 6A) on GyrB are compared to those exhibited by two noncompetitive binders (ADPNP and SD8 [Fig. 6B] and Novo and SD8 [Fig. 6C]).
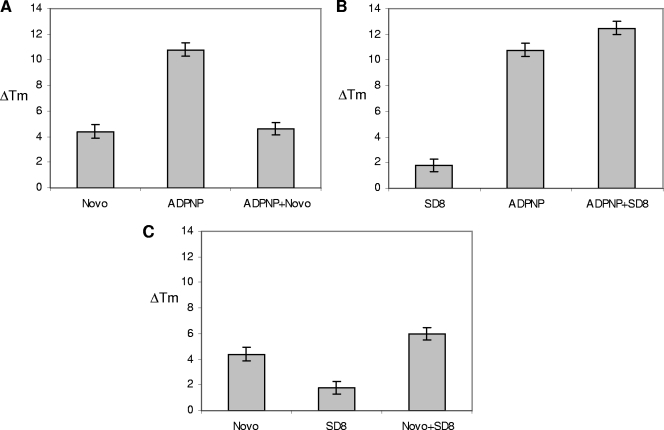
FIG. 6.
Increments of GyrB melting temperature (ΔT) resulting from the protein CD signal at 220 nm in 10 mM Tris-HCl (pH 7.5)-20 mM KCl-4 mM MgCl2 in the presence or the absence of different ligand (20 μM) combinations, as indicated.
When Novo was added to the GyrB-ADPNP complex, a displacement of the nonhydrolyzable ATP analogue was observed. Indeed, the protein melting profile recorded in the presence of the two ligands was essentially identical to the one corresponding to the GyrB-Novo complex. Experiments performed with a constant ADPNP concentration in the presence of various amounts of Novo showed that nucleotide displacement was directly related to the coumarin concentration and that it was complete at a 1:1 ADPNP/Novo ratio. In contrast, when ADPNP was added to the GyrB-SD8 complex, a GyrB thermal shift larger than the shifts obtained in the presence of the same concentrations of the single ligands was observed, which indicates an additive effect. The same was true for the Novo-SD8 system. In fact, by monitoring the effect of SD8 on the Novo-GyrB complex we were not able to observe coumarin displacement using up to a 10-fold excess of SD8, a range that includes equipotent drug concentrations. It should be noted that CFX was not able to displace any of the other ligands tested on GyrB. In fact, the melting profile of the ligand-enzyme mixture was not affected by the presence of the quinolone. The complete set of results is summarized in Table S1 in the supplemental material.
SD8 exhibits a unique GyrB proteolysis pattern.
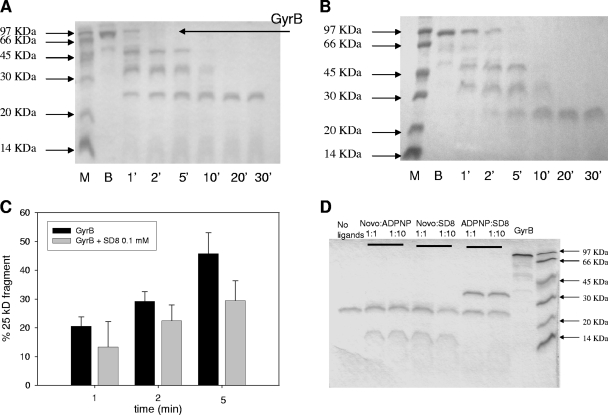
To further explore the GyrB-drug binding process, we analyzed the GyrB proteolysis pattern produced by trypsin and thermolysin in the presence or the absence of the drugs tested. Previous work showed that the tryptic digestion of GyrB initially produces the 43- and 47-kDa domains. GyrB43 is subsequently cleaved to a 33-kDa product that corresponds to the N-terminal domain, which is further extensively digested, whereas the 47-kDa product is rapidly digested to produce a main 25-kDa fragment that corresponds to the C-terminal sequence (2, 14).
A kinetic analysis of the GyrB digestion pattern in the presence of the test drugs showed a modulation of the proteolytic pattern (Fig. 7A and B). In the presence of all test derivatives, the rate of disappearance of the full-length enzyme to produce subdomains GyrB43 and GyrB47 remained almost unchanged (see Fig. S3 in the supplemental material). This further supports the lack of extensive protein rearrangement, as shown by CD titrations. ADPNP stabilized the 33-kDa N-terminal domain, as evidenced by the presence of this fragment even after 30 min of incubation. Novo led to the formation of a 13-kDa fragment that, again, derives from digestion of the 43-kDa N-terminal domain (2, 14). Neither ADPNP nor Novo affected the digestion of the 47-kDa C-terminal domain; however, in the presence of SD8, the appearance of the 25-kDa proteolytic fragment was postponed. Although this effect was subtle, it supports a drug interaction with the C-terminal domain. A quantitative evaluation of the effects of SD8 on GyrB is reported in Fig. 7C.
FIG. 7.
Kinetics of GyrB (3 μg) cleavage by trypsin in the absence (A) or the presence (B) of 100 μM SD8. Lanes M, molecular mass markers; lanes B, GyrB in the absence of trypsin. (C) Quantitative determination of the 25 kDa GyrB fragment formation; (D) GyrB (3 μg) cleavage patterns after 30 min of incubation with trypsin in the absence or the presence of the ligand (0.1 or 1 mM) combinations.
When the protein was incubated with two-ligand combinations, SD8 did not affect the typical cleavage pattern produced by ADPNP or Novo up to a 10-fold molar excess, consistent with the presence of distinct binding sites on the B subunit (Fig. 7D). Comparable results (a distinct proteolytic pattern and no competition between SD8 and the other ligands) were observed by using thermolysin as the cleaving enzyme (data not shown).
Inhibition of gyrase by SD8 is not affected by the presence of Novo or ADPNP.
All ligands tested are known to exhibit different mechanisms of interference with the catalytic activity of DNA gyrase. In particular, ADPNP and Novo compete for ATP binding and hydrolysis; hence, they efficiently reduce enzyme-mediated DNA supercoiling, whereas they do not significantly affect the enzymatic DNA relaxation activity (36). In contrast, SD8 efficiently inhibits gyrase-mediated DNA relaxation and supercoiling (7, 26). As a consequence, starting from a supercoiled plasmid, the efficiency of the relaxation process is related only to the activity of SD8, with the other ligands not contributing to the modulation of this reaction. These differences in the mechanisms of action allowed us to investigate the effects produced by drug combinations. In particular, we evaluated whether ADPNP and Novo could reduce the inhibitory activity of SD8. By applying these experimental protocols, we never observed a significant reduction in the inhibitory activity of SD8 as a function of the presence of Novo or ADPNP in solution (see Fig. S4 in the supplemental material).
SD8 prevents DNA from binding to DNA gyrase.
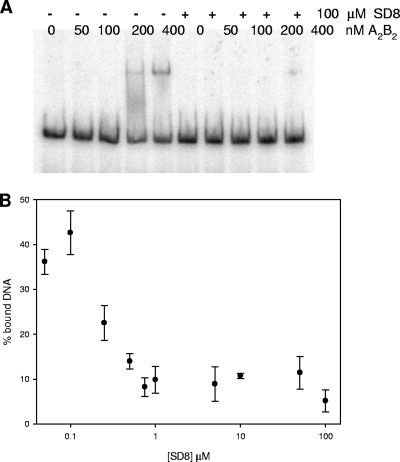
Previous studies indicated that SD8 impairs gyrase binding to DNA (7). To confirm this, we determined the extent of Gyr-DNA complex formation by a gel shift assay (Fig. 8) using a pBR322 fragment (161 bp) containing the known gyrase hot spot at position 990 (18). When SD8 was added to the Gyr-DNA reaction mixture, the formation of the DNA-protein complex was prevented in a drug concentration-dependent fashion. Interestingly, the amount of simocyclinone required to abolish DNA binding to the enzyme was in the low micromolar range, which is fully comparable to the amount necessary to suppress enzymatic activity in vitro. Spectroscopic evidence suggests that this effect is not due to a binary interaction of the drug with DNA (data not shown). This indicates that the SD8-Gyr complex is unable to appropriately recognize its DNA substrate.
FIG. 8.
Effect of SD8 on gyrase-DNA complex formation. DNA was incubated with increasing protein concentrations in the presence or the absence of 100 μM SD8 (A). (B) The percentage of DNA bound by DNA gyrase (400 nM) as a function of the SD8 concentration in the reaction mixture. The buffer was made up of 10 mM Tris-HCl (pH 7.5), 20 mM KCl, and 4 mM MgCl2.
DISCUSSION
Recently, simocyclinone D8 has attracted significant interest, as it represents a new antibiotic with an apparently novel mechanism of action that could be exploited for the rational design of potent chemotherapeutic agents (26, 29). In this connection, a detailed understanding of the molecular basis of the interaction of SD8 with its target is worthy of investigation. Recent work suggests that the antibiotic binds to the A subunit of the enzyme (6, 7). The results of our melting experiments with the N-terminal GyrA59 domain are fully consistent with this. An interesting feature of the binding process described above is represented by its S-shaped dependence upon the SD8 concentration (Fig. 3B). This indicates that under our experimental conditions, at least two SD8 molecules interact cooperatively per GyrA subunit. Interestingly, the conformational transition window compares well with the SD8 concentrations needed to impair enzymatic activity, which suggests a correlation between drug-induced conformational change and enzyme inhibition.
Unexpectedly, additional binding of SD8 was seen on GyrB and, specifically, on the GyrB47 domain, which does not contain the ATP- and coumarin-binding sites. Considering that SD8, like Novo, contains an aminocoumarin ring, an overlapping binding site common to the two drugs might have been predicted. However, this is not the case, as confirmed by the lack of reciprocal displacement in competition experiments performed by the use of three different experimental approaches (protein thermal stability, proteolysis, and activity assays). This is consistent with the lack of interference of SD8 with the intrinsic ATPase activity of gyrase (7). It is worth recalling that in Novo the l-noviosyl sugar is implicated in competitive binding with the ATP adenine ring (17). Instead, SD8 contains a d-olivose deoxyhexose function, and additionally, its distance from the aminocoumarin moiety is changed in comparison to that of Novo. These major differences likely justify a change in the drug binding preference. In fact, the protein thermal stability and proteolysis assay data indicate that the SD8 binding site is totally unprecedented. A previous investigation failed to find evidence for an interaction of SD8 with GyrB (7). Indeed, surface plasmon resonance titrations were performed by using the GyrB43 domain only, which we confirmed did not contain any drug-binding sites. Isothermal titration calorimetry (ITC) titrations were performed with the full-length GyrB subunit. In this case, the lack of apparent binding might be related to a low reaction enthalpy. Additionally, at the high protein concentration required by the ITC technique (16 to 22 μM), GyrB aggregation can occur (4), conceivably preventing drug recognition.
The two SD8 target domains on gyrase identified (GyrA59 and GyrB47) are known to be involved in DNA binding and protein-protein interactions. Although we do not know the exact location of the drug within GyrB47, the interaction could occur at the protein-protein interface, likely affecting the enzyme's local structure to render binding of DNA more difficult. Accordingly, experimental evidence confirmed that addition of the drug interferes with A2B2 recognition of DNA, thus impairing the proper generation of the cleavage complex and progression of the catalytic cycle.
However, the two sites are not equally effective in drug binding, the GyrA site exhibits a substantially higher affinity for SD8, as indicated by the melting experiments. At the moment, we do not have evidence indicating whether these two binding sites might be simultaneously operative. In any event, the assembly of GyrA and GyrB in the active form could possibly help to generate a more stable drug-protein complex. In this connection, protein thermal stabilization by SD8 is more notable on the reconstituted enzyme than on the isolated constituent subunits (Fig. 2 to 4), which suggests the potential cooperation of the two interaction events.
In conclusion, SD8 is not only characterized by a novel mechanism of action, but it may also represent the first DNA gyrase inhibitor directed toward two different target sites. The fact that simocyclinone is also active against cancer cell lines and inhibits human topoisomerase II (29) suggests that its mode of action is shared among several members of this family of enzymes, which gives an opportunity to better understand the general features of drug-mediated enzyme poisoning and inhibition, as well as to rationally design new active drug prototypes.
Supplementary Material
Acknowledgments
We gratefully acknowledge the financial support from the University of Padova (grant CPDA078422/07 to C.S.) and by AIRC, Milan, Italy (to M.P.). Work in A.M.'s laboratory was supported by the BBSRC (United Kingdom).
Footnotes
Published ahead of print on 26 October 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Ali, J. A., A. P. Jackson, A. J. Howells, and A. Maxwell. 1993. The 43-kilodalton N-terminal fragment of the DNA gyrase B protein hydrolyzes ATP and binds coumarin drugs. Biochemistry 32:2717-2724. [DOI] [PubMed] [Google Scholar]
- 2.Ali, J. A., G. Orphanides, and A. Maxwell. 1995. Nucleotide binding to the 43-kilodalton N-terminal fragment of the DNA gyrase B protein. Biochemistry 34:9801-9808. [DOI] [PubMed] [Google Scholar]
- 3.Appelbaum, P. C., and P. A. Hunter. 2000. The fluoroquinolone antibacterials: past, present and future perspectives. Int. J. Antimicrob. Agents 16:5-15. [DOI] [PubMed] [Google Scholar]
- 4.Blandamer, M. J., B. Briggs, P. M. Cullis, A. P. Jackson, A. Maxwell, and R. J. Reece. 1994. Domain structure of Escherichia coli DNA gyrase as revealed by differential scanning calorimetry. Biochemistry 33:7510-7516. [DOI] [PubMed] [Google Scholar]
- 5.Drlica, K., M. Malik, R. J. Kerns, and X. Zhao. 2008. Quinolone-mediated bacterial death. Antimicrob. Agents Chemother. 52:385-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards, M. J., R. H. Flatman, L. A. Mitchenall, C. E. Stevenson, T. B. K. Le, H. P. Fiedler, A. R. McKay, T. A. Clarke, M. J. Buttner, D. M. Lawson, and A. Maxwell. A crystal structure of the bifunctional antibiotic, simocyclinone D8, bound to DNA gyrase. Science, in press. [DOI] [PubMed]
- 7.Flatman, R. H., A. J. Howells, L. Heide, H. P. Fiedler, and A. Maxwell. 2005. Simocyclinone D8, an inhibitor of DNA gyrase with a novel mode of action. Antimicrob. Agents Chemother. 49:1093-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galm, U., J. Schimana, H. P. Fiedler, J. Schmidt, S. M. Li, and L. Heide. 2002. Cloning and analysis of the simocyclinone biosynthetic gene cluster of Streptomyces antibioticus Tu 6040. Arch. Microbiol. 178:102-114. [DOI] [PubMed] [Google Scholar]
- 9.Gellert, M., K. Mizuuchi, M. H. O'Dea, and H. A. Nash. 1976. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl. Acad. Sci. U. S. A. 73:3872-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gellert, M., K. Mizuuchi, M. H. O'Dea, H. Ohmori, and J. Tomizawa. 1979. DNA gyrase and DNA supercoiling. Cold Spring Harbor Symp. Quant. Biol. 43(Pt 1):35-40. [DOI] [PubMed] [Google Scholar]
- 11.Gormley, N. A., G. Orphanides, A. Meyer, P. M. Cullis, and A. Maxwell. 1996. The interaction of coumarin antibiotics with fragments of DNA gyrase B protein. Biochemistry 35:5083-5092. [DOI] [PubMed] [Google Scholar]
- 12.Heddle, J. G., F. M. Barnard, L. M. Wentzell, and A. Maxwell. 2000. The interaction of drugs with DNA gyrase: a model for the molecular basis of quinolone action. Nucleosides Nucleotides Nucleic Acids 19:1249-1264. [DOI] [PubMed] [Google Scholar]
- 13.Holzenkampfer, M., M. Walker, A. Zeeck, J. Schimana, and H. P. Fiedler. 2002. Simocyclinones, novel cytostatic angucyclinone antibiotics produced by Streptomyces antibioticus Tu 6040 II. Structure elucidation and biosynthesis. J. Antibiot. (Tokyo) 55:301-307. [DOI] [PubMed] [Google Scholar]
- 14.Kampranis, S. C., and A. Maxwell. 1998. Conformational changes in DNA gyrase revealed by limited proteolysis. J. Biol. Chem. 273:22606-22614. [DOI] [PubMed] [Google Scholar]
- 15.Laponogov, I., M. K. Sohi, D. A. Veselkov, X. S. Pan, R. Sawhney, A. W. Thompson, K. E. McAuley, L. M. Fisher, and M. R. Sanderson. 2009. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat. Struct. Mol. Biol. 16:667-669. [DOI] [PubMed] [Google Scholar]
- 16.Lee, C. 2008. Therapeutic challenges in the era of antibiotic resistance. Int. J. Antimicrob. Agents 32(Suppl. 4):S197-S199. [DOI] [PubMed] [Google Scholar]
- 17.Lewis, R. J., O. M. Singh, C. V. Smith, T. Skarzynski, A. Maxwell, A. J. Wonacott, and D. B. Wigley. 1996. The nature of inhibition of DNA gyrase by the coumarins and the cyclothialidines revealed by X-ray crystallography. EMBO J. 15:1412-1420. [PMC free article] [PubMed] [Google Scholar]
- 18.Lockshon, D., and D. R. Morris. 1985. Sites of reaction of Escherichia coli DNA gyrase on pBR322 in vivo as revealed by oxolinic acid-induced plasmid linearization. J. Mol. Biol. 181:63-74. [DOI] [PubMed] [Google Scholar]
- 19.Maxwell, A. 1999. DNA gyrase as a drug target. Biochem. Soc. Trans. 27:48-53. [DOI] [PubMed] [Google Scholar]
- 20.Maxwell, A. 1993. The interaction between coumarin drugs and DNA gyrase. Mol. Microbiol. 9:681-686. [DOI] [PubMed] [Google Scholar]
- 21.Maxwell, A., and D. M. Lawson. 2003. The ATP-binding site of type II topoisomerases as a target for antibacterial drugs. Curr. Top. Med. Chem. 3:283-303. [DOI] [PubMed] [Google Scholar]
- 22.Mulvey, M. R., and A. E. Simor. 2009. Antimicrobial resistance in hospitals: how concerned should we be? CMAJ 180:408-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noble, C. G., and A. Maxwell. 2002. The role of GyrB in the DNA cleavage-religation reaction of DNA gyrase: a proposed two metal-ion mechanism. J. Mol. Biol. 318:361-371. [DOI] [PubMed] [Google Scholar]
- 24.Nollmann, M., N. J. Crisona, and P. B. Arimondo. 2007. Thirty years of Escherichia coli DNA gyrase: from in vivo function to single-molecule mechanism. Biochimie 89:490-499. [DOI] [PubMed] [Google Scholar]
- 25.Oblak, M., M. Kotnik, and T. Solmajer. 2007. Discovery and development of ATPase inhibitors of DNA gyrase as antibacterial agents. Curr. Med. Chem. 14:2033-2047. [DOI] [PubMed] [Google Scholar]
- 26.Oppegard, L. M., B. L. Hamann, K. R. Streck, K. C. Ellis, H. P. Fiedler, A. B. Khodursky, and H. Hiasa. 2009. In vivo and in vitro patterns of the activity of simocyclinone D8, an angucyclinone antibiotic from Streptomyces antibioticus. Antimicrob. Agents Chemother. 53:2110-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palumbo, M., B. Gatto, G. Zagotto, and G. Palu. 1993. On the mechanism of action of quinolone drugs. Trends Microbiol. 1:232-235. [DOI] [PubMed] [Google Scholar]
- 28.Reece, R. J., and A. Maxwell. 1991. DNA gyrase: structure and function. Crit. Rev. Biochem. Mol. Biol. 26:335-375. [DOI] [PubMed] [Google Scholar]
- 29.Sadiq, A. A., M. R. Patel, B. A. Jacobson, M. Escobedo, K. Ellis, L. M. Oppegard, H. Hiasa, and R. A. Kratzke. 10 January 2009, posting date. Anti-proliferative effects of simocyclinone D8 (SD8), a novel catalytic inhibitor of topoisomerase II. Invest. New Drugs [Epub ahead of print.]. [DOI] [PMC free article] [PubMed]
- 30.Schimana, J., H. P. Fiedler, I. Groth, R. Sussmuth, W. Beil, M. Walker, and A. Zeeck. 2000. Simocyclinones, novel cytostatic angucyclinone antibiotics produced by Streptomyces antibioticus Tu 6040. I. Taxonomy, fermentation, isolation and biological activities. J. Antibiot. (Tokyo) 53:779-787. [DOI] [PubMed] [Google Scholar]
- 31.Schoeffler, A. J., and J. M. Berger. 2008. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 41:41-101. [DOI] [PubMed] [Google Scholar]
- 32.Sissi, C., A. Chemello, E. Vazquez, L. A. Mitchenall, A. Maxwell, and M. Palumbo. 2008. DNA gyrase requires DNA for effective two-site coordination of divalent metal ions: further insight into the mechanism of enzyme action. Biochemistry 47:8538-8545. [DOI] [PubMed] [Google Scholar]
- 33.Sissi, C., E. Marangon, A. Chemello, C. G. Noble, A. Maxwell, and M. Palumbo. 2005. The effects of metal ions on the structure and stability of the DNA gyrase B protein. J. Mol. Biol. 353:1152-1160. [DOI] [PubMed] [Google Scholar]
- 34.Sissi, C., E. Perdona, E. Domenici, A. Feriani, A. J. Howells, A. Maxwell, and M. Palumbo. 2001. Ciprofloxacin affects conformational equilibria of DNA gyrase A in the presence of magnesium ions. J. Mol. Biol. 311:195-203. [DOI] [PubMed] [Google Scholar]
- 35.Song, J. H. 2008. What's new on the antimicrobial horizon? Int. J. Antimicrob. Agents 32(Suppl. 4):S207-S213. [DOI] [PubMed] [Google Scholar]
- 36.Sugino, A., N. P. Higgins, P. O. Brown, C. L. Peebles, and N. R. Cozzarelli. 1978. Energy coupling in DNA gyrase and the mechanism of action of novobiocin. Proc. Natl. Acad. Sci. U. S. A. 75:4838-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugino, A., C. L. Peebles, K. N. Kreuzer, and N. R. Cozzarelli. 1977. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc. Natl. Acad. Sci. U. S. A. 74:4767-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Theobald, U., J. Schimana, and H. P. Fiedler. 2000. Microbial growth and production kinetics of Streptomyces antibioticus Tu 6040. Antonie Van Leeuwenhoek 78:307-313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.