Abstract
CEM-101 had MIC ranges of 0.002 to 0.016 μg/ml against macrolide-susceptible pneumococci and 0.004 to 1 μg/ml against macrolide-resistant phenotypes. Only 3 strains with erm(B), with or without mef(A), had CEM-101 MICs of 1 μg/ml, and 218/221 strains had CEM-101 MICs of ≤0.5 μg/ml. CEM-101 MICs were as much as 4-fold lower than telithromycin MICs against all strains. For Streptococcus pyogenes, CEM-101 MICs ranged from 0.008 to 0.03 μg/ml against macrolide-susceptible strains and from 0.015 to 1 μg/ml against macrolide-resistant strains. Against erm(B) strains, erythromycin, azithromycin, and clarithromycin MICs were 32 to >64 μg/ml, while 17/19 strains had telithromycin MICs of 4 to 16 μg/ml; CEM-101 MICs were 0.015 to 1 μg/ml. By comparison, erm(A) and mef(A) strains had CEM-101 MICs of 0.015 to 0.5 μg/ml, clindamycin and telithromycin MICs of ≤1 μg/ml, and erythromycin, azithromycin, and clarithromycin MICs of 0.5 to >64 μg/ml. Pneumococcal multistep resistance studies showed that although CEM-101 yielded clones with higher MICs for all eight strains tested, seven of eight strains had clones with CEM-101 MICs that rose from 0.004 to 0.03 μg/ml (parental strains) to 0.06 to 0.5 μg/ml (resistant clones); for only one erm(B) mef(A) strain with a parental MIC of 1 μg/ml was there a resistant clone with a MIC of 32 μg/ml, with no detectable mutations in the L4, L22, or 23S rRNA sequence. Among two of five S. pyogenes strains tested, CEM-101 MICs rose from 0.03 to 0.25 μg/ml, and only for the one strain with erm(B) did CEM-101 MICs rise from 1 to 8 μg/ml, with no changes occurring in any macrolide resistance determinant. CEM-101 had low MICs as well as low potential for the selection of resistant mutants, independent of bacterial species or resistance phenotypes in pneumococci and S. pyogenes.
Strains of Streptococcus pneumoniae resistant to macrolides, β-lactams, quinolones, and other agents are seen worldwide. Macrolide resistance is now predominant in some countries, such as Japan and Korea, most likely due to overuse of azithromycin and clarithromycin during the past 15 years. Macrolide resistance usually also occurs (although genetically unlinked) together with penicillin G resistance (8, 9, 22). Although all strains of group A streptococci remain β-lactam susceptible, macrolide resistance occurs, especially in Southern, Central, and Eastern Europe and Asia (15, 22, 23).
Although the pediatric conjugate vaccine has dramatically decreased the incidence of meningitis and bacteremia caused by most of the usual drug-resistant pneumococcal clones, recent papers have described the spread of multidrug-resistant pneumococcal strains with a serotype (19A), not included in the vaccine, which causes otitis media that is not amenable to treatment with any currently available Food and Drug Administration-approved antibiotic (9, 19, 24). The problem of drug-resistant pneumococci causing community-acquired respiratory infection, especially in children, is likely to worsen with the spread of this clone.
The introduction of telithromycin into the therapeutic armamentarium was, with the exception of erm(B) group A streptococci (which are naturally telithromycin resistant), intended to solve the problem of macrolide resistance in streptococci (2, 21, 22). However, safety issues have limited the clinical utility of this drug. Additionally, when the free area under the curve (AUC)/MIC ratio of telithromycin against macrolide-resistant pneumococci was examined carefully, even with low MICs, it could be seen that the number was not significantly above 25; thus, resistance was predicted, and this has indeed been the case, as evidenced by recent publications (25).
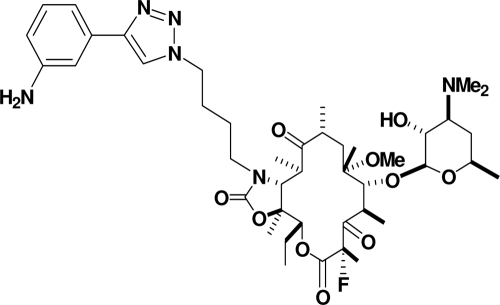
CEM-101 (Fig. 1) is a novel fluoroketolide containing an 11,12-carbamate-butyl-[1,2,3]-triazolyl-aminophenyl side chain. CEM-101 demonstrates enhanced potency compared to telithromycin, with activity against telithromycin-intermediate and telithromycin-resistant organisms (1, 7, 10-14, 17,18, 20, 26-29). In the current study, we have performed (i) MIC studies to compare the activity of CEM-101 to those of erythromycin, azithromycin, clarithromycin, telithromycin, clindamycin, penicillin G, amoxicillin-clavulanate, levofloxacin, and moxifloxacin against a spectrum of pneumococci and group A streptococci with different macrolide resistance phenotypes and genotypes, and (ii) single and multistep resistance studies to examine the ability of CEM-101 to select for resistant mutants of pneumococci and group A streptococci compared to those of telithromycin, azithromycin, clarithromycin, and clindamycin.
FIG. 1.
Structure of CEM-101.
MATERIALS AND METHODS
Bacteria and antimicrobials.
We tested 221 clinical pneumococcal strains by MIC testing. These comprised 50 macrolide-susceptible and 171 macrolide-resistant organisms. Macrolide-resistant strains all had defined genotypes and comprised strains with erm(B) (54 strains), mef(A) (51 strains), erm(B) plus mef(A) (31 strains), erm(A) (4 strains), and mutations in the L4 ribosomal protein (27 strains) and 23S rRNA (4 strains). These 221 strains also comprised 27 non-quinolone-susceptible phenotypes with defined quinolone resistance determinant regions (QRDRs) (levofloxacin MICs, 4 to 32 μg/ml) and the entire spectrum of penicillin G resistance phenotypes according to the latest Clinical and Laboratory Standards Institute (CLSI) oral penicillin V susceptibility classification (4). The 124 group A streptococci for which MICs were determined comprised 26 macrolide-susceptible and 98 macrolide-resistant strains. The latter comprised 19 strains with erm(B), 38 with mef(A), 40 with erm(A), and 1 with an L4 mutation. Because strains of both species studied were chosen for their macrolide resistance phenotypes, only susceptible strains were consistently recent (2003 to 2008) isolates; some resistant strains were isolated up to 5 years earlier (1998). Streptococcus pneumoniae ATCC 49619 was included as the quality control strain for each species and each run (5).
For resistance selection testing, one each of the following pneumococcal resistance phenotypes was tested: macrolide susceptible, erm(B) positive, mef(A) positive, erm(B) and mef(A) positive, erm(A) positive, and with mutations in ribosomal proteins (L4, L22) and 23S rRNA. Five strains of group A streptococci were tested; one each was macrolide susceptible, erm(B) positive, mef(A) positive, erm(A) positive, or had an L4 mutation. CEM-101 was obtained from Cempra Pharmaceuticals, Chapel Hill, NC, and other drugs were obtained either from their respective manufacturers or from Sigma Chemical, Inc., St. Louis, MO.
MIC determinations.
MICs were determined by the agar dilution technique, which, though not specifically recommended by the CLSI (5), has been in use in our research laboratory for >20 years (3, 6, 16). Mueller-Hinton agar (BD Diagnostics, Sparks, MD) supplemented with 5% sheep blood agar was used, with 104 CFU/spot and overnight incubation at 35°C in ambient air. The usual quality control strains were included in each run (5). For resistance selection, CLSI macrodilution (5) was used for MIC testing.
Mechanism of macrolide resistance.
All macrolide-resistant parental strains were tested for the presence of the erm(B), erm(A), mef(E), and mef(A) genes by PCR amplification (3, 5, 21, 22). All parental isolates and CEM-101-resistant clones (CEM-101 MIC, >1 μg/ml) were examined for the presence of mutations in the L4 and L22 ribosomal proteins and 23S rRNA (II and V domains) by using the primers and conditions described previously (2, 3, 6, 16, 21, 22). The nucleotide sequences were obtained by direct sequencing with a CEQ8000 genetic analysis system (Beckman Coulter, Fullerton, CA).
Multistep resistance selection.
Serial passages of each strain were performed daily in subinhibitory concentrations of all antimicrobials. In all cases, the broth medium was 1 ml per tube of cation-adjusted Mueller-Hinton broth (BD Diagnostics, Sparks, MD) plus 5% lysed horse blood. For each subsequent daily passage, an inoculum (10 μl) was taken from the tube at 1 to 2 dilutions below the MIC that matched the turbidity of a growth control tube. This inoculum was used to determine the next MIC. Daily passages were performed until a significant increase in the MIC (≥8 times) was obtained. A minimum of 14 passages were performed unless MICs of ≥32 μg/ml were obtained. The maximal number of passages was 50. The stability of the acquired resistance was ascertained by MIC determinations after 10 daily passages of the mutants on blood agar without antibiotics. The MIC of each compound for each resistant pneumococcal clone was determined by macrodilution. The identities of the mutants obtained and their respective parents were confirmed by pulsed-field gel electrophoresis (PFGE) at the end of the study. PFGE of SmaI-digested DNA was performed using a CHEF DR III apparatus (Bio-Rad, Hercules, CA) with the following run parameters: a switch time of 5 to 20 s and a run time of 16 h (3, 6, 16).
Single-step studies.
The frequency of spontaneous single-step mutations was determined by spreading suspensions (approximately 1010 CFU/ml) on Mueller-Hinton agar (BD Diagnostics, Sparks, MD) with 5% sheep blood at 2, 4, and 8 times the MIC (2×, 4×, and 8× MIC). After incubation at 35°C under 5% CO2 for 48 h, the frequency of resistance was calculated as the number of colonies per inoculum for which the MIC was at least 4 times higher than the MIC for the parental strain. Single-step studies were not performed with azithromycin, clarithromycin, clindamycin, and telithromycin for strains with MICs of ≥4 μg/ml (3, 6, 16).
RESULTS
The results of pneumococcal MIC testing are presented in Tables 1 and 2. As can be seen, CEM-101 MICs ranged from 0.002 to 0.015 μg/ml against macrolide-susceptible pneumococci and from 0.004 to 1 μg/ml against macrolide-resistant pneumococci (all phenotypes). Only 3 strains with erm(B) [with or without mef(A)] had CEM-101 MICs of 1.0 μg/ml, and 218/221 strains had CEM-101 MICs of ≤0.5 μg/ml. In contrast, corresponding telithromycin MICs ranged from 0.015 to 0.03 μg/ml for macrolide-susceptible strains and from 0.015 to 2 μg/ml for macrolide-resistant strains. CEM-101 MICs were as much as fourfold lower than telithromycin MICs against macrolide-susceptible and -resistant strains.
TABLE 1.
MICs of drugs against 221 pneumococcal strains
| Drug and type of strain testeda | MIC (μg/ml)b |
||
|---|---|---|---|
| Range | 50% | 90% | |
| Penicillin G | 0.008->16 | 1 | 4 |
| Penicillin S | 0.008-0.06 | 0.03 | 0.06 |
| Penicillin I | 0.125-1 | 0.5 | 1 |
| Penicillin R | 2->16 | 4 | 8 |
| Macrolide S | 0.015-8 | 1 | 2 |
| erm(B) | 0.03-16 | 1 | 4 |
| mef(A) | 0.008-4 | 0.125 | 4 |
| erm(A) | 0.03-0.03 | ||
| erm(B) mef(A) | 0.03-8 | 2 | 4 |
| L4 mutations | 1->16 | 4 | 16 |
| 23S rRNA mutations | 0.015-0.5 | ||
| Quinolone S | 0.008->16 | 1 | 4 |
| Quinolone R | 0.015-8 | 0.25 | 4 |
| CEM-101 | 0.002-1 | 0.03 | 0.25 |
| Penicillin S | 0.002-0.25 | 0.03 | 0.125 |
| Penicillin I | 0.002-0.25 | 0.03 | 0.25 |
| Penicillin R | 0.004-1 | 0.06 | 0.25 |
| Macrolide S | 0.002-0.015 | 0.008 | 0.015 |
| erm(B) | 0.004-1 | 0.03 | 0.5 |
| mef(A) | 0.008-0.25 | 0.03 | 0.125 |
| erm(A) | 0.008-0.015 | ----- | ----- |
| erm(B) mef(A) | 0.015-1 | 0.125 | 0.25 |
| L4 mutations | 0.03-0.125 | 0.06 | 0.125 |
| 23S rRNA mutations | 0.002-0.03 | ||
| Quinolone S | 0.002-1 | 0.03 | 0.25 |
| Quinolone R | 0.004-0.25 | 0.008 | 0.06 |
| Erythromycin | 0.03->64 | 64 | >64 |
| Penicillin S | 0.03->64 | 4 | >64 |
| Penicillin I | 0.03->64 | >64 | >64 |
| Penicillin R | 0.03->64 | >64 | >64 |
| Macrolide S | 0.03-0.25 | 0.06 | 0.125 |
| erm(B) | 16->64 | >64 | >64 |
| mef(A) | 1->64 | 4 | 32 |
| erm(A) | 2-4 | ||
| erm(B) mef(A) | 4->64 | >64 | >64 |
| L4 mutations | 4->64 | >64 | >64 |
| 23S rRNA mutations | 8->64 | ||
| Quinolone S | 0.03->64 | >64 | >64 |
| Quinolone R | 0.03->64 | 0.06 | >64 |
| Azithromycin | 0.06->64 | 16 | >64 |
| Penicillin S | 0.06->64 | 4 | >64 |
| Penicillin I | 0.06->64 | >64 | >64 |
| Penicillin R | 0.06->64 | >64 | >64 |
| Macrolide S | 0.06-0.25 | 0.125 | 0.0125 |
| erm(B) | >64->64 | >64 | >64 |
| mef(A) | 1->64 | 4 | 8 |
| erm(A) | 2-8 | ||
| erm(B) mef(A) | 2->64 | >64 | >64 |
| L4 mutations | 2->64 | >64 | >64 |
| 23S rRNA mutations | 32->64 | ||
| Quinolone S | 0.06->64 | >64 | >64 |
| Quinolone R | 0.06->64 | 0.125 | >64 |
| Clarithromycin | 0.125->64 | 8 | >64 |
| Penicillin S | 0.015->64 | 1 | >64 |
| Penicillin I | 0.03->64 | 16 | >64 |
| Penicillin R | 0.015->64 | 16 | >64 |
| Macrolide S | 0.015-0.06 | 0.03 | 0.06 |
| erm(B) | 4->64 | >64 | >64 |
| mef(A) | 0.5-32 | 2 | 4 |
| erm(A) | 0.25-0.5 | ||
| erm(B) mef(A) | 1->64 | >64 | >64 |
| L4 mutations | 1-32 | 16 | 32 |
| 23S rRNA mutations | 8-16 | ||
| Quinolone S | 0.015->64 | 16 | >64 |
| Quinolone R | 0.015->64 | 0.03 | >64 |
| Telithromycin | 0.015-2 | 0.06 | 0.5 |
| Penicillin S | 0.015-1 | 0.06 | 0.25 |
| Penicillin I | 0.015-1 | 0.06 | 0.5 |
| Penicillin R | 0.015-2 | 0.125 | 0.5 |
| Macrolide S | 0.015-0.03 | 0.03 | 0.03 |
| erm(B) | 0.03-2 | 0.06 | 1 |
| mef(A) | 0.03-0.5 | 0.125 | 0.25 |
| erm(A) | 0.03-0.06 | ||
| erm(B) mef(A) | 0.03-2 | 0.5 | 1 |
| L4 mutations | 0.06-0.25 | 0.125 | 0.25 |
| 23S rRNA mutations | 0.03-0.06 | ||
| Quinolone S | 0.015-2 | 0.125 | 0.5 |
| Quinolone R | 0.015-1 | 0.03 | 0.125 |
| Clindamycin | 0.015->64 | 0.06 | >64 |
| Penicillin S | 0.03->64 | 0.06 | >64 |
| Penicillin I | 0.03->64 | 0.125 | >64 |
| Penicillin R | 0.015->64 | 0.06 | >64 |
| Macrolide S | 0.015-0.06 | 0.03 | 0.06 |
| erm(B) | 0.06->64 | >64 | >64 |
| mef(A) | 0.03-0.125 | 0.06 | 0.06 |
| erm(A) | 0.125-0.25 | ||
| erm(B) mef(A) | 0.03->64 | 0.06 | >64 |
| L4 mutations | 0.03-0.125 | 0.06 | 0.125 |
| 23S rRNA mutations | 0.03-1 | ||
| Quinolone S | 0.015->64 | 0.06 | >64 |
| Quinolone R | 0.03-64 | 0.03 | 64 |
| Amoxicillin-clavulanate | 0.015-16 | 0.05 | 8 |
| Penicillin S | 0.015-0.125 | 0.03 | 0.06 |
| Penicillin I | 0.03-2 | 0.5 | 1 |
| Penicillin R | 0.125-16 | 2 | 8 |
| Macrolide S | 0.015-8 | 0.5 | 2 |
| erm(B) | 0.015-8 | 0.5 | 8 |
| mef(A) | 0.015-8 | 0.125 | 2 |
| erm(A) | 0.03-0.03 | ||
| erm(B) mef(A) | 0.03-16 | 2 | 8 |
| L4 mutations | 0.125-8 | 4 | 8 |
| 23S rRNA mutations | 0.03-0.06 | ||
| Quinolone S | 0.015-16 | 1 | 8 |
| Quinolone R | 0.015-4 | 0.5 | 2 |
| Levofloxacin | 0.06-32 | 1 | 8 |
| Penicillin S | 0.06-32 | 1 | 16 |
| Penicillin I | 1-32 | 1 | 2 |
| Penicillin R | 0.5-16 | 1 | 2 |
| Macrolide S | 1-32 | 1 | 16 |
| erm(B) | 0.5-32 | 1 | 2 |
| mef(A) | 0.5-8 | 1 | 2 |
| erm(A) | 1-1 | ||
| erm(B) mef(A) | 1-16 | 1 | 16 |
| L4 mutations | 0.5-16 | 1 | 2 |
| 23S rRNA mutations | 0.06-1 | ||
| Quinolone S | 0.06-2 | 1 | 2 |
| Quinolone R | 4-32 | 16 | 16 |
| Moxifloxacin | 0.125-8 | 0.25 | 2 |
| Penicillin S | 0.125-8 | 0.5 | 4 |
| Penicillin I | 0.125-4 | 0.25 | 0.5 |
| Penicillin R | 0.125-4 | 0.25 | 0.5 |
| Macrolide S | 0.125-8 | 0.25 | 4 |
| erm(B) | 0.125 | 0.25 | 0.5 |
| mef(A) | 0.125-4 | 0.25 | 0.5 |
| erm(A) | 0.25-0.5 | ||
| erm(B) mef(A) | 0.125-2 | 0.25 | 0.5 |
| L4 mutations | 0.125-4 | 0.25 | 0.5 |
| 23S rRNA mutations | 0.25-0.5 | ||
| Quinolone S | 0.125-1 | 0.25 | 0.5 |
| Quinolone R | 0.5-8 | 4 | 4 |
S, susceptible; I, intermediate; R, resistant. A total of 53 penicillin-susceptible, 63 penicillin-intermediate, 105 penicillin-resistant, 50 macrolide-susceptible, 54 erm(B), 51 mef(A), 4 erm(A), and 31 erm(B) mef(A) strains, 27 strains with L4 mutations, 4 strains with 23S rRNA mutations, 195 quinolone-susceptible strains, and 27 quinolone-resistant strains were tested.
50% and 90%, MICs at which 50% and 90% of isolates, respectively, are inhibited.
TABLE 2.
MIC50s and MIC90sa of pneumococcal strains with defined macrolide-resistant mechanisms
| Drug | MIC (μg/ml) for strains with the following macrolide-resistant mechanism (no. of strains): |
|||||||
|---|---|---|---|---|---|---|---|---|
|
erm(B) (54) |
mef(A) (51) |
erm(B) mef(A) (31) |
L4 mutations (27) |
|||||
| MIC50 | MIC90 | MIC50 | MIC90 | MIC50 | MIC90 | MIC50 | MIC90 | |
| CEM-101 | 0.03 | 0.5 | 0.03 | 0.125 | 0.125 | 0.25 | 0.06 | 0.125 |
| Erythromycin | >64 | >64 | 4 | 32 | >64 | >64 | >64 | >64 |
| Azithromycin | >64 | >64 | 4 | 8 | >64 | >64 | >64 | >64 |
| Clarithromycin | >64 | >64 | 2 | 4 | >64 | >64 | 16 | 32 |
| Telithromycin | 0.06 | 1 | 0.125 | 0.25 | 0.5 | 1 | 0.125 | 0.25 |
| Clindamycin | >64 | >64 | 0.06 | 0.06 | 0.06 | >64 | 0.06 | 0.125 |
| Amoxicillin-clavulanate | 0.5 | 8 | 0.125 | 2 | 2 | 8 | 4 | 8 |
| Levofloxacin | 1 | 2 | 1 | 2 | 1 | 16 | 1 | 2 |
| Moxifloxacin | 0.25 | 0.5 | 0.25 | 0.5 | 0.25 | 0.5 | 0.25 | 0.5 |
| Penicillin G | 1 | 4 | 0.125 | 4 | 2 | 4 | 4 | 16 |
MIC50 and MIC90, MICs at which 50% and 90% of isolates, respectively, are inhibited.
All group A streptococcal strains were penicillin G susceptible. MICs are presented in Tables 3 and 4. CEM-101 MICs were 0.008 to 0.03 μg/ml against macrolide-susceptible strains and 0.015 to 1 μg/ml against macrolide-resistant strains (all phenotypes). Telithromycin MICs were as much as fourfold higher than CEM-101 MICs. Importantly, 17/19 erm(B) strains were telithromycin resistant, with MICs between 4 and 16 μg/ml, while all had low CEM-101 MICs, similar to those of strains with other resistance phenotypes (range, 0.03 to 1 μg/ml).
TABLE 3.
MICs of drugs against 124 group A streptococci
| Drug and type of straina | MIC (μg/ml)b |
||
|---|---|---|---|
| Range (μg/ml) | 50% | 90% | |
| CEM-101 | 0.008-1 | 0.06 | 0.5 |
| Macrolide S | 0.008-0.03 | 0.015 | 0.03 |
| erm(B) | 0.03-1 | 0.5 | 1 |
| mef(A) | 0.06-0.25 | 0.125 | 0.25 |
| erm(A) | 0.016-0.5 | 0.03 | 0.125 |
| L4 mutations | 0.06 | ||
| Erythromycin | 0.03->64 | 16 | >64 |
| Macrolide S | 0.03-0.25 | 0.06 | 0.125 |
| erm(B) | >64->64 | >64 | >64 |
| mef(A) | 8-32 | 16 | 32 |
| erm(A) | 2->64 | 4 | >64 |
| L4 mutations | 2 | ||
| Azithromycin | 0.06->64 | 8 | >64 |
| Macrolide S | 0.06-0.25 | 0.125 | 0.25 |
| erm(B) | >64->64 | >64 | >64 |
| mef(A) | 0.5-16 | 8 | 8 |
| erm(A) | 2->64 | 16 | >64 |
| L4 mutations | 2 | ||
| Clarithromycin | 0.015->64 | 4 | >64 |
| Macrolide S | 0.015-0.06 | 0.03 | 0.06 |
| erm(B) | 32->64 | >64 | >64 |
| mef(A) | 0.5-8 | 4 | 8 |
| erm(A) | 0.25->64 | 2 | >64 |
| L4 mutations | 1 | ||
| Telithromycin | 0.03-16 | 0.125 | 8 |
| Macrolide S | 0.03-0.06 | 0.06 | 0.06 |
| erm(B) | 0.03-16 | 8 | 16 |
| mef(A) | 0.125-1 | 0.5 | 1 |
| erm(A) | 0.03-0.25 | 0.06 | 0.125 |
| L4 mutations | 0.06 | ||
| Amoxicillin-clavulanate | <0.015-0.125 | 0.03 | 0.03 |
| Macrolide S | 0.015-0.03 | 0.03 | 0.03 |
| erm(B) | <0.015-0.125 | <0.015 | 0.03 |
| mef(A) | <0.015-0.125 | 0.015 | 0.06 |
| erm(A) | <0.015-0.03 | 0.03 | 0.03 |
| L4 mutations | 0.03 | ||
| Levofloxacin | 0.5-2 | 0.5 | 1 |
| Macrolide S | 0.5-1 | 0.5 | 0.5 |
| erm(B) | 0.5-1 | 0.5 | 1 |
| mef(A) | 0.5-2 | 0.5 | 1 |
| erm(A) | 0.5-2 | 0.5 | 1 |
| L4 mutations | 0.5 | ||
| Moxifloxacin | 0.0125-0.5 | 0.25 | 0.25 |
| Macrolide S | 0.125-0.25 | 0.25 | 0.25 |
| erm(B) | 0.125-0.25 | 0.25 | 0.25 |
| mef(A) | 0.25-0.5 | 0.25 | 0.25 |
| erm(A) | 0.125-0.5 | 0.25 | 0.25 |
| L4 mutations | 0.25 | ||
| Penicillin G | <0.008-0.125 | 0.015 | 0.015 |
| Macrolide S | 0.008-0.015 | 0.015 | 0.015 |
| erm(B) | <0.008-0.125 | 0.015 | 0.015 |
| mef(A) | <0.008-0.125 | 0.015 | 0.03 |
| erm(A) | <0.008-0.015 | 0.015 | 0.015 |
| L4 mutations | 0.015 | ||
| Clindamycin | 0.03->64 | 0.125 | >64 |
| Macrolide S | 0.03-0.125 | 0.06 | 0.06 |
| erm(B) | 0.06->64 | >64 | >64 |
| mef(A) | 0.03-0.125 | 0.06 | 0.125 |
| erm(A) | 0.06-0.5 | 0.125 | 0.25 |
| L4 mutations | 0.06 | ||
S, susceptible. A total of 26 macrolide-susceptible, 19 erm(B), 38 mef(A), and 40 erm(A) strains, as well as 1 strain with L4 mutations, were tested.
50 and 90%, MICs at which 50% and 90% of isolates, respectively, are inhibited.
TABLE 4.
MIC50s and MIC90sa of group A streptococcal strains with defined macrolide-resistant mechanisms
| Drug | MIC (μg/ml) for strains with the following macrolide-resistant mechanism (no. of strains): |
|||||
|---|---|---|---|---|---|---|
|
erm(B) (19) |
mef(A) (38) |
erm(A) (40) |
||||
| MIC50 | MIC90 | MIC50 | MIC90 | MIC50 | MIC90 | |
| CEM-101 | 0.5 | 1 | 0.125 | 0.25 | 0.03 | 0.125 |
| Erythromycin | >64 | >64 | 16 | 32 | 4 | >64 |
| Azithromycin | >64 | >64 | 8 | 8 | 16 | >64 |
| Clarithromycin | >64 | >64 | 4 | 8 | 2 | >64 |
| Telithromycin | 8 | 16 | 0.5 | 1 | 0.06 | 0.125 |
| Clindamycin | >64 | >64 | 0.06 | 0.125 | 0.125 | 0.25 |
| Amoxicillin-clavulanate | <0.015 | 0.03 | 0.015 | 0.06 | 0.03 | 0.03 |
| Levofloxacin | 0.5 | 1 | 0.5 | 1 | 0.5 | 1 |
| Moxifloxacin | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 |
| Penicillin G | 0.015 | 0.015 | 0.015 | 0.03 | 0.015 | 0.015 |
MIC50 and MIC90, MICs at which 50% and 90% of isolates, respectively, are inhibited.
The results of pneumococcal multistep resistance selection studies are presented in Table 5. As can be seen for pneumococci, parental MICs (in micrograms per milliliter) were as follows: CEM-101, 0.004 to 1; azithromycin, 0.03 to 8; clarithromycin, 0.016 to 16; telithromycin, 0.004 to 0.5; clindamycin, 0.016 to 1. Four, two, and two strains with azithromycin, clarithromycin, and clindamycin MICs of ≥64 μg/ml, respectively, were not tested. CEM-101 MICs increased after 14 to 43 days for all eight strains tested. For seven strains, MICs rose from 0.004 to 0.03 μg/ml (parents) to 0.06 to 0.5 μg/ml (resistant clones) in 14 to 43 days. For the eighth strain, containing erm(B) plus mef(A), MICs rose from 1 μg/ml (parent) to 32 μg/ml (resistant clone) in 18 days. This CEM-101-resistant clone was subjected to sequencing analysis, which revealed no alterations from parental sequences in the L4 and L22 proteins and in domains II and V of 23S rRNA. Azithromycin produced resistant clones after 14 to 29 days for three of four strains, with MICs rising from 0.03 to 2 μg/ml (parents) to 0.5 to >64 μg/ml (resistant clones). Clarithromycin produced resistant clones after 14 to 49 days for five of six strains, with MICs rising from 0.03 to 16 μg/ml (parents) to 16 to >64 μg/ml (resistant clones). Telithromycin produced stable resistant clones after 14 to 38 days for five of eight strains tested, with MICs rising from 0.004 to 0.5 μg/ml (parents) to 0.06 to >64 μg/ml (resistant clones). Clindamycin produced resistant clones after 14 to 43 days for two of five strains, with MICs rising from 0.03 to 0.06 μg/ml (parents) to 0.25 to >64 μg/ml (resistant clones).
TABLE 5.
S. pneumoniae multistep selection resultsa
| Strain no. | Phenotype (resistance determinant)a | Drugb | Initial MIC (μg/ml) | Selected resistance |
Retest MICc after passages in subinhibitory concns of the following antibiotic and 10 antibiotic-free subcultures: |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| MIC (μg/ml) | No. of passages | CEM | AZI | CLA | TEL | CLI | ||||
| 1077 | Macrolide S | CEM | 0.008 | 0.06 | 43 | 0.06 | 0.03 | 0.06 | 0.06 | 0.03 |
| AZI | 0.03 | >64 | 29 | 0.03 | >64 | >64 | 0.125 | 4 | ||
| CLA | 0.016 | 0.008 | 50 | |||||||
| TEL | 0.004 | 0.25 | 15 | 0.06 | >64 | >64 | 0.25 | 8 | ||
| CLI | 0.016 | 4 | 49 | 0.06 | >64 | >64 | 0.06 | 8 | ||
| 24 | Macrolide R [erm(B)] | CEM | 0.004 | 0.06 | 14 | 0.06 | >64 | >64 | 0.125 | >64 |
| AZI | >64 | NTd | NT | |||||||
| CLA | >64 | NT | NT | |||||||
| TEL | 0.25 | 32 | 14 | 0.06 | >64 | >64 | 16 | >64 | ||
| CLI | >64 | NT | NT | |||||||
| 3665 | Macrolide R [mef(A)] | CEM | 0.03 | 0.5 | 14 | 0.5 | 16 | 4 | 0.25 | 0.03 |
| AZI | 8 | 16 | 50 | |||||||
| CLA | 2 | 16 | 26 | 0.06 | 8 | 8 | 0.06 | 0.03 | ||
| TEL | 0.125 | 2 | 14 | 0.03 | 4 | 2 | 0.25 | 0.03 | ||
| CLI | 0.016 | 0.03 | 50 | |||||||
| 1076 | Macrolide-R [erm(B) mef(A)] | CEM | 1 | 32 | 18 | 32 | >64 | >64 | 32 | >64 |
| AZI | >64 | NT | NT | |||||||
| CLA | >64 | NT | NT | |||||||
| TEL | 0.5 | >64 | 14 | 2 | >64 | >64 | >64 | >64 | ||
| CLI | 64 | NT | NT | |||||||
| 1635 | Macrolide R [erm(A)] | CEM | 0.008 | 0.06 | 32 | 0.125 | 4 | 1 | 0.03 | 0.03 |
| AZI | 2 | >64 | 14 | 0.004 | >64 | >64 | 0.004 | 0.03 | ||
| CLA | 0.5 | >64 | 49 | 0.008 | >64 | >64 | 0.016 | 0.25 | ||
| TEL | 0.004 | 0.008 | 50 | |||||||
| CLI | 0.06 | >64 | 14 | 0.004 | 4 | 0.5 | 0.008 | >64 | ||
| 2686 | Macrolide R (L4 mutation) | CEM | 0.03 | 0.5 | 22 | 1 | >64 | 32 | 0.25 | 0.03 |
| AZI | >64 | NT | NT | |||||||
| CLA | 8 | >64 | 14 | 0.03 | >64 | >64 | 0.06 | 0.03 | ||
| TEL | 0.06 | 0.5 | 25 | 0.016 | >64 | 16 | 0.5 | 0.03 | ||
| CLI | 0.03 | 0.125 | 50 | |||||||
| 7127 | Macrolide S (S20N in L4, A105V in L22) | CEM | 0.008 | 0.125 | 16 | 0.06 | 0.06 | 0.125 | 0.06 | 0.03 |
| AZI | 0.06 | 0.5 | 29 | 0.016 | 1 | 0.5 | 0.016 | 0.06 | ||
| CLA | 0.03 | 16 | 15 | 0.008 | >64 | 16 | 0.008 | 1 | ||
| TEL | 0.008 | 0.06 | 38 | 0.008 | 0.06 | 0.06 | 0.03 | 0.03 | ||
| CLI | 0.03 | 0.25 | 43 | 0.004 | 0.03 | 0.016 | 0.008 | 0.25 | ||
| 3009 | Macrolide R (23S rRNA mutation) | CEM | 0.016 | 0.25 | 20 | 0.25 | >64 | >64 | 0.06 | 1 |
| AZI | >64 | NT | NT | |||||||
| CLA | 16 | >64 | 25 | 0.03 | >64 | 64 | 0.06 | 1 | ||
| TEL | 0.016 | 0.03 | 50 | |||||||
| CLI | 1 | 2 | 50 | |||||||
S, susceptible; R, resistant.
CEM, CEM-101; AZI, azithromycin; CLA, clarithromycin; TEL, telithromycin; CLI, clindamycin.
Boldface indicates cross-reactivity.
NT, not tested.
For Streptococcus pyogenes (Table 6), parental MICs (μg/ml) were: CEM-101, 0.008 to 1; azithromycin, 0.06 to 4; clarithromycin, 0.03 to 4; telithromycin, 0.008 to 8; clindamycin 0.06. One strain with azithromycin, clarithromycin and clindamycin MICs >64 μg/ml was not tested. CEM-101 MICs increased after 18 to 43 days in 3/5 strains, rising from 0.03 to 1 μg/ml (parents) to 0.25 to 8 μg/ml (resistant clones). The resistant clone with a CEM-101 MIC of 8 μg/ml was subjected to sequencing analysis, which showed no changes in all genes (L4, L22 and II and V domain of 23S rRNA) tested. CEM-101 MICs for the remaining 2 clones did not go above 0.25 μg/ml when passages were continued for the maximum 50 days. Azithromycin had resistant clones after 5 to 35 days in 3/4 strains tested, with MICs rising from 0.06 to 4 μg/ml (parents) to 1 to >64 μg/ml (resistant clones). Clarithromycin had resistant clones after 6 days in 1/4 strains tested, with MICs rising from 0.5 μg/ml (parent) to >64 μg/ml (resistant clone). Telithromycin had resistant clones after 6 to 22 days in 2/5 strains tested, with MICs rising from 0.03 to 8 μg/ml (parents) to 0.25 to >64 μg/ml (resistant clones). Clindamycin had resistant clones after 34 to 43 days in 2/4 strains tested with MICs rising from 0.06 μg/ml (parents) to 0.5 to >64 μg/ml (resistant clones).
TABLE 6.
S. pyogenes multistep selection results
| Strain no. | Phenotype (resistance determinant)a | Antibioticb | Initial MIC (μg/ml) | Selected resistance |
Retest MIC (μg/ml)c after passages in subinhibitory concns of the following antibiotic and 10 antibiotic-free subcultures: |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| MIC (μg/ml) | No. of passages | CEM | AZI | CLA | TEL | CLI | ||||
| 2132 | Macrolide S | CEM | 0.008 | 0.016 | 50 | |||||
| AZI | 0.06 | 1 | 28 | 0.016 | 1 | 0.25 | 0.03 | 0.03 | ||
| CLA | 0.03 | 0.016 | 50 | |||||||
| TEL | 0.008 | 0.03 | 50 | |||||||
| CLI | 0.06 | 0.06 | 50 | |||||||
| 2368 | Macrolide R [erm(B)] | CEM | 1 | 8 | 18 | 8 | >64 | >64 | >64 | >64 |
| AZI | >64 | NTd | NT | |||||||
| CLA | >64 | NT | NT | |||||||
| TEL | 8 | >64 | 6 | 0.5 | >64 | >64 | >64 | >64 | ||
| CLI | >64 | NT | NT | |||||||
| 2094 | Macrolide R [erm(A)] | CEM | 0.03 | 0.25 | 43 | 0.25 | 4 | 8 | 0.5 | 0.06 |
| AZI | 4 | >64 | 5 | 0.016 | >64 | 1 | 0.03 | 0.06 | ||
| CLA | 0.5 | >64 | 6 | 0.016 | >64 | >64 | 0.03 | 0.06 | ||
| TEL | 0.03 | 0.25 | 22 | 0.03 | >64 | 8 | 0.125 | >64 | ||
| CLI | 0.06 | >64 | 34 | 0.03 | 16 | 1 | 0.03 | >64 | ||
| 2011 | Macrolide R [mef(A)] | CEM | 0.125 | 0.125 | 50 | |||||
| AZI | 4 | 32 | 35 | 0.06 | 16 | 4 | 0.25 | 0.06 | ||
| CLA | 4 | 8 | 50 | |||||||
| TEL | 0.5 | 1 | 50 | |||||||
| CLI | 0.06 | 0.06 | 50 | |||||||
| 237 | Macrolide R (L4 mutation) | CEM | 0.03 | 0.25 | 20 | 0.5 | 4 | 1 | 1 | 0.03 |
| AZI | 4 | 8 | 50 | |||||||
| CLA | 0.25 | 1 | 50 | |||||||
| TEL | 0.06 | 0.125 | 50 | |||||||
| CLI | 0.06 | 0.5 | 43 | 0.03 | 8 | 0.5 | 0.06 | 1 | ||
S, susceptible; R, resistant.
CEM, CEM-101; AZI, azithromycin; CLA, clarithromycin; TEL, telithromycin; CLI, clindamycin.
Boldface indicates cross-reactivity.
NT, not tested.
The results of single-step resistance selection studies for pneumococci are presented in Table 7. The same four comparators used in multistep selection were tested for their propensities to produce spontaneous mutations. Mutant selection frequencies for CEM-101 ranged from <2.0 × 10−10 to 6.8 × 10−7 at 2× MIC to <2.0 × 10−10 to 9.1 × 10−9 at 8× MIC. These comparators of CEM-101 had higher frequencies of resistance: telithromycin, 1.1 × 10−9 to 1.3 × 10−4 at 2× MIC to <1.5 × 10−10 to 4.8 × 10−6 at 8× MIC; clindamycin, <2.4 × 10−10 to 1.7 × 10−4 at 2× MIC to <1.2 × 10−10 to 5.6 × 10−7 at 8× MIC; and clarithromycin, <1.0 × 10−9 to 5.0 × 10−7 at 2× MIC to <1.2 × 10−10 to <3.1 × 10−9 at 8× MIC. A small number, three strains, were tested with azithromycin; mutant selection frequencies were <2.0 × 10−10 to 7.2 × 10−9 at 2× MIC to <1.9 × 10−10 to <2.0 × 10−10 at 8× MIC.
TABLE 7.
S. pneumoniae single-step mutation frequencies
| Strain no. | Phenotype (resistance determinant)a | Selecting drug | Frequency of mutation at: |
||
|---|---|---|---|---|---|
| 2× MIC | 4× MIC | 8× MIC | |||
| 1077 | Macrolide S | CEM-101 | <9.1 × 10−9 | <9.1 × 10−9 | <9.1 × 10−9 |
| Azithromycin | 7.2 × 10−9 | <1.9 × 10−10 | <1.9 × 10−10 | ||
| Clarithromycin | 1.1 × 10−9 | <1.2 × 10−10 | <1.2 × 10−10 | ||
| Telithromycin | 1.1 × 10−9 | <5.5 × 10−10 | <5.5 × 10−10 | ||
| Clindamycin | <3.8 × 10−10 | <3.8 × 10−10 | <3.8 × 10−10 | ||
| 24 | Macrolide R [erm(B)] | CEM-101 | 1.8 × 10−7 | <5.0 × 10−9 | <5.0 × 10−9 |
| Azithromycin | NTb | NT | NT | ||
| Clarithromycin | NT | NT | NT | ||
| Telithromycin | 1.3 × 10−4 | 2.5 × 10−6 | 1.7 × 10−6 | ||
| Clindamycin | NT | NT | NT | ||
| 3665 | Macrolide R [mef(A)] | CEM-101 | 6.8 × 10−7 | 1.4 × 10−7 | <4.5 × 10−10 |
| Azithromycin | NT | NT | NT | ||
| Clarithromycin | 5.0 × 10−7 | <5.0 × 10−10 | <5.0 × 10−10 | ||
| Telithromycin | 7.5 × 10−9 | 2.5 × 10−9 | <2.5 × 10−10 | ||
| Clindamycin | <2.4 × 10−10 | <2.4 × 10−10 | <2.4 × 10−10 | ||
| 1076 | Macrolide R [erm(B) mef(A)] | CEM-101 | <2.5 × 10−8 | 7.5 × 10−9 | 2.0 × 10−9 |
| Azithromycin | NT | NT | NT | ||
| Clarithromycin | NT | NT | NT | ||
| Telithromycin | 6.5 × 10−6 | 9.7 × 10−6 | 4.8 × 10−6 | ||
| Clindamycin | NT | NT | NT | ||
| 1635 | Macrolide R [erm(A)] | CEM-101 | 1.6 × 10−8 | <2.0 × 10−10 | <2.0 × 10−10 |
| Azithromycin | <2.0 × 10−10 | <2.0 × 10−10 | <2.0 × 10−10 | ||
| Clarithromycin | <3.1 × 10−9 | <3.1 × 10−9 | <3.1 × 10−9 | ||
| Telithromycin | <2.7 × 10−9 | <2.7 × 10−9 | <2.7 × 10−9 | ||
| Clindamycin | 7.3 × 10−7 | 5.5 × 10−7 | 5.6 × 10−7 | ||
| 2686 | Macrolide R (L4 mutation) | CEM-101 | <2.5 × 10−9 | <2.5 × 10−9 | <2.5 × 10−9 |
| Azithromycin | NT | NT | NT | ||
| Clarithromycin | NT | NT | NT | ||
| Telithromycin | 2.2 × 10−5 | 2.2 × 10−9 | <1.1 × 10−9 | ||
| Clindamycin | <1.2 × 10−9 | <1.2 × 10−9 | <1.2 × 10−9 | ||
| 7127 | Macrolide S (S20N in L4, A105V in L22) | CEM-101 | <2.0 × 10−10 | <2.0 × 10−10 | <2.0 × 10−10 |
| Azithromycin | <2.0 × 10−10 | <2.0 × 10−10 | <2.0 × 10−10 | ||
| Clarithromycin | <1.0 × 10−9 | <1.0 × 10−9 | <1.0 × 10−9 | ||
| Telithromycin | <5.0 × 10−9 | <5.0 × 10−9 | <5.0 × 10−9 | ||
| Clindamycin | <2.9 × 10−8 | <2.9 × 10−8 | <2.9 × 10−8 | ||
| 3009 | Macrolide R (23S rRNA mutation) | CEM-101 | <5.9 × 10−9 | <5.9 × 10−9 | <5.9 × 10−9 |
| Azithromycin | NT | NT | NT | ||
| Clarithromycin | NT | NT | NT | ||
| Telithromycin | 3.8 × 10−7 | <1.5 × 10−10 | <1.5 × 10−10 | ||
| Clindamycin | 1.7 × 10−4 | 7.3 × 10−9 | <1.2 × 10−10 | ||
S, susceptible; R, resistant.
NT, not tested.
The results of single-step resistance selection studies for S. pyogenes are presented in Table 8. As with the pneumococci, the four comparators used in multistep selection were tested for their propensities to produce spontaneous mutations. Mutant selection frequencies for CEM-101 ranged from <5.9 × 10−11 to 5.3 × 10−8 at 2× MIC to <5.9 × 10−11 to <5.3 × 10−10 at 8× MIC. The following comparators had higher frequencies of resistance than CEM-101: clindamycin, <7.7 × 10−11 to 2.1 × 10−7 at 2× MIC to < 7.7 × 10−11 to 1.1 × 10−7 at 8× MIC; clarithromycin, <1.0 × 10−10 to 1.7 × 10−7 at 2× MIC to <1.0 × 10−10 to 5.0 × 10−9 at 8× MIC. Mutant selection frequencies for telithromycin were similar to those for CEM-101: <8.3 × 10−11 to 7.7 × 10−8 at 2× MIC to <8.3 × 10−11 to <6.3 × 10−10 at 8× MIC. The mutation frequency for the one macrolide-sensitive strain tested with azithromycin was <1.0 × 10−10 at 2× and 8× MIC.
TABLE 8.
S. pyogenes single-step mutation frequencies
| Strain no. | Phenotype (resistance determinant)a | Selecting drug | Frequency of mutation at: |
||
|---|---|---|---|---|---|
| 2× MIC | 4× MIC | 8× MIC | |||
| 2132 | Macrolide S | CEM-101 | <1.0 × 10−10 | <1.0 × 10−10 | <1.0 × 10−10 |
| Azithromycin | <1.0 × 10−10 | <1.0 × 10−10 | <1.0 × 10−10 | ||
| Clarithromycin | <1.0 × 10−10 | <1.0 × 10−10 | <1.0 × 10−10 | ||
| Telithromycin | <8.3 × 10−11 | <8.3 × 10−11 | <8.3 × 10−11 | ||
| Clindamycin | <7.7 × 10−11 | <7.7 × 10−11 | <7.7 × 10−11 | ||
| 2368 | Macrolide R [erm(B)] | CEM-101 | <5.9 × 10−11 | <5.9 × 10−11 | <5.9 × 10−11 |
| Azithromycin | NTb | NT | NT | ||
| Clarithromycin | NT | NT | NT | ||
| Telithromycin | NT | NT | NT | ||
| Clindamycin | NT | NT | NT | ||
| 2094 | Macrolide R [erm(A)] | CEM-101 | 5.3 × 10−8 | 2.1 × 10−9 | <5.3 × 10−10 |
| Azithromycin | NT | NT | NT | ||
| Clarithromycin | 1.7 × 10−7 | 1.0 × 10−7 | 5.0 × 10−9 | ||
| Telithromycin | 7.7 × 10−8 | <1.5 × 10−10 | <1.5 × 10−10 | ||
| Clindamycin | 2.1 × 10−7 | 1.3 × 10−7 | 1.1 × 10−7 | ||
| 2011 | Macrolide R [mef(A)] | CEM-101 | 3.9 × 10−8 | <1.1 × 10−10 | <1.1 × 10−10 |
| Azithromycin | NT | NT | NT | ||
| Clarithromycin | NT | NT | NT | ||
| Telithromycin | 3.8 × 10−8 | <6.3 × 10−10 | <6.3 × 10−10 | ||
| Clindamycin | <1.0 × 10−10 | <1.0 × 10−10 | <1.0 × 10−10 | ||
| 237 | Macrolide-R (L4 mutation) | CEM-101 | <1.3 × 10−10 | <1.3 × 10−10 | <1.3 × 10−10 |
| Azithromycin | NT | NT | NT | ||
| Clarithromycin | <3.3 × 10−10 | <3.3 × 10−10 | <3.3 × 10−10 | ||
| Telithromycin | <2.0 × 10−10 | <2.0 × 10−10 | <2.0 × 10−10 | ||
| Clindamycin | <1.0 × 10−10 | <1.0 × 10−10 | <1.0 × 10−10 | ||
S, susceptible; R, resistant.
NT, not tested.
DISCUSSION
CEM-101 (Fig. 1) is a novel fluoroketolide that demonstrates enhanced potency compared to telithromycin, with activity against telithromycin-intermediate and telithromycin-resistant organisms (11). CEM-101 has shown significantly greater potency against phagocytized Staphylococcus aureus than telithromycin, azithromycin, and clarithromycin; CEM-101 was also about 50-fold and 100-fold more potent than azithromycin against phagocytized Listeria monocytogenes and Legionella pneumophila (17). CEM-101 exhibits the widest spectrum of activity against respiratory tract pathogens, including multidrug-resistant pneumococcus type 19A, compared to azithromycin, clarithromycin, erythromycin, telithromycin, clindamycin, and quinupristin-dalfopristin (11). CEM-101 is also potent against Chlamydia trachomatis, Chlamydophila pneumoniae (27), human mycoplasmas, and ureaplasmas (29), and MICs also point to clinical utility against most enterococci, gonococci, and Gram-positive anaerobes (1). CEM-101 is active against common organisms that cause gastroenteritis, such as Campylobacter jejuni, Salmonella spp., and Shigella spp., and is also active against Helicobacter pylori (12). CEM-101 has been shown to be more bactericidal against several Gram-positive species than telithromycin, with postantibiotic effects (in hours) of 2.3 to 6.1 and 3.7 to 5.3 against Gram-positive and -negative strains, respectively (10). CEM-101 has also demonstrated significant in vivo activity in a variety of murine infection models (20). Preliminary multistep studies have shown CEM-101 to have no or modest variation in MICs for one strain each of S. aureus and Enterococcus faecalis, and for two S. pneumoniae strains; low rates of spontaneous mutants were found in single-step experiments (13). At a projected human therapeutic dose of 400 to 500 mg, a Tmax (time to maximum concentration of the drug in serum) of 4 h and a t1/2 (half-life) of 5 to 6 h would be expected for CEM-101. Increases in the maximum concentration of the drug in serum (Cmax) and in the AUC were more than dose-proportional across the dose range administered (28).
In the current studies, CEM-101 yielded MICs that were usually a few dilutions lower than those of telithromycin against all resistance phenotypes of S. pneumoniae and S. pyogenes tested, including drug-resistant pneumococcus type 19A and erm(B)-positive S. pyogenes. Our results confirm previous findings cited above (11). CEM-101 yielded clones with higher MICs for all eight pneumococcal strains, but seven of the eight strains had clones with CEM-101 MICs of ≤0.5 μg/ml, and only for one erm(B) mef(A) strain with a parental MIC of 1 μg/ml was a resistant clone found with a MIC of 32 μg/ml. For two of the three resistant S. pyogenes CEM-101 clones [parental strains had the erm(A) gene or L4 mutations], MICs were 0.25 μg/ml, and only for the one strain with erm(B) did CEM-101 MICs rise from 1 to 8 μg/ml. Single-step studies also showed low yields of spontaneous mutations compared to those with other agents tested.
Based on pharmacokinetic findings reported from phase I clinical trials (28), recommendations for tentative CEM-101 susceptibility breakpoints have been set at ≤1 μg/ml as susceptible and ≥4 μg/ml as resistant; the tentative susceptibility breakpoint of ≤1 μg/ml is the same as that for telithromycin against streptococci (4).
The potent activity of CEM-101, and its low tendency to select for resistant mutants, against all streptococcal strains tested, irrespective of resistance phenotype, points to a promising clinical future for this compound, subject to pharmacokinetic/pharmacodynamic, toxicity, and animal infection model studies.
Acknowledgments
This study was supported by grants from Cempra Pharmaceuticals, Inc., Chapel Hill, NC.
Footnotes
Published ahead of print on 2 November 2009.
REFERENCES
- 1.Biedenbach, D. J., L. M. Deshpande, T. R. Fritsche, H. S. Sader, and R. N. Jones. 2008. Antimicrobial characterization of CEM-101: activity against enterococci, uncommon gram-positive pathogens, Neisseria gonorrhoeae, and anaerobes. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F1-3976.
- 2.Canu, A., B. Malbruny, M. Coquemont, T. A. Davies, P. C. Appelbaum, and R. Leclercq. 2002. Diversity of ribosomal mutations conferring resistance to macrolides, clindamycin, streptogramin, and telithromycin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark, C. L., K. Kosowska-Shick, L. M. Ednie, and P. C. Appelbaum. 2007. Capability of 11 antipneumococcal antibiotics to select for resistance by multistep and single-step methodologies. Antimicrob. Agents Chemother. 51:4196-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial susceptibility testing. Approved standard M100-S19. Nineteenth informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A7, 7th ed. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Davies, T. A., B. E. Dewasse, M. R. Jacobs, and P. C. Appelbaum. 2000. In vitro development of resistance to telithromycin (HMR 3647), four macrolides, clindamycin, and pristinamycin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:414-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heine, H. S., L. Miller, J. Bassett, and K. Holman. 2008. Antimicrobial activity of CEM-101, a new macrolide, tested against diverse collections of bacterial biowarfare/bioterrorism (BW/BT) agents. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F1-3980.
- 8.Jacobs, M. R., D. Felmingham, P. C. Appelbaum, and R. N. Gruneberg. 2003. The Alexander Project 1998-2000: susceptibility of pathogens isolated from community-acquired respiratory tract infection to commonly used antimicrobial agents. J. Antimicrob. Chemother. 52:229-246. [DOI] [PubMed] [Google Scholar]
- 9.Jacobs, M. R., C. E. Good, S. Bajaksouzian, and A. R. Windau. 2008. Emergence of Streptococcus pneumoniae serotypes 19A, 6C, and 22F and serogroup 15 in Cleveland, Ohio, in relation to introduction of the protein-conjugated pneumococcal vaccine. Clin. Infect. Dis. 47:1388-1395. [DOI] [PubMed] [Google Scholar]
- 10.Jones, R. N. 2008. Antimicrobial characterization of CEM-101: PAE, bactericidal activity and combinations. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F1-3981.
- 11.Jones, R. N., D. J. Biedenbach, P. R. Rhomberg, T. R. Fritsche, and H. S. Sader. 2008. Antimicrobial characterization of CEM-101 activity against 331 respiratory tract pathogens including multi-drug resistant pneumococcal serogroup 19A (MDR-19A) isolates. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F1-3975.
- 12.Jones, R. N., H. S. Sader, T. R. Fritsche, D. J. Biedenbach, and M. Castanheira. 2008. Antimicrobial potential application against species causing enteritis/gastroenteritis. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F1-3977.
- 13.Jones, R. N., P. R. Rhomberg, and H. S. Sader. 2008. Antimicrobial characterization of CEM-101: single step, selection by passaging and inducible resistances. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F1-3982.
- 14.Jones, R. N., T. R. Fritsche, H. S. Sader, D. J. Biedenbach, and P. R. Rhomberg. 2008. Assessment of CEM-101 susceptibility conditions and optimization of disk diffusion methods. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F1-3984.
- 15.Kaplan, E. L., D. R. Johnson, M. C. Del Rosario, and D. L. Horn. 1999. Susceptibility of group A beta-hemolytic streptococci to thirteen antibiotics: examination of 301 strains isolated in the United States between 1994 and 1997. Pediatr. Infect. Dis. J. 18:1069-1072. [DOI] [PubMed] [Google Scholar]
- 16.Kosowska-Shick, K., C. Clark, K. Credito, P. McGhee, B. Dewasse, T. Bogdanovich, and P. C. Appelbaum. 2006. Single- and multistep resistance selection studies on the activity of retapamulin compared to other agents against Staphylococcus aureus and Streptococcus pyogenes. Antimicrob. Agents Chemother. 50:765-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemaire, S., F. Van Bambeke, and P. M. Tulkens. 2009. Cellular accumulation and pharmacodynamic evaluation of the intracellular activity of CEM-101, a novel fluoroketolide, towards Staphylococcus aureus, Listeria monocytogenes, and Legionella pneumophila in human THP-1 macrophages. Antimicrob. Agents Chemother. 53:3734-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Llano-Sotelo, B., D. Klepacki, and A. S. Mankin. 2008. Binding and action of CEM-101, a new macrolide/ketolide, in development for treating infections with macrolide-resistant and macrolide-susceptible bacteria. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F-3983.
- 19.Moore, M. R., R. E. Gertz, Jr., R. L. Woodbury, G. A. Barkocy-Gallagher, W. Schaffner, C. Lexau, K. Gershman, A. Reingold, M. Farley, L. H. Harrison, J. L. Hadler, N. M. Bennett, A. R. Thomas, L. McGee, T. Pilishvili, A. B. Brueggemann, C. G. Whitney, J. H. Jorgensen, and B. Beall. 2008. Population snapshot of emergent Streptococcus pneumoniae serotype 19A in the United States, 2005. J. Infect. Dis. 197:1016-1027. [DOI] [PubMed] [Google Scholar]
- 20.Murphy, T. M., M. Gaffney, S. Little, A. M. Slee, and P. Fernandes. 2008. Evaluation of CEM-101, a novel macrolide, in murine infection models. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F1-3985.
- 21.Nagai, K., P. C. Appelbaum, T. A. Davies, L. M. Kelly, D. B. Hoellman, A. T. Andrasevic, L. Drukalska, W. Hryniewicz, M. R. Jacobs, J. Kolman, J. Miciuleviciene, M. Pana, L. Setchanova, M. K. Thege, H. Hupkova, J. Trupl, and P. Urbaskova. 2002. Susceptibilities to telithromycin and six other agents and prevalence of macrolide resistance due to L4 ribosomal protein mutation among 992 pneumococci from 10 Central and Eastern European countries. Antimicrob. Agents Chemother. 46:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagai, K., P. C. Appelbaum, T. A. Davies, L. M. Kelly, D. B. Hoellman, A. T. Andrasevic, L. Drukalska, W. Hryniewicz, M. R. Jacobs, J. Kolman, J. Miciuleviciene, M. Pana, L. Setchanova, M. K. Thege, H. Hupkova, J. Trupl, and P. Urbaskova. 2002. Susceptibility to telithromycin in 1,011 Streptococcus pyogenes isolates from 10 Central and Eastern European countries. Antimicrob. Agents Chemother. 46:546-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pérez-Trallero, E., C. Fernandez-Mazarrasa, C. Garcia-Rey, E. Bouza, L. Aguilar, J. Garcia-de-Lomas, and F. Baquero. 2001. Antimicrobial susceptibilities of 1,684 Streptococcus pneumoniae and 2,039 Streptococcus pyogenes isolates and their ecological relationships: results of a 1-year (1998-1999) multicenter surveillance study in Spain. Antimicrob. Agents Chemother. 45:3334-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pichichero, M. E., and J. R. Casey. 2007. Emergence of a multiresistant serotype 19A pneumococcal strain not included in the 7-valent conjugate vaccine as an otopathogen in children. JAMA 298:1772-1778. [DOI] [PubMed] [Google Scholar]
- 25.Rantala, M., M. Haanpera-Heikkinen, M. Lindgren, H. Seppala, P. Huovinen, and J. Jalava. 2006. Streptococcus pneumoniae isolates resistant to telithromycin. Antimicrob. Agents Chemother. 50:1855-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhomberg, P. R., J. E. Ross, and R. N. Jones. 2008. Proposed MIC quality control ranges for CEM-101 using the CLSI multi-laboratory M23-A2 study design. Abstr. 46th Intersci. Conf. Animicrob. Agents Chemother., abstr. D-2250.
- 27.Roblin, P. M., S. A. Kohlhoff, and M. R. Hammerschlag. 2008. In vitro activity of CEM-101, a new ketolide antibiotic against Chlamydia trachomatis and Chlamydia pneumoniae. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F1-3978. [DOI] [PMC free article] [PubMed]
- 28.Still, J. G., K. Clark, T. P. Degenhardt, D. Scott, P. Fernandes, and M. J. Gutierrez. 2008. Single oral dose pharmacokinetics and safety of CEM-101 in healthy subjects. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., poster F1-3972a. [DOI] [PMC free article] [PubMed]
- 29.Waites, K. B., D. M. Crabb, and L. B. Duffy. 2008. Comparative in vitro susceptibilities of a new investigational macrolide, CEM-101, against human mycoplasmas and ureaplasmas. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F1-3979. [DOI] [PMC free article] [PubMed]