Abstract
To identify factors associated with virological response (VR) to an etravirine (ETR)-based regimen, 243 patients previously treated with nonnucleoside reverse transcriptase inhibitors (NNRTIs) were studied. The impact of baseline HIV-1 RNA, CD4 cell count, past NNRTIs used, 57 NNRTI resistance mutations, genotypic sensitivity score (GSS) for nucleoside reverse transcriptase inhibitors (NRTIs) and protease inhibitors (PIs), and the number of new drugs used with ETR for the first time on the VR to an ETR regimen were investigated. Among the 243 patients, the median baseline HIV-1 RNA level was 4.4 log10 copies/ml (interquartile range [IQR], 3.7 to 4.9) and the median CD4 count was 175 cells/mm3 (IQR, 69 to 312). Patients had been previously exposed to a median of 6 NRTIs, 1, NNRTI, and 5 PIs. Overall, 82% of patients achieved a VR at month 2, as defined by a decrease of at least 1.5 log10 copies/ml and/or HIV-1 RNA level of <50 copies/ml. No difference in VR was observed between patients receiving or not a boosted PI in combination with ETR. Factors independently associated with a better VR to ETR were the number of drugs (among enfuvirtide, darunavir, or raltegravir) used for the first time in combination with ETR and the presence of the K103N mutation at baseline. Mutations Y181V and E138A were independently associated with poor VR, whereas no effect of the Y181C on VR was observed. In conclusion, ETR was associated with high response rates in NNRTI-experienced patients in combination with other active drugs regardless of the therapeutic class used.
Nonnucleoside reverse transcriptase inhibitors (NNRTIs) are frequently used components of combination antiretroviral therapy. However, drug resistance remains the primary cause of treatment failure and a single-amino-acid substitution in HIV-1 reverse transcriptase can confer cross-resistance to narrow-spectrum NNRTIs, restricting their use in treatment-experienced patients. Etravirine (ETR), or TMC125, is an expanded-spectrum NNRTI with a potent and broad in vitro activity against HIV-1, including virus with NNRTI resistance-associated mutations (1), and recently has been approved for clinical use in experienced patients. The molecular structure of ETR allows it to accommodate mutational changes in the binding pocket of the reverse transcriptase (RT) enzyme, even in the presence of significant mutations (25). Its efficacy and safety in treatment-experienced, NNRTI-resistant patients have been demonstrated in phase IIb and III trials (10, 13, 17). In the phase IIb C223 study, ETR treatment led to a significantly greater reduction in plasma viral load than control treatment at 24 weeks (17). In the large-phase III DUET-1 and −2 trials, ETR combined with other antiretroviral drugs significantly improved virological suppression at 24 weeks relative to placebo (10, 13). Resistance analyses from the DUET trials have first identified 13 mutations at 7 positions (V90I, A98G, L100I, K101E/P, V106I, V179D/F, Y181C/I/V, and G190A/S) associated with decreased virological responses (VR) to ETR; the concurrent presence of 3 or more of these were required to substantially reduce virological efficacy. Later, 4 mutations were added to this list of mutations (K101H, V179T, E138A, and M230L), resulting in a list of 17 ETR resistance-associated mutations (RAMs). Recently, a weighted score has been proposed comprising these 17 ETR RAMs based upon impact on response (weight factor), with some mutations having a weight of 3.0 (Y181I/V), 2.5 (L100I, K101P, Y181C, and M230L), 1.5 (V106I, V179F, E138A, and G190S), or 1.0 (V90I, A98G, K101E/H, V179D/T, and G190A) (28). The three resulting categories were defined on the basis of weighted mutation scores of 0 to 2, 2.5 to 3.5, and ≥4, leading to response rates of 74% (highest response), 52% (intermediate response), and 38% (reduced response), respectively, in the DUET trials. Beyond these resistance analyses in clinical trials, there are few data on the clinical and virological factors associated with ETR VR in clinical practice.
The aim of this study was to identify factors associated with virological response (VR) to ETR-containing regimens in NNRTI-experienced patients.
(This work was presented at the 16th Conference on Retroviruses and Opportunistic Infections, Montreal, Quebec, Canada, February 2009 [13a].)
MATERIALS AND METHODS
Patients and antiretroviral regimens.
Two hundred forty-three patients who experienced virological failure to an NNRTI were recruited to the study. All patients were treated with ETR (200 mg twice a day [b.i.d.]) with a background regimen comprising nucleoside reverse transcriptase inhibitors (NRTIs), protease inhibitor (PI), enfuvirtide (ENF), and/or raltegravir (RAL). Sociodemographic data, clinical data, and treatment histories were collected for all patients recruited. Inclusion criteria and all data were checked by the study monitor. Participating laboratories belong to the Agence Nationale de Recherches sur le SIDA (ANRS) AC11 network and participate in the ANRS quality control assessment of HIV-1 drug resistance sequencing (6).
Genotypic resistance testing.
The sequences of the protease and reverse transcriptase (RT) genes were determined at baseline in each laboratory using the ANRS consensus technique (http://www.hivfrenchresistance.org/), the Bayer TrueGene kit, the Abbott ViroSeq kit, or an in-house method. All protease and RT gene mutations were identified from the International AIDS Society—USA resistance testing panel (December 2008) (8).
Phylogenetic analyses.
Genetic subtypes were determined by phylogenetic tree analysis. The RT and protease sequences were aligned with sequences from reference strains representing all subtypes and circulating recombinant forms with the ClustalW program. Phylogenetic trees were constructed using the neighbor-joining method and Kimura two-parameter model. We evaluated 100 replicates by phylogenetic analysis. Among the 243 patients, 216 were infected with HIV-1 subtype B and 27 with HIV-1 subtype non-B (6 with CRF02_AG, 5 with CRF01_AE, 4 with CRF06_cpx, 6 with D, 1 with A, 1 with A1, 1 with CRF11, 1 with CRF14, and 1 with F1 and 1G).
Statistical methods.
Virological response (VR) was defined as a decrease of at least 1.5 log10 copies/ml and/or HIV-1 RNA level of <50 copies/ml at month 2. Association between 57 NNRTI resistance mutations and VR was studied using Fisher's exact test. This list of 57 mutations includes 44 NNRTI RAMs (V90I, A98G, L100I, K101E/P/Q, K103H/N/S/T, V106A/I/M, V108I, E138G/K/Q, V179D/E/F/G/I, Y181C/I/V, Y188C/H/L, V189I, G190A/C/E/Q/S, H221Y, P225H, F227C/L, M230I/L, P236L, K238N/T, and Y318F) identified from an extensive review of the existing literature on NNRTI resistance plus 13 mutations by addition of all mutations observed at NNRTI resistance amino acid positions (23, 28). All past genotypic resistance tests were taken into account in the analyses, and a mutation was considered as present if it was detected in baseline genotype or in at least one previous genotype.
The impact of baseline HIV-1 RNA, CD4 cell count, viral subtype (B versus non-B), past NNRTI used, genotypic sensitivity score (GSS) for PI, GSS for NRTI, the previous use of nevirapine (NVP) or efavirenz (EFV), having had an active boosted PI or not, and the number of new drugs associated with ETR for the first time on the VR to an ETR regimen was also investigated. Multivariate analysis (logistic model) was performed to search for independent predictive factors associated with VR to ETR. The statistical program used for analyses was SAS (version 9.0).
We used mutations present in the RT and protease gene and the ANRS algorithm (http://www.hivfrenchresistance.org/) to determine whether patients receiving a particular nucleoside reverse transcriptase inhibitor (NRTI) or protease inhibitor (PI), had resistant or susceptible virus strains.
RESULTS
Overall, 199/243 (82%) patients receiving an ETR-containing regimen displayed a VR at month 2. One hundred twenty patients had a viral load (VL) of <50 copies/ml, 179 had a decrease in VL of at least 1.5 log10 at month 2, and 100 patients had both criteria. Patients had a median of 4 genotypes (interquartile range [IQR], 2 to 6). The main characteristics of the study population are shown in Table 1. In the background regimen associated with ETR, the most frequent NRTIs prescribed were emtricitabine/lamivudine (FTC/3TC) (63%), tenofovir (TDF) (49%), and abacavir (ABC) (21%); the most frequent PI was darunavir (DRV) (79%). RAL was associated with ETR in 73% of cases and ENF in 23% of cases. Patients used for the first time ENF, DRV, and RAL in combination with ETR in 10%, 49%, and 61% of the cases, respectively.
TABLE 1.
Baseline characteristics of the study populationa
| Characteristicb | Result (IQR) |
|---|---|
| % of male patients | 82 |
| % of patients with subtype B | 89 |
| Median no. of plasma HIV-1 RNA log10 copies/ml | 4.4 (3.7-4.9) |
| Median CD4 cell count/mm3 | 175 (69-312) |
| Median no. of patients with previous antiretroviral treatment | |
| Antiretroviral drugs | 13 (11-15) |
| NRTIs | 6 (5-7) |
| NNRTIs | 1 (1-2) |
| PIs | 5 (4-6) |
| % of patients treated with with ENF | 60 |
| % of patients treated with RAL | 12 |
| % of patients with ETR cotreatment | |
| NRTIs + PI + RAL | 30 |
| PI + RAL | 19 |
| NRTIs + PI | 16 |
| NRTIs + PI + ENF | 8 |
| NRTIs + PI + ENF + RAL | 8 |
| NRTIs + RAL | 7 |
n = 243.
IQR, interquartile range; NRTIs, nucleoside reverse transcriptase inhibitors; NNRTIs, non-nucleoside reverse transcriptase inhibitors; PIs, protease inhibitors; ENF, enfuvirtide; RAL, raltegravir; ETR, etravirine.
Effect of NNRTI resistance mutations on virological response.
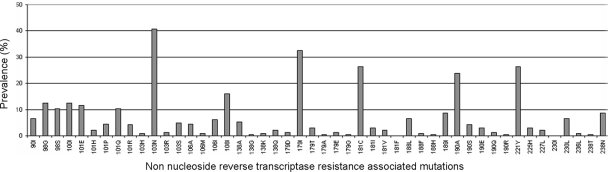
The prevalence of NNRTI resistance mutations among the viruses studied is displayed in Fig. 1. Among the list of 57 NNRTI RAMs, the following mutations were not detected in the population studied: K101N, K103T, V179F, Y188C, G190C, F227C, and Y318F.
FIG. 1.
Prevalence of the 57 nonnucleoside reverse transcriptase inhibitor resistance-associated mutations among the 243 isolates.
In univariate analysis, 4 NNRTI mutations were associated with a lower VR to ETR (P < 0.05)—Y181V, V179I, V106I, and Y188L—and the K103N mutation was associated with a higher VR (Table 2). A trend toward significance was found for the E138A mutation (P = 0.06), which was kept for further analyses because it belongs to the weighted score previously described. In the ETR weighted score defined by Vingerhoets et al., the Y181I and Y181V mutations had the highest weight, followed by L100I, K101P, Y181C, and M230L (28). In our analysis, although all of these mutations were present in the data set, only Y181V had a significant negative impact on VR to ETR.
TABLE 2.
Univariate analysis of the virological response to etravirine as a function of the presence of mutated or wild-type codons among the list of 57 nonnucleoside reverse transcriptase inhibitor resistance-associated mutations
| Position | No. (%) of responders/total |
P value | |
|---|---|---|---|
| Wild type | Mutation | ||
| Y181V | 198/238 (83) | 1/5 (20) | 0.00415 |
| K103N | 110/144 (76) | 89/99 (90) | 0.00694 |
| V179I | 142/164 (87) | 57/79 (72) | 0.00784 |
| V106I | 190/228 (83) | 9/15 (60) | 0.03477 |
| Y188L | 189/228 (83) | 10/16 (63) | 0.04800 |
| E138A | 191/230 (83) | 8/13 (62) | 0.06415a |
A trend toward significance was found for the E138A mutation, which was kept for further analyses.
Patients harboring viruses with the K103N mutation compared with patients not harboring the K103N mutation had fewer NNRTI mutations (median of 2 mutations versus 3 mutations; P = 0.054). They were also more frequently exposed to EFV in the past (77% versus 58%; P = 0.004) and less frequently exposed to NVP in the past (45% versus 63%; P = 0.008) and had no difference regarding the number of new drugs received in combination with ETR.
Although Y181C was not associated with a reduced VR, we compared patients harboring viruses with the Y181C mutation with patients not harboring the Y181C mutation and found that patients with viruses containing Y181C had more NNRTI mutations (median of 3 mutations versus 2 mutations; P < 0.0001). There was no difference in past exposure to EFV, but they were more frequently exposed to NVP in the past (73% versus 50%; P = 0.001) and no difference was observed in the number of new drugs received in combination with ETR.
Factors associated with the virological response. (i) Univariate analysis.
Baseline HIV-1 RNA, CD4 cell count, viral subtype (B versus non-B), GSS for PI, and GSS for NRTI were not associated with VR. There was no difference in VR between patients receiving an active boosted PI and those receiving a nonactive PI or no PI at all (84% versus 80%). The number of drugs received for the first time including DRV, RAL, ENF, and ETR was strongly associated with the VR (P < 0.001) (Fig. 2).
FIG. 2.
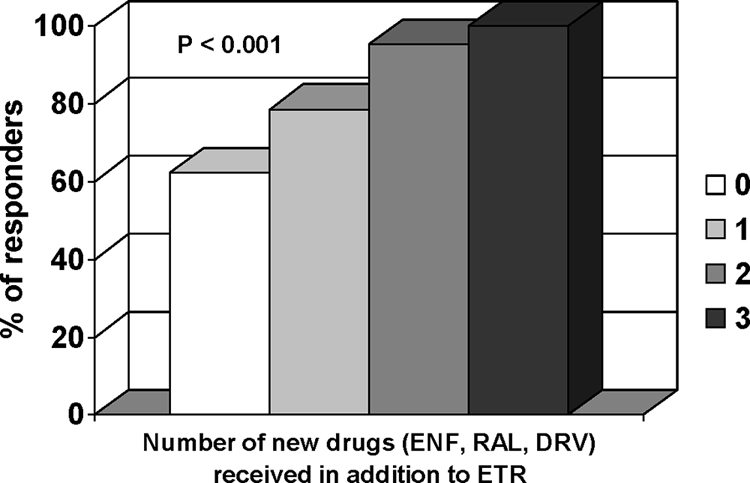
Virological response to etravirine (ETR) as a function of the number of drugs received for the first time, comprising darunavir (DRV), raltegravir (RAL), enfuvirtide (ENF), and etravirine.
Although the number of previous NNRTIs received in the past was not associated with VR to ETR, among patients exposed to only one NNRTI, the previous use of NVP rather than EFV was associated with a poorer response (77% versus 91%; P = 0.03). In these patients who received only NVP or EFV before the use of ETR, the number of NNRTI mutations was not statistically different.
(ii) Multivariate analysis.
The following variables were potentially be included in the final multivariate model: past NNRTI used, GSS for PIs and NRTIs, the number of new drugs associated with ETR for the first time, and mutations associated with VR in univariate analysis.
Four variables were retained in the final multivariate model as independently associated with the ETR VR: the number of new drugs used in combination with ETR for the first time and K103N with a better VR and mutations Y181V and E138A with a poorer VR (Table 3).
TABLE 3.
Factors associated with virological response to etravirine in multivariate analysisa
| Variable | OR | 95% CI | P value |
|---|---|---|---|
| Receiving 2 new drugs or more in combination with ETR | 8 | 2.6-24.5 | 0.0003 |
| Presence of Y181V | 0.05 | 0-0.6 | 0.02 |
| Presence of K103N | 2.4 | 1.1-5.4 | 0.03 |
| Presence of E138A | 0.23 | 0.06-0.9 | 0.03 |
OR, odds ratio; CI, confidence interval.
DISCUSSION
In this study, we observed a VR (defined as a decrease of at least 1.5 log10 copies/ml and/or HIV-1 RNA at <50 copies/ml at month 2) in 82% of the patients studied. The following NNRTI mutations were associated with a decreased VR to ETR: Y181V, V179I, V106I, Y188L, and E138A. The K103N mutation had no effect and was even associated with a better VR to ETR. In the multivariate analysis, some variables were retained as independently positively associated with the VR, including the number of new drugs used in combination with ETR for the first time and the K103N mutation, in addition to others negatively associated with the VR, such as mutations Y181V and E138A.
The VR was defined here as a decrease of at least 1.5 log10 copies/ml and/or HIV-1 RNA at <50 copies/ml at month 2, which is a criterion currently used in such studies (7, 14-16). Month 2 has been chosen as an endpoint because the impact of mutations on the response needs to be measured early after initiation of a new agent to avoid possible interference of drug discontinuation, dropout, toxicity, and adherence to the regimen in the VR.
From the 17 ETR RAMs involved in the weighted genotypic score, only 3 of them were associated in this study with a decreased VR to ETR in univariate analysis. A certain number of mutations had a very low frequency in our study, limiting the power to detect their potential effect on the virological response. For example, the following 8 mutations were found in <5% of patients: Y181I/V, K101P, M230L, G190S, K103H, and V179D/T. Of note, nine mutations (K101N, K103T, V179F, Y188C, G190C, F227C, Y318F, N348I, and N348T) were not found in our sample. The Y181V has been previously described to have a high weight (3.0) and both V106I and E138A a medium weight (1.5) on VR to ETR (28). Two mutations not previously included in the 17 ETR RAMs have been identified in this study: V179I and Y188L. In vitro selection experiments identified mutations selected by ETR that included known NNRTI-associated mutations L100I, Y181C, G190E, M230L, and Y318F and the novel mutations V179I and V179F (21, 26). Thus, it is understandable that V179I has been associated with a decreased VR to ETR since it can be selected in vitro by ETR. Moreover, it has been shown in the DUET trials that V179I can emerge upon virological failure in patients on an ETR-containing regimen (24). Y188L is a NNRTI resistance mutation already known that can be selected in case of EFV failure, and viruses with this mutation in the RT gene show reduced in vitro susceptibility to EFV and NVP (3). Moreover, Y188L was previously described in a study to have a potential role in ETR resistance (4).
K103N is the most commonly reported resistance mutation for NNRTIs arising from clinical use of this drug class (12). A number of structures of the RT K103N mutant have been reported (5, 11, 20). An early suggestion of how this mutation can give rise to resistance was based on the crystal structure of unliganded HIV-1 RT into which the K103N change was modeled. This work showed that a hydrogen bond could be made between the hydroxyl group of the Tyr188 side chain in the unliganded down position and the Asn103 amide group. Such an interaction would have the effect of stabilizing the apo-RT conformation and hence create an energy barrier to binding NNRTIs, thereby giving a reduction in potency. Modeling of ETR entry into the NNRTI using targeted molecular dynamics has led to the suggestion that hydrogen bonding from a cyano substituent may assist in breaking the Asn103-Tyr188 H-bond, thereby explaining the potency of this compound against the K103N mutant RT (22). In the DUET trials, it has been shown that K103N was not associated with a decreased VR (27). Our results confirm that the K103N mutation has no effect on VR to ETR and further suggest it may be associated with a better VR when present. Phenotypic studies have shown that ETR retains in vitro efficacy against K103N mutants, but there is no evidence of hypersusceptibility. The positive impact of the K103N mutation on ETR VR should be further investigated, especially the type of mutations associated with K103N that could explain a hypersensitivity to ETR, such as NRTI mutations (18).
It is noteworthy that certain baseline characteristics of our patients were similar to those of the DUET trial populations, such as the number of antiretroviral drugs received before starting the ETR regimen; the numbers of NRTIs, NNRTIs, and PIs received in the past; the CD4 cell count; and the plasma viral load. However, we did not identify strictly the same mutations impacting the ETR VR. Indeed, this may be due to the use of different patient populations, resistance methodologies, and statistical approaches. These different approaches are nonetheless interesting because this allows identification of mutations in a population that would not be present in another population, thus contributing to enlarging the knowledge of mutations implicated in resistance to a drug.
Our results confirm that previous exposure to NVP was associated with a poorer VR to ETR that is consistent with other studies showing that NVP rather than EFV was associated with an increased number of ETR mutations (9, 19). Thus, use of EFV may be less likely to lead to ETR resistance than that of NVP. In addition, it has been shown that the duration of initial NNRTI exposure was associated with an increasing likelihood of developing in vitro ETR resistance (9). Therefore, the choice and treatment duration of the initial NNRTI may affect the subsequent ETR response. Finally, the number of active drugs associated with ETR seems to be a factor strongly impacting the VR to ETR, as it was independently associated with the VR. Prior publications have already indicated the number of new agents to be a common predictor of success (2, 29). In our study, in patients receiving 3 active drugs in their regimen comprising ETR, 95% of patients were responders. This result, combined with the fact that the use of a boosted PI in combination with ETR was not associated with a better VR, suggests that the most important condition is the number of active drugs in the ETR regimen regardless of the therapeutic class. However, as the virological and clinical efficacy of ETR has been demonstrated in combination with boosted darunavir in the DUET trials, the use of ETR in combination with a boosted PI should be prescribed when possible.
In conclusion, in this population of NNRTI-experienced patients, ETR was associated with high response rates. Factors associated with a better VR to ETR were the number of new drugs (among RAL, DRV, or T20) used for the first time in combination with ETR and the presence of K103N. The use of a boosted PI in combination with ETR was not associated with a better VR, suggesting that the most important factor is the use of active drugs regardless of the therapeutic class. Mutations Y181V and E138A were independently associated with poor VR, whereas no effect on VR was observed with Y181C. The positive impact of K103N mutation on ETR VR should be further investigated, especially the type of mutations associated with K103N that could explain a hypersensitivity to ETR, such as NRTI mutations.
Acknowledgments
The research leading to these results has received funding from the Agence Nationale de Recherches sur le SIDA (ANRS), the European Community's Seventh Framework Programme (FP7/2007-2013) under the project “Collaborative HIV and Anti-HIV Drug Resistance Network (CHAIN)”—grant agreement no. 223131, and ARVD (Association de Recherche en Virologie et Dermatologie).
The members of the ANRS AC11 Resistance Study Group by location are as follows: Avicenne, C. Alloui; Besançon, D. Bettinger; Bordeaux, G. Anies and B. Masquelier; Brest, S. Vallet; Clermont-Ferrand, C. Henquell; Créteil, M. Bouvier-Alias; Fort de France, G. Dos Santos; Grenoble, A. Signori-Schmuck; Limoges, S. Rogez; Lyon, P. Andre, J. C. Tardy, and M. A. Trabaud; Marseille, C. Tamalet; Montpellier, B. Montes; Nice, J. Cottalorda; Paris-Bichat Claude Bernard, D. Descamps and F. Brun-Vézinet; Paris-HEGP, C. Charpentier; Paris-Necker, M. L. Chaix; Paris-Pitié-Salpêtrière, S. Fourati, A. G. Marcelin, V. Calvez, and P. Flandre; Paris-Saint Antoine, L. Morand-Joubert; Paris-Saint Louis, C. Delaugerre; Rennes, A. Ruffault and A. Maillard; Saint Etienne, T. Bourlet and H. Saoudin; Toulouse, J. Izopet; and Tours, F. Barin.
The members of the ANRS Clinical Centers by location are as follows: Avicenne, O. Bouchaud; Besançon, B. Hoen; Bordeaux, M. Dupon, P. Morlat, and D. Neau; Brest, M. Garré and V. Bellein; Clermont-Ferrand, C. Jacomet; Créteil, Y. Lévy and S. Dominguez; Fort de France, A. Cabié; Grenoble, P. Leclercq; Limoges, P. Weinbreck; Lyon, L. Cotte; Marseille, I. Poizot-Martin and I. Ravaud; Montpellier, J. Reynes; Nice, P. Dellamonica; Paris-Bichat Claude Bernard, P. Yeni and R. Landman; Paris-HEGP, L. Weiss and C. Piketty; Paris-Necker, J. P. Viard; Paris-Pitié-Salpêtrière, C. Katlama and A. Simon; Paris-Saint Antoine, P. M. Girard and J. L. Meynard; Paris-Saint Louis, J. M. Molina; Pointe à Pitre, M. T. Goeger-Sow and I. Lamaury; Rennes, C. Michelet, Saint Etienne, F. Lucht; Toulouse, B. Marchou and P. Massip; and Tours, J. M. Besnier.
Footnotes
Published ahead of print on 9 November 2009.
REFERENCES
- 1.Andries, K., H. Azijn, T. Thielemans, D. Ludovici, M. Kukla, J. Heeres, P. Janssen, B. De Corte, J. Vingerhoets, R. Pauwels, and M. P. de Bethune. 2004. TMC125, a novel next-generation nonnucleoside reverse transcriptase inhibitor active against nonnucleoside reverse transcriptase inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 48:4680-4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arazo Garces, P., and T. Omiste Sanvicente. 2008. Darunavir in patients with advanced HIV and multiresistance. The POWER, DUET and BENCHMRK studies. Enferm. Infecc. Microbiol. Clin. 26(Suppl. 10):23-31. (In Spanish.) [DOI] [PubMed] [Google Scholar]
- 3.Bacheler, L., S. Jeffrey, G. Hanna, R. D'Aquila, L. Wallace, K. Logue, B. Cordova, K. Hertogs, B. Larder, R. Buckery, D. Baker, K. Gallagher, H. Scarnati, R. Tritch, and C. Rizzo. 2001. Genotypic correlates of phenotypic resistance to efavirenz in virus isolates from patients failing nonnucleoside reverse transcriptase inhibitor therapy. J. Virol. 75:4999-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benhamida, J., C. Chappey, E. Coakley, and N. T. Parkin. 2008. Abstr. XVII Int. HIV Drug Resist. Workshop, abstr. 130.
- 5.Das, K., A. D. Clark, Jr., P. J. Lewi, J. Heeres, M. R. De Jonge, L. M. Koymans, H. M. Vinkers, F. Daeyaert, D. W. Ludovici, M. J. Kukla, B. De Corte, R. W. Kavash, C. Y. Ho, H. Ye, M. A. Lichtenstein, K. Andries, R. Pauwels, M. P. De Bethune, P. L. Boyer, P. Clark, S. H. Hughes, P. A. Janssen, and E. Arnold. 2004. Roles of conformational and positional adaptability in structure-based design of TMC125-R165335 (etravirine) and related non-nucleoside reverse transcriptase inhibitors that are highly potent and effective against wild-type and drug-resistant HIV-1 variants. J. Med. Chem. 47:2550-2560. [DOI] [PubMed] [Google Scholar]
- 6.Descamps, D., C. Delaugerre, B. Masquelier, A. Ruffault, A. G. Marcelin, J. Izopet, M. L. Chaix, V. Calvez, F. Brun-Vezinet, and D. Costagliola. 2006. Repeated HIV-1 resistance genotyping external quality assessments improve virology laboratory performance. J. Med. Virol. 78:153-160. [DOI] [PubMed] [Google Scholar]
- 7.Descamps, D., S. Lambert-Niclot, A. G. Marcelin, G. Peytavin, B. Roquebert, C. Katlama, P. Yeni, M. Felices, V. Calvez, and F. Brun-Vezinet. 2009. Mutations associated with virological response to darunavir/ritonavir in HIV-1-infected protease inhibitor-experienced patients. J. Antimicrob. Chemother. 63:585-592. [DOI] [PubMed] [Google Scholar]
- 8.Johnson, V. A., F. Brun-Vezinet, B. Clotet, H. F. Gunthard, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, and D. D. Richman. 2008. Update of the drug resistance mutations in HIV-1. Top. HIV Med. 16:138-145. [PubMed] [Google Scholar]
- 9.Lapadula, G., A. Calabresi, F. Castelnuovo, S. Costarelli, E. Quiros-Roldan, G. Paraninfo, F. Ceresoli, F. Gargiulo, N. Manca, G. Carosi, and C. Torti. 2008. Prevalence and risk factors for etravirine resistance among patients failing on non-nucleoside reverse transcriptase inhibitors. Antivir. Ther. 13:601-605. [PubMed] [Google Scholar]
- 10.Lazzarin, A., T. Campbell, B. Clotet, M. Johnson, C. Katlama, A. Moll, W. Towner, B. Trottier, M. Peeters, J. Vingerhoets, G. de Smedt, B. Baeten, G. Beets, R. Sinha, and B. Woodfall. 2007. Efficacy and safety of TMC125 (etravirine) in treatment-experienced HIV-1-infected patients in DUET-2: 24-week results from a randomised, double-blind, placebo-controlled trial. Lancet 370:39-48. [DOI] [PubMed] [Google Scholar]
- 11.Lindberg, J., S. Sigurdsson, S. Lowgren, H. O. Andersson, C. Sahlberg, R. Noreen, K. Fridborg, H. Zhang, and T. Unge. 2002. Structural basis for the inhibitory efficacy of efavirenz (DMP-266), MSC194 and PNU142721 towards the HIV-1 RT K103N mutant. Eur. J. Biochem. 269:1670-1677. [DOI] [PubMed] [Google Scholar]
- 12.Llibre, J. M., J. R. Santos, T. Puig, J. Molto, L. Ruiz, R. Paredes, and B. Clotet. 2008. Prevalence of etravirine-associated mutations in clinical samples with resistance to nevirapine and efavirenz. J. Antimicrob. Chemother. 62:909-913. [DOI] [PubMed] [Google Scholar]
- 13.Madruga, J. V., P. Cahn, B. Grinsztejn, R. Haubrich, J. Lalezari, A. Mills, G. Pialoux, T. Wilkin, M. Peeters, J. Vingerhoets, G. de Smedt, L. Leopold, R. Trefiglio, and B. Woodfall. 2007. Efficacy and safety of TMC125 (etravirine) in treatment-experienced HIV-1-infected patients in DUET-1: 24-week results from a randomised, double-blind, placebo-controlled trial. Lancet 370:29-38. [DOI] [PubMed] [Google Scholar]
- 13a.Marcelin, A. G., et al. 2009. 16th Conf. Retroviruses Opportunistic Infect., abstr. 645.
- 14.Marcelin, A. G., P. Flandre, J. M. Molina, C. Katlama, P. Yeni, F. Raffi, Z. Antoun, M. Ait-Khaled, and V. Calvez. 2008. Genotypic resistance analysis of the virological response to fosamprenavir-ritonavir in protease inhibitor-experienced patients in CONTEXT and TRIAD clinical trials. Antimicrob. Agents Chemother. 52:4251-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marcelin, A. G., B. Masquelier, D. Descamps, J. Izopet, C. Charpentier, C. Alloui, M. Bouvier-Alias, A. Signori-Schmuck, B. Montes, M. L. Chaix, C. Amiel, G. D. Santos, A. Ruffault, F. Barin, G. Peytavin, M. Lavignon, P. Flandre, and V. Calvez. 2008. Tipranavir-ritonavir genotypic resistance score in protease inhibitor-experienced patients. Antimicrob. Agents Chemother. 52:3237-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masquelier, B., K. L. Assoumou, D. Descamps, L. Bocket, J. Cottalorda, A. Ruffault, A. G. Marcelin, L. Morand-Joubert, C. Tamalet, C. Charpentier, G. Peytavin, Z. Antoun, F. Brun-Vezinet, and D. Costagliola. 2008. Clinically validated mutation scores for HIV-1 resistance to fosamprenavir/ritonavir. J. Antimicrob. Chemother. 61:1362-1368. [DOI] [PubMed] [Google Scholar]
- 17.Nadler, J. P., D. S. Berger, G. Blick, P. J. Cimoch, C. J. Cohen, R. N. Greenberg, C. B. Hicks, R. M. Hoetelmans, K. J. Iveson, D. S. Jayaweera, A. M. Mills, M. P. Peeters, P. J. Ruane, P. Shalit, S. R. Schrader, S. M. Smith, C. R. Steinhart, M. Thompson, J. H. Vingerhoets, E. Voorspoels, D. Ward, and B. Woodfall. 2007. Efficacy and safety of etravirine (TMC125) in patients with highly resistant HIV-1: primary 24-week analysis. AIDS 21:F1-F10. [DOI] [PubMed] [Google Scholar]
- 18.Picchio, G., J. Vingerhoets, N. T. Parkin, H. Azijn, and M. P. De Bethune. 2008. Abstr. XVII Int. HIV Drug Resistance Workshop, abstr. 23.
- 19.Poveda, E., C. Garrido, C. de Mendoza, A. Corral, J. Cobo, J. Gonzalez-Lahoz, and V. Soriano. 2007. Prevalence of etravirine (TMC-125) resistance mutations in HIV-infected patients with prior experience of non-nucleoside reverse transcriptase inhibitors. J. Antimicrob. Chemother. 60:1409-1410. [DOI] [PubMed] [Google Scholar]
- 20.Ren, J., J. Milton, K. L. Weaver, S. A. Short, D. I. Stuart, and D. K. Stammers. 2000. Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase. Structure 8:1089-1094. [DOI] [PubMed] [Google Scholar]
- 21.Rimsky, L. T., H. Azijn, I. Tirry, J. Vingerhoets, R. Mersch, G. Kraus, M. P. De Bethune, and G. Picchio. 2009. Abstr. XVIII Int. HIV Drug Resist. Workshop, abstr. 120.
- 22.Rodriguez-Barrios, F., J. Balzarini, and F. Gago. 2005. The molecular basis of resilience to the effect of the Lys103Asn mutation in non-nucleoside HIV-1 reverse transcriptase inhibitors studied by targeted molecular dynamics simulations. J. Am. Chem. Soc. 127:7570-7578. [DOI] [PubMed] [Google Scholar]
- 23.Tambuyzer, L., H. Azijn, L. T. Rimsky, J. Vingerhoets, P. Lecocq, G. Kraus, G. Picchio, and M. P. de Bethune. 2009. Compilation and prevalence of mutations associated with resistance to non-nucleoside reverse transcriptase inhibitors. Antivir. Ther. 14:103-109. [PubMed] [Google Scholar]
- 24.Tambuyzer, L., J. Vingerhoets, H. Azijn, B. Daems, G. Picchio, and M. P. De Bethune. 2008. Abstr. 6th Eur. HIV Drug Resist. Workshop, abstr. 47.
- 25.Udier-Blagovic, M., J. Tirado-Rives, and W. L. Jorgensen. 2003. Validation of a model for the complex of HIV-1 reverse transcriptase with nonnucleoside inhibitor TMC125. J. Am. Chem. Soc. 125:6016-6017. [DOI] [PubMed] [Google Scholar]
- 26.Vingerhoets, J., H. Azijn, E. Fransen, I. De Baere, L. Smeulders, D. Jochmans, K. Andries, R. Pauwels, and M. P. de Bethune. 2005. TMC125 displays a high genetic barrier to the development of resistance: evidence from in vitro selection experiments. J. Virol. 79:12773-12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vingerhoets, J., B. Clotet, M. Peeters, G. Picchio, L. Tambuyzer, K. Cao Van, G. De Smedt, B. Woodfall, and M. P. De Bethune. 2007. Abstr. 11th Eur. AIDS Conf., abstr. P7.3/05.
- 28.Vingerhoets, J., M. Peeters, H. Azijn, L. Tambuyzer, A. Hoogstoel, S. Nijs, M. P. De Bethune, and G. Picchio. 2008. Abstr. XVII Int. HIV Drug Resist. Workshop, abstr. 24.
- 29.Vray, M., J. L. Meynard, C. Dalban, L. Morand-Joubert, F. Clavel, F. Brun-Vezinet, G. Peytavin, D. Costagliola, and P. M. Girard. 2003. Predictors of the virological response to a change in the antiretroviral treatment regimen in HIV-1-infected patients enrolled in a randomized trial comparing genotyping, phenotyping and standard of care (Narval trial, ANRS 088). Antivir. Ther. 8:427-434. [DOI] [PubMed] [Google Scholar]