Abstract
In this paper we provide the first biochemical evidence of the existence of a family of structure-related antimicrobial peptides, the siderophore-microcins, in the Enterobacteriaceae family. We isolated and characterized two novel siderophore-microcins, MccM and MccH47, previously characterized through genetic studies. MccM and MccH47 were expressed from several Escherichia coli strains containing the microcin gene clusters. The spectra of their bactericidal activities were found to be restricted to some species of the Enterobacteriaceae. MccM and MccH47 were unable to inhibit the growth of strains carrying mutations in the fepA, cir, and fiu genes, which showed the requirement of the iron-catecholate receptors for their recognition. The MccM and MccH47 peptide moieties contain 77 and 60 residues, respectively, and are derived from the microcin precursors McmA and MchB, respectively. In addition, both peptides carried a C-terminal posttranslational modification containing a salmochelin-like siderophore moiety also found in MccE492 (X. Thomas et al., J. Biol. Chem., 279:28233-28242, 2004). Interestingly, when MccM was isolated from E. coli Nissle 1917, which lacks the two genes necessary for modification biosynthesis, it was devoid of posttranslational modification. Those two genes could be complemented by their homologues from the MccH47 gene cluster, thereby showing their functional interchangeability between at least two members of the siderophore-microcin family. Finally, from the sequence analysis of the MccE492 gene cluster, we hypothesized the existence of an additional member of the siderophore-microcin family. Therefore, we propose that the siderophore-microcin family contains five representatives.
Microcins (Mccs) are low-molecular-weight antimicrobial peptides secreted by members of the Enterobacteriaceae family and are involved in microbial competition within the intestinal tract. They are generally synthesized by the ribosomal pathway. A microcin classification in which class IIb microcins (microcins MccE492, MccH47, MccI47, and MccM) exhibit a conserved serine-rich C-terminal region has recently been proposed (9). While MccE492 was shown to carry at its C terminus a siderophore acquired by posttranslational modification (28), the other class IIb microcins remain uncharacterized to date.
In a previous study, we have shown that the unique posttranslational modification of MccE492 consists of a C-glucosylated linear trimer of N-(2,3 dihydroxybenzoyl)-l-serine (DHBS) linked to the C-terminal serine carboxylate (28). This modification is reminiscent of the catecholate siderophores, especially of salmochelin S4. Salmochelin S4 is derived from enterobactin (Ent), the cyclic trimer of DHBS, by addition of two β-d-glucose (Glc) moieties linked to the DHBS units through C-glucosidic bonds (1). Moreover, we have previously shown that Ent is a precursor for the biosynthesis of MccE492 posttranslational modification (29). Siderophores are generally synthesized by the nonribosomal pathway. Indeed, the Ent gene cluster is necessary for Ent biosynthesis, and genes in the iroA locus are required for the conversion of Ent into salmochelins (5, 16). Consequently, siderophore-microcin MccE492 can be considered to be both ribosomally (peptide moiety) and nonribosomally (enterobactin moiety) synthesized, with glucose being the linker between the two moieties. Nonetheless, MccE492 can also be isolated from bacterial culture supernatants as an unmodified peptide, which lacks the siderophore moiety (u-MccE492), and intermediate forms (u-McE492-Glc-DHBS and u-MccE492-Glc-DHBS2), which were shown to be degradation products (29).
Ten genes (mceABCDEFGHIJ) located on the bacterial chromosome of Klebsiella pneumoniae RYC492 (Fig. 1A) are required for MccE492 production, export, and posttranslational modification biosynthesis and the immunity of the producing strain (18), with four genes (mceC, mceD, mceI, and mceJ) being necessary for the acquisition of the siderophore moiety (22, 29). MceC and MceD are responsible for the C glucosylation of Ent and the cleavage of one of the trilactone ester bonds of Glc-Ent, respectively. The MceIJ complex is required for attachment of the siderophore moiety (linear Glc-Ent) to the C-terminal serine of the microcin precursor via an ester bond (23). The resulting posttranslational modification (siderophore moiety) enhances the antibacterial activity of MccE492, which is mainly directed against Escherichia coli, and MccE492 targets bacteria via the catecholate siderophore receptors (6, 28).
FIG. 1.
Genetic organization of class IIb microcin gene clusters. The name of the known or putative microcins produced by the gene cluster in each strain is indicated in parentheses. Genes are indicated by arrows whose orientations refer to the direction of gene transcription. Genes encoding microcin precursors are shown in yellow, while the genes required for self-immunity, microcin export, and posttranslational modification are shown in red, blue, and green, respectively. Genes with unknown function are shown in gray. Genes encoding homologous proteins in different gene clusters are colored by different shades of the same color. The names of the genes are indicated above or below each gene. (A) Genetic organization in five wild-type strains. In K. pneumoniae RYC492, genes with speculative functions are shown in gray, and the colors corresponding to the speculative functions are shown in stripes. Genes insC, tra-5, and insE, which encode transposases, are in brown. The names of the genes specifically required for the production of the MccE492, MccG492, MccH47, MccI47, and MccM precursors and immunity proteins are in green, brown, purple, blue, and red, respectively. Truncated genes in K. pneumoniae RYC492, E. coli H47, and E. coli Nissle 1917 are shown with slashes. (B) Genetic organization in recombinant plasmids pMM75 and pMM1.15 used for the production of MccM and MccH47, respectively. The kanamycin resistance cassette is shown in white, and inactivated genes are indicated by a cross. The names of the genes specifically involved in the production of MccM, MccH47, and MccI47 precursor and immunity proteins are in red, purple, and blue, respectively.
In order to identify potential novel siderophore-microcins, we have performed an in-depth analysis of the microcin gene clusters found in several microcinogenic strains (9). While several genetic studies support the existence of other siderophore-microcins (25), no biochemical evidence is available to date, with MccE492 being the only member of this family to have been isolated and characterized. We focused on MccM, MccH47, and MccI47, three microcins potentially carrying a posttranslational modification similar to that carried by MccE492. The gene clusters encoding these microcins are found in the genomes of E. coli H47, CA46, and CA58 and the probiotic E. coli Nissle 1917 (Fig. 1A), as well as in the pathogenicity island of uropathogenic E. coli (9). All of these strains were therefore analyzed for the production of siderophore-microcins.
Here we report for the first time on the isolation and structure characterization of MccM and MccH47, which are derived from the microcin precursors McmA and MchB, respectively. Both peptides were shown to carry a 831-Da posttranslational modification similar to that carried by MccE492, and the genes required for MccM and MccH47 posttranslational modification biosynthesis were shown to be interchangeable. Altogether our data provide biochemical evidence of the existence of a structure-related family of microcins. This family of siderophore-microcins gathers the class IIb microcins, namely, MccE492, MccM, and MccH47, as well as uncharacterized MccI47. Finally, a novel putative siderophore-microcin was identified in the corrected sequence of the MccE492 gene cluster in K. pneumoniae RYC492.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The strains and plasmids used for microcin production and the study of the mechanisms of action are described in Table 1. The bacteria used to determine the spectra of activity have been described previously (7). Recombinant plasmid pMM21 was constructed by inserting a 6.2-kb HindIII/EcoRI fragment containing mchA to mchX from E. coli H47 into the pBR322 vector containing the mchI to mcmM region (10.1 kb) from E. coli Nissle 1917 (F. Moreno, unpublished results). Microcin production was performed with wild-type strains Nissle 1917, CA46, and CA58 or the E. coli MC4100 recombinant strain transformed with plasmids. The plasmids used for the production of MccM and MccH47 were pMM75 and pMM1.15, respectively (Fig. 1B). Both plasmids were constructed from pMM21 by insertion of a 1.8-kb kanamycin resistance cassette inside mceB and mcmA, respectively (F. Moreno, unpublished results). In order to verify these constructions, plasmids pMM21 (20.2 kb), pMM75 (22 kb), and pMM1.15 (22 kb) were fully sequenced (Eurofins MWG Operon).
TABLE 1.
Bacterial strains and plasmids used for this study
| Strain or plasmid | Relevant features | Reference or source |
|---|---|---|
| E. coli strains | ||
| MC4100 | araD139 Δ(argF lac)U169rpsL relAflbBdeoC | Laboratory collection |
| RYC1000 | MC4100 Δrbs-7 recA56 gyrA | 12 |
| W3110 | F− IN(rrnD-rrnE)1 | Laboratory collection |
| KP1344 | W3110 (tonB::blaM) | 19 |
| W3110-6 | W3110 Δ(exbB-exbD) | S. P. Howard, unpublished data |
| H1443 | MC4100 aroB | 15 |
| H873 | H1443 fepA::Tn10 | K. Hantke, unpublished data |
| H1596 | H1443 fiu::Mud X | K. Hantke, unpublished data |
| H2222 | H1443 cir::Mud X | K. Hantke, unpublished data |
| H1728 | H1443 cir fiu::Mud X | 14 |
| H1875 | H1443 cir::Mud X fepA::Tn10 | 14 |
| H1877 | H1443 fepA::Tn10 fiu::Mud X | 14 |
| H1876 | H1728 fepA::Tn10 | 14 |
| CA46 CIP 105662 | Institut Pasteur collection | |
| CA58 CIP 105663 | Institut Pasteur collection | |
| Plasmids | ||
| pJAM229 | Derived from pHC79, carrying MccE492 gene cluster, Ampr | 30 |
| pMM21 | Carrying MccM, MccI47, and MccH47 gene clusters; Ampr | F. Moreno, unpublished data |
| pMM75 | Carrying MccM, MccI47, and MccH47 gene clusters; mchB inactivation; Ampr Kanr | F. Moreno, unpublished data |
| pMM1.15 | Carrying MccM, MccI47, and MccH47 gene clusters; mcmA inactivation; Ampr, Kanr | F. Moreno, unpublished data |
| pMcMi | Carrying mcmI from MccM gene cluster, Ampr | F. Moreno, unpublished data |
| pMcHi | Carrying mchI from MccH47 gene cluster, Cmr | F. Moreno, unpublished data |
MccE492 was expressed in E. coli MC4100(pJAM229), as described previously (28). For microcin production, E. coli MC4100 was freshly transformed with the plasmid harboring the microcin gene cluster of interest. The transformants were grown in M63 minimal medium supplemented with 2.5 g/liter glucose, 0.25 g/liter MgSO4, and 1 mg/liter thiamine. For E. coli Nissle 1917, CA46, and CA58 and E. coli MC4100(pJAM229) cultures, M63 medium was supplemented with 1 g/liter Bacto tryptone (BD Biosciences). The following antibiotics were used at the indicated concentrations: ampicillin and kanamycin at 50 μg/ml and chloramphenicol at 30 μg/ml. Cultures were routinely performed for 15 h at 37°C with vigorous shaking (250 rpm).
Microcin purification.
MccE492 was purified as described previously (28). MccM and MccH47 were purified as follows. Culture supernatants were separated from bacterial cells by centrifugation (10,000 × g, 15 min, 4°C) and were subjected to solid-phase extraction on a Sep-Pak C8 cartridge (Waters Corp.) preequilibrated with 0.1% aqueous trifluoroacetic acid (TFA). The cartridges were washed with 0.1% aqueous TFA prior to stepwise elution with 30%, 35%, 40%, 45%, and 50% acetonitrile (ACN) in 0.1% aqueous TFA at a flow rate of 10 ml/min. The detection of MccM- and MccH47-specific antibacterial activities in the Sep-Pak fractions was performed by radial diffusion assays, as described below. The 40% ACN Sep-Pak fraction containing MccM and the 50% ACN Sep-Pak fraction containing MccH47 were lyophilized and resuspended in 50% ACN in 0.1% aqueous TFA. The MccM-containing fraction was further purified by reversed-phase high-performance liquid chromatography (RP-HPLC) on an Inertsil ODS2 C18 column (5 μm, 4.6 by 250 mm; Interchim, France). Separation was performed at a flow rate of 1 ml/min with a linear gradient of 20 to 60% ACN in 0.1% aqueous TFA over 20 min. The fractions issued from the Sep-Pak cartridge and the HPLC column were collected by hand, according to their absorbance at 226 nm.
Antibacterial assays.
The microcin-susceptible strain (E. coli RYC1000) and the strains carrying the MccM or the MccH47 immunity gene on a plasmid [E. coli RYC1000(pMcMi) and E. coli RYC1000(pMcH1), respectively] were grown in Luria-Bertani (LB) medium to an optical density at 620 nm (OD620) of 0.3. Ten milliliters of M63-tryptone medium (containing 6.5 g/liter agar) was inoculated with 100 μl of bacteria, i.e., 107 CFU/ml. Petri dishes containing 40 ml M63 solid medium (20 g/liter agar) were overlaid with the bacterial suspension. After solidification, each fraction to be analyzed was spotted onto the overlay. After a 16-h incubation at 37°C, the plates were analyzed for the presence of inhibition halos. Fractions inhibitory to microcin-susceptible strain E. coli RYC1000 but not to microcin-resistant strains E. coli RYC1000(pMcMi) and RYC1000(pMcHi) were considered to contain MccM and MccH47, respectively. Fractions inhibitory to E. coli RYC1000, E. coli RYC1000(pMcMi), and E. coli RYC1000(pMcHi) but not to E. coli RYC1000(pMM21) were considered to contain MccI47. Time course bactericidal assays were performed for MccM and MccH47 by monitoring the culturability of E. coli W3110 exposed to microcins. Briefly, a bacterial suspension in exponential phase of growth was diluted in poor-broth medium (10 g/liter Bacto tryptone, 5 g/liter NaCl) to obtain a final OD620 of 0.001. Seventy-five microliters of this suspension was then incubated with 25 μl of the MccM- or MccH47-containing fraction or an equivalent volume of solvent (40% ACN in 0.1% aqueous TFA). At different times of incubation (0, 5, 10, 15, 30, 45, 60, and 120 min), each fraction was plated onto LB agar plates. The colonies were counted after 16 h of incubation at 37°C.
SDS-polyacrylamide gel electrophoresis (PAGE) and gel overlay.
An MccM-containing fraction was loaded onto three lanes of a 16.5% sodium dodecyl sulfate (SDS)-tricine polyacrylamide gel (27). After electrophoresis, the three lanes were cut out from the gel and treated separately. The first piece of gel, which contained the prestained protein marker (NE Biolabs) and the Mcc-containing fraction, was fixed and stained with Coomassie blue. The two other pieces of the gel were fixed overnight in 15% isopropanol-10% acetic acid, washed with deionized water for 4 h, and rinsed abundantly with sterile water. Every piece of gel was put into a sterile petri dish and overlaid with 40 ml M63 agar (0.7% [wt/vol] agar) containing 107 CFU/ml E. coli RYC1000, E. coli RYC1000(pMcMi), or E. coli RYC1000(pMcHi). The plates were incubated overnight at 37°C and examined for an Mcc-specific inhibition halo at the position of the Mcc migration band.
N-terminal sequencing.
Gel electrophoresis was performed as described above, and the proteins were subsequently transferred onto 0.1-μm-pore-size polyvinylidene difluoride membranes (Immobilon-PSQ; Millipore) for 1 h at 80 V in 25 mM Tris-190 mM glycine-20% ethanol-0.05% SDS buffer (pH 8.3). The membranes were stained (0.1% Coomassie blue in 1% acetic acid-40% methanol), destained (50% methanol), and dried. The band of interest was cut out for Edman degradation. N-terminal sequencing was performed on a Procise model protein sequencer (Applied Biosystems), according to the manufacturer's protocol.
Tryptic digestion.
Freeze-dried MccM (∼1.5 nmol) from the HPLC fraction was resuspended in 100 μl of 100 mM Tris-HCl (pH 7) and incubated with 20 μl of 1.3 μg/μl trypsin (Sigma) overnight at 37°C with vigorous shaking (250 rpm). The reaction was stopped by addition of 10 μl of 10% formic acid. The solution was then desalted on C18 tips (Omix; Varian) before mass spectrometry (MS) analysis.
Mass spectrometry.
Microcin-containing fractions were analyzed by matrix-assisted laser desorption ionization-time-of-flight MS (MALDI-TOF MS) on a Voyager-De-Pro MALDI-TOF mass spectrometer (Applied Biosystems), which was operated in the linear/positive mode and which used α-cyano-4-hydroxycinnamic acid as the matrix. The tryptic digests of MccM were analyzed by nanoelectrospray mass spectrometry on a qQ-TOF hybrid mass spectrometer (Q-Star Pulsar; Applied Biosystems) operated in the positive mode. Collision-induced dissociations were carried out on the triply charged ion of the N-terminal fragment of MccM, MccM(1-35) (m/z 1128), with a 35-V collision energy. Alternatively, the digests were analyzed by liquid chromatography (LC)-mass spectrometry on a Dionex U3000 micro-HPLC system connected to a Q-Star Pulsar Qq-TOF mass spectrometer operated in the positive mode. The separation was achieved on a Zorbax 300 SB C8 column (3.5 μm, 150 by 1 mm; Agilent). The elution gradient was 0 to 60% ACN in 0.1% formic acid over 20 min at a flow rate of 40 μl/min. Collision-induced dissociations were performed on the triply charged ions of the MccM(36-64) fragment (m/z 965) with a 30-V collision energy.
RESULTS
Production and purification of MccM and MccH47.
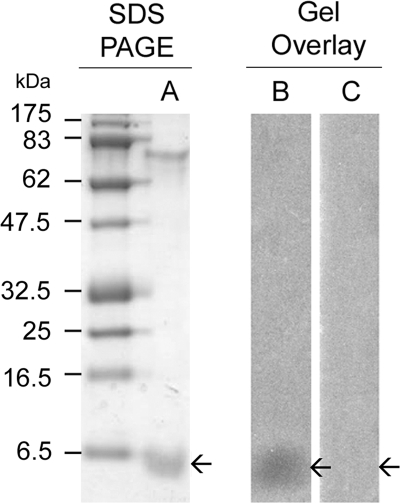
MccM and MccH47 were isolated from strain E. coli MC4100 transformed with pMM75 and pMM1.15, respectively. Both microcins were also isolated from wild-type strain E. coli Nissle 1917, CA46, or CA58 by a protocol similar to that described previously for MccE492 production (28). Whereas MccE492 was expressed in rather large amounts from plasmid pJAM229, 40 liters of culture supernatants of the MccM-producing strains was required for isolation, tryptic digestion, mass spectrometry analyses, and antibacterial assays. Both MccM and MccH47 were obtained by reversed-phase fractionation of the culture supernatants on Sep-Pak C8 cartridges. The fractions eluted with 30% to 50% ACN were submitted to antibacterial assays against a microcin-susceptible strain (E. coli RYC1000) and against various strains specifically resistant (immune) to MccM [E. coli RYC1000(pMcMi)], MccH47 [E. coli RYC1000(pMcHi)], and MccM/MccH47/MccI47 [E. coli RYC1000(pMM21)]. Fractions active against the susceptible strain but inactive against the MccM-immune strain contained MccM, while fractions inactive against the MccH47-immune strain contained MccH47. From these experiments, we concluded that MccM was recovered in 40% ACN Sep-Pak fractions from E. coli Nissle 1917, CA46, CA58, and MC4100(pMM75), whereas the 50% ACN Sep-Pak fractions from E. coli Nissle 1917, CA46, CA58, and MC4100(pMM1.15) contained MccH47. The 40% and 50% ACN Sep-Pak fractions were subjected to MS and SDS-PAGE. Gel overlay experiments revealed a single band with antibacterial activity in the 40% Sep-Pak fraction isolated from the E. coli Nissle 1917, CA46, CA58, and MC4100 harboring pMM75 (Fig. 2). From its molecular weight and MccM-specific activity, we concluded that the peptide band contained MccM. No MccH47-specific antibacterial activity could be detected by gel overlay in the 50% ACN Sep-Pak fractions from E. coli Nissle 1917, CA46, CA58, or MC4100 harboring pMM1.15, presumably due to the low level of expression of MccH47. MALDI-TOF MS analysis of the 40% and 50% ACN fractions showed ions at m/z 7284 and 4865, respectively. These ions corresponded to the calculated masses of MccM and MccH47, respectively, as deduced from the sequence of the microcin precursors McmA and MchB after cleavage at their expected maturation sites (see below). Neither the MccM nor the MccH47 Sep-Pak fractions led to the isolation of highly purified microcins. MccM-containing fractions could be further purified by RP-HPLC on a C18 column. N-terminal sequencing of MccM indicated a peptide purity of ∼90% (data not shown). In contrast, very small amounts of MccH47 could be isolated from the 50% ACN Sep-Pak fractions, prohibiting further purification by RP-HPLC. All attempts to isolate sufficient amounts of MccI47 from the Sep-Pak fractions were unsuccessful, even under iron-starvation culture conditions (25). Nevertheless, MALDI-TOF MS analysis of the 40% ACN fraction from the E. coli MC4100(pMM1.15) supernatant showed one ion at m/z 6275, which corresponds to the theoretical molecular mass of the putative MccI47.
FIG. 2.
SDS-PAGE and detection of MccM isolated from E. coli MC4100(pMM75) by gel overlay. Following separation on a 16.5% SDS-tricine polyacrylamide gel, the MccM-containing fraction was stained with Coomassie blue (lane A), subjected to antibacterial assay by gel overlay with the microcin-susceptible strain E. coli RYC1000 (lane B), and submitted to antibacterial assay by gel overlay with MccM-resistant strain E. coli RYC1000(pMcMi) (lane C). The molecular mass ladder (in the first unlabeled lane) was the prestained broad range protein marker (NE Biolabs). Arrows indicate the locations of the band corresponding to MccM.
MccM and MccH47 display bactericidal activity against phylogenetically related bacteria.
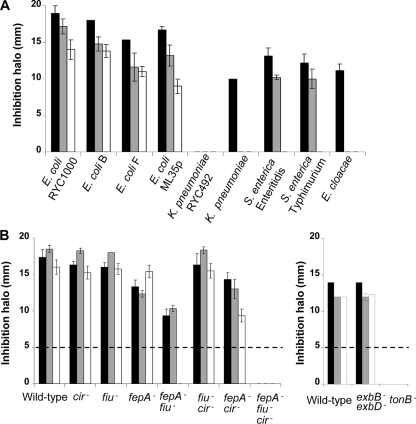
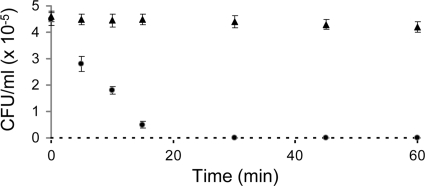
MccM and MccH47 were compared to MccE492 in terms of their antibacterial activities. The three microcins revealed similar spectra of activity (Fig. 3A). No activity against the gram-positive bacteria tested (Staphylococcus and Bacillus; data not shown) could be detected, and the E. coli strains were the most susceptible among the gram-negative bacteria tested (Fig. 3A). While MccM and MccE492 were less active against Salmonella than against E. coli, at the concentrations tested, MccH47 was inactive against Salmonella. MccE492 was the only microcin active against Enterobacter cloacae and Klebsiella pneumoniae. Like MccE492, MccM and MccH47 were bactericidal, as deduced from the culturability of microcin-treated E. coli (no countable CFU after a 30-min exposure to the microcins) (Fig. 4). Controls in the absence of microcins indicated no spontaneous loss of culturability.
FIG. 3.
Comparative antibacterial activities of MccE492 (black bars), MccM (gray bars), and MccH47 (white bars) against wild-type and mutant strains. Antibacterial activity is expressed by the size (in mm) of the inhibition halo. Values are presented as the means of three independent experiments with standard errors. (A) Antibacterial activity against gram-negative bacterial strains of different genera (E. coli, K. pneumoniae, Salmonella enterica, Enterobacter cloacae); (B) antibacterial activity against isogenic strains carrying mutations affecting the siderophore receptors FepA, Fiu, and Cir and associated proteins TonB, ExbB, and ExbD. The limit of detection (inhibition halo of 5 mm) is indicated by a broken line.
FIG. 4.
Kinetics of bacterial killing. MccM-containing fractions (•) or solvent (▴) were added to an exponential-phase culture of E. coli W3110. After different incubation times, the fractions were diluted and plated on LB agar. The surviving bacteria were counted (as the numbers of CFU/ml) after overnight incubation at 37°C. Error bars represent the standard deviations for three independent experiments. The reliable limit of detection (100 colonies/ml) is indicated by a broken line.
Siderophore receptors FepA, Fiu, Cir, and associated protein TonB are necessary for MccM and MccH47 antibacterial activity.
In order to identify the outer membrane receptors required for the recognition of MccM and MccH47, antibacterial assays were performed with strains carrying mutations in siderophore receptor genes. In previous studies, TonB and the catecholate siderophore receptors FepA, Fiu, and Cir were shown to be necessary for MccE492 recognition (7). We therefore compared the susceptibilities to MccE492, MccM, and MccH47 of wild-type and isogenic E. coli strains carrying mutations in the genes encoding FepA, Cir, and Fiu. We also examined the antibacterial activities of the three microcins against isogenic strains lacking the associated inner membrane proteins TonB, ExbB, and ExbD (Fig. 3B). For the three microcins, the lack of the Fiu or the Cir receptor did not significantly alter the antibacterial activities compared to the activity detected against wild-type strain E. coli H1443. In contrast, the diameters of the inhibition halos for MccM and MccE492 decreased significantly with fepA inactivation. When FepA and Fiu were nonfunctional, the inhibition halos were smaller than those observed with the wild-type strain, which suggests that of the three receptors Cir is the least efficient to promote microcin uptake. All three microcins were unable to inhibit the growth of the fepA fiu cir triple mutant and the tonB mutant. In contrast, deletion of both exbB and exbD had no noticeable effect.
The MccM amino acid sequence derives from the microcin precursor McmA sequence.
The MccM-containing HPLC fraction was subjected to SDS-PAGE and Edman degradation. A 17-residue N-terminal sequence (DGNDGQAELIAIGSLAG), which corresponds to the McmA sequence after removal of a 15-residue N-terminal sequence, could be determined for MccM. This cleavage occurs at a similar site in MccE492 (Fig. 5A). MccM was also sequenced by mass spectrometry from a tryptic digest. Two peptide fragments corresponding to the Asp1-Lys35 N-terminal region and the Val36-Arg64 central region of MccM were purified and analyzed by micro-LC-tandem MS (LC/MS/MS). As with Edman degradation, the complete sequences of both fragments obtained by MS/MS fragmentation (Fig. 5B) corresponded to the McmA sequence obtained after removal of the 15-residue leader peptide. Taken together, these results unambiguously indicate that the peptide moiety of MccM is a 7,283-Da and 77-residue peptide, whose sequence is given in Fig. 5B.
FIG. 5.
Amino acid sequences of siderophore-microcins. (A) Multiple-sequence alignment of the N- and C-terminal regions of class IIb microcin precursors. The alignments were performed with the ClustalW program. The names of the isolated or putative (in italics) microcins are indicated in parentheses. The numbers in angled brackets refer to the number of amino acids in the unaligned central regions. Dash indicates gaps. The amino acid sequences for all microcins except MceL (this study) are from the UniProt KB database. MceA (accession no. Q9Z4N4), MceL, MchB (accession no. P62530), McmA (accession no. Q83TS1), and MchS2 (accession no. Q712Q0) correspond to the MccE492, MccG492, MccH47, MccM, and MccI47 precursors, respectively. The known (MceA, McmA, MchB) or putative (MchS2, MceL) cleavage site of the microcin precursors is shown with a double-headed arrow. Letters on gray backgrounds highlight identical amino acids. (B) Amino acid sequences of the MccM, MccH47, and the putative MccI47 and MccG492 peptide moieties. For MccM, the amino acids determined by N-terminal sequencing are underlined. Arrows indicate trypsin cleavage sites. The amino acid sequence obtained by MS/MS fragmentation of the Asp1-Lys35 and the Val36-Arg64 tryptic peptides of MccM is indicated by a broken line. The sequences of MccI47 and MccG492 were deduced from the sequences of the microcin precursors MchS2 (UniProt KB accession no. Q712Q0) and MceL (this study), respectively.
MccM and MccH47 carry the same posttranslational modification as MccE492.
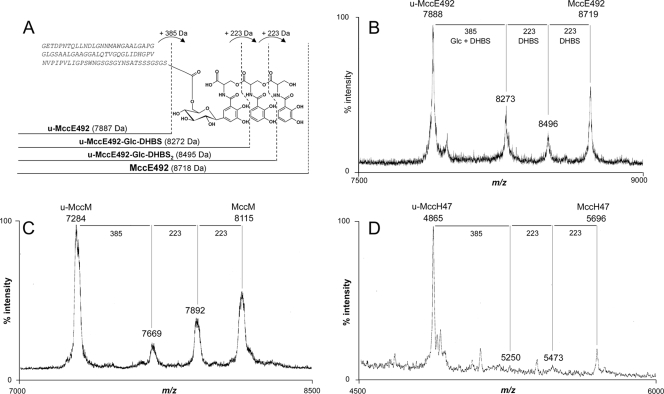
Mass spectrometry was used to determine whether MccM and MccH47 carry a posttranslational modification similar to that of MccE492 (Fig. 6A and B). MALDI-TOF MS analysis of the MccM produced by E. coli CA46 and CA58 revealed four ions at m/z 7284, 7669, 7892, and 8115 (Fig. 6C; Table 2). However, only the ion at m/z 7284, which corresponds to the MccM peptide moiety, was detected in an MccM preparation obtained from E. coli Nissle 1917 (Table 2). Similar experiments were carried out with MccH47. Ions at m/z 4865, 5250, 5473, and 5696 (Fig. 6D) were observed in an MccH47 preparation obtained from E. coli CA46 and CA58 and E. coli MC4100(pMM1.15) (Table 2). Again, as for MccM, when MccH47 was isolated from E. coli Nissle 1917, the only ion found corresponded to the MccH47 peptide moiety (m/z 4865) (Table 2), as was also observed with MccM. Careful analysis of the MALDI-TOF mass spectral data showed that the four ions found in the MccM (Fig. 6C) and the MccH47 (Fig. 6D) spectra displayed the same mass differences as those observed for MccE492 and its derivatives, u-MccE492, u-MccE492-Glc-DHBS, and u-MccE492-Glc-DHBS2 (Fig. 6A and B). These mass differences correspond to the peptide moiety carrying (i) one glucose molecule and one monomer of DHBS (385 Da), (ii) an additional monomer of DHBS (plus 223 Da), and (iii) a third monomer of DHBS (plus 223 Da), respectively.
FIG. 6.
Structures of the different forms of MccE492 and MALDI-TOF MS spectra of the siderophore-microcins. (A) MccE492 (8,718 Da) is a posttranslationally modified 84-residue peptide. The modification consists of a trimer of 2,3-dihydroxybenzoylserine (DHBS) linked via a C-glycosidic bond to a β-d-glucose (Glc) that is itself linked to Ser84 through an O-glycosidic bond. u-MccE492 (7,887 Da) is the unmodified 84-residue peptide. u-MccE492-Glc-DHBS (8,272 Da) and u-MccE492-Glc-DHBS2 (8,495 Da) correspond to intermediate forms carrying a β-d-glucose linked to one DHBS and two DHBS moieties, respectively. The amino acid sequence is indicated in italics. The mass differences between the different structures are shown at the top of the panel. (B) MALDI-TOF MS spectra showing the different ions corresponding to the unmodified, intermediate, and modified forms of MccE492 obtained from the 40% Sep-Pak fraction of E. coli MC4100 harboring pJAM229; (C) MALDI-TOF MS spectra showing the different ions corresponding to the unmodified, intermediate, and modified forms of MccM obtained from the 40% Sep-Pak fraction of E. coli MC4100 harboring pMM75; (D) MALDI-TOF MS spectra showing the different ions corresponding to the unmodified, intermediate, and modified forms of MccH47 obtained from the 50% Sep-Pak fraction of E. coli MC4100 harboring pMM1.15.
TABLE 2.
Micocins produced by the different strains used in this study
| Strain | Microcin | Microcin produceda |
|
|---|---|---|---|
| Unmodified form | Modified form | ||
| K. pneumoniae RYC492 | MccE492 | + | + |
| E. coli Nissle 1917 | MccM | + | − |
| MccH47 | + | − | |
| E. coli CA46 | MccM | + | + |
| MccH47 | + | + | |
| E. coli CA58 | MccM | + | + |
| MccH47 | + | + | |
| E. coli MC4100(pMM75) | MccM | + | + |
| E. coli MC4100(pMM1.15) | MccH47 | + | + |
+, isolated from the culture supernatant; −, not present in the culture supernatant.
We next examined the interchangeability of MchA-McmL and MchS1-McmK, the two pairs of homologous proteins responsible for the acquisition of the posttranslational modification and encoded by two different microcin gene clusters. MccM was therefore expressed in recombinant E. coli MC4100 harboring pMM75. This plasmid carries the microcin gene clusters from E. coli Nissle 1917 as well as the mchA and mchS1 genes from E. coli H47 (Fig. 1B). Again, the MALDI-TOF mass spectra showed ions at m/z 7284, 7669, 7892, and 8115 (Table 2). Altogether, these results indicate that the ions at m/z 7669, 7892, and 8115 correspond to u-MccM-Glc-DHBS, u-MccM-Glc-DHBS2, and MccM under its modified form, respectively. Similarly, the ions at m/z 5250, 5473, and 5696 can be assigned to u-MccH47-Glc-DHBS, u-MccH47-Glc-DHBS2, and MccH47 under its modified form, respectively. Although the existence of isomeric forms of the siderophore moiety cannot be ruled out, altogether our data indicate (i) that MccM and MccH47 carry the same posttranslational modification and (ii) that the modification is most likely the same as that in MccE492.
Sequence analysis of the MccE492 gene cluster from K. pneumoniae RYC492 reveals a novel putative siderophore-microcin.
Analysis of the recently corrected nucleotide sequence of K. pneumoniae RYC492 (GenBank accession no. AF063590) revealed that the orientation of mceFGHIJ is the reverse of that described previously (18). Thus, the MccE492 gene cluster has the same organization on K. pneumoniae chromosomal DNA and on plasmid pJAM229 (data not shown). Furthermore, we identified six novel open reading frames (ORFs; mceS2, mceS3, mceM, mceL, mceX, and mceK). MceX exhibits amino acid sequence identity with the MchX encoded by different microcin gene clusters (Fig. 1A) and probably displays the same unknown function. MceK is composed of a fusion of two sequences that exhibit identity with McmA and McmI on their N- and C-terminal regions, respectively. Interestingly, mcmA and mcmI are also truncated in a similar fashion in E. coli H47 (Fig. 1A). However, only one ORF is observed in K. pneumoniae RYC492, suggesting deletions of the K. pneumoniae chromosomal DNA. MceS2 and MceS3 have been annotated as peptides similar to MchS2 and MchS3, the MccI47 precursor and self-immunity protein, respectively, and are therefore a putative class IIb microcin and a self-immunity protein, respectively.
The amino acid sequences of class IIb microcins show two main features: (i) cleavage of an N-terminal leader peptide after a double glycine or a Gly-Ala motif and (ii) a C-terminal moiety rich in serine and glycine residues (Fig. 5A) (9). Nevertheless, MceS2 does not display either of these two characteristics, thus ruling out the hypothesis that it may be a novel microcin precursor. Besides, two other new genes, mceM and mceL, seemed to be particularly interesting. MceL is a putative 89-residue peptide. Its amino acid sequence exhibits the two mains features of class IIb microcins: (i) it displays a putative cleavage site at Gly15-Ala16 and a Ser- and Gly-rich C-terminal region (Fig. 5A), and (ii) it shows strong sequence identity at both ends with all other known siderophore-microcins (Fig. 5A). Furthermore, MALDI-TOF MS analysis of the 45% ACN fraction from the E. coli MC4100(pJAM229) supernatant showed one ion at m/z 7342, which is compatible with the theoretical molecular mass of the MceL precursor carrying a 831-Da posttranslational modification after removal of a 15-residue N-terminal sequence (Fig. 5A). Preceding rather than following the putative microcin gene mceL, a potential immunity gene, mceM, is found in the gene cluster. The sequence of its deduced amino acid, MceM, exhibits a number of residues similar to the numbers of MceB, MchB, and McmI, which confer immunity to MccE492, MccH47, and MccM, respectively, and contains putative transmembrane regions. Those two features are conserved in class IIb microcin self-immunity proteins (9). Therefore, MceM is most likely the self-immunity protein associated with MceL. Altogether, these data strongly support the existence of a novel siderophore-microcin encoded by K. pneumoniae RYC492 that we name MccG492 (Fig. 5B).
DISCUSSION
This article describes for the first time the isolation, purification, and characterization of MccM and MccH47. While the putative posttranslational modifications and functions of MccM, MccH47, and MccI47 have been the subject of many hypotheses, all of which relied on genetic studies, we provide here the first biochemical evidence that MccM and MccH47 carry the same posttranslational modification as MccE492. We demonstrate that among the microcins, MccM, MccH47, and MccE492 form a homogeneous family, the siderophore-microcins. We also show that several genes from the MccE492, MccM, MccH47, and MccI47 gene clusters are interchangeable and display similar functions.
MccM and MccH47 were produced from several wild-type and recombinant strains. As could be expected from the absence of biochemical data for these two peptides in the existing literature, both MccM and MccH47 proved to be difficult to isolate from culture supernatants, and only MccM could be obtained in sufficient amounts to be sequenced. Together with Edman sequencing, our electrospray ionization MS data demonstrated that MccM results from the cleavage of McmA between Gly15 and Asp16. Similarly, MccH47 was shown to result from the cleavage of MchB between Ala15 and Gly16. Thus, the MccM and MccH47 15-residue leader peptides are cleaved after the typical double-glycine or glycine-alanine motifs, as is the case for MccE492 and some bacteriocins (9, 17) (Fig. 5A). Edman degradation and MS/MS sequencing covered 83% of the MccM sequence, and the sequence agreed with the amino acid sequence deduced from the mcmA nucleotide sequence. All masses and sequences were in agreement with the amino acid sequences of MccM and MccH47 (Fig. 5B) deduced from the microcin precursor genes mcmA and mchB, respectively (9, 26).
Analysis of the different microcin gene clusters (Fig. 1A) revealed the presence of several homologous genes. The predicted amino acid sequences of MceC, MchA, and McmL exhibited 85% identity, while those of MceD, MchS1, and McmK exhibited 71% identity. Previous in vivo (29) and in vitro (22) experiments have shown that MceC and MceD are required for MccE492 posttranslational modification biosynthesis. Furthermore, MceI and MceJ, which are necessary for the linkage of the C-glucosylated linear Ent to the MceA C-terminal serine during MccE492 biosynthesis (23), showed 80% and 71% amino acid sequence identities with MchD and MchC, respectively. Thus, all genes encoding proteins homologous to MceC/MceD/MceI/MceJ would share the same functions. Interestingly, both MccM and MccH47 isolated from E. coli CA46 and CA58 and from recombinant E. coli MC4100 harboring pMM75 or pMM1.15, which includes the genes potentially required for the posttranslational modification biosynthesis, were shown here to carry a 831-Da posttranslational modification similar to that of MccE492. The microcins isolated from the E. coli Nissle 1917 culture supernatants were also found not to carry any posttranslational modification. In genomic island I from E. coli Nissle 1917, an iroA locus, which contains iroB and iroD homologous to mchA/mcmL and mchS1/mcmK, respectively, is located 14 kb downstream of the microcin gene cluster (13). The absence of posttranslational modification in E. coli Nissle 1917 demonstrates the necessity for mchA and mchS1 in the biosynthesis of this modification and supports the idea that iroB and iroD are not involved in MccM and MccH47 maturation. The key role of mchA and mchS1 is also supported by the isolation of siderophore-microcins (Table 2) from an E. coli MC4100 recombinant strain harboring pMM75, a plasmid which contains the microcin gene cluster from E. coli Nissle 1917 linked to mchA and mchS1 from E. coli H47. In this study we have also shown that MccH47 can be posttranslationally modified by either MchA/MchS1 in E. coli MC4100(pMM1.15) or McmL/McmK in E. coli CA46 and CA58 (Table 2). Altogether, the MS data and the findings of the complementation experiments show that MccM and MccH47 carry the same posttranslational modification as MccE492 and that this results from conserved enzyme machinery. Thus, the MchA and McmL proteins and the MchS1 and McmK proteins, which display the same potential roles as MceC and MceD, respectively, are required for MccM and MccH47 posttranslational modification biosynthesis, respectively. Although MccI47 has not been isolated to date, mass spectrometry and genetic data strongly suggest that this microcin carries a similar posttranslational modification.
Consequently, (i) MccE492, MccM, MccH47, and, most probably, MccI47 belong to the siderophore-microcin family; and (ii) the posttranslational modification requires MchA, McmL, and MceC, on the one hand, and MchS1, McmK, and MceD, on the other hand, all of which are most likely interchangeable and display the same function in the posttranslational modification biosynthesis. Thus, MchA/McmL/MceC and MchS1/McmK/MceD are enzymes potentially responsible for the C glucosylation of Ent and the cleavage of an ester bond of Glc-Ent, respectively. Similarly, we propose that MceJ/MchC and MceI/MchD, which are also required for posttranslational modification (22, 23), could be interchangeable, in agreement with the findings of Poey et al. (25). We speculate that this interchangeability might extend to all genes belonging to one of the five siderophore-microcin gene clusters located on K. pneumoniae RYC492 and E. coli CA46, CA58, H47, and Nissle 1917, except for genes encoding the microcin precursor and its self-immunity protein.
Studies of the mechanisms of action of MccM and MccH47 (this work) showed that, like MccE492, both microcins are recognized by the catecholate siderophore receptors. Genes encoding catecholate receptors are present on the genomes of E. coli (2, 21), Salmonella enterica serovar Typhimurium (20), as well as K. pneumoniae (11), all of which are susceptible to siderophore-microcins. The TonB protein associated with these receptors is also required for the antibacterial activities of MccM and MccH47, but ExbB and ExbD are not essential. It was previously demonstrated that TolQ and TolR can complement ExbB and ExbD, respectively, as regards E. coli susceptibility to group B colicins (3). Similarly, MccM and MccH47 could also use TolQ and TolR for their translocation when exbB and exbD are inactivated. Consequently, all the siderophore-microcins potentially use a “Trojan horse” mechanism (10) and are recognized as siderophores by outer membrane receptors.
Finally, the nucleotide sequence of K. pneumoniae RYC492 has recently been corrected, unveiling six novel ORFs, among which two may encode novel microcin precursors. Analysis of the N- and C-terminal sequences of MceS2 reveals that this peptide does not display the structural characteristics of the class IIb microcin precursors. Consequently, MceS2 is probably not a new class IIb microcin precursor. On the contrary, the N- and C-terminal sequences of MceL (Fig. 5A) exhibit the two features of class IIb microcins and MceM displays the characteristics of a self-immunity protein (see Results). Therefore, we propose that K. pneumoniae RYC492 produces a novel microcin, which we name MccG492, with a microcin precursor encoded by mceL and a self-immunity protein encoded by mceM. The genetic organization of MccE492/MccG492 seems to be similar to that of MccM/MccH47/MccI47, with specific genes encoding microcin precursors and self-immunity proteins and common genes for export and posttranslational modification acquisition. Both its C-terminal sequence and the location of the encoding gene in a class IIb microcin gene cluster suggest that MccG492 most probably carries the same posttranslational modification as all other class IIb siderophore-microcins.
Therefore, we provide here for the first time biochemical evidence of the existence of a family of structure-related microcins secreted by members of the Enterobacteriaceae family. The family of siderophore-microcins corresponds to class IIb of microcins described previously (9) and contains three members (MccE492, MccM, and MccH47), which have been isolated, purified, and characterized. Two additional putative members (Fig. 5B), which remain to be isolated (MccG492 and MccI47), most probably belong to this family. This is the first family of structure-related peptides among the antimicrobial peptides of gram-negative bacteria. In contrast, several families of bacteriocins, including lantibiotics and pediocin-like and two-peptide bacteriocins, each of which is exemplified by hundreds of peptides, have been described among the antimicrobial peptides of gram-positive bacteria (4, 8, 24).
The persistence of numerous genes encoding several microcins in a single gene cluster raises fundamental questions. Why do these bacteria synthesize several microcins which appear to have similar behavior? It is generally accepted that useless genes are eliminated from dynamic genomes. E. coli and K. pneumoniae do not appear to eliminate different genes encoding several siderophore-microcins. Each siderophore-microcin possibly has specific functions, but these remain to be elucidated.
Acknowledgments
We are grateful to Klaus Hantke and Felipe Moreno for the generous gifts of the E. coli strains. We thank Manon Vandervennet and Gérard Gastine for careful technical assistance with the bacteriology. We thank Florent Vandervennet for careful technical assistance with the microcin purification. We are also grateful to Alyssa Carré-Mlouka for providing helpful comments on the manuscript. We thank Paul Smith for the English revision. We thank Robert Thai for the N-terminal sequencing. We thank the mass spectrometry facility (Plate-Forme de Spectrométrie de Masse) at the National Museum of Natural History and Séverine Zirah for assistance with mass spectrometry.
Footnotes
Published ahead of print on 2 November 2009.
REFERENCES
- 1.Bister, B., D. Bischoff, G. J. Nicholson, M. Valdebenito, K. Schneider, G. Winkelmann, K. Hantke, and R. D. Sussmuth. 2004. The structure of salmochelins: C-glucosylated enterobactins of Salmonella enterica. Biometals 17:471-481. [DOI] [PubMed] [Google Scholar]
- 2.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 3.Braun, V., and C. Herrmann. 1993. Evolutionary relationship of uptake systems for biopolymers in Escherichia coli: cross-complementation between the TonB-ExbB-ExbD and the TolA-TolQ-TolR proteins. Mol. Microbiol. 8:261-268. [DOI] [PubMed] [Google Scholar]
- 4.Cotter, P. D., C. Hill, and R. P. Ross. 2005. Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3:777-788. [DOI] [PubMed] [Google Scholar]
- 5.Crosa, J. H., and C. T. Walsh. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66:223-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Destoumieux-Garzón, D., J. Peduzzi, X. Thomas, C. Djediat, and S. Rebuffat. 2006. Parasitism of iron-siderophore receptors of Escherichia coli by the siderophore-peptide microcin E492m and its unmodified counterpart. Biometals 19:181-191. [DOI] [PubMed] [Google Scholar]
- 7.Destoumieux-Garzón, D., X. Thomas, M. Santamaria, C. Goulard, M. Barthélémy, B. Boscher, Y. Bessin, G. Molle, A. M. Pons, L. Letellier, J. Peduzzi, and S. Rebuffat. 2003. Microcin E492 antibacterial activity: evidence for a TonB-dependent inner membrane permeabilization on Escherichia coli. Mol. Microbiol. 49:1031-1041. [DOI] [PubMed] [Google Scholar]
- 8.Drider, D., G. Fimland, Y. Hechard, L. M. McMullen, and H. Prevost. 2006. The continuing story of class IIa bacteriocins. Microbiol. Mol. Biol. Rev. 70:564-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duquesne, S., D. Destoumieux-Garzón, J. Peduzzi, and S. Rebuffat. 2007. Microcins, gene-encoded antibacterial peptides from enterobacteria. Nat. Prod. Rep. 24:708-734. [DOI] [PubMed] [Google Scholar]
- 10.Duquesne, S., V. Petit, J. Peduzzi, and S. Rebuffat. 2007. Structural and functional diversity of microcins, gene-encoded antibacterial peptides from enterobacteria. J. Mol. Microbiol. Biotechnol. 13:200-209. [DOI] [PubMed] [Google Scholar]
- 11.Fouts, D. E., H. L. Tyler, R. T. DeBoy, S. Daugherty, Q. Ren, J. H. Badger, A. S. Durkin, H. Huot, S. Shrivastava, S. Kothari, R. J. Dodson, Y. Mohamoud, H. Khouri, L. F. Roesch, K. A. Krogfelt, C. Struve, E. W. Triplett, and B. A. Methe. 2008. Complete genome sequence of the N2-fixing broad host range endophyte Klebsiella pneumoniae 342 and virulence predictions verified in mice. PLoS Genet. 4:e1000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Genilloud, O., M. C. Garrido, and F. Moreno. 1984. The transposon Tn5 carries a bleomycin-resistance determinant. Gene 32:225-233. [DOI] [PubMed] [Google Scholar]
- 13.Grozdanov, L., C. Raasch, J. Schulze, U. Sonnenborn, G. Gottschalk, J. Hacker, and U. Dobrindt. 2004. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J. Bacteriol. 186:5432-5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hantke, K. 1990. Dihydroxybenzoylserine—a siderophore for E. coli. FEMS Microbiol. Lett. 55:5-8. [DOI] [PubMed] [Google Scholar]
- 15.Hantke, K. 1983. Identification of an iron uptake system specific for coprogen and rhodotorulic acid in Escherichia coli K12. Mol. Gen. Genet. 191:301-306. [DOI] [PubMed] [Google Scholar]
- 16.Hantke, K., G. Nicholson, W. Rabsch, and G. Winkelmann. 2003. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc. Natl. Acad. Sci. U. S. A. 100:3677-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Håvarstein, L. S., D. B. Diep, and I. F. Nes. 1995. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 16:229-240. [DOI] [PubMed] [Google Scholar]
- 18.Lagos, R., M. Baeza, G. Corsini, C. Hetz, E. Strahsburger, J. A. Castillo, C. Vergara, and O. Monasterio. 2001. Structure, organization and characterization of the gene cluster involved in the production of microcin E492, a channel-forming bacteriocin. Mol. Microbiol. 42:229-243. [DOI] [PubMed] [Google Scholar]
- 19.Larsen, R. A., M. G. Thomas, and K. Postle. 1999. Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol. Microbiol. 31:1809-1824. [DOI] [PubMed] [Google Scholar]
- 20.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 21.Molloy, M. P., B. R. Herbert, M. B. Slade, T. Rabilloud, A. S. Nouwens, K. L. Williams, and A. A. Gooley. 2000. Proteomic analysis of the Escherichia coli outer membrane. Eur. J. Biochem. 267:2871-2881. [DOI] [PubMed] [Google Scholar]
- 22.Nolan, E. M., M. A. Fischbach, A. Koglin, and C. T. Walsh. 2007. Biosynthetic tailoring of microcin E492m: post-translational modification affords an antibacterial siderophore-peptide conjugate. J. Am. Chem. Soc. 129:14336-14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nolan, E. M., and C. T. Walsh. 2008. Investigations of the MceIJ-catalyzed posttranslational modification of the microcin E492 C-terminus: linkage of ribosomal and nonribosomal peptides to form “Trojan horse” antibiotics. Biochemistry 47:9289-9299. [DOI] [PubMed] [Google Scholar]
- 24.Oppegård, C., P. Rogne, L. Emanuelsen, P. E. Kristiansen, G. Fimland, and J. Nissen-Meyer. 2007. The two-peptide class II bacteriocins: structure, production, and mode of action. J. Mol. Microbiol. Biotechnol. 13:210-219. [DOI] [PubMed] [Google Scholar]
- 25.Poey, M. E., M. F. Azpiroz, and M. Laviña. 2006. Comparative analysis of chromosome-encoded microcins. Antimicrob. Agents Chemother. 50:1411-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodríguez, E., C. Gaggero, and M. Laviña. 1999. The structural gene for microcin H47 encodes a peptide precursor with antibiotic activity. Antimicrob. Agents Chemother. 43:2176-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 28.Thomas, X., D. Destoumieux-Garzón, J. Peduzzi, C. Afonso, A. Blond, N. Birlirakis, C. Goulard, L. Dubost, R. Thai, J. C. Tabet, and S. Rebuffat. 2004. Siderophore peptide, a new type of post-translationally modified antibacterial peptide with potent activity. J. Biol. Chem. 279:28233-28242. [DOI] [PubMed] [Google Scholar]
- 29.Vassiliadis, G., J. Peduzzi, S. Zirah, X. Thomas, S. Rebuffat, and D. Destoumieux-Garzón. 2007. Insight into siderophore-carrying peptide biosynthesis: enterobactin is a precursor for microcin E492 posttranslational modification. Antimicrob. Agents Chemother. 51:3546-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilkens, M., J. E. Villanueva, J. Cofré, J. Chnaiderman, and R. Lagos. 1997. Cloning and expression in Escherichia coli of genetic determinants for production of and immunity to microcin E492 from Klebsiella pneumoniae. J. Bacteriol. 179:4789-4794. [DOI] [PMC free article] [PubMed] [Google Scholar]