Abstract
A Pseudomonas fluorescens isolate (PF-1) resistant to carbapenems was recovered during an environmental survey performed with water from the Seine River (Paris). It expressed a novel Ambler class A carbapenemase, BIC-1, sharing 68 and 59% amino acid identities with β-lactamases SFC-1 from Serratia fonticola and the plasmid-encoded KPC-2, respectively. β-Lactamase BIC-1 hydrolyzed penicillins, carbapenems, and cephalosporins except ceftazidime and monobactams. The blaBIC-1 gene was chromosomally located and was also identified in two other P. fluorescens strains isolated from the Seine River 3 months later.
Pseudomonas fluorescens is a psychrotrophic bacterium that expresses a chromosomally encoded and inducible Ambler class C β-lactamase (10, 15). Carbapenemases (serine- or metallo-β-lactamases) remain the most common mechanism of resistance to carbapenems in Gram-negative organisms (22, 23). To date, most acquired metallo-β-lactamase (MBL)-encoding genes (blaIMP or blaVIM variants) have been reported from Pseudomonas aeruginosa and very rarely from P. fluorescens (8, 12). Class A carbapenemases are either chromosome encoded or plasmid encoded and remain rarely identified in Gram-negative organisms. Indeed, several class A carbapenemases are chromosomally encoded (NMC-A, SFC-1, SME-1 to -3, IMI-1), with the exception of the emerging KPC β-lactamases, which are plasmid encoded (24). The chromosomal location of the blaSFC-1 gene could be the result of a horizontal gene transfer into an environmental Serratia fonticola isolate (5). The blaSME genes from Serratia marcescens are presumed to be chromosomal. Genes encoding NMC-A and IMI β-lactamases have been found sporadically, either in clinical or in environmental isolates of Enterobacter cloacae and Enterobacter asburiae from rivers (2). The blaNMC-A gene was found chromosomally located in several clinical isolates. The blaIMI-1 gene was chromosomally located, whereas the blaIMI-2 genes were identified on plasmids. The imiR-imi-2 gene tandem in E. cloacae and E. asburiae appeared to be flanked by transposable elements (2). Class A carbapenemases such as GES-2, GES-5, and KPC-2 have been recently reported from P. aeruginosa (16) but not from other Pseudomonas species.
Here, we characterize a novel class A carbapenemase identified from a P. fluorescens environmental isolate. This identification occurred during a survey aimed to study the spread of multidrug-resistant Gram-negative organisms in the environment.
MATERIALS AND METHODS
Bacterial strains and plasmids.
P. fluorescens isolate PF-1 was recovered from a water sample from the Seine River in downtown Paris. Water samples (100 ml) were collected in sterile bottles in January 2009 and filtered through nitrocellulose membranes (0.45-μm pore size; Millipore), and the filters were resuspended in 1 ml of sterile water. Aliquots (0.1 ml) were plated on MacConkey agar plates supplemented with imipenem (1 μg/ml), whereas 10-fold dilutions (0.1 ml) were plated on antibiotic-free MacConkey agar plates for bacterial counts. Samples were collected and processed on the same day of collection. Isolate PF-1 was identified by sequencing of the 16S rRNA genes. Escherichia coli DH10B (Invitrogen, Life Technologies, Cergy-Pontoise, France) and P. aeruginosa PAO1 were used as hosts for cloning and transformation experiments as previously described (20). The kanamycin-resistant pBKCMV (Invitrogen, Life Technologies, Cergy-Pontoise, France) was used as the cloning vector. Bacterial cultures were grown in Trypticase soy (TS) broth at 37°C for 18 h unless indicated otherwise.
Antimicrobial agents and MIC determinations.
The antimicrobial agents and their sources have been described elsewhere (14). MICs were determined by the Etest (AB Biodisk, Solna, Sweden) technique and interpreted according to the recommendations of the Clinical and Laboratory Standards Institute (3).
Plasmid extraction and conjugation assays.
Extraction of natural plasmid DNAs from P. fluorescens isolate PF-1 was attempted as described previously (7). Recombinant plasmid DNA was prepared using Qiagen maxi columns (Qiagen, Courtaboeuf, France). Transfer of the imipenem resistance marker into E. coli DH10B and P. aeruginosa PAO1 was attempted by electroporation as described previously (20). Transformants were selected on ticarcillin (50 μg/ml)-containing TS agar plates (Sanofi-Diagnostics Pasteur).
Cloning experiments, recombinant plasmid analysis, and DNA sequencing.
All enzymes for DNA manipulations were used according to the recommendations of the supplier (GE Healthcare, Orsay, France). PCR amplification of the blaBIC-1 gene was performed by using the internal primers BIC-1A and BIC-1B (Table 1). The genetic context of the blaBIC-1 β-lactamase gene was further analyzed by using primers BIC-1inv-1 and BIC-1inv-2 (Table 1).
TABLE 1.
Nucleotide sequences of primers used for amplification and sequence analysis
Partially Sau3AI-restricted DNA was ligated into the BamHI-restricted pBKCMV plasmid and introduced into E. coli DH10B by electroporation as described previously (14). Recombinant plasmids were selected onto amoxicillin (50 μg/ml)- and kanamycin (30 μg/ml)-containing TS agar plates. The recombinant plasmid, pSau1, with the smallest Sau3AI insert was retained for further analysis. The recombinant plasmid pEco2 was obtained by ligation of EcoRI-digested genomic DNA from P. fluorescens PF-1 in the EcoRI-restricted plasmid pBKCMV followed by amoxicillin/kanamycin selection (14). Both strands of the cloned DNA inserts of recombinant plasmids were sequenced by using an Applied Biosystems sequencer (ABI 377). The nucleotide and deduced protein sequences were analyzed with software available over the Internet from the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST/BLAST/).
Southern hybridization and PFGE analysis.
The probes were labeled using the ECL labeling kit according to the manufacturer's recommendation (GE Healthcare, Orsay, France). To evaluate the genetic location of the blaBIC-1 gene, whole-cell DNA of P. fluorescens strain PF-1 was restricted with the I-CeuI restriction enzyme (New England Biolabs, Saint-Quentin-en-Yvelines, France) and analyzed by pulsed-field gel electrophoresis (PFGE) as described previously (4, 9). After a transfer onto a nylon membrane (20), DNA fragments were hybridized with PCR-generated probes for 16S and 23S rRNA genes and for the blaBIC-1 gene (Table 1). PFGE analysis of XbaI-restricted DNA from P. fluorescens isolates was done as previously described (4).
Protein analysis.
Multiple nucleotide and protein sequence alignments were carried out online using the program ClustalW, available over the Internet at the University of Cambridge (http://www.ebi.ac.uk/Tools/clustalW2/index.html). The β-lactamase extracts from cultures of P. fluorescens PF-1 and the E. coli DH10B strain harboring the pSau1 plasmid were subjected to analytical isoelectric focusing (IEF) analysis as described previously (14).
β-Lactamase purification.
β-Lactamase BIC-1 purification was carried out by ion-exchange chromatography. E. coli DH10B (pSau1) was grown overnight at 37°C in 2 liters of TS broth containing amoxicillin (50 μg/ml) and kanamycin (30 μg/ml). The bacterial suspension was resuspended and disrupted by sonication in 10 ml 100 mM sodium phosphate buffer (pH 7), cleared by ultracentrifugation, and dialyzed against 50 mM malonate (pH 6.1). The protein extracts obtained were loaded onto a preequilibrated S-Sepharose column (Amersham Pharmacia Biotech) with the same buffer. The β-lactamase recovered in the flowthrough was subsequently dialyzed against 20 mM bis-Tris buffer (pH 7.2), loaded onto a Q-Sepharose column preequilibrated with the same buffer, and eluted with a linear NaCl gradient (0 to 500 mM). The fractions containing the highest β-lactamase activity, as determined qualitatively using nitrocefin hydrolysis (Oxoid, Dardilly, France), were pooled and dialyzed overnight against 50 mM sodium phosphate buffer (pH 7). The protein content was measured by the Bio-Rad DC protein assay. The protein purification rate and the relative molecular mass of BIC-1 β-lactamase were estimated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) analysis (14). The signal peptide cleavage site was identified with the SignalP 3.0 server (http://www.cbs.dtu.dk/services/SignalP/).
Kinetic studies.
Kinetic measurements (kcat and Km) of purified β-lactamase BIC-1 were performed as described previously (11). The 50% inhibitory concentration (IC50) was determined for BIC-1 as the concentration of clavulanate, tazobactam, or sulbactam that reduced the hydrolysis rate of 100 μM benzylpenicillin by 50%, under conditions in which BIC-1 was preincubated with various concentrations of inhibitor for 3 min at 30°C, before the substrate was added.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper will appear in the GenBank nucleotide database under the accession no. GQ260093.
RESULTS
Susceptibility testing and IEF analysis.
MICs of β-lactams for P. fluorescens PF-1 showed resistance to amino- and carboxypenicillins, narrow- and broad-spectrum cephalosporins (except ceftazidime), moxalactam, aztreonam, and carbapenems (Table 2). The P. fluorescens PF-1 isolate did not present any other additional resistance to antibiotics. The addition of clavulanic acid did not modify significantly the resistance pattern (Table 2). IEF analysis revealed that strain PF-1 produced two β-lactamases with pI values of ca. 5.8 and 9, the latter pI value likely corresponding to the natural AmpC β-lactamase of P. fluorescens.
TABLE 2.
MICs of β-lactams for P. fluorescens PF-1, E. coli DH10B harboring recombinant plasmid pSauI, and the E. coli DH10B reference strain
| β-Lactam(s) | MIC (μg/ml) |
||
|---|---|---|---|
| P. fluorescens PF-1 | E. coli DH10B (pSauI)a | E. coli DH10B | |
| Amoxicillin | >256 | >256 | 4 |
| Amoxicillin + CLAb | >256 | 16 | 4 |
| Ticarcillin | >256 | >256 | 4 |
| Ticarcillin + CLA | >256 | 128 | 4 |
| Piperacillin | 4 | 256 | 1 |
| Cephalothin | >256 | 128 | 2 |
| Cefuroxime | >256 | 16 | 0.5 |
| Cefoxitin | >256 | 4 | 1 |
| Ceftazidime | 2 | 0.5 | 0.5 |
| Cefotaxime | >32 | 0.5 | 0.12 |
| Cefepime | >16 | 0.12 | 0.06 |
| Moxalactam | >256 | 0.12 | 0.12 |
| Aztreonam | >256 | 16 | 0.25 |
| Imipenem | >16 | 2 | 0.12 |
| Imipenem + CLA | >16 | 0.25 | 0.12 |
| Meropenem | >16 | 0.25 | 0.12 |
| Ertapenem | >16 | 0.25 | 0.12 |
E. coli DH10B(pSau1) expressed β-lactamase BIC-1 from P. fluorescens PF-1.
CLA, clavulanic acid at a fixed concentration of 4 μg/ml.
Cloning and sequencing of the β-lactamase gene.
Shotgun cloning resulted in the selection of an E. coli DH10B(pSau1) strain that expressed a clavulanic-acid-inhibited carbapenemase phenotype with resistance or reduced susceptibility to penicillins, cephalosporins, imipenem, and aztreonam. The addition of clavulanic acid partially restored the activities of amoxicillin and imipenem (Table 2). The MIC of imipenem for E. coli DH10B(pSau1) was 2 μg/ml, an 8-fold-higher value than that for E. coli DH10B. In addition, IEF analysis showed that E. coli DH10B(pSau1) produced a β-lactamase with a pI value of 5.8, identical to that identified in P. fluorescens PF-1 (data not shown).
Identification of β-lactamase BIC-1.
DNA sequence analysis of the 1,615-bp insert of pSau1 revealed an open reading frame (ORF) of 882 bp encoding a 294-amino-acid preprotein, named BIC-1 (Bicêtre carbapenemase), with a relative molecular mass of 29 kDa. The G+C content of this ORF was 53.8%. The signal peptide cleavage site was identified between the alanine and the glutamic acid residues at positions 24 and 25 (FA-ET) of BIC-1 (Fig. 1).
FIG. 1.
Amino acid sequence comparison of BIC-1 with those of SFC-1 from S. fonticola (5) and KPC-2 from K. oxytoca (27). Full stops indicate the gaps that were inserted to optimize the alignment, and dashes indicate residues identical to those of BIC-1. The numbering is according to the method described by Ambler et al. (1). The conserved domains of class A β-lactamases are underlined (6). The residues suggested to play a critical role in carbapenemase activity are marked by asterisks (13, 15). The arrow indicates the cleavage site for the leader peptide of BIC-1.
β-Lactamase BIC-1 contains four conserved motifs of class A serine β-lactamases: 70SSFK73, 130SDN133, 166EXXXN170, and 234KTG236 (6) (Fig. 1), where S70 is the active-site serine, S130, T235, and T237 are involved in H-bond interactions, and K234 is involved in salt-bridge interaction (6, 13). BIC-1 shares 68 and 59% amino acid identities with class A carbapenemases SFC-1 from Serratia fonticola (5) and the plasmid-encoded KPC-2 (27), respectively (Fig. 1). The amino acid sequences of class A carbapenemases aligned with that of BIC-1 showed that the cysteine residues at positions 69 and 238 are conserved whereas the His105 residue was conserved in β-lactamase BIC-1, as in SFC-1, SME-1, and NMC-A, but not in KPC-2 (Fig. 1).
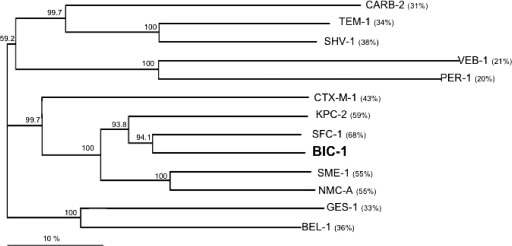
A dendrogram was generated from the amino acid sequence alignment of BIC-1 with main class A β-lactamases (21). It showed that BIC-1 is more closely related to the subgroup that includes SFC-1 and KPC-2 than to SME-1 and NMC-A (Fig. 2).
FIG. 2.
Dendrogram obtained for 13 representative class A β-lactamases by neighbor-joining analysis. The alignment used for the tree calculation was performed with the ClustalX program. Branch lengths are drawn to scale and are proportional to the number of amino acid changes. The distance along the vertical axis has no significance. Numbers in parentheses indicate percentages of amino acid identity with the β-lactamase BIC-1. The β-lactamases (GenBank accession numbers indicated) are CARB-2 (Q0370), TEM-1 (AAG47772), SHV-1 (AAD18054), VEB-1 (AAK14293), PER-1 (CAF18433), CTX-M-1 (CAJ01342), KPC-2 (AAO53443), SFC-1 (AAR06587), SME-1 (CAA82281), NMC-A (CAA79967), GES-1 (AAL82589), and BEL-1 (AAZ04368).
Biochemical properties of β-lactamase BIC-1.
The purification rate of BIC-1 was estimated to be >95% by SDS-PAGE analysis (data not shown). Kinetic parameters of the purified β-lactamase BIC-1 showed that it had a hydrolysis profile including penicillins, cephalosporins, carbapenems, and monobactams (Table 3). The highest kcat values were obtained with ampicillin, cephalothin, and cephaloridine, being approximately 4-fold higher than those for imipenem and piperacillin. Notably, kcat values for ticarcillin, cefotaxime, and oxacillin were approximately 2- to 3-fold lower than that for imipenem (Table 3). Also, BIC-1 showed no detectable activity against ceftazidime. The relatively weak catalytic efficiency (kcat/Km) of BIC-1 for most β-lactams resulted from a low affinity (high Km values) for most substrates. The highest affinities were found for ertapenem and meropenem, with Km values of 10 and 12 μM, respectively. The highest kcat/Km values were obtained for benzylpenicillin and cephalothin (660 and 500 mM−1·s−1, respectively), approximately 2- to 3-fold higher than those for piperacillin, cephaloridin, and ertapenem (210, 220 and 200 mM−1·s−1, respectively). Compared with the hydrolytic efficiencies of KPC-2, kcat/Km values obtained with BIC-1 were similar for most β-lactam substrates although substantially lower for cefuroxime and cefotaxime (Table 3).
TABLE 3.
Steady-state kinetic parameters of the β-lactamase BIC-1 and comparison of parameters obtained for β-lactamase KPC-2 (13)a
| β-Lactam | BIC-1 |
KPC-2 kcat/Km (mM−1·s−1) | ||
|---|---|---|---|---|
| kcat (s−1) | Km (μM) | kcat/Km (mM−1·s−1) | ||
| Benzylpenicillin | 31 | 48 | 660 | 1,000 |
| Ampicillin | 185 | 1,150 | 160 | 700 |
| Ticarcillin | 15 | 250 | 60 | ND |
| Piperacillin | 40 | 175 | 210 | 800 |
| Cephalothin | 250 | 550 | 500 | 750 |
| Cephaloridine | 170 | 770 | 220 | |
| Ceftazidime | ND | ND | ND | ND |
| Cefuroxime | 9b | >1,000 | <9 | 410 |
| Cefotaxime | 20 | 2,200 | 9 | 380 |
| Cefoxitin | 1 | 825 | 1 | |
| Aztreonam | 35b | >1,000 | <35 | 900 |
| Moxalactam | 3 | 1700 | 1 | |
| Oxacillin | 15 | 110 | 100 | |
| Imipenem | 50 | 300 | 170 | 230 |
| Meropenem | 1 | 12 | 80 | |
| Ertapenem | 2 | 10 | 200 | |
Data are means of results from three independent experiments. Standard deviations were within 10% of the means. ND, not determinable or no detectable hydrolysis (<0.01 s−1).
Data determined for the corresponding Km value (2,000 or 1,000 μM).
Inhibition studies as measured by IC50s showed that BIC-1 activity was weakly inhibited by clavulanic acid (IC50, 30 μM) and tazobactam (IC50, 6.5 μM). These values are in the same range as those found for KPC-1, with 10.5 and 0.37 μM for clavulanic acid and tazobactam, respectively (26).
Genetic environment of the blaBIC-1 gene.
A partial fragment of 4,135 bp of the ca. 10-kb insert of recombinant plasmid pEco2 was sequenced to identify the flanking sequences of the blaBIC-1 gene. Part of an ORF located 666 bp upstream of the ATG codon of blaBIC-1 encoded a 95-amino-acid peptide that had 58% identity with a truncated transposase of IS111A from Azotobacter vinelandii (GenBank accession no. ACO77208). Downstream of the blaBIC-1 gene, part of a truncated ORF was identified. Its deduced sequence shared 80% amino acid identity with 108 amino acids of a putative transposase of P. fluorescens SBW25 (GenBank accession no. CAY48720).
No plasmid was detected from P. fluorescens PF-1, and electrotransformation attempts using plasmid extracts from P. fluorescens PF-1 into E. coli DH10B and P. aeruginosa PAO1 as recipient strains failed to transfer any β-lactam resistance marker. The blaBIC-1-specific gene probe hybridized with an endonuclease I-CeuI-generated fragment of chromosomal DNA that hybridized also with the rRNA gene probe, revealing the chromosomal location of the blaBIC-1 gene in P. fluorescens PF-1 (data not shown).
Dissemination of blaBIC-1-like genes in water samples.
In order to evaluate the prevalence of the blaBIC-1-like genes, water samples were collected 2 months later from three distinct city sites, taking into consideration that the Seine River crosses Paris from southeast to northwest. Those samples, therefore, included the same collecting site (site 2), another site, located 2 km upstream (site 1), and another site, located 2 km downstream (site 3). PCR screening identified blaBIC-1-like genes in one (strain PF-2) out of nine isolates growing onto imipenem-containing agar plates from site 1, in one (strain PF-3) out of 18 isolates from site 2, and in none of the 17 isolates collected from site 3. Only BIC-1-positive isolates were identified by sequencing of the 16S rRNA genes. Isolates PF-2 and PF-3 were also identified as P. fluorescens, and PFGE analysis did not show any clonality between PF-1, PF-2, and PF-3 (data not shown).
DISCUSSION
Our study identified a novel chromosome-encoded class A carbapenemase from water samples collected in the Seine River, Paris. β-Lactamase BIC-1 shares the highest amino acid identity with class A carbapenemases SFC-1 from S. fonticola (5) and the plasmid-encoded KPC-2 (27). The cysteine residues conserved in class A carbapenemases at positions 69 and 238 may form a disulfide bond, as in KPC-2 and SFC-1 (13). Interestingly, the His105 residue was conserved in β-lactamase BIC-1, as in SFC-1, SME-1, and NMC-A, whereas KPC-2 possesses a tryptophan residue and SME-3, a tyrosine residue, at that position. Tyrosine and tryptophan seem to be the most common substitutions important for carbapenem hydrolysis (17).
The G+C content of this ORF (53.8%) differed from the expected range of the G+C content of Pseudomonas genes (ca. 60%), perhaps indicating a non-Pseudomonas origin. Moreover, BIC-1 was chromosomally encoded although it was not identified in all P. fluorescens isolates. This result suggests that the β-lactamase BIC-1 is not a feature of the P. fluorescens species, suggesting a mobile DNA structure.
Biochemical characterization of BIC-1 showed a significant hydrolysis of piperacillin, although the MIC of piperacillin for P. fluorescens was surprisingly low.
Several metallo-β-lactamases, such as VIM-2 from P. pseudoalcaligenes (18) and P. aeruginosa (19) and IMP-22 from P. fluorescens (12), were recovered from urban water samples. The number of different class A β-lactamases with carbapenemase activity is still limited, and the occurrence of most class A carbapenemases is at present sporadic (24), except for KPC enzymes. Only the class A carbapenemase IMI-2 in E. asburiae has been identified from U.S. rivers (2), and detection of a novel member of this class of carbapenemases in a P. fluorescens isolate recovered from the Seine River is intriguing. The blaBIC-1 gene is located on a possible transferable structure that may be the source of its dissemination in urban water. Multiple identification of blaBIC-1-positive P. fluorescens isolates in the same river may indicate a larger-than-expected spread of multidrug-resistant bacteria. Further work should be dedicated to the search for the spread of this novel broad-spectrum resistance determinant among human and animal isolates.
Acknowledgments
This work was funded by a grant from the Ministère de l'Education Nationale et de la Recherche (UPRES-EA 3539), Université Paris XI, Paris, France, and mostly by the European Community (DRESP2, LSHM-CT-2003-503-335 and TROCAR, HEALTH-F3-2008-223031) and by the INSERM.
Footnotes
Published ahead of print on 9 November 2009.
REFERENCES
- 1.Ambler, R. P., A. F. Coulson, J. M. Frère, J. M. Ghuysen, B. Joris, M. Forsman, R. C. Levesque, G. Tiraby, and S. G. Waley. 1991. A standard numbering scheme for the class A ß-lactamases. Biochem. J. 15:269-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubron, C., L. Poirel, R. J. Ash, and P. Nordmann. 2005. Carbapenemase-producing Enterobacteriaceae, U.S. rivers. Emerg. Infect. Dis. 11:260-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial susceptibility testing; 15th informational supplement. M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 4.Girlich, D., T. Naas, A. Leelaporn, L. Poirel, M. Fennewald, and P. Nordmann. 2002. Nosocomial spread of the integron-located veb-1-like cassette encoding an extended-spectrum beta-lactamase in Pseudomonas aeruginosa in Thailand. Clin. Infect. Dis. 34:603-611. [DOI] [PubMed] [Google Scholar]
- 5.Henriques, I., A. Moura, A. Alves, M. J. Saavedra, and A. Correia. 2004. Molecular characterization of a carbapenem-hydrolyzing class A ß-lactamase, SFC-1, from Serratia fonticola UTAD54. Antimicrob. Agents Chemother. 48:2321-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joris, B., P. Ledent, O. Dideberg, E. Fonze, J. Lamotte-Brasseur, J. A. Kelly, J. M. Ghuysen, and J.-M. Frère. 1991. Comparison of the sequences of class A β-lactamase and of the secondary structure elements of penicillin-recognizing proteins. Antimicrob. Agents Chemother. 35:2294-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kieser, T. 1984. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12:19-36. [DOI] [PubMed] [Google Scholar]
- 8.Koh, T. H., G. C. Wang, and L. H. Sng. 2004. IMP-1 and a novel metallo-ß-lactamase, VIM-6, in fluorescent pseudomonads isolated in Singapore. Antimicrob. Agents Chemother. 48:2334-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu, S. L., A. Hessel, and K. E. Sanderson. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. U. S. A. 90:6874-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michaux, C., J. Massant, F. Kerff, J.-M. Frère, J. D. Docquier, I. Vandenberghe, B. Samyn, A. Pierrard, G. Feller, P. Charlier, J. Van Beeumen, and J. Wouters. 2008. Crystal structure of a cold-adapted class C ß-lactamase. FEBS J. 275:1687-1697. [DOI] [PubMed] [Google Scholar]
- 11.Naas, T., W. Sougakoff, A. Casetta, and P. Nordmann. 1998. Molecular characterization of OXA-20, a novel class D β-lactamase, and its integron from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:2074-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pellegrini, C., P. S. Mercuri, G. Celenza, M. Galleni, B. Segatore, E. Sacchetti, R. Volpe, G. Amicosante, and M. Perilli. 2009. Identification of blaIMP-22 in Pseudomonas spp. in urban wastewater and nosocomial environments: biochemical characterization of a new IMP metallo-enzyme variant and its genetic location. J. Antimicrob. Chemother. 63:901-908. [DOI] [PubMed] [Google Scholar]
- 13.Petrella, S., N. Ziental-Gelus, C. Mayer, M. Renard, V. Jarlier, and W. Sougakoff. 2008. Genetic and structural insights into the dissemination potential of the extremely broad-spectrum class A ß-lactamase KPC-2 identified in an Escherichia coli strain and an Enterobacter cloacae strain isolated from the same patient in France. Antimicrob. Agents Chemother. 52:3725-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Philippon, L. N., T. Naas, A. T. Bouthors, V. Barakett, and P. Nordmann. 1997. OXA-18, a class D clavulanic acid-inhibited extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 41:2188-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pierrard, A., P. Ledent, J.-D. Docquier, G. Feller, C. Gerday, and J.-M. Frère. 1998. Inducible class C β-lactamases produced by psychrophilic bacteria. FEMS Microbiol. Lett. 161:311-315. [Google Scholar]
- 16.Poirel, L., J. D. Pitout, and P. Nordmann. 2007. Carbapenemases: molecular diversity and clinical consequences. Future Microbiol. 2:501-512. [DOI] [PubMed] [Google Scholar]
- 17.Queenan, A. M., W. Shang, P. Schreckenberger, K. Lolans, K. Bush, and J. Quinn. 2006. SME-3, a novel member of the Serratia marcescens SME family of carbapenem-hydrolyzing beta-lactamases. Antimicrob. Agents Chemother. 50:3485-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quinteira, S., H. Ferreira, and L. Peixe. 2005. First isolation of blaVIM-2 in an environmental isolate of Pseudomonas pseudoalcaligenes. Antimicrob. Agents Chemother. 49:2140-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quinteira, S., and L. Peixe. 2006. Multiniche screening reveals the clinically relevant metallo-ß-lactamase VIM-2 in Pseudomonas aeruginosa far from the hospital setting: an ongoing dispersion process? Appl. Environ. Microbiol. 72:3743-3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Swofford, D. L. 1989. PAUP (version 3.0): phylogenetic analysis using parsimony. Illinois Natural History Survey, Champaign, IL.
- 22.Toleman, M. A., D. Biedenbach, D. M. Bennet, R. N. Jones, and T. R. Walsh. 2005. Italian metallo-β-lactamases: a national problem? Report from the SENTRY and Antimicrobial Surveillance Programme. J. Antimicrob. Chemother. 55:61-70. [DOI] [PubMed] [Google Scholar]
- 23.Walsh, T. R., M. A. Toleman, L. Poirel, and P. Nordmann. 2005. Metallo-β-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walther-Rasmussen, J., and N. Høiby. 2007. Class A carbapenemases. J. Antimicrob. Chemother. 60:470-482. [DOI] [PubMed] [Google Scholar]
- 25.Yakupogullari, Y., L. Poirel, S. Bernabeu, A. Kizirgil, and P. Nordmann. 2007. Multidrug-resistant Pseudomonas aeruginosa isolate co-expressing extended-spectrum ß-lactamase PER-1 and metallo-ß-lactamase VIM-2 from Turkey. J. Antimicrob. Chemother. 61:221-222. [DOI] [PubMed] [Google Scholar]
- 26.Yigit, H., A. M. Queenan, G. J. Anderson, A. Domenech-Sanchez, J. W. Biddle, C. D. Steward, S. Alberti, K. Bush, and F. C. Tenover. 2001. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151-1161. (Erratum, 52:809.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yigit, H., A. M. Queenan, J. K. Rasheed, J. W. Biddle, A. Domenech-Sanchez, S. Alberti, K. Bush, and F. C. Tenover. 2003. Carbapenem-resistant strain of Klebsiella oxytoca harboring carbapenem-hydrolyzing beta-lactamase KPC-2. Antimicrob. Agents Chemother. 47:3881-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]