Abstract
Recent years have seen the rapid advancement of new therapeutic agents against hepatitis C virus (HCV) in response to the need for treatment that is unmet by interferon (IFN)-based therapies. Most antiviral drugs discovered to date are small molecules that modulate viral enzyme activities. In the search for highly selective protein-binding molecules capable of disrupting the viral life cycle, we have identified a class of anionic tetraphenylporphyrins as potent and specific inhibitors of the HCV replicons. Based on the structure-activity relationship studies reported herein, meso-tetrakis-(3,5-dicarboxy-4,4′-biphenyl) porphyrin was found to be the most potent inhibitor of HCV genotype 1b (Con1) replicon systems but was less effective against the genotype 2a (JFH-1) replicon. This compound induced a reduction of viral RNA and protein levels when acting in the low nanomolar range. Moreover, the compound could suppress replicon rebound in drug-treated cells and exhibited additive to synergistic effects when combined with protease inhibitor BILN 2061 or with IFN-α-2a. Our results demonstrate the potential use of tetracarboxyphenylporphyrins as potent anti-HCV agents.
Hepatitis C virus (HCV) exerts an increasingly heavy burden on global health care, and approximately 200 million people worldwide are infected (39). Chronically infected patients are often at risk for developing hepatic fibrosis, cirrhosis, and hepatocellular carcinoma (15). HCV is an enveloped virus that belongs to the Flaviviridae family, and seven recognized HCV genotypes and numerous subtypes have been identified. Genotype 1a is the most prevalent strain worldwide, and genotype 1b is predominant in Europe and North America, whereas genotype 2 is more prevalent in Asia (4, 33). The current standard of care, pegylated alpha interferon (IFN-α) combined with ribavirin, is plagued with adverse effects and has sustained viral response in less than half of the patients with genotype 1 infections (11, 20, 25). Therefore, more-effective and better-tolerated therapies are urgently needed, in particular for the treatment of nonresponders to IFN-based therapies.
The HCV genome, which is a single-stranded positive-sense RNA about 9.6 kb in length, encodes a polyprotein that is cleaved by viral and host proteases into structural (core, E1, E2, and possibly p7) and nonstructural proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) (4). The nonstructural proteins NS3 through NS5B assemble on the cytoplasmic membranes into a well-organized macromolecular machinery called the HCV replicase that is essential for the viral RNA replication (8, 14, 31). Until recently, the development of anti-HCV drugs had been hindered by the lack of a robust cell culture model. The establishment and optimization of the replicon systems have extensively widened our knowledge of the HCV replication and also proved a powerful tool for the discovery of novel agents that target the assembly and function of HCV replicase. HCV replicons are subgenomic constructs capable of autonomous replication in hepatoma cell lines, and the major viral components of the replicons consist of NS3 through NS5B (2, 23). Among these nonstructural proteins, viral protease NS3/4A and RNA-dependent RNA polymerase NS5B are the most extensively explored targets for anti-HCV drug development (for reviews, see references 6, 24, and 28). However, due to the error-prone nature of NS5B, mutational escapes could rapidly emerge under selective pressures from virus-specific inhibitors (35, 40). Other modalities under investigation include immune modulators and therapies targeting viral RNA.
Protein-protein interactions often involve substantial interfacial areas larger than 1,000 Å2 (34). Yet, selective targeting of a surface region in order to alter a protein's function or interaction with other biomolecules has not been extensively explored. In the current study, we have designed and synthesized a class of theoretical protein-binding molecules built on a porphyrin core, which is compatible with the biological milieu. The tetraphenylporphyrin (TPP) scaffold provides a sizable platform allowing hydrophobic interactions with the target surface, while charged peptidic appendages projected from the periphery support electrostatic interactions with complementary groups on the target(s). This contact with the large area may decrease the likelihood of high-level resistance developing in the targeted virus. We explored the potential of this class of compounds for antiviral activity against the HCV replicon systems. meso-Tetrakis-(3,5-dicarboxy-4,4′-biphenyl) porphyrin (compound 6) was found to be the most potent and selective inhibitor of HCV genotype 1b Con1 replicons (50% effective concentration [EC50], 0.024 ± 0.005 μM) with low cytotoxicity. While undertaking mechanistic studies to characterize the molecular target(s), we describe here the structure-activity relationships of tetraphenylporphyrin derivatives and the anti-HCV properties of compound 6, which is a proof-of-concept model for the development of proteomimetics in HCV drug discovery.
MATERIALS AND METHODS
Materials.
meso-Tetra(4-carboxyphenyl)porphine (compound 1) was purchased from Frontier Scientific, Inc. The synthesis of compounds 4, 6, and 8 were described in a previous publication (1), and the chemical synthesis procedures of the other analogues can be found in the supplemental material. 5,10,15,20-Tetrakis (4-(trimethylammonio)-phenyl)-21H,23H-porphine (TTMAPP) and 4,4′,4",4′"-(porphine-5,10,15,20-tetrayl) tetrakis (benzenesulfonic acid) tetrasodium salt hydrate (TPPS4) were purchased from Sigma-Aldrich. To protect photosensitive porphyrin compounds from degradation, compounds and compound-treated cell cultures were carefully shielded from light. NS3/4A protease inhibitor BILN 2061, developed by Boehringer Ingelheim (18), was a kind gift from Tsu-an Hsu from the National Health Research Institutes, Taiwan.
Cells.
The HCV genotype 1b (Con1 isolate) subgenomic replicon cell line with a luciferase reporter (Huh-luc/neo-ET) was kindly provided by Ralf Bartenschlager from the University of Heidelberg (37). Huh-luc/neo-ET cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1 mM nonessential amino acids, and 250 μg/ml of G418 (Invitrogen). Genotype 2a (JFH-1 isolate) subgenomic replicon cells (YSGR-JFH) were cultured in DMEM containing 10% fetal calf serum, 1 mM nonessential amino acids, and 400 μg/ml G418 (30). An additional genotype 1b (Con1) replicon cell line (429/BBix) was cultured in similar medium but with 3 μg/ml blasticidin (22).
Determination of antiviral activities.
Firefly (FF)-luciferase reporter activity was used to monitor the replication of HCV replicons in Huh-luc/neo-ET cells free from G418. Replicon cells were seeded at a density of 5 × 103 cells per well in 96-well plates. The following day, replicon cells were incubated in duplicates with dimethyl sulfoxide (DMSO) or serially diluted tetraphenylporphyrin (TPP) analogues at 37°C. Seventy-two hours after coincubation, cells were lysed with ice-cold passive lysis reagent after a phosphate-buffered-saline (PBS) wash, and the luciferase activity was measured with a luciferase assay kit (Promega) and a Tecan FARCyte luminometer (GE Healthcare) by following the manufacturers' instructions. The numbers of relative light units (RLU) were adjusted as percentage readings relative to the level for the compound-free controls, and the EC50 was determined from the dose-effect curve by nonlinear regression analysis using Origin 6.1 (OriginLab Software). TPP analogues were also screened in vitro against HIV-1 IIIB and hepatitis B virus (HBV) as previously described (26, 41). Briefly, 1 × 103 MT-2 cells per well were exposed in triplets to 0.1 50% tissue culture infective doses (TCID50)/cell of HIV-1 IIIB and cultured in the presence of compounds. The EC50s were estimated by MTT-based colorimetric quantitation of viral CPE after 5 days. Anti-HBV activities in 2.2.15 cells were evaluated by means of Southern DNA hybridization.
Determination of cytotoxicity and mitochondrial DNA (mtDNA) toxicity.
Exponentially growing Huh-7 cells were seeded at a density of 1 × 104 cells per well in a 24-well plate and incubated with TPP analogues for 3 days. Cells were fixed and stained with 0.5% methylene blue in 50% ethanol, followed by extensive washing. After the plates were air dried, cells were solubilized in 1% Sarkosyl and cell growth was determined from the extent of absorption by spectrophotometric measurements at 595 nm (Biotek Instruments). For compounds whose dark color interfered with 595-nm reading, cytotoxicity was measured by the CellTiter-Glo luminescence cell viability assay by following the manufacturer's instructions (Promega). Cytotoxicity in MT-2 cells was determined from colorimetric quantitation of uninfected MT-2 cells after 5 days of coincubation.
Quantitation of mitochondrial DNA content was performed as previously published (17). Briefly, CEM cell lysates were spotted onto Hybond paper by using a Miliford II slot blot apparatus (Schleicher & Schuell). mtDNA was detected with an mtDNA-specific probe and then reprobed with an Alu probe for an internal control. The autoradiographic bands were quantified with a scanning densitometer (Molecular Dynamics).
Impact of serum concentration in culture media on the antiviral activities of compound 6 and its analogues.
Similar to the HCV replicon assay, Huh-luc/neo-ET cells were incubated with serially diluted compounds in the presence of 5%, 10%, 20%, or 40% (vol/vol) FBS. Cells were harvested after 72 h for luciferase activity assays.
Quantification of HCV RNA and NS5A protein.
Huh-luc/neo-ET cells were seeded at a density of 1 × 105 cells per well in 6-well plate and treated with either the DMSO control or up to 250 nM compound 6. Cells were harvested after 24, 48, or 72 h of incubation and subjected to a luciferase activity assay, RNA quantification, and immunoblotting. Results from three independent repeats were averaged. The luciferase activity assay was done in triplets per experiment as aforementioned, and the RLU reading was normalized against the total protein level per sample determined from the Bradford assay.
Total RNA was isolated using an RNeasy minikit (Qiagen), and the RNA concentrations were measured by spectrophotometry (GE Healthcare) followed by dilution into 50 ng/μl. Replicon RNA was quantitated in triplicates by amplifying the HCV 5′ untranslated region (UTR) by using one-step quantitative real-time reverse transcriptase PCRs (qRT-PCR). Each 20-μl replicate contained 100 ng of total RNA, 100 nM probe (6-carboxyfluorescein [FAM]-5′-TATGAGTGTCGTACAGCCTCCAGG-3′-MGBNFQ [molecular-groove binding nonfluorescence quencher]; Applied Biosystems) and 200 nM forward and reverse primers (5′-CTTCACGCAGAAAGCGTCTA-3′ and 5′-CAAGCACCCTATCAGGCAGT-3′, respectively; W. M. Keck Facility, Yale University) (21), together with iScript reverse transcriptase and reaction mixture for one-step RT-PCR (Bio-Rad Laboratories). Reactions were run with the iCycler iQ RealTime thermocycler detection system (Bio-Rad Laboratories) as follows: 10 min at 50°C and 5 min at 95°C, followed by 42 cycles at 95°C for 15 s and 60°C for 30 min. Results were normalized against the β-actin mRNA levels in each sample (19).
For immunoblot analyses, cells were lysed in 100 μl of lysis/loading buffer (30 mM Tris, pH 6.8, 12.5% glycerol, 1% SDS, 5% β-mercaptol ethanol, and 0.01% bromophenol blue). Samples were electrophoresed by 8% SDS-PAGE and transferred onto a nitrocellulose membrane for 30 min at 15 V using a Trans-Blot semidry transfer apparatus (Bio-Rad Laboratories). The membrane was blocked with 5% nonfat dry milk in PBS for 1 h and probed by mouse monoclonal antibody (Ab) (7D4) specific for hepatitis C virus NS5A (Santa Cruz Biotechnology, Santa Cruz, CA) or monoclonal antibody specific for human α-tubulin (Sigma-Aldrich) at 4°C overnight, followed by washing in PBS with 0.2% Tween 20. After incubation with goat anti-mouse Ab (Sigma-Aldrich) for 1 h at room temperature, the membrane was washed extensively and detected by chemiluminescence procedures according to the manufacturer's instructions (PerkinElmer).
Reversibility of the action of compound 6 against the genotype 1b HCV replicon.
Huh-luc/neo-ET cells were seeded at a density of 2 × 105 cells per well in a 6-well plate and were incubated for 12 days with the DMSO control or up to 1 μM compound 6 in the absence of G418. Cells were split every three days when media and compounds were replenished, and samples were collected for RNA quantification. compound 6 was removed on day 12 when cells were split and cultured in the presence of 250 μg/ml of G418. Replicon cells were continuously monitored for another 12 days, during which cells were split and sampled whenever they reached confluence. Cell viability was measured by a CellTiter-Glo luminescence cell viability assay by following the manufacturer's procedures (Promega), and the HCV RNA was quantitated by qRT-PCR and normalized as described above.
Activity of compound 6 against genotype 2a (JFH-1) replicons.
In parallel to Huh-luc/neo-ET cells, 1 × 105 YSGR-JFH cells per well were incubated with the DMSO control, compound 6, or recombinant human IFN-α-2a (Pestka Biomedical Laboratories) in 6-well plates. Cells were harvested after 72 h of coincubation, and HCV RNAs were quantitated by qRT-PCR and normalized as described above.
Combination studies.
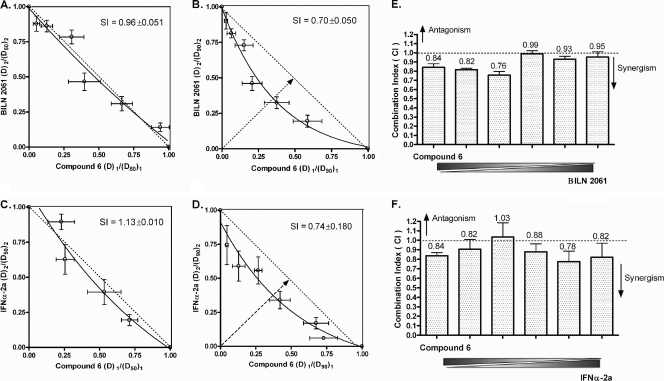
Huh-luc/neo-ET cells were seeded at a density of 5 × 103 cells per well in 96-well plates. On the following day, a mixture of two components (compound 6 with IFN-α-2a or compound 6 with BILN 2061) was applied in serial dilution and hence kept at a constant ratio. A total of eight different mixtures were assayed in duplicates such that the potency ratio of the two compounds ranged from emphasizing drug 1/deemphasizing drug 2 to deemphasizing drug 1/emphasizing drug 2. Seventy-two hours after coincubation, cells were harvested for the luciferase activity assay and the median effect equation was used for dose-effect analysis. The doses of drug 1 and drug 2 required to inhibit HCV replication by x% when used alone were denoted (Dx)1 and (Dx)2, respectively, whereas the apparent isoeffective doses needed to achieve x% inhibition when used in combination were denoted (D)1 and (D)2, respectively. The (D)1/(Dx)1 ratios were plotted against (D)2/(Dx)2 in antiviral isobolograms, in which the hypotenuse represents the line of additivity. If the experimental isobole bows below the hypotenuse, the combination is considered to be synergistic; if the isobole bows above the hypotenuse, antagonism is suggested. Synergy index (SI) values were calculated as the fractional distance from the origin to the intersection of the isobole and hypotenuse, with the total distance (half the length of the hypotenuse) designated 1.00. The smaller the SI value, the stronger the degree of synergism. Another parameter for quantitative determination of combination is the combination index [CI; calculated as (D)1/(Dx)1 + (D)2/(Dx)2]. CI values within the range of 1 ± 0.1 indicate additivity. CI values above or below the boundary suggest antagonism or synergism, respectively (5, 13, 32). The P value was determined by two-way analysis of variance (ANOVA), using GraphPad Prism 4.0.
RESULTS
Structure-activity relationships of the tetraphenylporphyrin analogues.
From a small library of porphyrin analogues that we initially explored for antiviral application, compound 2 emerged as a micromolar inhibitor of HCV replicons in vitro and provided the first insight into the development of meso-tetrakis-phenylporphyrin (TPP) derivatives as anti-HCV agents (Table 1). Greater-than-7-fold improvement in the anti-HCV EC50, together with a decrease of cytotoxicity and less effect on mitochondrial DNA synthesis, was observed in its synthetic precursor compound 1, in which the aspartic acid side chains were replaced with more-rigid and planar carboxylic acids. Substitution of the carboxylic acids in compound 1 with sulfonic acids as in TPPS4 led to complete loss of anti-HCV activity in vitro (EC50, 51.24 ± 8.577 μM). Reversing the charge of functional groups in the example of TTMAPP also compromised the activity against HCV (EC50, 3.58 ± 0.208 μM). The structures of the TPP analogues along with synthesis intermediates can be found in Fig S1 in the supplemental material.
TABLE 1.
Structure-activity relationships of anionic tetraphenylporphyrin analoguesa
Values shown are means ± SD for three independent experiments. “*” indicates that there were not enough compounds to complete the toxicity studies.
With the goal of improving hydrophobic surface recognition, we extended each peptidic appendage by one phenyl ring, giving rise to the family of meso-tetrakis-biphenylporphyrins (TBPs) that included compounds 4 and 5. The distance from opposite para positions of the phenyl groups was thus extended from approximately 15.5 Å in TPPs to 24.0 Å in TBPs, thereby increasing the total recognition area by 100 Å2 (34). Expansion of the surface area from compound 2 to compound 5 was accompanied by a 12-fold improvement in anti-HCV EC50. However, this was not the case in the comparison of compound 1 to its larger homolog compound 4, which showed reduced antiviral activity.
We then increased the total negative charges of the peripheral groups from four to eight in order to enhance electrostatic interactions with potentially complementary regions on the target(s). While compound 3 was relatively ineffective against HCV replicons, compound 6 proved to be the most potent nanomolar inhibitor in our study, with an EC50 of 0.024 ± 0.0051 μM, which represented a 75-fold improvement over the level for the lead, compound 2. Given the extremely low cytotoxicity in naïve Huh-7 cells, compound 6 offers a favorable selective index in culture (50% cytotoxic concentration for cell growth [CC50]/EC50) of over 2,000. Additionally, compound 6 did not alter the amount of mitochondrial DNAs. No changes in cellular morphology were observed in compound 6-treated replicon cells, confirming the lack of subtle cytotoxicity. Removal of one carboxylic acid from the meta position on each phenyl ring gave rise to compound 9, with decreased antiviral activity, suggesting that the projections of all eight negative charges are indispensable for potent inhibition of HCV replicons. As the functional groups became bulkier and more flexible in the cases of compounds 7 and 8, the antiviral activity was substantially decreased. Overall, there was no evidence of increased cytotoxicity over time. To study if metallation of the porphyrin rings can influence the antiviral function, we synthesized zinc, copper, and iron conjugates of compound 6 and compared their EC50s. The results suggested that metallation of the porphyrin core did not significantly alter the anti-HCV activity (see Table S1 in the supplemental material).
In addition to anti-HCV SAR, we also examined the activities of the nine TPP analogues against HBV and HIV-1 IIIB to establish antiviral specificity (Table 1). None of the compounds were able to inhibit HBV, whereas analogues bearing tetrabiphenylporphyrin motifs (compounds 5 to 8) exhibited micromolar activity against HIV-1 IIIB.
Compound 6 suppressed viral macromolecules in genotype 1b replicon cells in a time-dependent and dose-dependent manner.
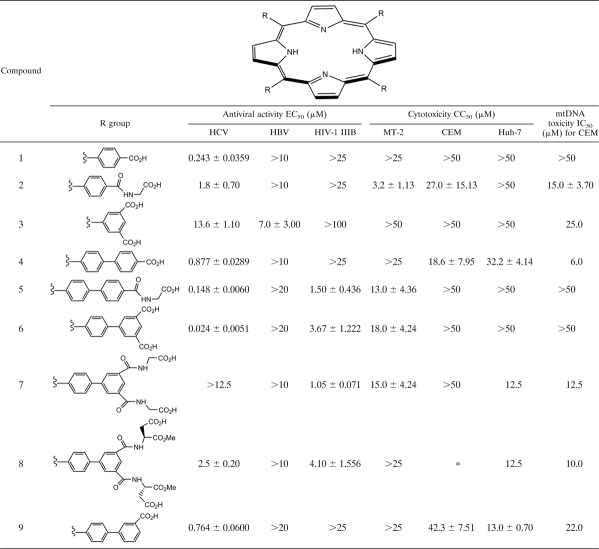
The anti-HCV activities of compound 6 in Huh-luc/neo-ET cells were characterized by studying different viral parameters: the luciferase reporter activity, the HCV RNA level, and the protein level of NS5A (Fig. 1). The replicon luciferase activity was markedly inhibited by compound 6 in a dose-dependent manner, and the EC50s decreased with incubation time, indicating improved efficacy. The 24-h and 48-h EC50s of compound 6 were 57.8 nM and 19.2 nM, respectively; the 72-h EC50 was 17.6 nM, a >3-fold improvement compared with the level observed following 24 h of incubation (Fig. 1A).
FIG. 1.
Time- and dose-dependent reduction of viral parameters in genotype 1b (Con1) replicon cells induced by compound 6. Huh-luc/neo-ET cells were incubated with serially diluted compound 6 for 24, 48, or 72 h. Results are expressed as percentages of the level for mock-treated controls. (A) Reduction of reporter luciferase activity, which indirectly reflects the replication level of HCV replicons. (B) Reduction of HCV RNA level normalized against the mRNA level of human β-actin. (C) Reduction of the NS5A protein level as in one experiment. (D) NS5A protein level quantitated and normalized against the protein level of α-tubulin (means ± standard deviations [SD] of results from three independent experiments).
Since luciferase activity indirectly reflects the overall level of viral replication, we expected the HCV RNA to be suppressed in a similar fashion following exposure to the inhibitor. Quantitative amplification of the HCV 5′ UTR demonstrated that compound 6 indeed led to a reduction of the HCV RNA level in replicon cells in a time- and dose-dependent manner (Fig. 1B). The relative HCV RNA level was expressed as a percentage of the level for the mock-treated control. The 24-h and 48-h EC50s of compound 6 were estimated to be 67.8 nM and 36.2 nM, respectively; the 72-h EC50 was 29.2 nM, a >2-fold improvement compared with the level observed following 24 h of incubation. When the viral protein amount was quantitated by Western blot analyses, a similar reduction of the NS5A protein level in drug-treated replicon cells was observed (Fig. 1C and D).
The antiviral activities were confirmed for 429/BBix (EC500.014 μM), a Huh-7.5 cell line carrying the genotype 1b replicon that confers blasticidin resistance and does not carry the luciferase reporter gene (see Fig. S2 in the supplemental material). In addition, we confirmed that compound 6 did not affect the luciferase reporter activities in FF-luciferase expression systems that carried NF-κB, transforming growth factor β (TGF-β), and the AP-1 response element (data not shown). Together, this ruled out the possibility that compound 6 exerted its action by interacting with nonviral components of the replicon, i.e., FF-luciferase and neomycin resistance genes and their gene products.
Effect of serum concentrations in culture media on the anti-HCV activity of compound 6 and its analogues.
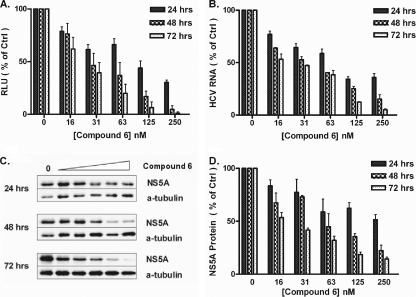
Sequestration of compounds by serum proteins could decrease the availability of the free agent but might also improve the uptake of hydrophobic derivatives. Hence, we studied the effects of serum protein binding on the antiviral activity of compounds 1, 4, and 6 by using different amounts of FBS in the media. Huh-luc/neo-ET cells were incubated with compound 1, 4, or 6 for 72 h, and the anti-HCV activity was evaluated by measuring the reduction of reporter luciferase activities. Up to 40% (vol/vol) FBS in media did not alter the luciferase activity of Huh-luc/neo-ET cells. In all cases, the EC50s increased with the percentage of FBS, which could reflect a decrease of available compounds due to sequestration by serum proteins. Extrapolation of the plot of EC50s versus serum amount provided an estimate of the theoretical EC50 values at 0% FBS (Table 2). Comparison of the relative fold changes in EC50 with increasing percentages of FBS showed that compound 1 was more affected by serum binding than compound 6. Compound 4 appeared to be least affected by serum concentration, and the fold increase was nonlinear unlike those observed for the other two analogues (Fig. 2).
TABLE 2.
Effects of serum concentrations in culture media on the anti-HCV EC50s of compounds 1, 4, and 6a
| Compound | EC50 (μM) for indicated FBS concn |
||||
|---|---|---|---|---|---|
| 0% (theoretical) | 5% | 10% | 20% | 40% | |
| 1 | 0.0482 | 0.124 ± 0.008 | 0.243 ± 0.036 | 0.513 ± 0.069 | 0.948 ± 0.089 |
| 4 | 0.3800 | 0.633 ± 0.176 | 0.877 ± 0.029 | 1.210 ± 0.056 | 1.483 ± 0.015 |
| 6 | 0.0065 | 0.013 ± 0.003 | 0.024 ± 0.005 | 0.039 ± 0.005 | 0.069 ± 0.011 |
Values shown are means ± SD for three independent experiments.
FIG. 2.
Effect of serum binding on the EC50s of compounds 1, 4, and 6. The fold changes in EC50 were plotted against percentages of serum by using the EC50 at 5% FBS as the 1-fold level (means ± SD of results from three independent experiments). The antiviral activities of compounds 1 and 6 decreased linearly with increasing serum and also differed significantly from each other at the 95% confidence intervals. Change in the EC50s of compound 4, which was the least affected by serum binding, was nonlinear. The P value was determined by a two-way ANOVA test, using GraphPad Prism 4.0.
The activity of compound 6 against genotype 1b (Con1) replicon was reversible, but longer treatment with higher dosages could prevent viral rebound.
The goal of anti-HCV treatment is to completely eliminate the virus from infected cells. In order to assess the reversibility of the antiviral action and the possibility of replicon clearance, we incubated Huh-luc/neo-ET cells with increasing concentrations of compound 6 for 12 days, free from G418 (Fig. 3). On day 12, compound 6 was removed and 250 μg/ml G418 was reintroduced, and the replicon RNA levels and cell growth were monitored for another 12 days. Due to the inhibition of viral replication by compound 6, cells that have lower levels of replicons should become more sensitive to G418. Therefore, the percentage of cells killed reflected the percentage of cells “cured” by compound 6.
FIG. 3.
Compound 6 could prevent the rebound of genotype 1b (Con1) replicons. Replicon cells were incubated with increasing concentrations of compound 6 for 12 days, free from G418. At the end of the incubation, compound 6 was removed and 250 μg/ml G418 was reintroduced. (A) The level of HCV RNA in cells was quantitated by qRT-PCR. RNA copy number per μg of total RNA was expressed as a ratio relative to the level for the mock-treated controls. During the rebound period, replicon cells incubated with 300 nM and 1 μM compound 6 were not confluent enough for sampling. (B) Cell viability was shown on the log10 scale as a percentage of the level for mock-treated controls. Replicon cells that were treated with 300 nM and 1 μM compound 6 were no longer viable by day 21.
Up to 1 μM compound 6 did not cause significant toxicity in cells during the 12-day treatment. A steady decrease in viability was initially observed when replicon cells were cocultured with G418, followed by a gradual rebound (Fig. 3B). Cells that were treated with higher concentrations of compound 6 experienced a significant delay in rebound. Some (8.5%) of the cells treated with 50 nM compound 6 survived 6 days after the removal of inhibitor, and replicon-positive cells slowly rebounded to 13.1% after a lapse of another 6 days. Only 0.007% of the cells treated with 100 nM compound 6 survived under the selective pressure of G418 by the end of the experiment; no rebound was observed, and the replicon RNA level fell beneath the detection limit once. Cells that were exposed to 300 nM and 1 μM compound 6 were no longer viable 9 days after coincubation with G418, which indicated a complete “cure.” Concentrations of compound 6 of 300 nM and above induced approximately 4.5-log10 reductions in the HCV RNA levels after 12 days of exposure. HCV RNA level in cells treated with lower concentrations of compound 6 rebounded faster than those in cells treated with higher dosages (Fig. 3A).
In a separate experiment, we treated the replicon cells with compound 6 for 9 days, followed by a 15-day rebound period in the presence of G418. Only 0.4% of the cells treated with 300 nM compound 6 survived with a lack of rebound, and 1 μM compound 6 achieved a complete “cure” (data not shown).
Genotype 2a (JFH-1) replicon cells were more resistant to both compound 6 and IFN-α-2a.
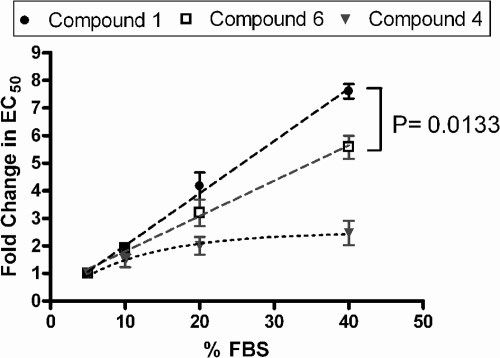
In contrast to Huh-luc/neo-ET replicon cells, YSGR-JFH, a genotype 2a JFH-1 isolate replicon cell line, was more resistant to treatment with compound 6 as well as IFN-α-2a. Replicon cells of the two genotypes were incubated with various concentrations of compound 6 or IFN-α-2a for 72 h, and the HCV RNA level was quantitated by qRT-PCR. The antiviral EC50 of compound 6 against YSGR-JFH replicon cells was 1.38 ± 0.148 μM, which was over 57-fold higher than the EC50 for Huh-luc/neo-ET replicons (Fig. 4A). The antiviral EC50s of IFN-α-2a were 2.39 ± 1.765 IU/ml and 25.99 ± 4.119 IU/ml in Huh-luc/neo-ET and YSGR-JFH replicon cells, respectively, representing an approximately 11-fold difference (Fig. 4B). YSGR-JFH replicon cells were also less responsive to compound 1, as expected (EC50, 1.9 μM).
FIG. 4.
Compared with genotype 1b (Con1) replicons, genotype 2a (JFH-1) replicons were more resistant to both compound 6 and IFN-α-2a. Genotype 1b (Con1) replicon cells (Huh-luc/neo-ET) and genotype 2a (JFH-1) replicon cells (YSGR-JFH) were incubated with increasing concentrations of compound 6 (A) or IFN-α-2a (B). Cells were harvested 72 h after incubation. The HCV RNA level was quantitated by qRT-PCR and expressed as a percentage of the level for mock-treated controls (mean ± SD of results from three independent experiments).
Compound 6 exhibited additive to synergistic effects when combined in vitro with BILN 2061 or IFN-α-2a.
The development of more-effective and nontoxic combinations of therapeutic agents has become an important goal in the management of HCV infection. We assessed the combination of compound 6 and established anti-HCV agents with respect to their antiviral activities when used alone. In a classical isobologram, the synergy index (SI) represents the fractional distance from the origin to where the isobole and hypotenuse intersect. Hence, an SI value of >1 indicates antagonism and an SI of <1 synergism. The smaller the SI value, the higher the degree of synergism. The intersection also represents the optimum potency ratio [theoretically, (D)1/(Dx)1 = (D)2/(Dx)2] for achievement of the highest degree of synergy at a given effect level when the isobole is of symmetrical distribution. As shown in Fig. 5A and C, the combination of compound 6 with BILN 2061 or IFN-α-2a was near-additive at the EC50, with SI values around 1.00. Synergistic effect became more apparent at the 90% inhibition level when the SI value was around 0.70, and the two isoboles differed significantly, as indicated by a P value of <0.0001 (Fig. 5B and D). The combination of compound 6 with BILN 2061 (optimum potency ratio, ≈0.34:0.36) was slightly more synergistic than the combination with IFN-α-2a (optimum potency ratio, ≈0.38:0.37). Calculation of the combination index [CI = (D)1/(Dx)1 + (D)2/(Dx)2] (5) provided a similar conclusion on drug combination: CI90 values of compound 6 in combination with BILN 2061 (Fig. 5E) or IFN-α-2a (Fig. 5F) confirmed the additive to synergistic interactions between the compounds.
FIG. 5.
Antiviral isobologram and CI90 plot of compound 6 in combination with BILN 2061 or IFN-α-2a in vitro. Huh-luc/neo-ET cells were coincubated for 72 h with various concentrations of drug 1 or 2 alone or the two in combination at different potency ratios. (A, C) Ratios of the apparent EC50 of each drug in combination over its EC50 when applied alone were plotted against each other in isobolograms. The hypotenuse represents the linear additive response to the action of two therapeutic agents. Isoboles that bow below the hypotenuse indicate synergism, and isoboles that bow above the hypotenuse indicate antagonism. Experimental data points on the isobole represent a combination that inhibits the HCV replication by 50% and is hence isoeffective with the line of additivity. The EC90 isobolograms of compound 6 in combination with BILN 2061 or IFN-α-2a are shown in panels B and D, respectively (means ± SD of results from at least four independent experiments). Different degrees of synergism/antagonism are expected at different effect levels. The combination indices (CI) at the 90% effect level for the combination of compound 6 with BILN 2061 and IFN-α-2a are plotted in panels E and F, respectively. Mean CI90 values for each dose ratio are indicated above the bars. A CI value of 1 ± 0.1 suggests additivity, as indicated by the dashed line. CI values below and above the boundary indicate synergism and antagonism, respectively.
DISCUSSION
In recent decades, efforts in medicinal chemistry have been concentrated in the development of small molecule inhibitors that are selective and high-affinity binders of active sites in the protein cavities, with the goal of disrupting protein-protein or protein-ligand interactions. In contrast, the protein exterior surfaces frequently employed in specific recognition during intermolecular interactions have been less explored. Specific targeting of such large interfacial areas with their complex topological distribution of hydrophobic, polar, and charged residues can potentially be achieved by molecules that mimic protein surface structures. Porphyrins, peptidocalixarenes, and α-helical mimetics are examples of macromolecules that have been designed to bind to the protein surface and modulate protein-protein interactions (for a review, see reference 9). Porphyrins are attractive macrocyclic scaffolds due to their intrinsic compatibility with the biological milieu, and their physicochemical properties along with synthesis procedures are also well documented. The photoinactivation of viruses by tetrapyrroles has been widely studied. Porphyrins and metalloporphyrins have also demonstrated light-independent activity in the micromolar range against HIV and vaccinia virus (7, 38). In particular, anionic tetrapyrroles, including sulfonated porphyrins, such as metallo-TPPS4, were shown to inhibit HIV-1 infection by blocking cell fusion induced by the envelope protein and also possibly by disruption of gp120-CD4 binding (38). Interestingly, an uncharged molecule, TPP[2,6-(OH)2], was equally active against vaccinia virus, suggesting that the interaction between charged groups may not be the sole basis for its antiviral activity (3). Exploration of the 4-fold symmetry of porphyrin derivatives is best illustrated in the rational design of tetraphenylporphyrins to reversibly block the conductance of voltage-gated potassium channels, which are homotetrameric molecules essential for numerous cellular functions. As synthetic mimics of peptide toxins, these cationic porphyrins appear to bind the channel pore and also mediate polyvalent interactions with the conserved acidic residues on the channel subunits (12).
For the development of HCV enzyme-specific therapies, viral protease NS3/4A and RdRP NS5B are the most intensely exploited targets. Successful examples of small-molecule inhibitors include the protease inhibitors telaprevir (VX-950) and boceprevir (SCH503034), the nucleoside polymerase inhibitor R7128, and the nonnucleoside NS5B inhibitor VCH-222. The macrocyclic inhibitor of NS3, BILN 2061, despite being suspended in clinical development, is a proof-of-principle peptidomimetic compound that was designed to mimic the conformation of substrate-based hexapeptides bound to NS3 and is active both in vitro and in vivo (18, 36). In the present study, we report the development of tetracarboxyphenylporphyrins for feasible interaction with biomolecules involved in HCV replication. This class of tetraphenylporphyrins (TPPs) offers a rigid scaffold capable of forming hydrophobic interactions with protein exteriors or solvent-exposed shallow clefts. The binding of the synthetic ligands could be further strengthened through electrostatic interactions with the cationic groups on the targets. The inherent 4-fold symmetry of TPPs can potentially lead to simultaneous binding to several components/subunits of a heteromeric or homomeric complex. The structural features of TPPs could be of particular interest in antiviral drug discovery, because the virus would require multisite mutations (possibly spanning more than one target protein) to become highly resistant, an event with significantly lower probability than single-site mutation, which is often sufficient for conferring resistance to small-molecule inhibitors.
Based on a lead, compound 2, that exhibited micromolar activity against HCV replicons, we explored TPP analogues with different structural features in search of a selective inhibitor active in the low nanomolar range. The following key factors were taken into consideration during our structural optimization: (i) surface area; (ii) charge, size, and flexibility of peptidic appendages; (iii) the projection of functional groups relative to the hydrophobic core; and finally, (iv) solubility and serum sequestration. An interesting feature of tetraphenylporphyrin (TPP) and tetrabiphenylporphyrin (TBP) derivatives is that the first phenyl ring is perpendicular to the porphyrins core but that the second phenyl ring lies perpendicular to the first ring and in the same plane as the porphyrin core. Consequently, compounds 1 and 4, for example, represent a completely different projection of anionic appendages. As shown in Table 1, structure-activity relationship (SAR) analysis revealed that the most optimum structure for activity against HCV in vitro is that of an octaanionic tetrabiphenylporphyrin, compound 6 (EC50, 0.024 ± 0.0051 μM), which represented a 75-fold improvement in EC50 over the lead compound and is comparable to the levels for other anti-HCV agents developed to date. Moreover, the carboxylic acids could not be replaced with sulfonate, trimethylammonium, the more flexible aspartic acid, or bulkier moieties, nor could the number of negative charges be decreased; all of these courses led to reductions in activity. Metallated derivatives of compound 6 demonstrated anti-HCV activity similar to that of the parent compound, suggesting that contact with the porphyrin core does not contribute toward anti-HCV activity or that compound 6 itself becomes metallated upon entering the cells. Expansion of the hydrophobic surface area improved antiviral efficacy except in the cases of compounds 1 to 4, which may be due in part to their different projections of anionic groups and their differences in serum binding. Sequestration by serum has the potential to decrease the availability of free drug but may also improve its solubility and promote uptake into the hepatocytes. Compound 1 appeared to have the highest degree of binding to serum proteins, and its anti-HCV EC50 increased linearly with the percentage of serum in the medium (Fig. 2). In contrast, compound 4 has the lowest degree of serum association, which could hinder its uptake. Sharing the same hydrophobic core, compound 6, however, benefits from a greater number of electrostatic interactions that could help toward uptake into cells. The activity of compound 6 against HCV replicons was confirmed by the suppression of viral RNA and the protein levels of two independent genotype 1b (Con1) replicons established in Huh 7 and Huh 7.5 cells, respectively (Fig. 1; see also Fig. S1 in the supplemental material). If compound 6 disrupts the assembly of the HCV replication complex, active replicases that have already assembled prior to drug treatment could continue viral replication until turnover, which may explain the initial increase of antiviral potency over time as shown in Fig. 1.
Compounds 1, 4, 5, 6, and 9 are selective inhibitors of HCV in vitro and are relatively inactive against DNA virus HBV and RNA virus HIV-1 IIIB. Although compounds 5 and 6 and their bulkier derivatives showed micromolar inhibition of HIV-1 IIIB comparable to that observed for tetraporphines that are under development as microbicides, there was no correlation between the trends of anti-HCV and anti-HIV efficacy; therefore, it is unlikely that the two types of antiviral activities share the same mechanism of action. The micromolar anti-HIV activities of compounds 5 to 8 suggested that the tetrabiphenylporphyrin scaffold and flexibility but not the bulkiness of the peptidic appendages may be favorable to the inhibition of HIV-1 IIIB. Hamilton et al. had previously shown that tetracarboxyphenylporphyrin derivatives bind cytochrome c. Compounds 1, 4, 6, and 8 were found to bind cytochrome c, with Kd values of 0.95 ± 0.25, 17 ± 0.84, 1.5 ± 0.17, and 1.7 ± 0.097 μM, respectively (1, 16), but this property did not correlate with the SAR in the present study. We treated Huh-luc/neo-ET cells with up to 1 μM compound 6 for 9 days, during which the media were replenished every 3 days and the cells were passaged once. Live cells were stained with the ratiometric indicator JC-1 (Invitrogen) in order to measure the mitochondrial potential using confocal microscopy. Compared with mock-treated control, compound 6 did not affect the mitochondrial membrane potential, unlike the classical uncoupler valinomycin (Calbiochem). In light of the extremely low toxicity on cells and particularly on the amount of mitochondrial DNA, it is unlikely that the potent anti-HCV activity of compound 6 is mediated through cytochrome c binding.
Compared with subgenomic genotype 1b (Con1) replicons, genotype 2a (JFH-1) replicons appeared to be more resistant to IFN-α-2a, with an 11-fold difference in anti-HCV EC50, in accordance with the literature (27). Con1 and JFH-1 isolates differ significantly in their replicase coding regions. Surprisingly, the genotype 2a (JFH-1) replicon was also more resistant to compound 6, and the anti-HCV EC50 fell into the micromolar range, with the activity being 57 times less effective than that against genotype 1b (Con1) replicons. The HCV RNA levels were similar between the two cell lines, indicating that differences in replication capacity could not be the major contributing factor. Such different sensitivities toward compound 6 suggests the possible involvement of a cellular interactor of the viral replicase, which, however, could be dispensable for the replication of genotype 2a (JFH-1) replicons. Besides the significant impact of genetic variability on drug sensitivity, the observed differential responses to IFN-α-2a and compound 6 between the two subgenomic replicons could be correlated. HCV is known to suppress host immune responses, and reduction of viral load restores the production of IFN-α/β and related antiviral signaling pathways (10, 29). Therefore, the antiviral activity exerted by compound 6 could be augmented through the action of revived host defenses and the IFN amplification loops. Such amplification could be more significant in genotype 1b (Con1) replicon-containing cells because of their intrinsic IFN sensitivity, and this potential “dual inhibition” could be masked in cells harboring genotype 2a (JFH-1) replicons.
Unlike treatment of HIV infection, HCV therapy can lead to complete eradication of virus in a significant proportion of patients. We demonstrated that the antiviral activity of compound 6 was irreversible if the treatment period is sufficiently long and if the dosages are adequate (Fig. 3). Moreover, the longer the treatment was, the further the delay in viral rebound was. Whether the percentage of replicon-positive cells that survived the treatment under high concentrations of compound 6 could have involved the development of resistance remains to be addressed. The limitation of the replicon model, however, could be the relationship between the viral load per cell and the sensitivity of host cells to G418. If the HCV replicon falls below a threshold level enough to subject hepatocytes to Geneticin toxicity, the remaining replicon is beneath the detection limit because of decreased cell viability. On the other hand, Geneticin selectively amplifies replicon-positive cells above the threshold. We have also carried out rebound studies in the absence of Geneticin; however, the HCV RNA levels in mock- and drug-treated cells were all reduced with time due to the lack of selective pressure.
As in HIV management, combination therapy is an important focal point in the development of anti-HCV agents. Optimum combination of drugs with different mechanisms of action should improve efficacy with a wider therapeutic window and reduced viral resistance. It is important that the combination should produce at least additive effects with no antagonism. Our in vitro synergy studies showed that the combination of compound 6 with BILN 2061 or with IFN-α-2a was additive to synergistic at the effect levels studied, more so at 90% inhibition (Fig. 5). According to the antiviral isobolograms, approximately equipotent combinations of compound 6 and BILN 2061 (∼0.350 EC90) or compound 6 and IFN-α-2a (∼0.375 EC90) were sufficient to inhibit HCV replication by 90%. The difference between the degrees of synergism at the 50% and 90% response level illustrated how the nature of drug-drug interactions may vary depending on the dose ratio in combination and on the endpoints of choice (5, 32).
The antiviral specificity and genotypic selectiveness of compound 6 suggest that the compound could be targeting the viral replicase. Whether the binding of octa-anionic tetrabiphenylporphyrin to viral protein blocks the interaction with HCV genome, other proteins in the replicase or with host factors are under investigation. If the synthetic agent targets highly conserved sequences that are essential for viral replication, mutations at these hot spots should have decreased probability and, as a result, it could be difficult for the virus to develop high-level resistance. In vitro resistance characterization and transient replication assays will be employed in future research to elucidate the mechanism of action of compound 6 and to establish its cross-resistance profiles involving known inhibitors of HCV NS3/4A as well as NS5B. While undertaking detailed mechanistic studies, we present here the proof-of-concept design and antiviral results for compound 6, which show great potential as a potent and selective inhibitor of HCV.
Supplementary Material
Acknowledgments
We thank Ralf Bartenschlager (University of Heidelberg) for generously providing us with the Huh-luc/neo-ET cell line. We thank Tsu-an Hsu (National Health Research Institutes) for kindly providing BILN 2061 for our drug combination studies. We also thank Mingjun Huang (Achillion Pharmaceuticals) for providing many helpful suggestions. Finally, we thank Sourav Saha, Elijah Paintsil, and Wing Lam (Yale University) for their help with the project.
This work was partly supported by grant AI-073299 from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health. Y.-C. Cheng is a fellow of the National Foundation for Cancer Research.
Footnotes
Published ahead of print on 9 November 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Aya, T., and A. D. Hamilton. 2003. Tetrabiphenylporphyrin-based receptors for protein surfaces show sub-nanomolar affinity and enhance unfolding. Bioorg. Med. Chem. Lett. 13:2651-2654. [DOI] [PubMed] [Google Scholar]
- 2.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 3.Chen-Collins, A. R., D. W. Dixon, A. N. Vzorov, L. G. Marzilli, and R. W. Compans. 2003. Prevention of poxvirus infection by tetrapyrroles. BMC Infect. Dis. 3:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choo, Q. L., K. H. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, R. Medina-Selby, and P. J. Barr. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 88:2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chou, T.-C. 1991. The median-effect principle and the combination index for quantitation of synergism and antagonism, p. 61-102. In D. C. Rideout and T.-C. Chou (ed.), Synergism and antagonism in chemotherapy. Academic Press, Inc., San Diego, CA.
- 6.De Francesco, R., and G. Migliaccio. 2005. Challenges and successes in developing new therapies for hepatitis C. Nature 436:953-960. [DOI] [PubMed] [Google Scholar]
- 7.Dixon, D. W., A. F. Gill, L. Giribabu, A. N. Vzorov, A. B. Alam, and R. W. Compans. 2005. Sulfonated naphthyl porphyrins as agents against HIV-1. J. Inorg. Biochem. 99:813-821. [DOI] [PubMed] [Google Scholar]
- 8.El-Hage, N., and G. Luo. 2003. Replication of hepatitis C virus RNA occurs in a membrane-bound replication complex containing nonstructural viral proteins and RNA. J. Gen. Virol. 84:2761-2769. [DOI] [PubMed] [Google Scholar]
- 9.Fletcher, S., and A. D. Hamilton. 2005. Protein surface recognition and proteomimetics: mimics of protein surface structure and function. Curr. Opin. Chem. Biol. 9:632-638. [DOI] [PubMed] [Google Scholar]
- 10.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 11.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 12.Gradl, S. N., J. P. Felix, E. Y. Isacoff, M. L. Garcia, and D. Trauner. 2003. Protein surface recognition by rational design: nanomolar ligands for potassium channels. J. Am. Chem. Soc. 125:12668-12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greco, W. R., H. S. Park, and Y. M. Rustum. 1990. Application of a new approach for the quantitation of drug synergism to the combination of cis-diamminedichloroplatinum and 1-beta-D-arabinofuranosylcytosine. Cancer Res. 50:5318-5327. [PubMed] [Google Scholar]
- 14.Hardy, R. W., J. Marcotrigiano, K. J. Blight, J. E. Majors, and C. M. Rice. 2003. Hepatitis C virus RNA synthesis in a cell-free system isolated from replicon-containing hepatoma cells. J. Virol. 77:2029-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoofnagle, J. H. 1997. Hepatitis C: the clinical spectrum of disease. Hepatology 26:15S-20S. [DOI] [PubMed] [Google Scholar]
- 16.Jain, R. K., and A. D. Hamilton. 2000. Protein surface recognition by synthetic receptors based on a tetraphenylporphyrin scaffold. Org Lett. 2:1721-1723. [DOI] [PubMed] [Google Scholar]
- 17.Lam, W., C. Chen, S. Ruan, C. H. Leung, and Y. C. Cheng. 2005. Expression of deoxynucleotide carrier is not associated with the mitochondrial DNA depletion caused by anti-HIV dideoxynucleoside analogs and mitochondrial dNTP uptake. Mol. Pharmacol. 67:408-416. [DOI] [PubMed] [Google Scholar]
- 18.Lamarre, D., P. C. Anderson, M. Bailey, P. Beaulieu, G. Bolger, P. Bonneau, M. Bos, D. R. Cameron, M. Cartier, M. G. Cordingley, A. M. Faucher, N. Goudreau, S. H. Kawai, G. Kukolj, L. Lagace, S. R. LaPlante, H. Narjes, M. A. Poupart, J. Rancourt, R. E. Sentjens, R. St. George, B. Simoneau, G. Steinmann, D. Thibeault, Y. S. Tsantrizos, S. M. Weldon, C. L. Yong, and M. Llinas-Brunet. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186-189. [DOI] [PubMed] [Google Scholar]
- 19.Leung, C. H., S. P. Grill, W. Lam, Q. B. Han, H. D. Sun, and Y. C. Cheng. 2005. Novel mechanism of inhibition of nuclear factor-kappa B DNA-binding activity by diterpenoids isolated from Isodon rubescens. Mol. Pharmacol. 68:286-297. [DOI] [PubMed] [Google Scholar]
- 20.Lindahl, K., L. Stahle, A. Bruchfeld, and R. Schvarcz. 2005. High-dose ribavirin in combination with standard dose peginterferon for treatment of patients with chronic hepatitis C. Hepatology 41:275-279. [DOI] [PubMed] [Google Scholar]
- 21.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 22.Lindenbach, B. D., B. M. Pragai, R. Montserret, R. K. Beran, A. M. Pyle, F. Penin, and C. M. Rice. 2007. The C terminus of hepatitis C virus NS4A encodes an electrostatic switch that regulates NS5A hyperphosphorylation and viral replication. J. Virol. 81:8905-8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lohmann, V., F. Körner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 24.Manns, M. P., G. R. Foster, J. K. Rockstroh, S. Zeuzem, F. Zoulim, and M. Houghton. 2007. The way forward in HCV treatment—finding the right path. Nat. Rev. Drug Discov. 6:991-1000. [DOI] [PubMed] [Google Scholar]
- 25.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M.-H. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 26.Mellors, J. W., G. E. Dutschman, G. J. Im, E. Tramontano, S. R. Winkler, and Y. C. Cheng. 1992. In vitro selection and molecular characterization of human immunodeficiency virus-1 resistant to non-nucleoside inhibitors of reverse transcriptase. Mol. Pharmacol. 41:446-451. [PubMed] [Google Scholar]
- 27.Miyamoto, M., T. Kato, T. Date, M. Mizokami, and T. Wakita. 2006. Comparison between subgenomic replicons of hepatitis C virus genotypes 2a (JFH-1) and 1b (Con1 NK5.1). Intervirology 49:37-43. [DOI] [PubMed] [Google Scholar]
- 28.Pereira, A. A., and I. M. Jacobson. 2009. New and experimental therapies for HCV. Nat. Rev. Gastroenterol. Hepatol. 6:403-411. [DOI] [PubMed] [Google Scholar]
- 29.Pflugheber, J., B. Fredericksen, R. Sumpter, Jr., C. Wang, F. Ware, D. L. Sodora, and M. Gale, Jr. 2002. Regulation of PKR and IRF-1 during hepatitis C virus RNA replication. Proc. Natl. Acad. Sci. U. S. A. 99:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phan, T., R. K. Beran, C. Peters, I. Lorenz, and B. D. Lindenbach. 2009. Hepatitis C virus NS2 protein contributes to virus particle assembly via opposing epistatic interactions with the E1-E2 glycoprotein and NS3-NS4A enzyme complexes. J. Virol. 83:8379-8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinkert, D., R. Bartenschlager, and V. Lohmann. 2005. Quantitative analysis of the hepatitis C virus replication complex. J. Virol. 79:13594-13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rideout, D. C. 1991. The median-effect principle and the combination index for quantitation of synergism and antagonism, p. 3-60. In D. C. Rideout and T.-C. Chou (ed.), Synergism and antagonism in chemotherapy. Academic Press, Inc., San Diego, CA.
- 33.Simmonds, P., J. Bukh, C. Combet, G. Deléage, N. Enomoto, S. Feinstone, P. Halfon, G. Inchauspé, C. Kuiken, G. Maertens, M. Mizokami, D. G. Murphy, H. Okamoto, J.-M. Pawlotsky, F. Penin, E. Sablon, T. Shin-I, L. J. Stuyver, H.-J. Thiel, S. Viazov, A. J. Weiner, and A. Widell. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962-973. [DOI] [PubMed] [Google Scholar]
- 34.Stites, W. E. 1997. Protein-protein interactions: interface structure, binding thermodynamics, and mutational analysis. Chem. Rev. 97:1233-1250. [DOI] [PubMed] [Google Scholar]
- 35.Trozzi, C., L. Bartholomew, A. Ceccacci, G. Biasiol, L. Pacini, S. Altamura, F. Narjes, E. Muraglia, G. Paonessa, U. Koch, R. De Francesco, C. Steinkuhler, and G. Migliaccio. 2003. In vitro selection and characterization of hepatitis C virus serine protease variants resistant to an active-site peptide inhibitor. J. Virol. 77:3669-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsantrizos, Y. S. 2004. The design of a potent inhibitor of the hepatitis C virus NS3 protease: BILN 2061—from the NMR tube to the clinic. Biopolymers 76:309-323. [DOI] [PubMed] [Google Scholar]
- 37.Vrolijk, J. M., A. Kaul, B. E. Hansen, V. Lohmann, B. L. Haagmans, S. W. Schalm, and R. Bartenschlager. 2003. A replicon-based bioassay for the measurement of interferons in patients with chronic hepatitis C. J. Virol. Methods 110:201-209. [DOI] [PubMed] [Google Scholar]
- 38.Vzorov, A. N., D. W. Dixon, J. S. Trommel, L. G. Marzilli, and R. W. Compans. 2002. Inactivation of human immunodeficiency virus type 1 by porphyrins. Antimicrob. Agents Chemother. 46:3917-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wasley, A., and M. J. Alter. 2000. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin. Liver Dis. 20:1-16. [DOI] [PubMed] [Google Scholar]
- 40.Yang, W., Y. Zhao, J. Fabrycki, X. Hou, X. Nie, A. Sanchez, A. Phadke, M. Deshpande, A. Agarwal, and M. Huang. 2008. Selection of replicon variants resistant to ACH-806, a novel hepatitis C virus inhibitor with no cross-resistance to NS3 protease and NS5B polymerase inhibitors. Antimicrob. Agents Chemother. 52:2043-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeo, H., Y. Li, L. Fu, J. L. Zhu, E. A. Gullen, G. E. Dutschman, Y. Lee, R. Chung, E. S. Huang, D. J. Austin, and Y. C. Cheng. 2005. Synthesis and antiviral activity of helioxanthin analogues. J. Med. Chem. 48:534-546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.