Abstract
The recently discovered antibacterial compound alaremycin, produced by Streptomyces sp. A012304, structurally closely resembles 5-aminolevulinic acid, the substrate of porphobilinogen synthase. During the initial steps of heme biosynthesis, two molecules of 5-aminolevulinic acid are asymmetrically condensed to porphobilinogen. Alaremycin was found to efficiently inhibit the growth of both Gram-negative and Gram-positive bacteria. Using the newly created heme-permeable strain Escherichia coli CSA1, we are able to uncouple heme biosynthesis from bacterial growth and demonstrate that alaremycin targets the heme biosynthetic pathway. Further studies focused on the activity of alaremycin against the opportunistic pathogenic bacterium Pseudomonas aeruginosa. The MIC of alaremycin was determined to be 12 mM. Alaremycin was identified as a direct inhibitor of recombinant purified P. aeruginosa porphobilinogen synthase and had a Ki of 1.33 mM. To understand the molecular basis of alaremycin's antibiotic activity at the atomic level, the P. aeruginosa porphobilinogen synthase was cocrystallized with the alaremycin. At 1.75-Å resolution, the crystal structure reveals that the antibiotic efficiently blocks the active site of porphobilinogen synthase. The antibiotic binds as a reduced derivative of 5-acetamido-4-oxo-5-hexenoic acid. The corresponding methyl group is, however, not coordinated by any amino acid residues of the active site, excluding its functional relevance for alaremycin inhibition. Alaremycin is covalently bound by the catalytically important active-site lysine residue 260 and is tightly coordinated by several active-site amino acids. Our data provide a solid structural basis to further improve the activity of alaremycin for rational drug design. Potential approaches are discussed.
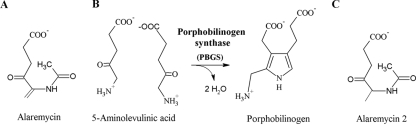
Modified tetrapyrroles are complex macromolecules and the most abundant pigments found in nature. Tetrapyrroles such as hemes and chlorophyll are essential prosthetic groups involved in numerous electron transport chains for energy recovery in essentially all forms of life. The biosynthetic pathways of tetrapyrroles are correspondingly highly conserved (16, 30), making heme biosynthesis an attractive target for antibacterial drug discovery and application (24). In fact, the tetrapyrrole biosynthetic pathway serves both as a source for the production of antibiotics such as asukamycin (25) and as a target for antibiotics, as in the case of gabaculin (1, 19). Recently, the antibiotic alaremycin was isolated from the culture broth of the actinomycete Streptomyces sp. A012304. Its structure, determined to be 5-acetamido-4-oxo-5-hexenoic acid (2) (Fig. 1A), is related to that of 5-aminolevulinic acid (ALA), the first common precursor molecule of all tetrapyrroles. The enzyme porphobilinogen (PBG) synthase (PBGS; EC 4.2.1.24) asymmetrically condenses two such molecules of ALA to generate porphobilinogen (26, 27) (Fig. 1B). The activity of alaremycin against Escherichia coli is enhanced by the presence of ALA, implying that its antimicrobial activity derives from the inhibition of PBGS (2). During tetrapyrrole biosynthesis, four molecules of porphobilinogen are fused to generate the first tetrapyrrole, uroporphyrinogen III. Thereafter, the biosynthetic pathways of heme and chlorophyll separate from those of vitamin B12, siroheme, and factor F430 due to distinct modifications of the initial tetrapyrrole skeleton (30).
FIG. 1.
(A) Alaremycin (5-acetamido-4-oxo-5-hexenoic acid). (B) Enzyme reaction of PBGS. PBGS catalyzes the first common step in tetrapyrrole biosynthesis involving the asymmetric condensation of two 5-aminolevulinic acid molecules to form the monopyrrolic porphobilinogen. (C) Alaremycin 2 (5-acetamido-4-oxo-5-hexanoic acid).
PBGSs from a range of organisms have been investigated biochemically and structurally, providing in-depth insights into the catalysis of PBGS (4, 7-10). The two ALA substrate molecules occupy two distinct binding sites in PBGS, referred to as the A and the P sites, to indicate that they contribute to porphobilinogen an acetic acid and a propanoic acid side chain, respectively. The ALA A and P sites are covalently bound through a stable Schiff base with lysine residues 205 and 260, respectively (12). Although PBGSs from different organisms differ with respect to the metal ion requirements and localization, the amino acid sequences are highly conserved between bacteria, archaea, and eukaryotes. Whereas human PBGS is Zn2+ dependent (17), catalysis by PBGS from the opportunistic human pathogen Pseudomonas aeruginosa is metal independent but retains a structural Mg2+ site near the active site (9, 13).
Many organisms, including bacteria, produce dedicated antimicrobial compounds to ward off infections or competing microbes. Streptomyces sp. A012304 hence produces the compound alaremycin, thereby preventing growth in neighboring microorganisms. Strategies to counteract the effect of alaremycin on Streptomyces sp. A012304 itself have so far not been described. We have investigated the molecular basis for the antimicrobial activity of alaremycin by analyzing its activity against Escherichia coli, Pseudomonas aeruginosa, Bacillus subtilis, Bacillus megaterium, Streptomyces coelicolor, and Streptomyces avermitilis. As PBGS is the molecular target of alaremycin, we studied the impact of the antibiotic on recombinant Mg2+-stimulated P. aeruginosa PGBS and Zn2+-dependent Methanosarcina barkeri PBGS. Furthermore, alaremycin was cocrystallized with the PBGS from P. aeruginosa, and its was structure solved at a resolution of 1.75 Å to describe its mode of PBGS inactivation at the atomic level.
MATERIALS AND METHODS
Materials.
ALA was purchased from Merck (Darmstadt, Germany), porphobilinogen was purchased from Porphyrin Products (Logan, UT), and other chemicals were purchased from Sigma-Aldrich (Hamburg, Germany). Oligonucleotides were purchased from Metabion (Planegg-Martinsried, Germany), Protino Ni-IDA resin was purchased from Machery-Nagel (Düren, Germany), and ethyl methanesulfonate was purchased from ABCR (Karlsruhe, Germany).
Bacterial strains and growth conditions.
To determine the antibacterial effect of alaremycin, P. aeruginosa PAO1 was grown in AB minimal medium; Bacillus subtilis JH642, S. coelicolor (DMS 40233), and S. avermitilis (DSM 46492) were grown in Spizizen minimal medium; and E. coli CSA1 was grown in M9 minimal medium containing 20 μM hemin (20). B. megaterium (DSM 319) was grown in MOPSO [3-(N-morpholino)-2-hydroxypropanesulfonic acid] minimal medium. To quantify the activity of PBGS in cell extracts, S. avermitilis was cultured in 2% oat meal, pH 7.2, at 30°C and 200 rpm for 4 days. S. coelicolor was grown in 0.4% glucose-0.4% yeast extract-1% malt extract, pH 7.2, at 30°C and 200 rpm for 4 days. Streptomyces sp. A012304 was grown in seed medium (4% glucose, 1% dry bouillon, 0.3% soybean meal, 0.3% CaCO3, pH 7.0) at 30°C and 200 rpm for 2 days. All other bacterial strains were grown in LB medium at 37°C for 20 h.
Isolation of alaremycin.
For alaremycin production, Streptomyces sp. A012304 was cultivated in production medium containing 6% dextrin, 2% yeast extract, 0.3% NaCl, 0.3% CaCO3, 0.1% dry bouillon, and 0.1% K2HPO4, pH 7.0, at 30°C and 200 rpm for 4 days. Alaremycin was isolated as described previously (2). It was shown to be at least 98% pure, as analyzed by nuclear magnetic resonance (NMR) spectroscopy (2).
Determination of MICs.
MICs were determined by microdilution techniques: twofold dilutions (1 to 100 mg/liter) of alaremycin in double-distilled water were placed in microtiter plates; and 108 cells/ml exponentially growing B. subtilis, B. megaterium, P. aeruginosa, S. coelicolor, S. avermitilis, E. coli DH10b, and E. coli CSA1 were added. E. coli strain CSA1 was incubated in the presence and the absence of 50 μg/ml hemin. For all strains, at least four independent curves of growth in the presence of different alaremycin concentrations over 24 h were recorded. The MIC was determined as the lowest alaremycin concentration that inhibited growth for 5 h.
Expression vectors for recombinant P. aeruginosa and Methanosarcina barkeri PBGS genes.
M. barkeri hemB was amplified from chromosomal DNA by PCR with primers 5′-CCGGAATTCCGGATGTTTCCAGATGTCAGGTTAAG-3′ and 5′-CCGCTCGAGCGGTTACTTCAACATGCGGGCAGC-3′, which contained EcoRI and XhoI sites, respectively. The resulting 975-bp PCR product was digested with EcoRI and XhoI and inserted into pET32a (Novagen, Madison, WI) to create pET32aMbhemB. Vector pGEXhemB, which contained P. aeruginosa PBGS was kindly provided by N. Frankenberg-Dinkel (University of Bochum, Bochum, Germany) (10).
Protein production and purification.
Recombinant P. aeruginosa PBGS was produced and purified as described previously (10). Protein integrity was analyzed by mass spectrometry and Western blot analysis. Recombinant M. barkeri PBGS was produced by using E. coli strain BL21(DE3)RIL (Stratagene, Heidelberg, Germany) in LB medium at 37°C and 180 rpm. At an optical density at 578 nm of 0.7, protein production was induced by 150 μM isopropyl-1-thio-ß-d-galactopyranoside (IPTG). Cells were cultivated overnight at 17°C and 150 rpm, harvested, washed with buffer A (50 mM Tris HCl, pH 8.5, 300 mM NaCl, 10 mM ZnCl2), and resuspended in a minimal volume of buffer A. After cell disruption by sonication (HD 2070; Bandelin) and centrifugation, (100,000 × g for 45 min), the soluble fraction was applied to Protino Ni-IDA agarose (Machery-Nagel, Düren, Germany) and washed and the PBGS was eluted with 300 mM imidazole in buffer A. Further purification steps involved anion-exchange chromatography on a DEAE-Sepharose column (PBGS was eluted with 200 mM NaCl in buffer A) and gel permeation chromatography on a 30-ml Superdex 200 HR 10/30 column (0.5 ml/min; GE Healthcare) in buffer A. The overall yield was ∼9 mg of M. barkeri PBGS per liter of culture.
Enzyme activity assay.
PBGS activity was quantified by a modified Ehrlich's test, based on the reaction between the product PBG with 4-(dimethylamino)-benzaldehyde (10, 15). The Michaelis-Menten constant (Km), the maximal velocity (Vmax), and the catalytic constant (kcat) were determined by measuring the constant rate of PBG formation for 0 to 10 mM ALA and iteratively optimized Lineweaver-Burk plots by using the SigmaPlot (version 8.0) and Enzyme Kinetics (version 1.1) programs. The catalytic efficiency (kcat) was obtained by dividing Vmax by the enzyme concentration. The activity of PBGS in cell extracts was determined by adjusting protein concentrations to an A280 of 5 with buffer K1 (100 mM bis-Tris-propane, pH 8.5). As a control, cell extracts were inactivated by heating to 95°C for 10 min, which resulted in negligible background activity. Reactions without additional ALA were used to determine the concentration of cellular PBG in the cell extracts.
Determination of inhibition constants.
P. aeruginosa PBGS at 2 μg/ml and M. barkeri PBGS at 25 μg/ml were diluted to a final absorbance of 5 at λ280 with kinetic buffer K1 (100 mM bis-Tris-propane, pH 8.5). ALA (40 mM) and alaremycin solutions were prepared in buffer K1. For P. aeruginosa PBGS, 10 mM MgCl2 was added, and for M. barkeri PBGS, 10 mM ZnCl2 was added. Protein, buffer, and alaremycin (0 to 10 mM) were mixed and incubated at 37°C for 10 min. Longer incubation times did not affect enzyme inhibition. Substrate (5 mM) was added to start the reaction. The PBGS-catalyzed reaction was stopped at times of between 1 and 60 min by adding equivalent volumes of stop reagent (50% trichloroacetic acid, 100 mM HgCl2) to the reaction mixture. After centrifugation (at 5,000 × g for 3 min), the supernatant was treated with an equivalent amount of Ehrlich's reagent (0.4 g 4-dimethylamino benzaldehyde in 10 ml acetic acid and 10 ml HClO4). After 15 min of incubation at room temperature, the product was quantified by measurement of the absorbance at 555 nm (ɛ = 60,200 M−1 cm−1). The alaremycin concentration that inhibited the enzyme activity by 50% (IC50) was determined for both P. aeruginosa and M. barkeri PGBSs. As inhibition was time independent, kinetic inhibition constants (kis) were determined by the use of Michaelis-Menten kinetics.
Crystallization and structure determination.
Recombinant P. aeruginosa PBGS (20 mg/ml) was mixed with 5 mM alaremycin (both in 100 mM Tris-HCl, pH 7.5, and in 10 mM MgCl2). The purity of the alaremycin was at least 98%, as determined by NMR spectroscopy (2). The crystallization conditions were identified by using the Crystal Screen and Crystal Screen 2 programs (Hampton Research, Aliso Viejo, CA) in 96-well sitting-drop racks (Douglas Instruments) with 100 μl reservoir solution and drops of 3 μl of reservoir solution and 3 μl of protein solution. P. aeruginosa PBGS crystals (space group I422, a = b = 84 Å, c = 159 Å) grew in 12 h at 17°C in 100 mM HEPES, pH 7.5-200 mM MgCl2-30% polyethylene glycol 400 (PEG 400). The crystals were cryoprotected by 20% (vol/vol) PEG 400 and were flash-cooled in liquid N2.
The data were collected at 100 K and at beam line BL1 (Bessy, Berlin, Germany) and were indexed, integrated, and scaled by using the HKL2000 program (23). Data statistics are listed in Table 1. The structure of P. aeruginosa PBGS in complex with alaremycin was solved by molecular replacement by using the Phaser program (5, 6) and the A chain of a related crystal (Protein Data Bank code 2WOQ). The structure was refined by restrained refinement with TLS protocols (22). For manual adjustments and quality control the graphics program Coot was used (7). Molecular depictions were prepared by using the PyMOL program (www.pymol.org).
TABLE 1.
Data collection and refinement statistics
| Parameter | Value |
|---|---|
| Crystallographic data | |
| Space group | I422 |
| Cell dimensions, a, c (Å) | 84.4, 158.5 |
| Total no. of reflections (>1σ) | 401,885 |
| Unique reflections | 29,316 |
| Resolution range (Å)b | 28-1.75 (1.78-1.75) |
| Completeness (%)b | 100 (99.9) |
| Redundancyb | 13.7 (8.6) |
| Rmergeb,c | 6.9 (50.4) |
| I/σIb | 46.0 (5.3) |
| Temp factor from Wilson plot (Å2) | 18.7 |
| Refinement | |
| Resolution range (Å)b | 25.3-1.75 (1.80-1.75) |
| No. of reflectionsa,b | 27,821 (2,006) |
| R factor (%), Rfree (%)b,d | 14.6 (21.5), 17.8 (25.4) |
| No. of non-H protein atoms/monomer | 2,703 |
| No. of water molecules/monomer | 177 |
| No. of Mg2+ ions/monomer | 4 |
| Average B factor (Å2) | 16.1 |
| Ramachandran plot,e preferred, allowed, outliers (%/no.) | 97.0/292, 2.7/8, 0.3/1 |
| RMS deviation from ideality: bond lengths (Å), bond angles (degrees)f | 0.015, 1.49 |
| Estimated overall coordinate error, based on maximum likelihood (Å) | 0.06 |
| Protein Data Bank code | 2WOQ |
>1σ, where σ is the average standard deviation.
Value for shell of highest resolution in parentheses.
Rmerge = 100[(Σhj |Ihj − Ih|)/(Σhj Ihj)] for all observations Ihj contributing to Ih, where Rmerge is the R-factor for reflections classified as “observed” in a given resolution shell, Ihj is the intensity of the jth observation of reflection h, I/σI is the average intensity of a group of reflections divided by the average standard deviation of the same group of reflections, and Rfree is the R-factor based on a test set consisting of 10% of reflections included from refinement.
Five percent of the data were omitted from the refinement.
Determined by use of the program Coot (7).
Determined by use of the program Refmac (21). RMS, root mean square.
Protein structure accession number.
The atomic coordinates have been deposited in the Protein Data Bank under accession number 2WOQ.
RESULTS AND DISCUSSION
Rationale for experimental approach.
Alaremycin was recently shown to inhibit the growth of an E. coli laboratory strain (2). To determine the general applicability of alaremycin as an antibiotic, a range of bacterial species was exposed to the compound. Representatives of Gram-positive bacteria included B. megaterium and B. subtilis, and representatives of Gram-negative bacteria included P. aeruginosa and E. coli DH10b. Alaremycin-producing and non-alaremycin-producing Streptomyces strains were analyzed for their intrinsic immunity to the antibiotic. To identify the cellular target of alaremycin activity, heme-permeant E. coli strain CSA1 was created to test alaremycin independently of intrinsic heme biosynthesis. The alaremycin inhibition of PBGS was determined in cell extracts for the species listed above and of the enzymes purified from P. aeruginosa and M. barkeri. The PBGS from P. aeruginosa was also used for biochemical characterization as well as for cocrystallization with alaremycin and crystal structure determination.
Heme biosynthesis is the target of alaremycin activity.
E. coli laboratory strains have generally lost their ability to import heme from the surrounding medium (29). We have created an E. coli strain capable of heme uptake by treating hemA-negative E. coli strain EV61 (28), which normally requires ALA for growth, with ethyl methanesulfonate to induce chemical mutagenesis (20, 21). Colonies were screened for their ability to grow in ALA-free, heme-containing medium. Our isolated heme-permeant hemA-negative strain was designated E. coli CSA1. If heme biosynthesis is the target of alaremycin activity, the addition of heme to E. coli CSA1 should allow the strain to overcome the growth-inhibitory phenotype. The treatment of E. coli CSA1 with alaremycin (≤40 mg/ml) in the presence and the absence of ALA completely inhibited bacterial growth. The addition of additional heme rendered E. coli CSA1 alaremycin insensitive. This implicates heme biosynthesis as the target of alaremycin.
Alaremycin inhibits the growth of Gram-negative and -positive bacteria.
The MICs of alaremycin for several Gram-positive and -negative bacteria ranged from 4 mM to over 20 mM (Table 2). For example, for E. coli, the MIC for alaremycin was 4 mM. Eight and 14 mM alaremycin inhibited the growth of S. coelicolor and S. avermitilis in complex medium, whereas the producer strain, Streptomyces sp. A012304, was not inhibited by alaremycin at concentrations of up to 20 mM.
TABLE 2.
MICsa
| Isolate | MIC |
PBGS synthase activity in cell extracts (% PBGS activity) |
||
|---|---|---|---|---|
| mM | mg/liter | 5 mM alaremycin | 10 mM alaremycin | |
| Escherichia coli DH10b | 4 | 740 | 30 | <1 |
| Pseudomonas aeruginosa | 12 | 2,220 | 92 | 25 |
| Bacillus subtilis | 14 | 2,590 | 61 | 30 |
| Bacillus megaterium | 4 | 740 | 3 | <1 |
| Streptomyces coelicolor | 8 | 1,480 | 77 | 22 |
| Streptomyces avermitilis | 8 | 1,480 | 68 | <1 |
| Escherichia coli CSA1 | 10 | 1,850 | 36 | <1 |
| Streptomyes sp. A012304 | >20 | >3,700 | 90 | 79 |
The MICs for bacterial growth and the residual PBGS activity in cell extracts inhibited by alaremycin were determined as described in the text. The PBGS activity in cell extracts of each strain without alaremycin was set equal to 100%. The PBGS activities after the addition of alaremycin are relative values. The standard errors were estimated to be between 10 and 15%.
Quantifying PBGS inhibition by alaremycin in cell extracts.
As alaremycin is structurally related to ALA (the substrate of PBGS), we analyzed the effects of 0, 5, and 10 mM alaremycin on PBGS activity in cell extracts of S. coelicolor, S. avermitilis, Streptomyces sp. A012304, E. coli, B. megaterium, B. subtilis, P. aeruginosa, E. coli DH10b, and E. coli CSA1. Surprisingly, the trend of alaremycin inhibition of PBGS in cell extracts was distinct from the trend of alaremycin inhibition of the PBGS from the corresponding organisms (Table 2). Thus, 5 mM alaremycin essentially abolished the PBGS activity of B. megaterium cell extracts, whereas the S. coelicolor (22%), P. aeruginosa (25%), and B. subtilis (30%) cell extracts retained residual PBGS activity even in the presence of 10 mM alaremycin. The differences in the alaremycin inhibitory activities between intact organisms (MICs) and cell extracts (PBGSs) observed for P. aeruginosa were presumably due to the various permeabilities and uptake capacities of the individual organisms. Interestingly, Streptomyces sp. A012304 cell extracts retained 78% PBGS activity in the presence of 10 mM alaremycin, implying that the PBGS from this species possesses significant immunity against this antibiotic.
Determination of inhibitory effect on recombinant PBGS.
To unambiguously identify the molecular target of alaremycin, we tested its effect on recombinant, purified PBGS from P. aeruginosa (metal independent) and M. barkeri (Zn2+ dependent). The kinetic parameters (Table 3) of P. aeruginosa PBGS are essentially as published previously (11). The specific activity of purified native M. barkeri PBGS is 0.39 μmol min−1 mg−1 (3), while the Km of 0.07 μM is quite low. To quantify the potency of the inhibitor, IC50s were determined as described previously (2) and are listed in Table 3. No time dependence of inhibition was observed. The ki for the P. aeruginosa PBGS was 1.33 mM, and that for the M. barkeri PBGS was 1.51 mM.
TABLE 3.
Kinetic parameters of P. aeruginosa and M. barkeri PBGSs and IC50s for alaremycina
| Parameter | P. aeruginosa | M. barkeri |
|---|---|---|
| General catalytic properties | ||
| Km(μM) | 0.32 | 0.07 |
| kcat (s−1) | 16.1 | 0.012 |
| kcat/Km (μM−1s−1) | 50.3 | 0.17 |
| Inhibition by alaremycin | ||
| IC50 (mM) | 2.1 | 2.2 |
| ki (mM) | 1.33 | 1.51 |
The Michaelis-Menten constant (Km) and the kcat and the kcat/Km values for the PBGSs were determined from substrate velocity plots by measuring the constant velocity formation of porphobilinogen from ALA over a substrate concentration range of from 1 to 10 mM. Values were determined by the use of iterative, curve-fitting Lineweaver-Burk plots (SigmaPlot program, version 8.0; Enzyme Kinetics program, version 1.1). For inhibition studies, 0 to 40 mM alaremycin was added to the enzyme activity test. The standard errors of the results were between 5 and 10%.
Crystal structure of the P. aeruginosa PBGS-alaremycin complex.
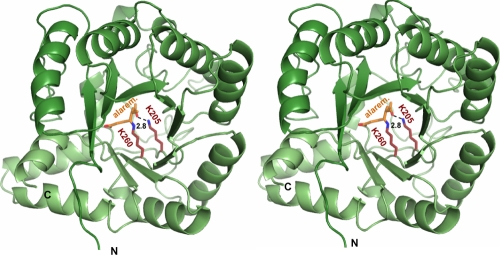
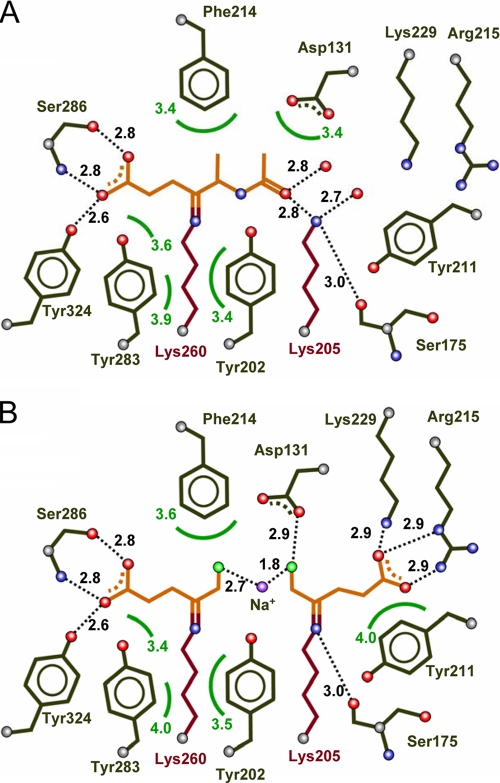
P. aeruginosa PBGS was cocrystallized with alaremycin to elucidate its mode of inhibition in detail. The structure was solved and refined to a resolution of 1.75 Å. The overall structure is unchanged from that presented in earlier reports (10, 13), despite a change in the space group from P4212 (one dimer per asymmetric unit) to I422 (one monomer per asymmetric unit). Interestingly, the antibiotic is observed to bind as 5-acetamido-4-oxo-5-hexanoic acid in the active site of PBGS, a reduced derivative of 5-acetamido-4-oxo-5-hexenoic acid. We correspondingly refer to this compound as alaremycin 2 (Fig. 1C). Alaremycin 2, which is located in the active site (Fig. 2) and whose electron density is clearly defined, is coordinated by the P-site Lys260 via a Schiff base to its C-4 atom. This resembles substrate ALA binding to the P site and has a binding mode comparable to that of the inhibitor 5-fluorolevulinic acid (Fig. 3) (9, 14). Inhibitor binding is stabilized by two hydrogen bonds from the antibiotic carboxylate to Ser286, as described for 5-hydroxylevulinic acid, a substrate analogue, in the P site of P. aeruginosa PBGS (12). Alaremycin 2 thus mimics the P-site substrate ALA rather than the product PBG. The Schiff base bond of a second A-site substrate ALA with Lys205 is not mirrored by alaremycin 2 coordination. Instead, the amino group of the A-site Lys205 hydrogen bonds to the C-7 keto group of alaremycin 2. Inhibitor binding is further stabilized by hydrogen bonds to water molecules that in turn form hydrogen bonds with Gln233, Asp127, and Ser175. An Mg2+ in the active site (12) is coordinated by Asp139, Asp131, and Asp 176. The part of the antibiotic that blocks the A site of the active site has a conformation similar to that previously observed for inhibitors mimicking the intermediate of the condensation reaction (18). The methyl group attached to the nonplanar C-5 carbon atom of alaremycin 2 is coordinated only by weak van der Waals interactions (≥3.9 Å) in the active site of PBGS, implying that the modification to alaremycin may be of limited relevance for its antibiotic function. The conversion of alaremycin (characterized by NMR [2]) to alaremycin 2 (observed in the active site of PBGS by X-ray crystallography) appears to occur during crystallization by an as yet unknown mechanism. However, the inhibitory features of the compound are not affected.
FIG. 2.
Stereo image of the catalytic domain of Pseudomonas aeruginosa PBGS in complex with alaremycin 2 (alarem.). Alaremycin 2 binds to Lys260 through a Schiff base, while its keto-oxygen forms a hydrogen bond to Lys205 (A site).
FIG. 3.
Comparison of the binding modes of alaremycin 2 (A) and the two 5-fluorolevulinc acid (FLA) substrate analogue molecules (B) in the PBGS catalytic site. The two FLA substrate molecules and the alaremycin 2 inhibitor are indicated in orange, the conserved lysine residues of the P and A sites are indicated in violet, amino acid residues involved in substrate recognition are indicated in black, and hydrogen bonding is indicated as dotted lines. The two FLA molecules are covalently bound to Lys260 (P site) and Lys205 (A site) by Schiff bases linked by Na+. The natural inhibitor alaremycin 2 is bound by a Schiff base bond to Lys260 (P site) but additionally partly blocks the A site.
Alaremycin as a lead compound for antibiotic development.
Alignment of the amino acid sequences of PBGSs from a range of bacteria demonstrated a high degree of conservation at the catalytic site. Correspondingly, the amino acid residues in contact with alaremycin 2 are conserved in many bacteria, which explains the broad antibiotic activity of the compound. It also provides a structural basis for rational drug design to further improve its specificity and affinity. Clearly, the observed alaremycin 2 IC50 of 2.1 mM for the P. aeruginosa PBGS (Table 3) would need to be further improved prior to any clinical applications. Previous investigations of the catalytic mechanism of P. aeruginosa PBGS by the use of synthetic inhibitors provide a rational basis for this (12). These inhibitors, with a (carbon) backbone of 11 atoms, bear ether, amine, thioether, sulfoxide, and sulfonyl groups at the central position, position 6. All were cocrystallized with P. aeruginosa PBGS. The amine and thioether derivatives revealed IC50s of about 0.3 mM. Their coordination by PGBS differs somewhat from that of alaremycin, largely on the basis of the fact that alaremycin is shorter than the other inhibitors. Thus, inter alia, it lacks the A-site carboxyl group. The extension of alaremycin to a chain length of 11 atoms and restoration of the terminal carboxyl group would be one way of increasing the overall binding affinity. As described here, alaremycin is unable to utilize the second Schiff base coordination by Lys205, even though it possesses a carbonyl group at position 7, positioned within 2.8 Å of the lysine amine group. The geometry is, however, such that a Schiff base bond would require the introduction of a carbonyl group at carbon 8 in the potentially optimized inhibitor, which could restore this interaction. Finally, the synthetic amine and thioether derivatives that have been described partly serve as substrates for the enzyme, leading to C—C bond formation between C-3 and C-8, mirroring the ring closure via the aldol addition of the A-side C-3 to the P-side C-8 of the natural substrate and leading to a dead-end product. Larger and charged atoms in position 6, such as sulfoxide and sulfonyl, prevent this reaction. In contrast, alaremycin 2, which has an amine group in this position, could provide a good starting point for the synthesis of a novel inhibitory derivative, since the product of natural PBGS catalysis also carries an amino group in this position. Overall, the crystal structure of the P. aeruginosa PBGS-alaremycin complex provides a solid basis for the rational development of novel antibiotic compounds targeted to bacterial heme biosynthesis.
Acknowledgments
We gratefully acknowledge the synchrotron beam time at BL1, Bessy.
This work was supported by the Deutsche Forschungsgemeinschaft (grant JA 470-7-3).
Footnotes
Published ahead of print on 12 October 2009.
REFERENCES
- 1.Anderson, C. M., and J. C. Gray. 1991. Effect of gabaculine on the synthesis of heme and cytochrome f in etiolated wheat seedlings. Plant Physiol. 96:584-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Awa, Y., N. Iwai, T. Ueda, K. Suzuki, S. Asano, J. Yamagishi, K. Nagai, and M. Wachi. 2005. Isolation of a new antibiotic, alaremycin, structurally related to 5-aminolevulinic acid from Streptomyces sp. A012304. Biosci. Biotechnol. Biochem. 69:1721-1725. [DOI] [PubMed] [Google Scholar]
- 3.Bhosale, S., D. Kshirsagar, P. Pawar, T. Yeole, and D. Ranade. 1995. Purification and characterization of 5-aminolevulinic acid dehydratase from Methanosarcina barkeri. FEMS Microbiol. Lett. 127:151-155. [Google Scholar]
- 4.Breinig, S., J. Kervinen, L. Stith, A. S. Wasson, R. Fairman, A. Wlodawer, A. Zdanov, and E. K. Jaffe. 2003. Control of tetrapyrrole biosynthesis by alternate quaternary forms of porphobilinogen synthase. Nat. Struct. Biol. 10:757-763. [DOI] [PubMed] [Google Scholar]
- 5.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 6.Collaborative Computational Project Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 7.Erskine, P. T., E. Norton, J. B. Cooper, R. Lambert, A. Coker, G. Lewis, P. Spencer, M. Sarwar, S. P. Wood, M. J. Warren, and P. M. Shoolingin-Jordan. 1999. X-ray structure of 5-aminolevulinic acid dehydratase from Escherichia coli complexed with the inhibitor levulinic acid at 2.0 Å resolution. Biochemistry 38:4266-4276. [DOI] [PubMed] [Google Scholar]
- 8.Erskine, P. T., N. Senior, S. Awan, R. Lambert, G. Lewis, I. J. Tickle, M. Sarwar, P. Spencer, P. Thomas, M. J. Warren, P. M. Shoolingin-Jordan, S. P. Wood, and J. B. Cooper. 1997. X-ray structure of 5-aminolaevulinate dehydratase, a hybrid aldolase. Nat. Struct. Biol. 4:1025-1031. [DOI] [PubMed] [Google Scholar]
- 9.Frankenberg, N., P. T. Erskine, J. B. Cooper, P. M. Shoolingin-Jordan, D. Jahn, and D. W. Heinz. 1999. High resolution crystal structure of a Mg2+-dependent porphobilinogen synthase. J. Mol. Biol. 289:591-602. [DOI] [PubMed] [Google Scholar]
- 10.Frankenberg, N., D. W. Heinz, and D. Jahn. 1999. Production, purification, and characterization of a Mg2+-responsive porphobilinogen synthase from Pseudomonas aeruginosa. Biochemistry 38:13968-13975. [DOI] [PubMed] [Google Scholar]
- 11.Frankenberg, N., D. Jahn, and E. K. Jaffe. 1999. Pseudomonas aeruginosa contains a novel type V porphobilinogen synthase with no required catalytic metal ions. Biochemistry 38:13976-13982. [DOI] [PubMed] [Google Scholar]
- 12.Frere, F., M. Nentwich, S. Gacond, D. W. Heinz, R. Neier, and N. Frankenberg-Dinkel. 2006. Probing the active site of Pseudomonas aeruginosa porphobilinogen synthase using newly developed inhibitors. Biochemistry 45:8243-8253. [DOI] [PubMed] [Google Scholar]
- 13.Frere, F., H. Reents, W. D. Schubert, D. W. Heinz, and D. Jahn. 2005. Tracking the evolution of porphobilinogen synthase metal dependence in vitro. J. Mol. Biol. 345:1059-1070. [DOI] [PubMed] [Google Scholar]
- 14.Frere, F., W. D. Schubert, F. Stauffer, N. Frankenberg, R. Neier, D. Jahn, and D. W. Heinz. 2002. Structure of porphobilinogen synthase from Pseudomonas aeruginosa in complex with 5-fluorolevulinic acid suggests a double Schiff base mechanism. J. Mol. Biol. 320:237-247. [DOI] [PubMed] [Google Scholar]
- 15.Gacond, S., F. Frere, M. Nentwich, J. P. Faurite, N. Frankenberg-Dinkel, and R. Neier. 2007. Synthesis of bisubstrate inhibitors of porphobilinogen synthase from Pseudomonas aeruginosa. Chem. Biodivers. 4:189-202. [DOI] [PubMed] [Google Scholar]
- 16.Heinemann, I. U., M. Jahn, and D. Jahn. 2008. The biochemistry of heme biosynthesis. Arch. Biochem. Biophys. 474:238-251. [DOI] [PubMed] [Google Scholar]
- 17.Jaffe, E. K. 2003. Investigations on the metal switch region of human porphobilinogen synthase. J. Biol. Inorg. Chem. 8:176-184. [DOI] [PubMed] [Google Scholar]
- 18.Jaffe, E. K. 2004. The porphobilinogen synthase catalyzed reaction mechanism. Bioorg. Chem. 32:316-325. [DOI] [PubMed] [Google Scholar]
- 19.May, T. B., J. A. Guikema, R. L. Henry, M. K. Schuler, and P. P. Wong. 1987. Gabaculine inhibition of chlorophyll biosynthesis and nodulation in Phaseolus lunatus L. Plant Physiol. 84:1309-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McConville, M. L., and H. P. Charles. 1979. Mutants of Escherichia coli K12 permeable to haemin. J. Gen. Microbiol. 113:165-168. [DOI] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1992. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 22.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 24.Padmanaban, G., and P. N. Rangarajan. 2000. Heme metabolism of Plasmodium is a major antimalarial target. Biochem. Biophys. Res. Commun. 268:665-668. [DOI] [PubMed] [Google Scholar]
- 25.Petricek, M., K. Petrickova, L. Havlicek, and J. Felsberg. 2006. Occurrence of two 5-aminolevulinate biosynthetic pathways in Streptomyces nodosus subsp. asukaensis is linked with the production of asukamycin. J. Bacteriol. 188:5113-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shemin, D., and C. S. Russel. 1953. Delta-aminolevulinic acid, its role in the biosynthesis of porphyrins and purins. J. Am. Chem. Soc. 75:4873-4875. [Google Scholar]
- 27.Shoolingin-Jordan, P. M., and K. M. Cheung. 1999. The biosynthesis of heme, vol. 4. Springer, London, United Kingdom.
- 28.Verkamp, E., and B. K. Chelm. 1989. Isolation, nucleotide sequence, and preliminary characterization of the Escherichia coli K-12 hemA gene. J. Bacteriol. 171:4728-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wandersman, C., and I. Stojiljkovic. 2000. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr. Opin. Microbiol. 3:215-220. [DOI] [PubMed] [Google Scholar]
- 30.Warren, M. J., and A. G. Smith. 2008. Tetrapyrroles: birth, life and death. Landes Bioscience, Springer Science and Business Media, New York, NY.