Abstract
Antifolate drugs have an important role in the treatment of malaria. Polymorphisms in the genes encoding the dihydrofolate reductase and dihydropteroate synthetase enzymes cause resistance to the antifol and sulfa drugs, respectively. Rwanda has the highest levels of antimalarial drug resistance in Africa. We correlated the efficacy of chlorproguanil-dapsone plus artesunate (CPG-DDS+A) and amodiaquine plus sulfadoxine-pyrimethamine (AQ+SP) in children with uncomplicated malaria caused by Plasmodium falciparum parasites with pfdhfr and pfdhps mutations, which are known to confer reduced drug susceptibility, in two areas of Rwanda. In the eastern province, where the cure rates were low, over 75% of isolates had three or more pfdhfr mutations and two or three pfdhps mutations and 11% had the pfdhfr 164-Leu polymorphism. In the western province, where the cure rates were significantly higher (P < 0.001), the prevalence of multiple resistance mutations was lower and the pfdhfr I164L polymorphism was not found. The risk of treatment failure following the administration of AQ+SP more than doubled for each additional pfdhfr resistance mutation (odds ratio [OR] = 2.4; 95% confidence interval [CI] = 1.01 to 5.55; P = 0.048) and each pfdhps mutation (OR = 2.1; 95% CI = 1.21 to 3.54; P = 0.008). The risk of failure following CPG-DDS+A treatment was 2.2 times higher (95% CI = 1.34 to 3.7) for each additional pfdhfr mutation, whereas there was no association with mutations in the pfdhps gene (P = 0.13). The pfdhfr 164-Leu polymorphism is prevalent in eastern Rwanda. Antimalarial treatments with currently available antifol-sulfa combinations are no longer effective in Rwanda because of high-level resistance.
Antifolate drugs have an important role in the treatment of Plasmodium falciparum malaria and are among the most widely available drugs. Antifols and sulfonamides target the de novo folate synthesis pathway of the parasite, inhibiting the activities of two key enzymes, dihydrofolate reductase (DHFR) and dihydropteroate synthetase (DHPS), respectively, and thereby interrupting DNA synthesis. When they are used in combinations, DHFR and DHPS inhibitors act synergistically. Sulfadoxine-pyrimethamine (SP) is the most commonly used antifolate for the treatment of malaria, but its efficacy varies widely because of resistance, which has spread rapidly in many regions where malaria is endemic. Antifolate drug resistance results from polymorphisms in the genes encoding the DHFR and DHPS enzymes. Mutations in the pfdhfr gene and in the corresponding gene in Plasmodium vivax are acquired sequentially, with each mutation conferring a stepwise reduction in susceptibility (11, 12). This progression is less clearly delineated for mutations in the pfdhps gene. Under drug pressure, antifolate resistance can be selected rapidly both in vivo and in vitro (17, 24).
The relative importance of different mutation patterns in predicting therapeutic responses remains a subject of debate, particularly when there is significant background immunity to augment antimalarial drug effects (11, 15). In Africa, treatment failure following SP treatment of falciparum malaria is often attributed to mutations at codons 108, 51, and 59 of the pfdhfr gene product (referred to as the triple mutant, which has Asn-108/Ile-51/Arg-59 mutations) and point mutations at codon 437 and/or 540 of the pfdhps gene product (referred to as the double mutant, which has Gly-437/Glu-540 mutations) (14, 29). A significantly higher level of resistance to antifolates is associated with a mutation at codon 164 of the pfdhfr gene product (Ile instead of Leu). This almost invariably occurs in addition to the mutations at codons 108, 51, and 59; and thus, parasites with the 164-Leu mutation are usually called quadruple mutants (10, 25). Until recently, quadruple mutants were confined to parts of Asia and South America (16). In 1997, Watkins et al. (30) predicted that selection of this fourth mutation would compromise the clinical usefulness of the more potent antifolate combination chlorproguanil-dapsone (CPG-DDS), designed primarily for use in Africa. East Africa has historically had more resistant P. falciparum parasites than other areas of Africa. The resistance patterns in that region often act as harbingers of the spread of resistance to the remainder of the continent.
Between 2001 and 2006, various artemisinin-based combination treatments were tested in Rwanda to identify the best treatment for uncomplicated malaria to be adopted as the first-line therapy in that country (8, 13, 26, 27). In 2006, the efficacy and safety of CPG-DDS plus artesunate (CPG-DDS+A) were compared with the efficacy and safety of amodiaquine plus SP (AS+SP) (6, 7).
The aim of the present study was to evaluate the clinical efficacies of these two antifolate combinations, CPG-DDS+A and AQ+SP, in relation to drug resistance mutations in the pfdhfr and pfdhps genes in Rwanda, a country with high levels of antimalarial drug resistance. Although neither combination is now used in Rwanda, at the time we carried out this study, SP was still recommended for the intermittent preventive treatment of malaria (IPT) during pregnancy and other antifolates had been suggested to be possible treatments for malaria in Rwanda (28).
MATERIALS AND METHODS
P. falciparum isolates.
The treatment responses in this large antimalarial drug trial (ClinicalTrials.gov identifier, NCT00461578) have been described previously (6). Briefly, we tested the efficacy and safety of CPG-DDS+A compared to AQ+SP for the treatment of uncomplicated P. falciparum malaria at two different sites in Rwanda. The trial had an open-label design; 800 children, aged 6 to 59 months, with uncomplicated malaria were randomized to receive either AQ+SP (n = 400) or CPG-DDS+A (n = 400). The patients were hospitalized for 4 days for directly observed treatment and appropriate management, including blood transfusion, if necessary, and were followed up weekly until day 28 after treatment. If the child had a second episode of parasitemia within 28 days, blood samples from the first and second episodes were used to genotype the parasite strains by using msp-1 and msp-2 to distinguish between new infections and recrudescences. Clinical and parasitological outcomes were recorded according to WHO guidelines (31), and new infections and other study withdrawals (e.g., because of drug-induced adverse events and protocol deviations) were censored at the time of withdrawal. For further genotyping, blood was collected on Whatman filter paper.
Parasite pfdhfr and pfdhps analysis.
The blood samples on filter paper were taken to Bangkok, Thailand, for molecular analysis. DNA extraction was done with a QIamp blood kit (Qiagen, Hilden, Germany). Alleles at residues 51, 59, 108, and 164 of the pfdhfr gene and at residues 436, 437, 540, 581, and 613 of the pfdhps gene were identified by PCR-restriction fragment length polymorphism analysis (5) and single-nucleotide primer extension and detection of fluorescent products on a capillary sequencer (21). These two techniques can occasionally give discordant results; therefore, both the pfdhfr and the pfdhps genes in 100 samples were randomly amplified and the results were confirmed by the sequencing technique, with identical results.
We genotyped the isolates in all blood samples taken at enrollment (day 0). Isolates in samples from patients who experienced a treatment failure were genotyped on the day that the recrudescence was detected. After day 7, recrudescences were confirmed by PCR (of the msp-1 and msp-2 genes). Only samples from patients included in the per protocol analysis were analyzed.
Statistical analysis. (i) Univariate analysis.
The frequencies of the different genotypes and genotype patterns were compared between patients who failed treatment (the total treatment failure [TTF] group) and those who cleared the infection (the adequate clinical and parasitological response [ACPR] group) by using the χ2 and Fisher's exact two-tailed tests. The cure rate for a defined parasite genotype was calculated as the number of isolates with a defined mutation from patients who were cured (the ACPR group) divided by the total number of isolates with that mutation (the ACPR and TTF groups).
(ii) Multivariate analysis.
By assuming that the failure rate increases as a result of increasing numbers of resistance-associated mutations in the pfdhfr and pfdhps genes (without further defining the relationship between the increase in the rate of resistance and the number of additional mutations in either gene), a logistic regression model for the probability of failure with two continuous covariates, one coding for the pfdhfr mutations and the other coding for the pfdhps mutations, was fitted for each treatment; and their interaction was tested. A site covariate was also added to the model. The possibility that the effects of mutations on failure rates were different between the two sites was evaluated by including an interaction term. By fitting mutations as continuous covariates, we estimated an average change in the probability of treatment failure with each increase in the number of pfdhfr or pfdhps mutations. This representation was selected since the many different possible combinations of pfdhfr and pfdhps mutations meant that despite the large size of the study population, there was a limited power for comparisons between all the different individual mutation combinations.
In these analyses, only patients with 28 days of follow-up were included. Failure was defined as early treatment failure or failure on day 7 or later PCR-confirmed recrudescence.
Analyses were performed with STATA (version 10) software (Stata Corp., College Station, TX).
RESULTS
Distribution and frequencies of pfdhfr haplotypes.
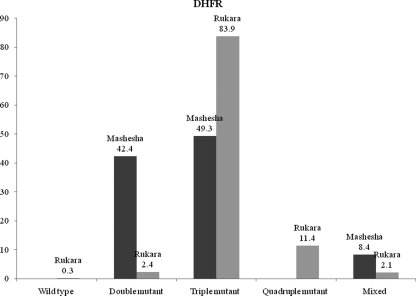
A total of 725 blood samples were successfully genotyped for polymorphisms in the pfdhfr gene. Four codons were found to be polymorphic: codons 51 (Asn → Ile), 59 (Cys → Arg), 108 (Ser → Asn), and 164 (Ile → Leu). No mutations at codon 16 (Ala→Val) were detected (results are shown in Fig. 1).
FIG. 1.
Distribution and frequency of pfdhfr genotypes at the two sites (samples were collected before treatment).
In Rukara, eastern Rwanda, 83.9% of the isolates analyzed were triple mutants (Asn-108/Ile-51/Arg-59), whereas 49.3% of the isolates in Mashesha, western Rwanda, analyzed were triple mutants (P < 0.001). The 164-Leu mutation (pfdhfr mutations 108-Asn/51-Ile/59-Arg/164-Leu) was found only in Rukara and was detected at a frequency of 11.4%; none of the Mashesha isolates harbored this mutation.
In Mashesha, 42.4% of the isolates were double mutants (108-Asn/51-Ile), whereas in Rukara, only 2.4% of the isolates were double mutants (P < 0.001). The wild-type gene was found in only one sample from Rukara. The remaining isolates were from patients with mixed infections: 2.1% and 8.4% in Rukara and Mashesha, respectively.
Distribution and frequencies of pfdhps haplotypes.
A total of 725 blood samples were successfully genotyped for polymorphisms in the pfdhps gene. Four codons in the pfdhps gene product were polymorphic: codons 436 (Ser → Ala), 437 (Ala → Gly), 540 (Lys → Glu), and 581 (Ala → Gly). Mutations at codon 613 (Ala→Ser) were not detected (results are shown in Fig. 2).
FIG. 2.
Distribution and frequency of pfdhps genotypes at the two sites (samples were collected before treatment).
In Rukara, 36.2% of the isolates analyzed were double pfdhps mutants (Gly-437/Glu-540 or Glu-540/Gly-581) and 56.4% were triple pfdhps mutants (Gly-437/Glu-540/Gly-581). The remaining samples were from patients with infections caused by single mutants (0.8%), the wild type (1.3%), or mixed mutants (5.3%).
In Mashesha, 47.0% of the isolates analyzed were double mutants and 24.4% were triple mutants. The remaining samples were from patients with infections caused by single mutants (3.2%), the wild type (10.3%), or mixed mutants (15.2%).
As for the pfdhfr genotypes, the pfdhps genotype frequency distributions were significantly different between sites, with more resistance mutations detected in isolates from Rukara (P < 0.001).
Overall, the codon 437 (Gly) mutation was present in 97% of the isolates in Rukara and 80% of the isolates in Mashesha. Fifteen isolates were wild type at codon 437 (Ala) and mutated at codon 436 (Ala), 540 (Glu), or 581 (Gly), suggesting that a mutation at codon 437 is not always necessary for the other mutations to be selected. A mutation at codon 436 (Ala) was detected in only 3% (n = 22) of the samples analyzed, and in 11 isolates 436-Ala was detected in isolation, supporting the hypothesis that serine and alanine at codon 436 are alternative wild types. A mutation at codon 581 (Gly) was present in 60% of the samples in Rukara and 29% in Mashesha. In four isolates, this mutation was observed in isolation.
pfdhfr and pfdhps together.
Considering mutations in the pfdhfr and pfdhps genes together, in Rukara, the prevalence of quintuple mutants (pfdhfr triple mutants and pfdhps double mutants) was 31.3%, whereas 46.5% of the isolates were triple mutants for both pfdhfr and pfdhps. The pfdhfr triple mutants were found with wild-type or single mutations in the pfdhps gene in only 2% of isolates. The 164 (Leu) pfdhfr mutants occurred only with the pfdhps double or triple mutants.
In Mashesha, the distribution was more heterogeneous. The pfdhfr double and triple mutants were associated with all pfdhps genotypes. Quadruple mutant (pfdhfr double mutant and pfdhps double mutant) comprised 20.5% of the isolates. Quintuple mutants comprised 23.1% of the isolates, and only 14.2% of the isolates were triple mutants at both the pfdhfr and the pfdhps genes.
Pre- and posttreatment genotype frequencies.
In Rukara, treatment failures were more likely to carry parasites with a pfdhfr quadruple-mutation genotype than those who cleared the infection in either treatment arm: for AQ+SP, 13.3% versus 4.2% (P = 0.047); for CPG-DDS+A, 21.4% versus 9.2% (P = 0.03). Patients with treatment failures following AQ+SP treatment were also more likely than those who cleared their infections to carry parasites with a pfdhps triple-mutation genotype: 65.8% versus 48.6% (P = 0.02). No differences in the prevalence of isolates that were carriers of pfdhfr triple or pfdhps double mutations were observed when the pretreatment and the recrudescent parasitemia groups were compared.
In Mashesha, patients who remained parasitemic following CPG-DDS+A treatment were more likely to carry parasites with a triple pfdhfr mutation than those who cleared the infection (60.87% versus 39.34%; P = 0.012). Similarly, in the AQ+SP arm the prevalence of the pfdhfr triple-mutation genotype was higher in the posttreatment group, although this difference was not significant (66.7% versus 51.3%; P = 0.184). There were no differences in the prevalence of pfdhfr double mutants or mixed infections or in the prevalence of the different pfdhps genotypes when the pretreatment and the recrudescent parasitemia groups in either treatment arms were compared.
Relationship of cure rates with genotypes.
Considering only the pfdhfr gene, irrespective of any mutations in pfdhps, the cure rate following treatment with CPG-DDS+A in Rukara was 100% for patients with double mutant infections, 72% for patients with triple mutant infections, and 50.0% for patients with quadruple mutant infections (Table 1). In Mashesha, the cure rates for patients with double mutant and triple mutant infections were 90.6% and 86.1%, respectively (Table 2).
TABLE 1.
Prevalence of pfdhfr and pfdhps genotypes by treatment arm and treatment outcome in Rukara
| pfdhfr mutant | pfdhps mutant | CPG-DDS+A |
AQ+SP |
||||
|---|---|---|---|---|---|---|---|
| ACPR group | TTF group | Cure rate (%) | ACPR group | TTF group | Cure rate (%) | ||
| Wild type | Double | 0 | 0 | 1 | 0 | 100 | |
| Double | Double | 2 | 0 | 100 | 2 | 1 | 66.7 |
| Triple | 0 | 0 | 1 | 2 | 33.3 | ||
| Mixed | 0 | 0 | 0 | 1 | 0 | ||
| All double | 2 | 0 | 100 | 3 | 4 | 42.3 | |
| Triple | Wild type/single | 4 | 2 | 66.7 | 2 | 0 | 100 |
| Double | 47 | 14a | 77.0 | 23 | 34 | 40.4 | |
| Triple | 54 | 26 | 67.5 | 31 | 64 | 32.6 | |
| Mixed | 6 | 1 | 85.7 | 6 | 2 | 75.0 | |
| All triple | 111 | 43 | 72.0 | 62 | 100 | 38.2 | |
| Quadruple | Double | 3 | 4 | 42.9 | 2 | 3 | 40.0 |
| Triple | 7 | 7 | 50.0 | 1 | 13 | 7.1 | |
| Mixed | 2 | 1 | 66.7 | 0 | 0 | ||
| All quadruple | 12 | 12 | 50.0 | 3 | 16 | 15.8 | |
| Mixed | Triple | 5 | 1 | 83.3 | 1 | 0 | 100 |
| Mixed | 1 | 0 | 100 | 0 | 0 | ||
| All mixed | 6 | 1 | 85.7 | 1 | 0 | 100 | |
| Total | 131 | 56 | 70 | 120 | |||
One patient in this group received a blood transfusion.
TABLE 2.
Prevalence of pfdhfr and pfdhps genotypes by treatment arm and treatment outcome in Mashesha
| pfdhfr mutant | pfdhps mutant | CPG-DDS+A |
AQ+SP |
||||
|---|---|---|---|---|---|---|---|
| ACPR group | TTF group | Cure rate (%) | ACPR group | TTF group | Cure rate (%) | ||
| Double | Wild type/single | 9 | 1a | 90.0 | 14 | 0 | 100.0 |
| Double | 34 | 11a | 75.6 | 22 | 4a | 84.6 | |
| Triple | 14 | 5 | 73.7 | 13 | 0 | 100.0 | |
| Mixed | 8 | 1 | 88.9 | 9 | 2 | 81.8 | |
| All double | 65 | 18 | 78.3 | 58 | 6 | 90.6 | |
| Triple | Wild type/single | 6 | 4 | 60.0 | 10 | 1 | 90.9 |
| Double | 26 | 14 | 65.0 | 34 | 6 | 85.0 | |
| Triple | 11 | 7 | 61.1 | 25 | 6 | 80.6 | |
| Mixed | 5 | 3 | 62.5 | 12 | 0 | 100.0 | |
| All triple | 48 | 28 | 63.2 | 81 | 13 | 86.1 | |
| Mixed | Wild type/single | 1 | 0 | 100.0 | 1 | 0 | 100.0 |
| Double | 5 | 0 | 100.0 | 6 | 1 | 85.7 | |
| Triple | 0 | 0 | 3 | 0 | 100.0 | ||
| Mixed | 3 | 0 | 100.0 | 9 | 0 | 100.0 | |
| All mixed | 9 | 0 | 100 | 19 | 1 | 95 | |
| Total | 122 | 46 | 72.6 | 158 | 20 | 88.8 | |
One patient in each group received a blood transfusion.
Following treatment with AQ+SP in Rukara, the cure rate was 38.2% for patients with pfdhfr triple mutant infections but it was only 15.8% for patients with pfdhfr quadruple mutant infections (Table 1). In Mashesha, 90.6% of those with double mutant infections and 86.1% of those with triple mutant infections were cured (Table 2).
A model was fitted to assess the contribution of mutations in the pfdhps gene within each category of pfdhfr mutations. The analysis showed that the risk of treatment failure following treatment with AQ+SP more than doubled for each unit increase in the number of pfdhfr resistance mutations (odds ratio [OR] = 2.4; 95% confidence interval [CI] = 1.01 to 5.55; P = 0.048) and also doubled for each unit increase in the number of pfdhps resistance mutations (OR = 2.1; 95% CI = 1.21 to 3.54; P = 0.008) (Fig. 3). The effects of the mutations in the two genes were additive (P = 0.39 in the test for interaction). The effect of study site was independent of the mutations present, and after adjustment for the category of mutations, the risk of treatment failure was 13 times higher in Rukara than in Mashesha (OR = 13.4; 95% CI = 6.62 to 26.93; P < 0.001).
FIG. 3.
Relationship between probability of failure and pfdhfr and pfdhps genotypes at the two sites after treatment with AQ+SP (blue, predicted probability of failure; green, observed failure rate; brown, 95% CI around the observed rate; W, wild type, S, single mutant; D, double mutant; T, triple mutant).
The risk of failure following treatment with CPG-DDS+A also doubled for each unit increase in the number of pfdhfr mutations (OR = 2.23; 95% CI = 1.34 to 3.7; P = 0.002), but the risk of failure was not related to the number of mutations in the pfdhps gene (P = 0.13). There was a difference between sites, but it did not reach statistical significance (P < 0.10) (Fig. 4).
FIG. 4.
Relationship between probability of failure and pfdhfr and pfdhps genotypes at the two sites after treatment with CPG-DDS+A (blue, predicted probability of failure; green, observed failure rate; brown, 95% CI around the observed rate; W, wild type; S, single mutant; D, double mutant; T, triple mutant).
DISCUSSION
The responses to antimalarial treatment regimens containing SP and CPG-DDS were poor in Rwandan children with acute malaria because of the high prevalence of polymorphisms in the genes coding for the DHFR and DHPS enzymes in Plasmodium falciparum. The distribution and relative frequency of alleles differed significantly between the two study areas.
At Rukara, in the eastern province, the parasite population showed a high level of resistance, and more than 75% of the isolates had three mutations in the pfdhfr gene and two or three mutations in the pfdhps gene. Importantly, the pfdhfr 164-Leu polymorphism was present in 11% of the samples analyzed.
At Mashesha, in the western province, the treatment responses were better and the prevalence of pfdhfr triple mutants together with pfdhps double and triple mutants was lower (less than 40%). The pfdhfr I164L polymorphism was not found in Mashesha. The parasite genotype mix there was more heterogeneous than at the site with high levels of resistance. Of relevance, the only other place in Africa where a high frequency (14%) of the genotype with a polymorphism at position 164 in pfdhfr has been reported is Kabale in Uganda (16), which is close to the eastern Rwanda border. The genotype with a polymorphism at position 164 in pfdhfr has been reported at lower frequencies in Kenya (18), Malawi (1), and Central Africa (19) and from travelers returning from Ghana and Kenya (9). The quadruple mutant is prevalent at higher frequencies in Southeast Asia and South America, where SP is no longer effective. These data confirm that eastern Rwanda and adjacent Uganda are a focus of high-level antifol resistance in Africa.
Whereas there is a general consensus on the relative contributions of different pfdhfr mutations to antifol resistance on the basis of the results of in vitro tests and examination of the biochemical properties of the isolated enzyme, the relative importance of different mutations in the pfdhps gene to sulfonamide and sulfone resistance and the therapeutic responses is less clear. In most of the samples analyzed in this study, the polymorphic site at codon 436 of the pfdhps gene product was not mutated (Ala), and in 11 samples the alanine-to-serine mutation was found in isolation, supporting the hypothesis that this polymorphism is an alternative wild type (2, 23).
The mutation at codon 581 (Gly) in the pfdhps gene product is thought to confer high levels of resistance and was found in >40% of the samples at the site with high levels of resistance (Rukara) and >27% of the samples at the site with intermediate levels of resistance (Mashesha). This prevalence is similar to that in nearby Uganda (16), although it appears to be infrequent in the rest of Africa. In a small number of isolates, the mutation at codon 581 (Gly) was found in isolation. Alternatively, it has been proposed that this mutation and the mutation at codon 613 have a compensatory function and for this reason are rarely seen in isolation but generally follow the occurrence of mutations at codon 437 (11).
We also found isolates which were wild type at codon 437 (Ala) and mutated at codons 436 (Ala), 540 (Glu), or 581 (Gly). It has been suggested that mutation 437-Ala is necessary for the other mutations to be selected, analogous to the mutation at codon 108 in the pfdhfr gene product. An alternative hypothesis is that mutations at codon 436, 437, or 540 could be a first step for sulfa drug resistance, as all individual mutations confer some degree of resistance to sulfonamides (11). No mutations at codon 613 in the pfdhps gene product or at codon 16 of the pfdhfr gene product were found in Rwanda.
As expected, there was evidence of selection for parasites with the pfdhfr quadruple-mutation genotype following treatment with both study drug combinations, whereas the pfdhfr triple mutant was selected only after CPG-DDS+A treatment. In Tanzania, the pfdhfr triple mutant was found to be significantly more common after treatment with CPG-DDS than before treatment (4), but this was not confirmed in Kenya (22). No changes in genotype proportions were observed for the pfdhps gene following CPG-DDS+A treatment, suggesting that CPG-DDS does not place significant selection pressure on this gene, irrespective of its effect on pfdhfr, as observed previously (3, 4, 22). In contrast, there was selection for the pfdhps triple-mutation genotype after treatment with AQ+SP in Rukara but not in Mashesha. This difference may be explained by the better efficacy of AQ at the latter site.
The clinical efficacy of CPG-DDS+A treatment was significantly higher than that of AQ+SP in Rukara (cure rates, 70.5% and 38.1%, respectively; P < 0.001), whereas the opposite was observed in Mashesha (cure rates, 73.3% and 87.8%, respectively; P < 0.001). This was because of the marked difference in the efficacy of AQ+SP (P < 0.001) and was correlated with the difference in the numbers of point mutations in the pfdhfr and pfdhps genes in P. falciparum. In particular, the cure rate was affected by acquisition of a third mutation in the pfdhps gene (at codon 581) and the fourth mutation in the pfdhfr gene (at codon 164).
AQ appears to have contributed more to the outcome in Mashesha than in Rukara, suggesting a higher level of resistance at the latter site. The contribution of artesunate should have been similar at both sites, but the different relationships observed between pfdhfr and pfdhps mutations and the therapeutic responses to CPG-DDS+A at the two sites suggests that this needs to be examined further. Although the 3-day artesunate regimen is not sufficient to effect a cure by itself, it must have contributed significantly to the combination, as CPG-DDS alone had a very low level of efficacy; by day 28, more than 50% of patients had a recurrent infection (7). In Africa, CPG-DDS has been shown to be clinically effective against SP-resistant parasite strains, including the triple mutant (3, 15, 20). However, in Thailand, in an area of multidrug resistance where the mutation at codon 164 occurs in approximately 25% of the P. falciparum infections, the efficacy of CPG-DDS (chlorproguanil given at a lower dose of 1.4 mg/kg of body weight) was poor (32). However, a more recent study in Thailand (14) showed that a 3-day regimen of proguanil (8 mg/kg/day)-dapsone plus artesunate in adults with acute malaria provided an overall cure rate similar to that in Rwanda, although isolates from all treatment failures carried the mutation at codon 164.
The genotyping of the pfdhfr and pfdhps genes for mutations associated with antifolate resistance has helped explain the low levels of efficacy of antimalarial drugs observed in Rwanda in the past years (8, 13, 26, 27). The reasons why there is such a large difference in the level of resistance between sites still need to be understood, as the observed differences in the allele frequencies are not linked to the intensity of previous SP treatment for infections caused by P. falciparum, which has been homogeneous across the country in the past. At present, arthemether-lumefantrine is the first-line treatment for uncomplicated malaria. Following the findings of high-level resistance in the present study, IPT in pregnancy with SP has temporarily been stopped by the National Malaria Control Program in Rwanda, with the decision awaiting for final approval by the Ministry of Health. Antimalarial treatments containing SP and/or amodiaquine are not recommended in Rwanda, and the use of other antifolate combinations needs to be carefully evaluated.
Acknowledgments
The Wellcome Trust of Great Britain, the Wellcome Trust Intermediate Fellowship (grant 066439/Z/01/2), and the Thailand Research Fund and the Commission on Higher Education provided financial support for this study.
K.C., A.U., and C.I.F. performed the data collection. K.S. and C.I.F. performed the statistical analysis. M.I. and S.N. performed the laboratory analysis. C.I.F. prepared the manuscript. A.D., N.P.D., and N.J.W. contributed to data analysis and manuscript preparation.
We have no conflicts of interest to declare.
Footnotes
Published ahead of print on 19 October 2009.
REFERENCES
- 1.Alker, A. P., V. Mwapasa, A. Purfield, S. J. Rogerson, M. E. Molyneux, D. D. Kamwendo, E. Tadesse, E. Chaluluka, and S. R. Meshnick. 2005. Mutations associated with sulfadoxine-pyrimethamine and chlorproguanil resistance in Plasmodium falciparum isolates from Blantyre, Malawi. Antimicrob. Agents Chemother. 49:3919-3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basco, L. K., and P. Ringwald. 1998. Molecular epidemiology of malaria in Yaounde, Cameroon. II. Baseline frequency of point mutations in the dihydropteroate synthase gene of Plasmodium falciparum. Am. J. Trop. Med. Hyg. 58:374-377. [DOI] [PubMed] [Google Scholar]
- 3.Curtis, J., M. T. Duraisingh, and D. C. Warhurst. 1998. In vivo selection for a specific genotype of dihydropteroate synthetase of Plasmodium falciparum by pyrimethamine-sulfadoxine but not chloroproguanil-dapsone treatment. J. Infect. Dis. 177:1429-1433. [DOI] [PubMed] [Google Scholar]
- 4.Curtis, J., C. Maxwell, F. H. M. Msuya, S. Mkongewa, A. Alloueche, and D. C. Warhurst. 2002. Mutations in dhfr in Plasmodium falciparum infections selected by chlorproguanil-dapsone treatment. J. Infect. Dis. 186:1861-1864. [DOI] [PubMed] [Google Scholar]
- 5.Duraisingh, M. T., J. Curtis, and D. C. Warhurst. 1998. Plasmodium falciparum: detection of polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes by PCR and restriction digestion. Exp. Parasitol. 89:1-8. [DOI] [PubMed] [Google Scholar]
- 6.Fanello, C. I., C. Karema, P. Avellino, G. Bancone, A. Uwimana, S. J. Lee, U. d'Alessandro, and D. Modiano. 2008. High risk of severe anaemia after chlorproguanil-dapsone + artesunate antimalarial treatment in patients with G6PD (A−) deficiency. PLoS One 3:e4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fanello, C. I., C. Karema, D. Ngamije, A. Uwimana, V. Ndahindwa, C. Van Overmeir, W. Van Doren, J. Curtis, and U. D'Alessandro. 2008. A randomised trial to assess the efficacy and safety of chlorproguanil/dapsone + artesunate for the treatment of uncomplicated Plasmodium falciparum malaria. Trans. R. Soc. Trop. Med. Hyg. 102:412-420. [DOI] [PubMed] [Google Scholar]
- 8.Fanello, C. I., C. Karema, W. van Doren, C. Van Overmeir, D. Ngamije, and U. D'Alessandro. 2007. A randomised trial to assess the safety and efficacy of artemether-lumefantrine (Coartem) for the treatment of uncomplicated Plasmodium falciparum malaria in Rwanda. Trans. R. Soc. Trop. Med. Hyg. 101:344-350. [DOI] [PubMed] [Google Scholar]
- 9.Farnert, A., K. Tengstam, I. B. Palme, U. Bronner, M. Lebbad, G. Swedberg, and A. Bjorkman. 2002. Polyclonal Plasmodium falciparum malaria in travelers and selection of antifolate mutations after proguanil prophylaxis. Am. J. Trop. Med. Hyg. 66:487-491. [DOI] [PubMed] [Google Scholar]
- 10.Foote, S. J., D. Galatis, and A. F. Cowman. 1990. Amino acids in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum involved in cycloguanil resistance differ from those involved in pyrimethamine resistance. Proc. Natl. Acad. Sci. U. S. A. 87:3014-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gregson, A., and C. V. Plowe. 2005. Mechanisms of resistance of malaria parasites to antifolates. Pharmacol. Rev. 57:117-145. [DOI] [PubMed] [Google Scholar]
- 12.Imwong, M., S. Pukrittakayamee, S. Looareesuwan, G. Pasvol, J. Poirreiz, N. J. White, and G. Snounou. 2001. Association of genetic mutations in Plasmodium vivax dhfr with resistance to sulfadoxine-pyrimethamine: geographical and clinical correlates. Antimicrob. Agents Chemother. 45:3122-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karema, C., C. I. Fanello, C. van Overmeir, J. P. van Geertruyden, W. van Doren, D. Ngamije, and U. D'Alessandro. 2006. Safety and efficacy of dihydroartemisinin/piperaquine (Artekin) for the treatment of uncomplicated Plasmodium falciparum malaria in Rwandan children. Trans. R. Soc. Trop. Med. Hyg. 100:1105-1111. [DOI] [PubMed] [Google Scholar]
- 14.Krudsood, S., M. Imwong, P. Wilairatana, S. Pukrittayakamee, A. Nonprasert, G. Snounou, N. J. White, and S. Looareesuwan. 2005. Artesunate-dapsone-proguanil treatment of falciparum malaria: genotypic determinants of therapeutic response. Trans. R. Soc. Trop. Med. Hyg. 99:142-149. [DOI] [PubMed] [Google Scholar]
- 15.Kublin, J. G., F. K. Dzinjalamala, D. D. Kamwendo, E. M. Malkin, J. F. Cortese, L. M. Martino, R. A. Mukadam, S. J. Rogerson, A. G. Lescano, M. E. Molyneux, P. A. Winstanley, P. Chimpeni, T. E. Taylor, and C. V. Plowe. 2002. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J. Infect. Dis. 185:380-388. [DOI] [PubMed] [Google Scholar]
- 16.Lynch, C., R. Pearce, H. Pota, J. Cox, T. A. Abeku, J. Rwakimari, I. Naidoo, J. Tibenderana, and C. Roper. 2008. Emergence of a dhfr mutation conferring high-level drug resistance in Plasmodium falciparum populations from southwest Uganda. J. Infect. Dis. 197:1598-1604. [DOI] [PubMed] [Google Scholar]
- 17.Martin, D. C., and J. D. Arnold. 1968. The effect of parasite populations on the curative action of pyrimethamine. Trans. R. Soc. Trop. Med. Hyg. 62:379-384. [DOI] [PubMed] [Google Scholar]
- 18.McCollum, A. M., A. C. Poe, M. Hamel, C. Huber, Z. Zhou, Y. P. Shi, P. Ouma, J. Vulule, P. Bloland, L. Slutsker, J. W. Barnwell, V. Udhayakumar, and A. A. Escalante. 2006. Antifolate resistance in Plasmodium falciparum: multiple origins and identification of novel dhfr alleles. J. Infect. Dis. 194:189-197. [DOI] [PubMed] [Google Scholar]
- 19.Menard, D., D. Djalle, F. Yapou, A. Manirakiza, and A. Talarmin. 2006. Frequency distribution of antimalarial drug-resistant alleles among isolates of Plasmodium falciparum in Bangui, Central African Republic. Am. J. Trop. Med. Hyg. 74:205-210. [PubMed] [Google Scholar]
- 20.Mutabingwa, T., A. Nzila, E. Mberu, E. Nduati, P. Winstanley, E. Hills, and W. Watkins. 2001. Chlorproguanil-dapsone for treatment of drug-resistant falciparum malaria in Tanzania. Lancet 358:1218-1223. [DOI] [PubMed] [Google Scholar]
- 21.Nair, S., A. Brockman, L. Paiphun, F. Nosten, and T. J. Anderson. 2002. Rapid genotyping of loci involved in antifolate drug resistance in Plasmodium falciparum by primer extension. Int. J. Parasitol. 32:852-858. [DOI] [PubMed] [Google Scholar]
- 22.Nzila, A. M., E. K. Mberu, E. Nduati, A. Ross, W. M. Watkins, and C. H. Sibley. 2002. Genetic diversity of Plasmodium falciparum parasites from Kenya is not affected by antifolate drug selection. Int. J. Parasitol. 32:1469-1476. [DOI] [PubMed] [Google Scholar]
- 23.Nzila, A. M., E. K. Mberu, J. Sulo, H. Dayo, P. A. Winstanley, C. H. Sibley, and W. M. Watkins. 2000. Towards an understanding of the mechanism of pyrimethamine-sulfadoxine resistance in Plasmodium falciparum: genotyping of dihydrofolate reductase and dihydropteroate synthase of Kenyan parasites. Antimicrob. Agents Chemother. 44:991-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paget-McNicol, S., and A. Saul. 2001. Mutation rates in the dihydrofolate reductase gene of Plasmodium falciparum. Parasitology 122:497-505. [DOI] [PubMed] [Google Scholar]
- 25.Peterson, D. S., W. K. Milhous, and T. E. Wellems. 1990. Molecular basis of differential resistance to cycloguanil and pyrimethamine in Plasmodium falciparum malaria. Proc. Natl. Acad. Sci. U. S. A. 87:3018-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rwagacondo, C. E., C. Karema, V. Mugisha, A. Erhart, J. C. Dujardin, C. Van Overmeir, P. Ringwald, and U. D'Alessandro. 2004. Is amodiaquine failing in Rwanda? Efficacy of amodiaquine alone and combined with artesunate in children with uncomplicated malaria. Trop. Med. Int. Health 9:1091-1098. [DOI] [PubMed] [Google Scholar]
- 27.Rwagacondo, C. E., F. Niyitegeka, J. Sarushi, C. Karema, V. Mugisha, J. C. Dujardin, C. Van Overmeir, J. van den Ende, and U. D'Alessandro. 2003. Efficacy of amodiaquine alone and combined with sulfadoxine-pyrimethamine and of sulfadoxine pyrimethamine combined with artesunate. Am. J. Trop. Med. Hyg. 68:743-747. [PubMed] [Google Scholar]
- 28.Sagara, I., S. Rulisa, W. Mbacham, I. Adam, K. Sissoko, H. Maiga, O. B. Traore, N. Dara, Y. T. Dicko, A. Dicko, A. Djimde, F. H. Jansen, and O. K. Doumbo. 2009. Efficacy and safety of a fixed dose artesunate-sulphamethoxypyrazine-pyrimethamine compared to artemether-lumefantrine for the treatment of uncomplicated falciparum malaria across Africa: a randomized multi-centre trial. Malar. J. 8:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sibley, C. H., J. E. Hyde, P. F. Sims, C. V. Plowe, J. G. Kublin, E. K. Mberu, A. F. Cowman, P. A. Winstanley, W. M. Watkins, and A. M. Nzila. 2001. Pyrimethamine-sulfadoxine resistance in Plasmodium falciparum: what next? Trends Parasitol. 17:582-588. [DOI] [PubMed] [Google Scholar]
- 30.Watkins, W. M., E. K. Mberu, P. A. Winstanley, and C. V. Plowe. 1997. The efficacy of antifolate antimalarial combinations in Africa: a predictive model based on pharmacodynamic and pharmacokinetic analyses. Parasitol. Today 13:459-464. [DOI] [PubMed] [Google Scholar]
- 31.WHO. 2003. Assessment and monitoring of antimalarial drug efficacy for the treatment of uncomplicated P. falciparum malaria. Report WHO/HTM/RBM/2003.50. WHO, Geneva, Switzerland.
- 32.Wilairatana, P., D. E. Kyle, S. Looareesuwan, K. Chinwongprom, S. Amradee, N. J. White, and W. M. Watkins. 1997. Poor efficacy of antimalarial biguanide-dapsone combinations in the treatment of acute, uncomplicated, falciparum malaria in Thailand. Ann. Trop. Med. Parasitol. 91:125-132. [PubMed] [Google Scholar]