Abstract
The fitness costs associated with high-level fluoroquinolone resistance were examined for phenotypically and genotypically characterized ciprofloxacin-resistant Salmonella enterica serotype Enteritidis mutants (104-cip and 5408-cip; MIC, >32 μg/ml). The stability of the fluoroquinolone resistance phenotype in both mutants was investigated to assess whether clones with better fitness could emerge in the absence of antibiotic selective pressure. Mutants 104-cip and 5408-cip displayed altered morphology on agar and by electron microscopy, reduced growth rates, motility and invasiveness in Caco-2 cells, and increased sensitivity to environmental stresses. Microarray data revealed decreased expression of virulence and motility genes in both mutants. Two clones, 104-revert and 1A-revertC2, with ciprofloxacin MICs of 3 and 2 μg/ml, respectively, were recovered from separate lineages of 104-cip after 20 and 70 passages, respectively, on antibiotic-free agar. All fitness costs, except motility, were reversed in 104-revert. Potential mechanisms associated with reversal of the resistance phenotype were examined. Compared to 104-cip, both 104-revert and 1A-revertC2 showed decreased expression of acrB and soxS but still overexpressed marA. Both acquired additional mutations in SoxR and ParC, and 1A-revertC2 acquired two mutations in MarA. The altered porin and lipopolysaccharide (LPS) profiles observed in 104-cip were reversed. In contrast, 5408-cip showed no reversal in fitness costs and maintained its high-level ciprofloxacin resistance for 200 passages on antibiotic-free agar. In conclusion, high-level ciprofloxacin resistance in S. Enteritidis is associated with fitness costs. In the absence of antibiotic selection pressure, isolates may acquire mutations enabling reversion to an intermediate-level ciprofloxacin resistance phenotype associated with less significant fitness costs.
Salmonella enterica serotype Enteritidis is one of the most common causes of food-borne salmonellosis worldwide. Historically, S. Enteritidis has remained susceptible to most antibiotics, unlike more-common serotypes, such as Salmonella enterica serotypes Typhimurium, Virchow, Newport, and Hadar, in which resistance to a wide range of antimicrobial agents is common (16, 29). A number of reports have documented an increasing prevalence of nalidixic acid resistance among nontyphoidal Salmonella isolates, in particular S. Enteritidis (5, 26, 42). Such isolates typically show decreased susceptibility to ciprofloxacin (MIC, 0.12 to 1.0 μg/ml), although the MICs are within the susceptible range of the interpretive criteria of the Clinical and Laboratory Standards Institute (CLSI) (12). There is increasing evidence that fluoroquinolone therapy for infections caused by Salmonella strains with reduced susceptibility to fluoroquinolones may result in treatment failure (28, 34, 44). To date, high-level fluoroquinolone resistance remains relatively uncommon in Salmonella isolates, compared with that in other Enterobacteriaceae. However, the emergence and clonal spread of fluoroquinolone-resistant Salmonella enterica serotype Typhimurium DT204 strains were observed in the 1990s, and these strains presently reoccur in serotypes such as S. Typhimurium (22), S. Choleraesuis (9), and S. Schwarzengrund (31).
Well-documented mechanisms associated with the development of high-level fluoroquinolone resistance include mutations that reduce the affinity for the antibiotic targets DNA gyrase and/or DNA topoisomerase IV, active efflux due to overproduction of the AcrAB-TolC efflux pump, and plasmid-mediated protection of target topoisomerases (17, 20). The contribution of the decreased membrane permeability resulting from altered porin expression and lipopolysaccharide (LPS) profiles to quinolone resistance is currently unclear (19, 27, 35).
Mutations in antibiotic target genes and overexpression of multidrug resistance (MDR) efflux pumps have been associated with fitness costs, including reduced growth rates and virulence, which may limit the survival of resistant strains in the absence of antibiotic selective pressure (1, 3, 23, 39, 47). However, stabilization of resistance can occur through the development of compensatory mutations that restore fitness without loss of the original level of resistance (2). In contrast to the wealth of information available on the mechanisms leading to high-level fluoroquinolone resistance in Salmonella, few studies to date have investigated the fitness costs associated with this phenotype (18, 45). Data from these studies suggest that mechanisms that confer high-level ciprofloxacin resistance in Salmonella have a prohibitive fitness cost and may thus limit the emergence and spread of highly resistant clones in the absence of antibiotic selection.
In a previous study, we genotypically and phenotypically characterized two in vitro-selected ciprofloxacin-resistant S. Enteritidis mutants (32). The aim of this study was to assess their growth characteristics, colony morphology, motility, invasiveness, and acid and osmotic tolerance and to investigate whether clones with better fitness could emerge following serial transfer in the absence of antibiotic selective pressure. We examined the global gene expression in both mutants to investigate potential molecular mechanisms associated with the observed fitness costs. Mechanisms associated with the reversal of one of the mutants to a low-level ciprofloxacin resistance phenotype associated with lesser fitness costs were also investigated.
MATERIALS AND METHODS
Bacterial strains used.
The parent S. Enteritidis strains 104 and 5408, displaying high-level nalidixic acid resistance and reduced susceptibility to ciprofloxacin and harboring a single gyrA mutation (D87Y), were isolated from poultry and a human, respectively. Quinolone-resistant mutants 104-cip and 5408-cip were obtained from their isogenic parents after seven selection steps on tryptone soya agar (TSA; Oxoid, New Hampshire, United Kingdom) with increasing concentrations of ciprofloxacin (0.25 to 16 μg/ml; Sigma-Aldrich, Ireland), as previously described (32). Two clones, 104-revert and 1A-revertC2, were derived from separate lineages of 104-cip after 20 and 70 passages, respectively, on antibiotic-free TSA (Table 1). The reference strain S. Enteritidis PT4 NCTC 13349 was also included in this study.
TABLE 1.
Phenotypic and genotypic characteristics of the S. Enteritidis strains used in this study
| Strain | MIC (μg/ml)a |
Mutation(s) in target structureb |
Mutation(s) in regulator of AcrAB-TolCb |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NAL | CIP | GyrA | GyrB | ParC | ParE | SoxR | SoxS | MarA | RamR | |
| 104 | >256 | 0.25 | D87Y | * | * | * | * | * | * | * |
| 104-cip | >256 | >32 | D87Y, S83F | * | * | * | R20H | E52K | * | * |
| 104-revert | >256 | 4 | D87Y, S83F | * | D79N | * | E16G, R20H | E52K | * | * |
| 1A-revertC2 | >256 | 2 | D87Y, S83F | * | D79N | * | R20H, R133C | E52K | I111N, H115Q | * |
| 5408 | >256 | 0.25 | D87Y | * | * | * | * | * | * | * |
| 5408-cip | >256 | >32 | D87Y | E466D | * | V461G | * | * | * | G25A |
| NCTC 13349 | 8 | 0.032 | * | * | * | * | * | * | * | * |
Values represent means of results from 3 separate determinations. NAL, nalidixic acid; CIP, ciprofloxacin.
D, aspartic acid; Y, tyrosine; S, serine; F, phenylalanine; E, glutamic acid; G, glycine; R, arginine; H, histidine; K, lysine; N, asparagine; I, isoleucine; C, cysteine; Q, glutamine; A, alanine; *, wild-type allele (no mutation).
Stability of the resistance phenotype and molecular subtyping of isolates.
Ciprofloxacin-resistant mutants were serially passaged on TSA in the absence of ciprofloxacin for 200 generations. Cultures were tested for their quinolone resistance every 3 days. Individual isolates were subtyped by pulsed-field gel electrophoresis (PFGE). PFGE was performed following XbaI digestion of genomic DNA according to the standard 1-day PulseNet protocol of the Centers for Disease Control and Prevention (CDC) (http://pulsenetinternational.org/protocols/protocols.asp) (6).
Bacterial growth curves.
Bacterial growth was monitored by measuring optical density at 600 nm (OD600) in tryptone soya broth (TSB; Oxoid, New Hampshire, United Kingdom). Briefly, all bacteria were initially cultured on TSA at 37°C for 18 h, and an isolated colony was then inoculated into 10 ml TSB. A 1-ml aliquot of the overnight culture was inoculated into a conical flask containing 100 ml TSB and incubated in a shaking incubator aerobically at 37°C. Absorbance readings were taken every hour for 8 h and then after 24 h with a spectrophotometer (Biomate 5; Thermospectronic, Cambridge, United Kingdom).
Electron microscopy.
Bacterial cells were grown to stationary phase in Luria-Bertani (LB) broth (Difco, NJ), harvested by centrifugation at 1,968 × g for 10 min, and resuspended in phosphate-buffered saline (PBS; Sigma-Aldrich, Ireland) and 2.5% glutaraldehyde (fixative) at a concentration of approximately 108 cells per ml. Five microliters of the cell suspension was pipetted onto Formvar carbon-stabilized copper grids and incubated for 2 min. Negative staining was performed with 5 μl 2% phosphotungstic acid for 1 min. The grids were examined using a JEOL JEM-2000 FX microscope under standard operating conditions.
Antimicrobial susceptibility testing.
MICs for nalidixic acid, ciprofloxacin, ampicillin, chloramphenicol, tetracycline, and sulfamethoxazole-trimethoprim were determined by an Etest with Mueller-Hinton agar (Difco) by following the manufacturer's instructions (AB-Biodisk, Solna, Sweden). The MICs were evaluated by using the breakpoints of the CLSI (11). Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as quality control organisms. All antibiotics were supplied by Oxoid.
Phenotype microarray.
The ability of the bacterial strains to respond to environmental stresses (pH and osmotic) were examined by the use of OmniLog Biolog phenotype microarrays (Biolog, Inc., Hayward, CA). Briefly, bacteria were grown on blood agar overnight at 37°C. Colonies were picked with a sterile cotton swab and suspended in 10 ml IF-0a (Biolog), and the cell density was adjusted to an OD600 of 0.035 with a spectrophotometer (Biomate 5; Thermospectronic, Cambridge, United Kingdom). A 750-μl aliquot of this cell suspension was added to 150 ml IF-10 (Biolog). Microtiter plates (PM-9 and PM-10) were inoculated with 100 μl of cell suspension per well and then incubated at 37°C for 48 h in an OmniLog incubator and monitored continuously for color changes in the wells. Kinetic data were analyzed with OmniLog PM software.
Swim motility assay.
One microliter of an overnight culture was spotted in the middle of a swim plate (LB broth and 0.1% gellan gum [Sigma]) and allowed to dry for 1 h at room temperature. Plates were incubated at 30°C overnight. The region of visible colony spread on the agar was measured with a ruler (mm) (14, 37).
Swarm motility assay.
Bacterial cells were grown overnight in LB broth, harvested by centrifugation, washed once, and resuspended at a 50-fold concentration. Two microliters of this cell suspension was spotted onto a motility plate (LB broth and 0.2% gellan gum) and incubated at 30°C overnight. The region of visible colony spread on the agar was measured as described above (14, 37).
Adherence and invasion assays.
Caco-2 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 20% (vol/vol) fetal bovine serum (FBS), 1% penicillin-streptomycin solution, 1% (vol/vol) minimal nonessential amino acids in Triton X-100, and 1% fungizone (250 μg/ml amphotericin B) in a humidified atmosphere of 5% CO2 at 37°C. (All reagents were supplied by Biosciences, Dublin, Ireland.) Caco-2 cells (5 × 105 cells/0.5 ml) were seeded into 24-well plates and cultured for approximately 15 days. Cell medium was changed three times a week. An overnight culture of bacteria was diluted 1 in 100 in LB broth and allowed to grow to mid-log phase. The bacterial cells were collected by centrifugation (1,968 × g for 10 min) and resuspended in DMEM at a concentration of approximately 108 bacteria per ml. The Caco-2 cells in the 24-well plates were washed twice in PBS, serum-free medium was then added, and cells were allowed to equilibrate for 2 h in 5% CO2 at 37°C. The serum-free medium was removed, and the Caco-2 cells were incubated with the bacterial inoculum for 1 h in 5% CO2 at 37°C. For the adhesion assay, after infection, the Caco-2 cells were washed four times with PBS before disruption with a 1-ml volume of PBS containing 1% Triton X-100 (Sigma-Aldrich) at room temperature for 5 min. The number of bacteria per well was determined by plating out appropriate dilutions of this final suspension onto TSA. The adhesion level is reported as the number of adhered bacteria minus the number of invaded bacteria. For the invasion assay, after the bacteria were allowed to adhere to the monolayers, wells were washed twice with serum-free medium, replaced with serum-free medium containing gentamicin sulfate (50 μg/ml; Sigma-Aldrich), and incubated for 30 min to kill all external bacteria. The medium was then removed, and the cells were washed twice with PBS to remove any residual gentamicin sulfate before lysis with 1% Triton X-100. The number of bacteria invaded was estimated by plating serial dilutions. Adhesion and invasion assays were performed on three separate occasions, with four wells per assay used for each isolate.
PCR amplification and sequencing of the QRDRs of quinolone target genes and the local and global regulators of acrAB-tolC.
Genomic DNA was extracted from overnight cultures in TSB (Oxoid) at 37°C by using a Wizard genomic DNA purification kit (Promega, Madison, WI). The quinolone resistance-determining regions (QRDRs) of gyrA, gyrB, parC, and parE and the local (acrR) and global regulators of acrAB-tolC (ramA, ramR, marORAB, and soxRS) were amplified and sequenced as previously described (32).
Expression analysis of efflux transporter gene acrB and global regulators marA, soxS, ramA, and rob.
RNA extraction and real-time quantification of RNA templates by real-time one-step RT-PCR were carried out as previously described (32). Relative gene expression was calculated using the ΔΔCT method (25).
LPS and porin analyses.
LPS and porin analyses were carried out as previously described (32). Briefly, LPS was prepared from whole bacteria by proteolytic digestion of cellular proteins with proteinase K and separated by SDS-PAGE, and the bands were visualized by silver staining. For porin analysis, total cell proteins were separated by SDS-PAGE, electrotransferred onto nitrocellulose membranes, and probed with polyclonal antibodies (1:2,000 dilution) directed against denatured OmpF porin or with F4 polyclonal antibody directed against the L3 internal loop of E. coli porins. These antibodies directed against denatured OmpF and the L3 internal porin loop recognize the denatured enterobacterial porins, including Salmonella F and D porins (40). The detection of antigen-antibody complexes was performed with alkaline phosphatase-conjugated AffinitiPure goat anti-rabbit IgG antibodies.
Gene expression microarray.
RNA was extracted from six separate cultures (biological replicates) of each parent and mutant strain. A separate microarray analysis was performed with each biological replicate.
Microarray design and manufacture.
The nonredundant multiserotype microarray contained 5,660 PCR products that currently cover 95% of all genes in the genomes of Salmonella serovar Typhimurium strain LT2, S. Typhimurium strain SL1344, S. Typhi strain CT18, S. Typhi strain Ty2, S. Paratyphi A strain SARB42, and S. Enteritidis strain PT4. The array represents each gene in a separate spot, deposited in 50% dimethyl sulfoxide (DMSO) onto the amino silane-modified surfaces of bar-coded Corning Ultra-GAPS glass slides (catalogue no. 40015; Corning). Each glass slide contained triplicate identical arrays.
RNA labeling and hybridization.
cDNA probes were labeled with Cy3 and Cy5 dye-linked dUTP by direct incorporation during reverse transcription from total RNA to cDNA, as described in the protocol available at http://cmgm.stanford.edu/pbrown/protocols/4_Ecoli_RNA.txt, with minor modifications. Fifty micrograms of total RNA and 2.4 μg of random hexamers were resuspended in 30 ml of water, and subsequently, the amounts and volumes of all components were doubled compared to those used in the Brown protocol. Furthermore, 2 μl of RNasin (F. Hoffmann, La Roche, Ltd., Basel, Switzerland) was added to the reverse transcription mixture, and the reaction mixture was incubated at 42°C for 2 h. After the first hour of incubation, a further 2 μl of Superscript II reverse transcriptase was added. Probes were purified using a QIAquick PCR purification kit (Qiagen, Valencia, CA) and eluted in 1 mM Tris-HCl, pH 8.0. Subsequently, all probes were dried down and resuspended in 10 μl sterile water.
Hybridization and data acquisition.
Probes were hybridized to the Salmonella array overnight in 25% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% (vol/vol) SDS at 42°C by using a hybridization chamber (Corning, New York, NY) submerged in a water bath. Protocols suggested by the manufacturer for hybridizations in formamide buffer were applied for prehybridization, hybridization, and posthybridization processing. Microarrays were scanned with a ScanArray Lite Laser scanner (PerkinElmer Life and Analytical Sciences, Waltham, MA), using ScanArray Express 3.0 software, or with a GenePix 4100A scanner (Molecular Devices, Sunnyvale, CA) and GenePix Pro software.
Data analysis.
Signal intensities were quantified using the QuantArray 3.0 software package (Packard BioChip Technologies, Billerica, MA). Spots were analyzed by adaptive quantitation and subsequently statistically analyzed using WebArray (46). The following parameters were used: background subtraction was performed using the “half” method, print-tip Loess normalization was employed within arrays, and scale normalization was used between arrays.
Real-time RT-PCR versus microarray analysis.
Gene expression analyses of five differentially transcribed regulators of S. Enteritidis were assessed by real-time reverse transcriptase PCR (RT-PCR) (as described above) and compared to gene expression microarray results.
Statistical analysis.
Data are presented as means ± standard deviations (SD). Statistical comparisons were made using the two-tailed Student t test. A P value less than 0.05 was considered significant.
Microarray data accession number.
The Salmonella data from this study have been deposited in NCBI's Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE15024.
RESULTS
Stability of the high-level ciprofloxacin resistance phenotype and subtyping of isolates.
The separate lineages of 104-cip reverted to intermediate levels of ciprofloxacin resistance (MICs of 4 μg/ml [104-revert] and 2 μg/ml [1A-revert C2]) after 20 and 70 passages on antibiotic-free agar, respectively, and maintained this phenotype throughout the rest of this study (Table 1). In contrast, 5408-cip maintained its high-level ciprofloxacin resistance phenotype (MIC, ≥32 μg/ml) for 200 passages in the absence of antibiotic. All isolates showed identical PFGE restriction profiles (data not shown).
Growth and morphology of ciprofloxacin-resistant mutants.
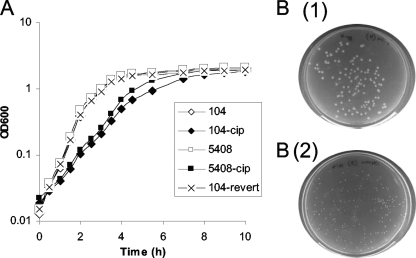
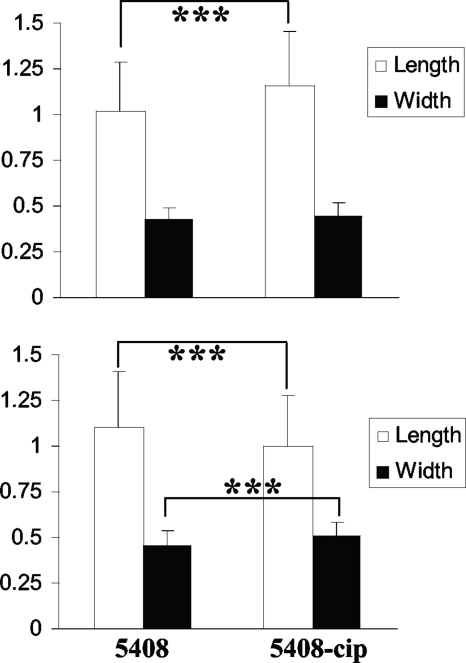
Mutant strains 104-cip and 5408-cip demonstrated lower growth rates and formed smaller colonies than their respective parent strains (Fig. 1). The growth profiles of both parent strains were identical to that of the reference strain (data not shown). The growth profiles of 104-revert and 1A-revertC2 were the same as that of their parent strain, 104. The two reverted mutants formed colonies similar in size to those formed by their isogenic parents. Electron microscopy analysis (Fig. 2) revealed that 104-cip rods were significantly longer (1.16 ± 0.3 μm versus 1.02 ± 0.27 μm; P < 0.0005) and wider (0.45 ± 0.07 μm versus 0.43 ± 0.07 μm; P < 0.005) than those of the parent strain. Rods of the mutant 5408-cip were significantly shorter (1.00 ± 0.28 μm versus 1.10 ± 0.31 μm; P < 0.0005) and wider (0.51 ± 0.08 μm versus 0.46 ± 0.08 μm; P < 0.0005) than those of its parent strain. After 200 passages on agar in the absence of antibiotic, 5408-cip continued to form small colonies and had a reduced growth rate in broth (data not shown).
FIG. 1.
Growth profiles of S. Enteritidis wild-type strains (104 and 5408), their isogenic ciprofloxacin-resistant mutants (104-cip and 5408-cip), and the reverted mutant (104-revert) (A) and colony morphology of parents [B (1)] and isogenic mutants [B (2)] (B). Both 104-cip and 5408-cip formed small colonies on agar. 104-revert formed colonies similar to those of the parent strains. For purposes of clarity, 1A-revertC2 and the S. Enteritidis reference strain (S100) are not included in the growth curve below. Both showed growth profiles similar to those of 104, 5408, and 104-revert.
FIG. 2.
Comparison of the widths and lengths of wild-type S. Enteritidis strains (104 and 5408) and their isogenic ciprofloxacin-resistant mutants (104-cip and 5408-cip) as determined by electron microscopy. Results are expressed as means ± SD for 400 measurements for individual Salmonella rods from 6 different fields. Statistical comparisons were made with unpaired Student t tests. **, P < 0.005; ***, P < 0.0005.
Susceptibility of mutants to environmental stresses and antibiotics.
Both 104-cip and 5408-cip displayed increased pH and osmotic susceptibilities compared to their isogenic parents in the phenotype microarrays. The mutant 104-cip had increased susceptibility to 5 to 6.5% sodium chloride; 3% sodium formate; 5 to 11% sodium lactate; 5% urea; 100 mM sodium nitrite; and pHs 4.5, 9.5, and 10.0. The 5408-cip mutant showed increased susceptibility to 3 to 6.5% sodium chloride; 5 to 6% potassium chloride; 3 to 6% sodium formate; 2 to 6% urea; 4 to 10% sodium lactate; 20 to 50 mM sodium benzoate, pH 5.2; 10 to 100 mM sodium nitrite; and pHs 4.5, 9.5, and 10.0. Both 104-cip and 5408-cip displayed a lack of acid and alkali habituation through production of decarboxylases and deaminases and a lack of ability to accumulate osmolytes. Both reverted mutants showed pH and osmotic susceptibilities similar to those of the parent strain. We previously reported that 104-cip showed decreased susceptibility to tetracycline, ampicillin, and chloramphenicol and that 5408-cip showed decreased susceptibility to sulfamethoxazole-trimethoprim, ampicillin, and chloramphenicol (32). Compared to 104-cip, 104-revert and 1A-revertC2 displayed increased susceptibility to tetracycline, ampicillin, and chloramphenicol (Table 2).
TABLE 2.
Antimicrobial susceptibility phenotypes of S. Enteritidis strains
| Strain | MIC (μg/ml)a |
|||
|---|---|---|---|---|
| TC | TS | AM | CL | |
| 104 | 1 | 0.125 | 1 | 2 |
| 104-cip | 16 | 0.25 | 16 | 128 |
| 104-revert | 2 | 0.064 | 4 | 8 |
| 1A-revertC2 | 0.50 | 0.032 | 0.50 | 2 |
| 5408 | 1 | 0.064 | 1 | 4 |
| 5408-cip | 2 | 0.25 | 4 | 16 |
Values represent means of results from 3 separate determinations. AM, ampicillin; CL, chloramphenicol; TC, tetracycline; TS, trimethoprim-sulfamethoxazole.
Mutant motility, adherence, and invasion.
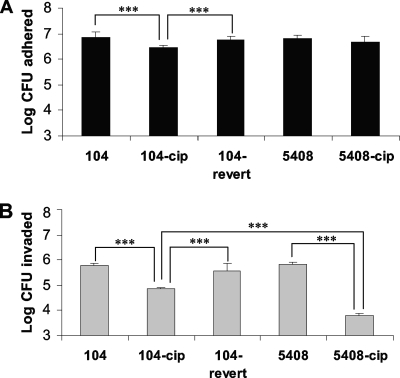
The mutated 104-cip, 104-revert, and 5408-cip strains showed 66, 90, and 93% reductions in swim motility and 36, 36, and 46% reductions in swarm motility, respectively, compared to their parent strains. The parent strains showed swim and swarm motilities identical to those of the reference strain. The mutant 104-cip showed significantly lower levels of adherence to Caco-2 monolayers than its isogenic parent (104) (6.47 ± 0.08 versus 6.84 ± 0.22 log CFU adhered; P < 0.0005). The 104-revert strain showed adherence levels similar to those observed for 104. Strains 5408 and 5408-cip adhered in similar numbers (Fig. 3). Mutants 104-cip and 5408-cip were significantly (P < 0.0005) less invasive in Caco-2 cells than their parent strains. The log numbers of CFU invaded in 104-cip and 5408-cip were 4.84 ± 0.05 and 3.78 ± 0.09, respectively, compared to the log numbers of CFU invaded by their respective parents (5.76 ± 0.1 and 5.8 ± 0.11, respectively). Strain 104-revert showed levels of invasion in Caco-2 cells similar (5.58 ± 0.3 log CFU) to those observed for its parent strain (Fig. 3). Both 104 and 5408 showed levels of adherence and invasion similar to those observed for the S. Enteritidis reference strain (data not shown).
FIG. 3.
Adherence (A) and invasion (B) of Caco-2 monolayers by S. Enteritidis wild-type strains (104 and 5408), their isogenic ciprofloxacin-resistant mutants (104-cip and 5408-cip), and the reverted mutant (104-revert). Results are expressed as mean ± SD log numbers of CFU invaded in 12 wells from three separate experiments. Statistical comparisons were made with unpaired Student t tests. ***, P < 0.0005.
Genotypic characteristics of the mutants.
In our previous study, we genotypically characterized mutants 104-cip and 5408-cip (32). The mutant 104-cip harbored two GyrA (D87Y and S83F) mutations, and 5408-cip harbored single GyrA (D87Y), GyrB (E466D), and ParE (V461G) mutations. Both mutants overexpressed acrB. The global regulator genes soxS and marA were overexpressed in 104-cip, and ramA was overexpressed in 5408-cip. Mutations were found in SoxR (R20H) and in SoxS (E52K) in 104-cip and in RamR (G25A) in 5408-cip (Tables 1 and 3). In this study, both 104-revert and 1A-revertC2 showed decreased expression of acrB and soxS compared to 104-cip but retained increased expression of marA, albeit at a lower level of expression than that in 104-cip. Expression of ramA and rob in both reverted mutants was similar to that in 104-cip (Table 3). Both reverted isolates maintained the previously identified double GyrA mutations (S83F and D87Y), the single SoxS mutation (E52K), and the single SoxR mutation (R20H). An additional mutation was found in SoxR in 104-revert (E16G) and in 1A-revertC2 (R113C). A mutation was observed in ParC (D79N) in both reverted isolates. 1A-revertC2 acquired two mutations in MarA (I111N and H115Q) (Table 1).
TABLE 3.
Gene expression analyses of S. Enteritidis isolates by real-time RT-PCR
| Strain | Fold change in gene expressiona |
||||
|---|---|---|---|---|---|
| acrB | soxS | marA | ramA | rob | |
| 104-cip | 6.1 ± 1.5 | 26.1 ± 4.0 | 8.9 ± 0.6 | 1.2 ± 0.2 | −4.6 ± 0.2 |
| 104-revert | −1.9 ± 0.4 | −5.0 ± 0.9 | 3.0 ± 0.4 | −1.45 ± 0.1 | −1.75 ± 0.5 |
| 1A-revertC2 | 1.5 ± 0.3 | 1.7 ± 0.1 | 6.6 ± 1.1 | 1.6 ± 0.2 | −2.2 ± 0.2 |
| 5408-cip | 5.4 ± 1.6 | −3.4 ± 0.5 | 1.3 ± 0.2 | 33.7 ± 4.0 | −2.3 ± 1.0 |
RNA was extracted from cultures growing at log phase in LB broth. Gene expression data represent means ± SD of results from 3 independent total RNA extractions. Changes in gene expression are relative to the levels for the respective parental strains.
LPS and porin profiles.
We previously examined the LPS and porin profiles of 104-cip and 5408-cip (32). The LPS profile of 104-cip showed significant loss of short and intermediate O-chain LPSs compared to the levels for its parent strain (data not shown). The 104-cip mutant showed decreased production of OmpF (Fig. 4). No changes were observed in the LPS or OmpF profile of 5408-cip. Both 104-revert and 1A-revertC2 gained short and intermediate O-chain LPSs (data not shown), and 104-revert showed increased OmpF production relative to 104-cip (Fig. 4).
FIG. 4.
The detection of porins was carried out using the polyclonal antibodies directed against denatured OmpF porin (A) or the F4 polyclonal antibody directed against the L3 internal loop of E. coli porins (B). Lanes 1, 104; lanes 2, 104-revert; lanes 3, 104-cip; lanes 4, 5408; lanes 5, 5408-cip; lanes 6, S. Enteritidis NCTC 13349. Arrows indicate the migration of F and D porins, respectively.
Gene expression microarray.
Microarray data revealed that 64 genes were differentially regulated in 104-cip and 97 genes in 5408-cip compared to what was observed for their respective parent strains. Two detailed tables representing the differentially regulated genes in both mutants can be found in the supplemental material. Activated and repressed genes were classified into clusters of orthologous groups (COGs) as defined at http://www.ncbi.nlm.nih.gov/COG/grace/fiew.cgi (Table 4). Several genes involved in energy production and conservation (31 genes), carbohydrate transport and metabolism (24 genes), and amino acid transport and metabolism (19 genes) were differentially regulated in both mutants (Table 4). Decreased expression of outer membrane porins was observed in ompF and ompW in 5408-cip and in ompC in 104-cip, along with an increased expression level of ompX in 5408-cip. A total of 26 genes involved in cell motility (including fimA, fliCHGTZ, motAB, cheAWMRBYZ, and flgLKFEDCN) were downregulated in 5408-cip. In contrast, only 3 genes (flgED and orgA) were decreased in 104-cip. Both mutants showed downregulation of Salmonella pathogenicity island 1 (SPI-1) genes involved in invasion (invJIGF, hilCD, and prgKIH) (included in the no-COG category). Among the genes that were upregulated in 104-cip, a number were SoxS regulated, including acnA, fpr, ribA, sodA, lpxC, and soxS itself. Increased expression of the efflux genes acrB and acrA was detected in 104-cip and 5408-cip. Comparisons of real-time RT-PCR and microarray results (Table 5) showed that there were similar trends occurring between the two methods for acrB and rob in the two mutants and for soxS in 104-cip.
TABLE 4.
Classification of regulated genes in ciprofloxacin-resistant mutants according to COGs
| Categorya | No. of genes regulated |
|||
|---|---|---|---|---|
| 104-cip activated | 104-cip repressed | 5408-cip activated | 5408-cip repressed | |
| Energy production and conversion | 11 | 8 | 5 | 7 |
| Carbohydrate transport and metabolism | 3 | 7 | 0 | 14 |
| Amino acid transport and metabolism | 4 | 3 | 2 | 10 |
| Coenzyme transport and metabolism | 2 | 0 | 0 | 0 |
| Inorganic ion transport and metabolism | 1 | 0 | 1 | 0 |
| Translation, ribosomal structure, and biogenesis | 0 | 0 | 3 | 0 |
| Transcription | 1 | 0 | 2 | 3 |
| Replication, recombination, and repair | 1 | 0 | 0 | 0 |
| Defense mechanisms | 2 | 0 | 2 | 0 |
| Signal transduction mechanisms | 0 | 1 | 0 | 1 |
| Cell wall/membrane/ envelope biogenesis | 1 | 1 | 0 | 3 |
| Cell motility | 0 | 3 | 0 | 26 |
| Posttranslational modification, protein turnover, and chaperones | 3 | 1 | 0 | 1 |
| General function prediction only | 1 | 0 | 1 | 4 |
| Function unknown | 2 | 0 | 3 | 1 |
| No COGb | 0 | 8 | 1 | 7 |
| Total | 32 | 32 | 20 | 77 |
Genes were classified according to COGs as defined at http://www.ncbi.nlm.nih.gov/COG/grace/fiew.cgi.
Repressed genes in 104-cip and 5408-cip were all SPI-1 genes. The activated gene in 5408-cip is the outer membrane protease gene ompX.
TABLE 5.
Gene expression analyses of select transcriptional regulators of S. Enteritidisa
| Strain | Change in gene expression (P)b |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
acrB |
soxS |
marA |
ramA |
rob |
||||||
| RT-PCR | Array | RT-PCR | Array | RT-PCR | Array | RT-PCR | Array | RT-PCR | Array | |
| 104-cip | 6.1 ± 1.5 | 2.22 (2.4E−11) | 26.1 ± 4.0 | 1.19 (3.5E−05) | 8.9 ± 0.6 | 0.15 (0.07) | 1.2 ± 0.2 | 0.16 (0.5) | −4.6 ± 0.2 | −0.8 (3.2E−06) |
| 5408-cip | 5.4 ± 1.6 | 1.53 (1.8E−06) | −3.4 ± 0.5 | −0.15 (0.25) | 1.3 ± 0.2 | 0.17 (0.03) | 33.7 ± 4.0 | 0.9 (1.1E−03) | −2.3 ± 1.0 | −0.34 (2.2E−03) |
RNA was extracted from cultures growing at log phase in LB. RT-PCR data represent means ± SD of results from 3 independent total RNA extractions. Changes in gene expression are relative to the levels for the respective parental strains.
Array data were gathered from six biological replicates. Analysis was done using WebArray (46), employing the print-tip Loess and scale methods for within- and between-array normalizations, respectively. The cutoff values for genes downregulated and upregulated in the ciprofloxacin-resistant mutants (log2 ciprofloxacin-resistant-mutant/wild-type ratio values) were <1 (P < 0.01) and >1.0 (P < 0.01), respectively. Log2 ciprofloxacin-resistant-mutant/wild-type ratio values with P values greater than 0.01 are marked in italics to indicate lower confidence levels.
DISCUSSION
S. Enteritidis is the predominant serotype associated with egg-borne salmonellosis in humans. This serotype is invasive in poultry and therefore has the potential to contaminate eggs by trans-ovarian transmission following colonization of the intestinal tract (43). In this study, the fitness costs associated with high-level ciprofloxacin resistance in S. Enteritidis isolates, including reduced growth rates, altered morphology, decreased motility, and invasiveness in Caco-2 cells and increased susceptibility to environmental stresses, such as pH and osmotic stimuli, all could negatively affect intestinal colonization, thereby limiting the spread of resistant clones.
Microarray data revealed consistent downregulation both in mutants of invasion genes found within SPI-1, which encodes a type III secretion system (TTSS), and in flagellar biosynthesis genes, which correlated with the observed phenotypes of decreased motility and epithelial cell invasiveness. Invasion, preceded by adherence, is a highly coordinated activity, involving fimbriae, flagella, chemotaxis, LPS, and the TTSS (4, 21, 24). The greater reductions in motility and invasiveness of 5408-cip than in those of 104-cip correlated with the significantly decreased expression of a larger number of genes involved in flagellar biosynthesis and chemotaxis in this isolate. The possibility that the altered LPS profile in 104-cip may also have influenced its adherence and invasiveness cannot be excluded. Microarray data from this study showed a shift in metabolic activity to support the energy requirements associated with the overexpression of AcrAB in both ciprofloxacin-resistant mutants. In particular, the expression levels of a number of enzymes of the Krebs cycle were upregulated, consistent with the channeling of reducing power to the terminal electron transport system.
Mechanisms widely associated with high-level fluoroquinolone resistance, such as multiple topoisomerase mutations and overexpression of MDR efflux pumps, have been associated with fitness costs (1, 3, 23, 39). However, the mechanisms by which these resistance traits cause fitness burdens, such as reduced growth rates and virulence, have not been elucidated. Decreased growth in high-level fluoroquinolone resistance isolates harboring multiple topoisomerase mutations has been associated with decreased DNA supercoiling (3, 23). Decreased supercoiling indicates the presence of a less efficient DNA gyrase (15) which could in turn slow replication and thereby affect growth. Growth defects could also result from the inopportune efflux of nutrients and metabolic intermediates by overexpressed MDR efflux pumps (30). However, evidence to support this hypothesis is currently lacking. Expression of genes associated with bacterial virulence, such as invasion and flagellar genes, is influenced by changes in DNA supercoiling (13) and is activated by quorum sensing (10, 41). As efflux pumps have been shown to expel quorum-sensing signals (7, 33), it is reasonable to speculate that altered quorum-sensing signal homeostasis, resulting from increased AcrAB activity and/or altered DNA supercoiling, may have contributed to the decreased virulence of isolates in this study. Currently, we are investigating the effects of the topoisomerase mutations on supercoiling.
No restoration of growth rates or decrease in the level of resistance was observed in 5408-cip after 200 passages in the absence of antibiotic. In contrast, 104-cip reverted to an intermediate-level ciprofloxacin resistance phenotype (104-revert) after 20 passages, with reversal of all fitness costs except motility. The ability of 104-cip to revert to a lower-level resistance phenotype was confirmed by the recovery of another reverted clone (1A-revertC2) 70 generations after a separate lineage of 104-cip was put into serial transfer. Expression of acrB was decreased in both 104-revert and 1A-revertC2, providing convincing evidence that overexpression of this pump makes a significant contribution to the fitness burden of fluoroquinolone resistance. The decreased expression of acrB in both reverted isolates was associated with decreased expression of the global regulator soxS. It would be interesting to speculate that the additional SoxR mutations acquired by the reverted mutants (E16G in 104-revert and R113C in 1A-revertC2) were responsible for the observed decrease in expression of soxS and the associated decrease in acrB expression, even in the genetic background of elevated marA expression. A mutation at position E16 in the helix-turn-helix region of the SoxR protein in E. coli has previously been reported, and it is thought that alterations in this region of the protein could disrupt DNA binding (8).
Reversal to an intermediate-level ciprofloxacin resistance phenotype was also associated with restoration of normal OmpF expression and LPS profiles. Although we have shown that these membrane alterations do not make a significant contribution to the resistance phenotype in 104-cip (32), the possibility that they may have contributed to the fitness burden of ciprofloxacin resistance in this isolate cannot be excluded. OmpF is controlled by the mar and sox regulons (36, 38). The normal expression of OmpF in 104-revert, despite elevated expression of marA, may be a consequence of the decreased expression of soxS and the greater role played by this regulator in antibiotic resistance in 104-cip (32). The significance of the acquired marA mutations in 1A-revertC2 is currently unclear.
Interestingly, both reverted mutants acquired the same mutation in parC (D79N). Kugelberg et al. reported that after serial passage in laboratory medium, the fitness of slow-growing fluoroquinolone-resistant P. aeruginosa isolates with a gyrA mutation and decreased DNA supercoiling was increased by a compensatory mutation(s) that restored supercoiling to normal levels. However, no mutations were found in any of the genes expected to affect supercoiling (23). The possibility that this parC mutation may contribute to fitness restoration by compensating for the potential negative effects of resistance-associated topoisomerase mutations on global supercoiling remains to be explored.
In conclusion, this study demonstrates that high-level ciprofloxacin resistance in S. Enteritidis in vitro-derived mutants is associated with fitness costs. Evolution in the absence of antibiotic selective pressure may result in mutational events favoring a reversion to a lower-level resistance phenotype associated with lesser fitness costs, rather than the acquisition of compensatory mutations that would maintain resistance while ameliorating the fitness burden.
The fitness costs of high-level ciprofloxacin resistance, in the absence of evidence of compensatory evolution, may account for the lack of emergence and spread of highly resistant S. Enteritidis clones along the farm-to-fork continuum to date. The lack of fitness costs observed in isolates showing high-level nalidixic acid resistance and decreased susceptibility to ciprofloxacin may be a contributory factor in the widespread dissemination of this phenotype. Infection with these first-step mutants could lead to generation of second-step mutants rapidly after fluoroquinolone therapy, which could ascend to dominance during the course of treatment and potentially result in treatment failure.
Supplementary Material
Acknowledgments
We thank David Cottell and Cormac O'Connell, Electron Microscopy Laboratory, University College Dublin (UCD), for technical support with the electron microscope. We also thank Evelyn Murphy and Kevin McMahon (UCD) for the use of their tissue culture facility and technical training. We thank Jarlath Nally, Avril Monaghan, and Emily Mulheirn (UCD) for technical support during the SDS-PAGE experiments and for the use of their scanner. We thank Niall Mullane and Steven O'Brien (UCD) for PFGE analysis and Mike Asher (USDA) for technical assistance during microarray experiments.
This work was supported in part by COST Action BM0701 ATENS. S.P. and M.M. were supported by National Institutes of Health grant 1R01AI075093.
Footnotes
Published ahead of print on 16 November 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Alonso, A., G. Morales, R. Escalante, E. Campanario, L. Sastre, and J. L. Martinez. 2004. Overexpression of the multidrug efflux pump SmeDEF impairs Stenotrophomonas maltophilia physiology. J. Antimicrob. Chemother. 53:432-434. [DOI] [PubMed] [Google Scholar]
- 2.Andersson, D. I. 2006. The biological cost of mutational antibiotic resistance: any practical conclusions? Curr. Opin. Microbiol. 9:461-465. [DOI] [PubMed] [Google Scholar]
- 3.Bagel, S., V. Hullen, B. Wiedemann, and P. Heisig. 1999. Impact of GyrA and ParC mutations on quinolone resistance, doubling time, and supercoiling degree of Escherichia coli. Antimicrob. Agents Chemother. 43:868-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baxter, M. A., and B. D. Jones. 2005. The fimYZ genes regulate Salmonella enterica serovar Typhimurium invasion in addition to type 1 fimbrial expression and bacterial motility. Infect. Immun. 73:1377-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrique-Mas, J. J., C. Papadopoulou, S. J. Evans, A. Wales, C. J. Teale, and R. H. Davies. 2008. Trends in phage types and antimicrobial resistance of Salmonella enterica serovar Enteritidis isolated from animals in Great Britain from 1990 to 2005. Vet. Rec. 162:541-546. [DOI] [PubMed] [Google Scholar]
- 6.CDC. 2007. One-day (24-48 h) standardized laboratory protocol for molecular subtyping of Escherichia coli O157:H7, non-typhoidal Salmonella serotypes, and Shigella sonnei by pulsed field gel electrophoresis (PFGE). Centers for Disease Control and Prevention, Atlanta, GA. http://www.cdc.gov/pulsenet/protocols/ecoli_salmonella_shigella_protocols.pdf.
- 7.Chan, Y. Y., H. S. Bian, T. M. Tan, M. E. Mattmann, G. D. Geske, J. Igarashi, T. Hatano, H. Suga, H. E. Blackwell, and K. L. Chua. 2007. Control of quorum sensing by a Burkholderia pseudomallei multidrug efflux pump. J. Bacteriol. 189:4320-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chander, M., L. Raducha-Grace, and B. Demple. 2003. Transcription-defective soxR mutants of Escherichia coli: isolation and in vivo characterization. J. Bacteriol. 185:2441-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu, C. H., T. L. Wu, L. H. Su, C. Chu, J. H. Chia, A. J. Kuo, M. S. Chien, and T. Y. Lin. 2002. The emergence in Taiwan of fluoroquinolone resistance in Salmonella enterica serotype Choleraesuis. N. Engl. J. Med. 346:413-419. [DOI] [PubMed] [Google Scholar]
- 10.Choi, J., D. Shin, and S. Ryu. 2007. Implication of quorum sensing in Salmonella enterica serovar Typhimurium virulence: the luxS gene is necessary for expression of genes in pathogenicity island 1. Infect. Immun. 75:4885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.CLSI/NCCLS. 2004. Performance standards for antimicrobial susceptibility testing. CLSI document M100-S14. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 12.Crump, J. A., T. J. Barrett, J. T. Nelson, and F. J. Angulo. 2003. Reevaluating fluoroquinolone breakpoints for Salmonella enterica serotype Typhi and for non-Typhi salmonellae. Clin. Infect. Dis. 37:75-81. [DOI] [PubMed] [Google Scholar]
- 13.Dorman, C. J., and C. P. Corcoran. 2009. Bacterial DNA topology and infectious disease. Nucleic Acids Res. 37:672-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dowd, S. E., K. Killinger-Mann, M. Brashears, and J. Fralick. 2008. Evaluation of gene expression in a single antibiotic exposure-derived isolate of Salmonella enterica typhimurium 14028 possessing resistance to multiple antibiotics. Foodborne Pathog. Dis. 5:205-221. [DOI] [PubMed] [Google Scholar]
- 15.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frost, J. A., A. Kelleher, and B. Rowe. 1996. Increasing ciprofloxacin resistance in Salmonella in England and Wales 1991-1994. J. Antimicrob. Chemother. 37:85-91. [DOI] [PubMed] [Google Scholar]
- 17.Giraud, E., S. Baucheron, and A. Cloeckaert. 2006. Resistance to fluoroquinolones in Salmonella: emerging mechanisms and resistance prevention strategies. Microbes Infect. 8:1937-1944. [DOI] [PubMed] [Google Scholar]
- 18.Giraud, E., A. Cloeckaert, S. Baucheron, C. Mouline, and E. Chaslus-Dancla. 2003. Fitness cost of fluoroquinolone resistance in Salmonella enterica serovar Typhimurium. J. Med. Microbiol. 52:697-703. [DOI] [PubMed] [Google Scholar]
- 19.Giraud, E., A. Cloeckaert, D. Kerboeuf, and E. Chaslus-Dancla. 2000. Evidence for active efflux as the primary mechanism of resistance to ciprofloxacin in Salmonella enterica serovar typhimurium. Antimicrob. Agents Chemother. 44:1223-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hopkins, K. L., R. H. Davies, and E. J. Threlfall. 2005. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int. J. Antimicrob. Agents 25:358-373. [DOI] [PubMed] [Google Scholar]
- 21.Ibarra, J. A., and O. Steele-Mortimer. 2009. Salmonella—the ultimate insider. Salmonella virulence factors that modulate intracellular survival. Cell. Microbiol. 11:1579-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Izumiya, H., K. Mori, T. Kurazono, M. Yamaguchi, M. Higashide, N. Konishi, A. Kai, K. Morita, J. Terajima, and H. Watanabe. 2005. Characterization of isolates of Salmonella enterica serovar Typhimurium displaying high-level fluoroquinolone resistance in Japan. J. Clin. Microbiol. 43:5074-5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kugelberg, E., S. Lofmark, B. Wretlind, and D. I. Andersson. 2005. Reduction of the fitness burden of quinolone resistance in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 55:22-30. [DOI] [PubMed] [Google Scholar]
- 24.La Ragione, R. M., W. A. Cooley, P. Velge, M. A. Jepson, and M. J. Woodward. 2003. Membrane ruffling and invasion of human and avian cell lines is reduced for aflagellate mutants of Salmonella enterica serotype Enteritidis. Int. J. Med. Microbiol. 293:261-272. [DOI] [PubMed] [Google Scholar]
- 25.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 26.Meakins, S., I. S. Fisher, C. Berghold, P. Gerner-Smidt, H. Tschape, M. Cormican, I. Luzzi, F. Schneider, W. Wannett, J. Coia, A. Echeita, and E. J. Threlfall. 2008. Antimicrobial drug resistance in human nontyphoidal Salmonella isolates in Europe 2000-2004: a report from the Enter-net International Surveillance Network. Microb. Drug Resist. 14:31-35. [DOI] [PubMed] [Google Scholar]
- 27.Miro, E., C. Verges, I. Garcia, B. Mirelis, F. Navarro, P. Coll, G. Prats, and L. Martinez-Martinez. 2004. Resistance to quinolones and beta-lactams in Salmonella enterica due to mutations in topoisomerase-encoding genes, altered cell permeability and expression of an active efflux system. Enferm. Infecc. Microbiol. Clin. 22:204-211. (In Spanish.) [DOI] [PubMed] [Google Scholar]
- 28.Molbak, K., D. L. Baggesen, F. M. Aarestrup, J. M. Ebbesen, J. Engberg, K. Frydendahl, P. Gerner-Smidt, A. M. Petersen, and H. C. Wegener. 1999. An outbreak of multidrug-resistant, quinolone-resistant Salmonella enterica serotype Typhimurium DT104. N. Engl. J. Med. 341:1420-1425. [DOI] [PubMed] [Google Scholar]
- 29.Molbak, K., P. Gerner-Smidt, and H. C. Wegener. 2002. Increasing quinolone resistance in Salmonella enterica serotype Enteritidis. Emerg. Infect. Dis. 8:514-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikaido, H. 1998. Multiple antibiotic resistance and efflux. Curr. Opin. Microbiol. 1:516-523. [DOI] [PubMed] [Google Scholar]
- 31.Olsen, S. J., E. E. DeBess, T. E. McGivern, N. Marano, T. Eby, S. Mauvais, V. K. Balan, G. Zirnstein, P. R. Cieslak, and F. J. Angulo. 2001. A nosocomial outbreak of fluoroquinolone-resistant Salmonella infection. N. Engl. J. Med. 344:1572-1579. [DOI] [PubMed] [Google Scholar]
- 32.O'Regan, E., T. Quinn, J. M. Pages, M. McCusker, L. Piddock, and S. Fanning. 2009. Multiple regulatory pathways associated with high-level ciprofloxacin and multidrug resistance in Salmonella enterica serovar Enteritidis: involvement of RamA and other global regulators. Antimicrob. Agents Chemother. 53:1080-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearson, J. P., C. Van Delden, and B. H. Iglewski. 1999. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 181:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piddock, L. J., D. J. Griggs, M. C. Hall, and Y. F. Jin. 1993. Ciprofloxacin resistance in clinical isolates of Salmonella typhimurium obtained from two patients. Antimicrob. Agents Chemother. 37:662-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piddock, L. J., V. Ricci, I. McLaren, and D. J. Griggs. 1998. Role of mutation in the gyrA and parC genes of nalidixic-acid-resistant Salmonella serotypes isolated from animals in the United Kingdom. J. Antimicrob. Chemother. 41:635-641. [DOI] [PubMed] [Google Scholar]
- 36.Pomposiello, P. J., and B. Demple. 2000. Identification of soxS-regulated genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:23-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramos, J. L., and R. C. Levesque. 2006. Pseudomonas: molecular biology of emerging issues, vol. 4. Springer, Berlin, Germany.
- 38.Randall, L. P., and M. J. Woodward. 2002. The multiple antibiotic resistance (mar) locus and its significance. Res. Vet. Sci. 72:87-93. [DOI] [PubMed] [Google Scholar]
- 39.Sanchez, P., J. F. Linares, B. Ruiz-Diez, E. Campanario, A. Navas, F. Baquero, and J. L. Martinez. 2002. Fitness of in vitro selected Pseudomonas aeruginosa nalB and nfxB multidrug resistant mutants. J. Antimicrob. Chemother. 50:657-664. [DOI] [PubMed] [Google Scholar]
- 40.Simonet, V., M. Mallea, D. Fourel, J. M. Bolla, and J. M. Pages. 1996. Crucial domains are conserved in Enterobacteriaceae porins. FEMS Microbiol. Lett. 136:91-97. [DOI] [PubMed] [Google Scholar]
- 41.Sperandio, V., A. G. Torres, and J. B. Kaper. 2002. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 43:809-821. [DOI] [PubMed] [Google Scholar]
- 42.Stevenson, J. E., K. Gay, T. J. Barrett, F. Medalla, T. M. Chiller, and F. J. Angulo. 2007. Increase in nalidixic acid resistance among non-Typhi Salmonella enterica isolates in the United States from 1996 to 2003. Antimicrob. Agents Chemother. 51:195-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thiagarajan, D., A. M. Saeed, and E. K. Asem. 1994. Mechanism of transovarian transmission of Salmonella Enteritidis in laying hens. Poult. Sci. 73:89-98. [DOI] [PubMed] [Google Scholar]
- 44.Vasallo, F. J., P. Martin-Rabadan, L. Alcala, J. M. Garcia-Lechuz, M. Rodriguez-Creixems, and E. Bouza. 1998. Failure of ciprofloxacin therapy for invasive nontyphoidal salmonellosis. Clin. Infect. Dis. 26:535-536. [DOI] [PubMed] [Google Scholar]
- 45.Wang, Y. P., L. Li, J. Z. Shen, F. J. Yang, and Y. N. Wu. 2009. Quinolone-resistance in Salmonella is associated with decreased mRNA expression of virulence genes invA and avrA, growth and intracellular invasion and survival. Vet. Microbiol. 133:328-334. [DOI] [PubMed] [Google Scholar]
- 46.Xia, X., M. McClelland, and Y. Wang. 2005. WebArray: an online platform for microarray data analysis. BMC Bioinformatics 6:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, Q., O. Sahin, P. F. McDermott, and S. Payot. 2006. Fitness of antimicrobial-resistant Campylobacter and Salmonella. Microbes Infect. 8:1972-1978. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.