Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) infections are complicated by the ability of the organism to grow in surface-adhered biofilms on a multitude of abiotic and biological surfaces. These multicellular communities are notoriously difficult to eradicate with antimicrobial therapy. Cells within the biofilm may be exposed to a sublethal concentration of the antimicrobial due to the metabolic and phenotypic diversity of the biofilm-associated cells or the protection offered by the biofilm structure. In the present study, the influence of a sublethal concentration of tigecycline on biofilms formed by an epidemic MRSA-16 isolate was investigated by transcriptome analysis. In the presence of the drug, 309 genes were upregulated and 213 genes were downregulated by more than twofold in comparison to the levels of gene regulation detected for the controls not grown in the presence of the drug. Microarray data were validated by real-time reverse transcription-PCR and phenotypic assays. Tigecycline altered the expression of a number of genes encoding proteins considered to be crucial for the virulence of S. aureus. These included the reduced expression of icaC, which is involved in polysaccharide intercellular adhesin production and biofilm development; the upregulation of fnbA, clfB, and cna, which encode adhesins which attach to human proteins; and the downregulation of the cap genes, which mediate the synthesis of the capsule polysaccharide. The expression of tst, which encodes toxic shock syndrome toxin 1 (TSST-1), was also significantly reduced; and an assay performed to quantify TSST-1 showed that the level of toxin production by cells treated with tigecycline decreased by 10-fold (P < 0.001) compared to the level of production by untreated control cells. This study suggests that tigecycline may reduce the expression of important virulence factors in S. aureus and supports further investigation to determine whether it could be a useful adjunct to therapy for the treatment of biofilm-mediated infections.
Staphylococcus aureus is one of the gram-positive pathogens that is the most frequently recovered from patients with hospital-acquired infections, and approximately 45% of S. aureus strains isolated in the United Kingdom are resistant to methicillin. Methicillin-resistant Staphylococcus aureus (MRSA) infections can range from those of the skin and surgical sites to more serious deep-seated conditions, such as bacteremia and endocarditis (27). This diversity is partly due to the arsenal of virulence factors that the staphylococcus employs to allow successful infection of the host. Virulence factors are instrumental in the induction and persistence of infection (15) and can be involved in such steps as binding to the cell surface, evasion of the host immune response, and host tissue invasion (16). One important virulence trait utilized by this organism is the ability to form biofilms on damaged tissues and implanted biomaterials (11).
Biofilm-mediated infections are particularly difficult to resolve, as biofilm-associated cells display a greater tolerance to antimicrobials than equivalent free-floating bacterial populations (25, 38). Cells within the biofilm display phenotypic diversity and variable levels of metabolic activity due to the heterogeneous diffusion of water and nutrients through the biofilm structure (21, 41). The architecture and chemical composition of the biofilm and extracellular matrix can also limit the penetration of antimicrobials into the biofilm core, exposing cells in this area to a subinhibitory concentration of the agent (30). Studies have shown that subinhibitory concentrations of some antibiotics influence the expression of key virulence factors in S. aureus (5, 19, 39), which may alter the pathogenesis of infection.
One novel antibiotic recently approved for the treatment of complicated skin and intra-abdominal infections is the glycylcycline tigecycline (13, 36). These types of infections may have a biofilm component; therefore, the aim of this study was to use microarray technology to examine the impact of a sublethal concentration of tigecycline on gene expression in biofilm-associated cells of a clinical isolate of MRSA. This study reveals the effect of this drug on cellular processes and virulence factor expression and provides evidence as to whether tigecycline could be useful for the treatment of biofilm-mediated infections caused by MRSA strains.
MATERIALS AND METHODS
Strains and antibiotics.
The S. aureus isolate (isolate 784) used in this study was obtained from the Scottish MRSA Reference Laboratory (Stobhill Hospital, Glasgow, United Kingdom). Isolate 784 is an epidemic MRSA-16 (EMRSA-16) strain which was recovered in 2006 from a patient with an infected wound and is one of the most commonly isolated genotypes in the Scottish health care system. This isolate was selected because of its multidrug-resistant phenotype and ability to form fully established biofilms (37). It was cultured on blood agar (Oxoid, Basingstoke, United Kingdom) and stored in Microbank storage vials at −70°C. Prior to each assay, the isolate was freshly subcultured on brain heart infusion agar (BHIA; Oxoid). The tigecycline used in this study was kindly provided by Wyeth Pharmaceuticals (Philadelphia, PA). The MIC of tigecycline was determined as described previously (38) by the broth dilution method described by the British Society for Antimicrobial Chemotherapy (BSAC) (1). The antibiotic was freshly prepared before each experiment in brain heart infusion (BHI) broth (Oxoid) at a working concentration of 4× MIC (0.24 μg/ml). This concentration did not kill biofilm-associated cells (99.8% of the cells in the biofilm survived treatment) (38).
Biofilm growth.
One colony of the EMRSA-16 isolate was used to inoculate 5 ml of BHI broth and was grown in an orbital shaker at 200 rpm for 18 h at 37°C. The culture was then adjusted to an optical density equivalent to 1 × 105 CFU/ml, 20 μl was used to inoculate 15 ml of BHI broth in a 25-cm2 polystyrene tissue culture flask (Sarstedt, Leicester, United Kingdom), and the broth was incubated for 24 h at 37°C to allow a mature biofilm to develop. Flasks were selected to provide a large surface area to support biofilm growth. Biofilms were prepared in triplicate on four concurrent days, producing a total of 12 replicate biofilms. A panel of 12 biofilms was grown for use as the tigecycline-free controls, and a panel of 12 biofilms was grown and then treated with tigecycline diluted in freshly prepared BHI broth at 4× MIC.
Treatment of MRSA biofilms with tigecycline.
Following biofilm growth, the supernatant was removed from each biofilm, 10 ml of tigecycline (4× MIC) in BHI broth was added to the flask at an amount sufficient to cover the entire biofilm (BHI broth was added to the drug-free biofilms), and the biofilm was incubated for 1 h on a rocking platform at 37°C. Following the challenge, the antibiotic solution was removed from the flask and the biofilm was rinsed three times with phosphate-buffered saline (PBS). The medium was also removed from the control drug-free biofilms, and these were rinsed in the same way to remove loosely attached planktonic cells.
RNA isolation.
After the biofilms were rinsed, each biofilm was immediately exposed to 1 ml of RNA Protect (Qiagen, Crawley, United Kingdom), in preparation for RNA isolation from biofilm-associated cells. The drug-free and tigecycline-treated biofilm cells were scraped from the surface of each tissue culture flask with a cell scraper (Fisherbrand, Loughborough, United Kingdom) and placed in the RNA Protect solution. The sessile cell suspension was then removed from each flask, transferred to a microcentrifuge tube, and incubated in the RNA Protect solution for 5 min at room temperature to stabilize the mRNA. The cell suspensions were centrifuged at 12,000 × g for 2 min to pellet the cells, and the RNA Protect solution was removed. Each pellet was then treated with 50 mM EDTA (pH 8), 1 mg/ml lysozyme, and 20 mg/ml lysostaphin and was incubated at 37°C for 30 min to digest the cell wall. The cell suspension was then combined with 1 ml of Trizol (Invitrogen, Paisley, United Kingdom), and the cells were subjected to mechanical lysis by using 100 μl of 0.1-mm zirconium beads and the FastPrep system (Qbiogene, Irvine, CA) at a speed of 6.0 m/s three times for 40 s each time. RNA was extracted from the lysate according to the Trizol manufacturer's instructions. Contaminating DNA was removed from the sample by using a Turbo DNA-free kit (Ambion, Warrington, United Kingdom), and the RNA sample was purified by using an RNeasy minikit (Qiagen). The quantity and integrity of the RNA were determined by using a NanoDrop 1000 apparatus (Thermo Scientific, Wilmington, DE) and a Bioanalyzer 2100 apparatus (Agilent Technologies, Stockport, United Kingdom), respectively. Triplicate RNA samples were pooled to make four independent RNA samples with an acceptable concentration for use in the microarray. The RNA samples were aliquoted and stored at −70°C until they were required.
Staphylococcus aureus microarray.
The S. aureus microarray employed in this study is a whole-genome microarray generated by the Bacterial Microarray Group at St. George's, University of London (BμG@S SAv1.1.0). It consists of 3,623 gene-specific PCR products representing every open reading frame predicted from the first seven complete S. aureus genome sequencing projects (43). The majority of the strains included in the array are genotype EMRSA-16. The array design is available in the BμG@Sbase database (accession number A-BUGS-17; http://bugs.sgul.ac.uk/A-BUGS-17) and also the ArrayExpress database (accession number A-BUGS-17).
Microarray hybridization and processing.
For each condition, four RNA samples were prepared from independently pooled cultures (from four sets of triplicate biofilms). The RNA was reverse transcribed to labeled cDNA and hybridized in competition with labeled genomic DNA from isolate 784 (control) in a common reference experimental design. The RNA was hybridized in competition with a DNA standard to allow a direct comparison with the findings of future experiments with the same isolate under different experimental conditions or with different antimicrobial agents. DNA was isolated from an 18-h culture of isolate 784 grown in BHI broth by use of a DNeasy blood and tissue genomic DNA extraction kit (Qiagen), according to the manufacturer's instructions. The RNA and DNA samples were fluorescently labeled and hybridized as described previously (40). Briefly, 2 μg of total RNA was labeled by reverse transcription (RT) with random hexamer primers and Cy5 dCTP (GE Healthcare, Chalfont St. Giles, United Kingdom). In parallel, 1 μg of genomic DNA was labeled with Cy3 dCTP (GE Healthcare) by using random primers and the DNA polymerase I large (Klenow) fragment. The Cy3-labeled DNA and Cy5-labeled cDNA samples were then copurified through a Qiagen MinElute column (Qiagen), mixed with hybridization solution (4× SSC [1× SSC is 0.15 M NaCl plus 0.015 sodium citrate], 0.3% sodium dodecyl sulfate [SDS]) and denatured at 95°C for 2 min. The labeled sample was loaded onto a prehybridized (3.5× SSC, 0.1% SDS, 10 mg/ml bovine serum albumin) microarray under two LifterSlips (22 by 22 mm; Erie Scientific, Portsmouth, United Kingdom), sealed in a humidified hybridization cassette (Corning, Loughborough, United Kingdom), and hybridized overnight by immersion in a water bath at 65°C for 16 to 20 h. The slides were washed once in 400 ml 1× SSC-0.06% SDS at 65°C for 2 min and twice in 400 ml 0.06× SSC for 2 min.
Microarray data analysis.
The microarrays were scanned with an Affymetrix 428 scanner, and the signal intensity data were extracted with the BlueFuse for microarrays (version 3.5) program (BlueGnome, Cambridge, United Kingdom). The intensity data were also postprocessed in BlueFuse to exclude both control data and low-confidence data (P < 0.1) prior to normalization by the global Lowess method. Further analysis of the normalized data was undertaken by use of the GeneSpring (version 7.3.1) program (Agilent Technologies, Santa Clara, CA). For each pair of conditions, genes were initially filtered by determination of the fold change in expression to select those genes with a greater than twofold change in expression level between conditions and were then analyzed by one-way analysis of variance (P < 0.05, with Benjamini and Hochberg false discovery rate correction) to select those genes that were statistically shown to be differentially expressed.
Validation of microarray experiment by real-time RT-PCR.
Twelve genes that were up- or downregulated by more than twofold in the presence of tigecycline were selected to validate the data generated from the microarray study by real-time RT-PCR. These genes were selected because they encoded a range of virulence factors, including those associated with biofilm formation, cell adhesion, toxin production, and capsule synthesis. The genes selected were as follows: capC, capG, clfB, clpC, cna, fnbA, icaC, isaA, mnhG, sasF, ssaA, and tst (Table 1).
TABLE 1.
Sequences of oligonucleotide primers used for RT-PCR
| Primer | Gene | Oligonucleotide primer sequence (5′-3′) |
|---|---|---|
| capC F | capC | CATCCAGAGCGGAATAAAGC |
| capC R | capC | GTGTTATGCGCATCTGAACC |
| capG F | capG | CAAGGCCTGAAATCATTCGT |
| capG R | capG | CGCAATAATATTCCCCATCG |
| clfB F | clfB | TTGCCGCCATAAATGTGTTA |
| clfB R | clfB | TCACCACAAACGATTTCCAA |
| clpC F | clpC | GGTCATGATGATGGTGGACA |
| clpC R | clpC | ACTTGAACCACCGAATCCAG |
| cna F | cna | AAAGCGTTGCCTAGTGGAGA |
| cna R | cna | AGTGCCTTCCCAAACCTTTT |
| fnbB F | fnbB | TTCTGCATGACCTTCTGCA |
| fnbB R | fnbB | AACTTGGAAAAATGGCGTTG |
| gyrB F | gyrB | TTATGGTGCTGGGCAAATACA |
| gyrB R | gyrB | CACCATGTAAACCACCAGATA |
| icaC F | icaC | CTTGGGTATTTGCACGCATT |
| icaC R | icaC | GCAATATCATGCCGACACCT |
| isaA F | isaA | ACAGCTGCGTTGATTTGTTG |
| isaA R | isaA | CTGCAGGTGCTACTGGTTCA |
| mnhG F | mnhG | TCGCAACGATTAATTGCATA |
| mnhG R | mnhG | CGCCCTAGCAGCTATAGGATT |
| sasF F | sasF | CGTCCTCGTCACTTTGTTGA |
| sasF R | sasF | CGAAAACAGCATCGCAAATA |
| ssaA F | ssaA | AATGGCCGTTCAATCTCAAG |
| ssaA R | ssaA | GCGTTAGCCCAGTTACTTGCATTGC |
| tst F | tst | GTAAGCCCTTTGTTGCTTGC |
| tst R | tst | CTGATGCTGCCATCTGTGTT |
In addition, RNA was also extracted from one other EMRSA-16 clinical isolate and one EMRSA-15 isolate (the most common genotype isolated in the Scottish health care system), and real-time RT-PCR was performed with the primer sets described in Table 1 to determine whether the expression of the selected genes is similar in genetically related isolates and different clinical isolates of MRSA and to further validate the results of the microarray study.
Real-time RT-PCR.
cDNA was synthesized by using Moloney murine leukemia virus reverse transcriptase (Invitrogen) from 200 ng of each of the original four replicate sets of RNA samples used in the microarray study, according to the manufacturer's instructions. For each of the 12 genes selected, the real-time amplification was performed with 400 ng cDNA, 12.5 μl of SYBR GreenER qPCR SuperMix (Invitrogen), 1 μl of forward primer (200 nM), 1 μl of reverse primer (200 nM) (Table 1), and RNase- and DNase-free water added to a final volume of 25 μl. The real-time PCR was carried out by using the DNA Engine Opticon system (Bio-Rad, Hemel Hemstead, United Kingdom) by using the following cycle parameters: 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and then 60°C for 60 s. All real-time RT-PCRs were performed in triplicate for each of the four replicate sets of RNA samples from control biofilm cells and biofilm cells treated with tigecycline. The gyrB gene was used as an internal control housekeeping gene to normalize the levels of expression between samples. The real-time RT-PCR data were analyzed by the 2−ΔΔCT method (24).
Characterization of biological function.
The results of the microarray study were further validated by performing assays to determine the biological function in cells treated with tigecycline. The levels of production of one secreted protein (toxic shock syndrome toxin 1 [TSST-1]) and structural proteins (capsule) involved in virulence in this organism were determined.
Evaluation of capsule production by antibody agglutination.
The biofilms were grown for 24 h and treated with tigecycline, as described above. Control drug-free biofilms were also prepared. Drug-treated and control biofilm cells were scraped from the surface of the flasks and placed into 1 ml of 50 mM Tris buffer with 2 mM MgSO4. The cells were then treated with 0.04 mg/ml lysostaphin and 10 μl of both DNase and RNase for 4 h at 37°C with shaking at 200 rpm. The cell debris was pelleted by centrifugation at 13, 000 × g, and the cell lysates were removed. The amount of capsule produced by the biofilm cells in the presence and the absence of tigecycline was determined by rocket immunoelectrophoresis with rabbit antisera raised to S. aureus capsule type 8 (CP8) (IM 95-122; Nabi Biopharmaceuticals, Rockville, MD) and capsule type 8 polysaccharide standards.
Quantification of TSST-1 production.
Cells from drug-free biofilms and biofilms treated with tigecycline were scraped from the surface of the flask and the contents were transferred to 20-ml centrifuge tubes. The cells were harvested by centrifugation at 1,000 × g for 20 min at 4°C, and the presence of TSST-1 was detected in the supernatant by use of a toxic shock toxin-reversed passive latex agglutination toxin detection kit (Oxoid), according to the manufacturer's instructions. Briefly, 25-μl doubling dilutions of the sample were made in the V-shaped wells of a 96-well plate (Fisher Scientific, Loughborough, United Kingdom). A 25-μl aliquot of latex beads coated in antibody to TSST-1 was then added to each well. A positive TSST-1 control and a negative control (diluent only) were added to each plate. The plate was incubated at 20°C for 24 h without agitation, and each well was examined for agglutination. The assay was performed in triplicate for each sample.
Statistical analysis of results.
Prism software (GraphPad Software, Inc.) was used to analyze the data produced in each experiment. The standard deviation between replicate samples was calculated, and the two-tailed unpaired t test was performed to determine statistical significance.
Microarray data accession numbers.
Fully annotated microarray data have been deposited in the BμG@Sbase database (accession number E-BUGS-81; http://bugs.sgul.ac.uk/E-BUGS-81) and also the ArrayExpress database (accession number E-BUGS-81).
RESULTS
Global gene expression in MRSA biofilm-associated cells treated with tigecycline.
The biofilms formed by isolate EMRSA-16 were examined by scanning electron microscopy, and a multilayered three-dimensional structure was confirmed. In tigecycline-treated biofilms, analysis of the transcriptome by use of the S. aureus microarrays showed that 309 genes were significantly upregulated by more than twofold and that 213 genes were downregulated by more than twofold in comparison to the levels of gene expression in cells within drug-free biofilms (http://bugs.sgul.ac.uk/E-BUGS-81). These differentially expressed genes belonged to various functional categories, including those involved with macromolecule metabolism, cell envelope biogenesis, cell processes, and global regulatory functions (Table 2). Twelve genes were selected from among the genes that were either upregulated (six genes) or downregulated (six genes) following antibiotic challenge as representatives of genes encoding virulence factors with different functions. The expression of these genes was examined by real-time RT-PCR with the RNA retained from the microarray experiment to validate the findings of this study (Table 2). The results of the real-time RT-PCR showed concordance with the data generated from the microarray experiment, and the modulation of gene expression in all cases was confirmed.
TABLE 2.
Selected genes that displayed altered expression in the presence of tigecycline, as determined by microarray analysis and real-time RT-PCR
| Functional group | Gene | Protein | Fold change in expression measured by: |
|
|---|---|---|---|---|
| Microarray analysis | Real-time RT-PCR | |||
| Protein synthesis, ribosomes | rpsH | 30S ribosomal protein S8 | 14.86 | |
| rplN | 50S ribosomal protein L14 | 13.76 | ||
| rplB | 50S ribosomal protein L2 | 13.38 | ||
| rplD | 50S ribosomal protein L4 | 13.06 | ||
| rplJ | 50S ribosomal protein L10 | 12.75 | ||
| rplC | 50S ribosomal protein L3 | 12.06 | ||
| rplK | 50S ribosomal protein L11 | 11.03 | ||
| rpsE | 30S ribosomal protein S5 | 11.01 | ||
| rpmI | 50S ribosomal protein L35 | 10.51 | ||
| Protein synthesis, translation and modification | fus | Elongation factor G | 10.51 | |
| tuf | Elongation factor Tu | 7.41 | ||
| tsf | Elongation factor Ts | 3.23 | ||
| infC | Initiation factor C | 6.29 | ||
| infA | Initiation factor A | 4.77 | ||
| infB | Initiation factor B | 2.35 | ||
| rho | Transcription termination factor | 2.15 | ||
| Cell envelope, surface proteins and antigens | capB | Capsular polysaccharide synthesis enzyme | −2.08 | |
| cap8H | Capsular polysaccharide synthesis enzyme | −2.18 | ||
| capD | Capsular polysaccharide synthesis enzyme | −2.19 | ||
| capG | Capsular polysaccharide synthesis enzyme | −2.30 | −1.01 | |
| capF | Capsular polysaccharide synthesis enzyme | −2.66 | ||
| capC | Capsular polysaccharide synthesis enzyme | −3.14 | −1.75 | |
| icaC | Intercellular adhesion protein C | −2.06 | −2.23 | |
| Cell envelope, surface anchored membrane exported lipoproteins | fnbA | Fibronectin-binding protein | 2.30 | |
| fnbB | Fibronectin-binding protein | 5.34 | 3.03 | |
| clfB | Fibrinogen- and keratin 10-binding surface anchored protein | 4 | 7.5 | |
| sasF | Putative surface anchored protein | −2.30 | −1.32 | |
| cna | Collagen adhesion precursor | 2.10 | 3.42 | |
| sdrD | Fibrinogen-binding protein | −2.46 | ||
| Cell envelope, exported lipoproteins | sstD | Lipoprotein | 3.27 | |
| sbi | IgG-binding protein | 2.39 | ||
| ssaA | Secretory antigen precursor | 2.84 | 2.04 | |
| isaA | Immunodominant antigen A | 5.08 | 7.68 | |
| Cell envelope-peptidoglycan | dltA | d-Alanine-d-alanyl carrier protein ligase | 2.63 | |
| dltB | Putative activated d-alanine transport protein | 2.02 | ||
| dltC | d-Alanyl carrier protein | 2.29 | ||
| pbp2 | Penicillin-binding protein 2 | 3.91 | ||
| DNA, replication and repair | nusG | Transcription antitermination protein | 4.34 | |
| rpoC | DNA-directed RNA polymerase protein | 3.28 | ||
| topA | DNA topoisomerase I | 2.26 | ||
| recU | Putative recombination protein | 4.38 | ||
| dnaG | DNA primase | 2.19 | ||
| polA | DNA polymerase I | −2.11 | ||
| Global regulatory functions | mecI | Methicillin resistance regulatory protein MecI | −2.39 | |
| clpC | Putative stress response-related Clp ATPase | −2.44 | −1.14 | |
| sarA | Staphylococcal accessory regulator A | 2.37 | ||
| saeR | Response regulator protein | 2.02 | ||
| gapR | Glycolytic operon regulator | 9.12 | ||
| dinR | DNA damage-inducible repressor | −3.31 | ||
| tcaR | MarR family regulatory protein | 2.79 | ||
| icaR | ica operon transcriptional regulator | −3.48 | ||
| Cellular processes, transport | mnhF | Na+/H+ antiporter subunit | 3.27 | |
| mnhE | Na+/H+ antiporter subunit | 3.15 | ||
| mnhG | Na+/H+ antiporter subunit | 2.85 | 5.16 | |
| mnhD | Na+/H+ antiporter subunit | 1.93 | ||
| Adaptation to atypical conditions | sodA | Superoxide dismutase | −2.59 | |
| clpB | Putative ATPase subunit of an ATP-dependent protease | −2.49 | ||
| asp23 | Alkaline shock protein 23 | −8.61 | ||
| cspB | Cold shock protein | 5.59 | ||
| Toxin production | tst | TSST-1 | −2.39 | −1.51 |
Effect of tigecycline on protein synthesis within biofilm-associated cells.
In biofilm-associated cells challenged with tigecycline, 47 genes encoding ribosomal proteins were upregulated by 2.4- to 13.7-fold (Table 2). Eighteen genes encoded proteins belonging to the Rps family of 30S ribosomal proteins, 7 genes encoded 50S ribosomal proteins belonging to the Rpm family, and 22 genes encoded proteins belonging to the Rpl family of 50S ribosomal proteins. In the tigecycline-treated biofilms, the initiation factors infA, infB, and infC, which are essential for the instigation of translation, were also upregulated by 2.34- to 6.23-fold; and the levels of expression of genes encoding elongation factors G, Tu, and Ts increased by 10.5-, 7.4-, and 3.23-fold, respectively.
Effect of tigecycline on synthesis of adhesins.
In biofilm-associated cells, tigecycline exposure influenced the expression of genes encoding important virulence factors involved in the adhesion of S. aureus cells to human proteins. In our study, fnbA, which encodes FnbA, a protein that binds to fibronectin and, to a lesser extent, to fibrinogen (42), was upregulated by 2.3-fold in cells treated with tigecycline; and the level of expression of the closely related fnbB was upregulated by 5.34-fold (Table 2). The level of transcription of clfB, which encodes fibrinogen-binding protein ClfB, was also higher (4-fold) in tigecycline-treated biofilms than in the drug-free controls, as was the level of expression of cna, which encodes an adhesin that binds to collagen, a major component of connective tissue (upregulated by 2.10-fold).
Influence of tigecycline on biofilm-associated proteins.
When biofilms were exposed to tigecycline, the level of expression of some genes linked to biofilm development was reduced. The level of transcription of icaC, which plays a vital role in polysaccharide intercellular adhesin (PIA) synthesis and biofilm structure, was reduced by 2.06-fold in treated cells in comparison to the level of transcription in biofilm cells in drug-free medium (Table 2). The other genes in the ica operon also displayed a downward trend in expression (less than a twofold change), but this was not statistically significant. The sasF gene, which encodes a putative surface anchored protein (SasF) with significant homology to the biofilm-associated protein SasG, was also downregulated by 2.3-fold in the presence of tigecycline.
Influence of tigecycline on capsule synthesis.
In the presence of tigecycline, the expression of genes (capB, capC, capD, capF, capG, and cap8H) encoding the capsule polysaccharide synthesis enzymes CapB, CapC, CapD, CapF, CapG, and Cap8H decreased by 2.08-, 3.14-, 2.19-, 2.66-, 2.30-, and 2.18-fold, respectively. The production of capsule type 8 was assessed by rocket immunoelectrophoresis; however, the amount of capsule produced by cells growing in drug-free and tigecycline-treated biofilms was below the detection limit of this assay.
Influence of tigecycline on TSST-1 production.
The presence of a sublethal concentration of tigecycline also reduced the level of expression of tst, which encodes TSST-1, by 2.39-fold in biofilm-associated cells. A phenotypic latex agglutination assay for quantification of the level of TSST-1 produced in cells treated with the antibiotic also showed that the level of toxin production in treated biofilms was significantly reduced (by 10-fold; P < 0.001) in comparison to the level produced by untreated biofilm-associated cells (Fig. 1).
FIG. 1.
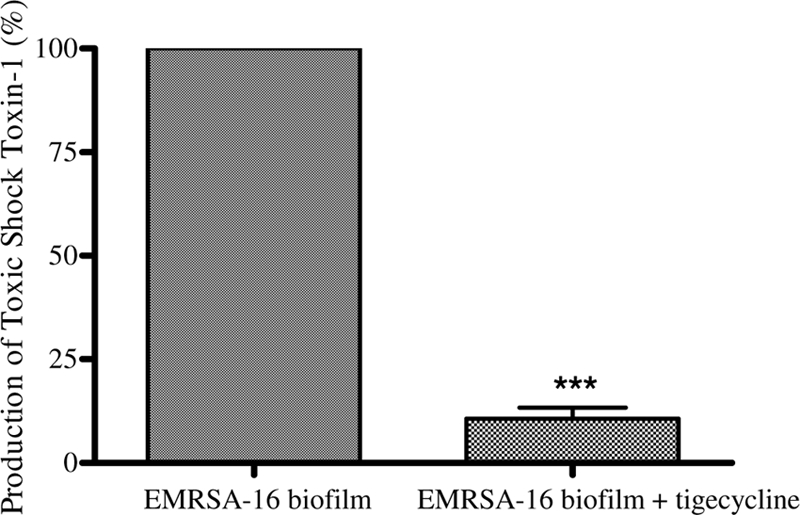
Production of TSST-1 in control biofilm-associated cells and those treated with a sublethal concentration of tigecycline, as measured by an antibody agglutination assay. Error bars represent the difference between replicate samples. ***, P < 0.001.
Expression of virulence-associated genes in other MRSA clinical isolates.
The expression of 12 genes that were up- or downregulated by more than twofold in the microarray experiment by a further two clinical isolates of MRSA was determined by real-time RT-PCR. One of these was an EMRSA-16 isolate with the same genotype as the isolate originally used in the microarray study; and the other was an EMRSA-15 isolate; which is the most commonly isolated genotype in the Scottish health care system. Following exposure to tigecycline, the levels of expression of ssaA, isaA, and cna by the second EMRSA-16 isolate increased by 1.72-fold, 3.7-fold, and 1.2-fold, respectively (Table 3). The levels of expression of capC, capG, clpC, clfB, icaC, tst, sasF, mnhG, and fnbB decreased by 1.09-, 1.96-, 19.87-, 1.18-, 1.6-, 5.52-, 3.53-, 6.99-, and 1.53-fold, respectively. For the EMRSA-15 isolate, exposure to tigecycline reduced the levels of expression of capC, capG, clpC, clfB, icaC, sasF, and mnhG by 1.64-, 1.58-, 1.23-, 1.03- 2.64-, 1.86-, and 1.64-fold, respectively. The presence of the antibiotic stimulated the expression of ssaA, isaA, cna, and fnbB, upregulating the genes by 1.17-, 1.09-, 1.55-, and 1.22-fold. This isolate did not carry the tst gene.
TABLE 3.
Expression of virulence-associated genes in different clinical isolates of MRSA
| Gene | Fold change in gene expression measured by RT-PCR |
||
|---|---|---|---|
| EMRSA-16 isolate 1 | EMRSA-16 isolate 2 | EMRSA-15 isolate | |
| capC | −1.75 | −1.09 | −1.64 |
| capG | −1.01 | −1.96 | −1.58 |
| clfB | 7.5 | −1.18 | −1.03 |
| clpC | −1.14 | −19.87 | −1.23 |
| cna | 3.42 | 1.2 | 1.55 |
| fnbB | 3.03 | −1.53 | 1.22 |
| icaC | −2.23 | 1.6 | −2.64 |
| isaA | 7.68 | 3.7 | 1.09 |
| mnhG | 5.16 | −6.99 | −1.64 |
| sasF | −1.32 | −3.53 | −1.86 |
| ssaA | 2.04 | 1.72 | 1.17 |
| tst | −1.51 | −5.52 | Gene not present |
DISCUSSION
The transcriptome analysis performed in this study has shown that a sublethal concentration of tigecycline promotes the expression of initiation factors, elongation factors, and ribosomal proteins. The drug elicits its antimicrobial effect on bacteria by binding to the 30S ribosomal subunit, which blocks the entry of amino-acyl tRNA molecules into the A site of the ribosome, which prevents the incorporation of amino acid residues into the elongating peptide chain and inhibits protein synthesis (3, 33). The upregulation of genes encoding essential components of the protein synthesis pathway may be part of the bacterial stress response in an attempt to withstand the antimicrobial challenge. These findings are consistent with the proposed mechanism of action of tigecycline (3, 4). As well as affecting the synthesis of proteins in biofilm-associated cells, the presence of tigecycline also altered the expression of a number of genes encoding key virulence factors involved in the pathogenesis of S. aureus.
Biofilm development is one aspect which enhances the virulence of S. aureus and which allows this organism to establish infection and persist in the host. The crucial first step in biofilm development is cellular attachment to biological or abiotic surfaces. In cases of infection, bacterial pathogens can adhere to components of the extracellular matrix (ECM) of host tissues to initiate colonization. Adherence to these cells is mediated by protein adhesins of the microbial surface components recognizing adhesive matrix molecules (MSCRAMM) family (15). S. aureus can express several MSCRAMMs that enable the bacteria to attach to human proteins, including fibronectin, fibrinogen, and collagen, and help establish infection (15). Devices such as stents, prosthetic joints, and artificial heart valves become coated in proteins, such as fibronectin and fibrinogen, conditioning the surface for bacterial attachment and posing an increased risk of biofilm-mediated infection.
Among the adhesins that have been well characterized in S. aureus are the fibronectin-binding proteins FnbA and FnbB, the structurally related fibrinogen binding proteins ClfA and ClfB, and the collagen adhesin Cna. Fibronectin-binding proteins are required for bacterial adhesion to fibronectin-coated surfaces; and they mediate the adhesion of S. aureus to human epithelial cells, endothelial cells, and fibroblasts (12). FnbA and FnbB have been significantly implicated in invasive diseases such as endocarditis, osteomyelitis, septic arthritis, and nosocomial pneumonia (28, 31). In this study fnbA, which encodes FnbA, a protein that binds to fibronectin and, to a lesser extent, fibrinogen (42), is upregulated by 2.30-fold in biofilm cells treated with tigecycline; and the level of expression of the closely related gene fnbB is upregulated by 5.34-fold. Blickwede et al. (2005) reported similar findings and found that fnbB was upregulated by twofold in S. aureus strain Newman exposed to a sublethal concentration clindamycin, which is also an inhibitor of protein synthesis (6). This effect may vary between clinical isolates. In our study the level of expression of fnbB increased (by 1.22-fold) in a clinical isolate of EMRSA-15 exposed to tigecycline but decreased (by 1.53-fold) in a second EMRSA-16 clinical isolate exposed to the drug. In S. aureus, the regulation of the fnb genes is complex. It has been reported that under the control of agr, fnbA displays reduced levels of expression in vivo (44); however, in vitro the fnb genes have displayed enhanced expression by SarA (encoded by sarA) through promoter binding (8). In our study, the level of expression of sarA increased by 2.37-fold in the presence of tigecycline, and this may have directly influenced the enhanced transcription of fnbA and fnbB. If tigecycline stimulates the expression of sarA, it will have a knock-on effect on the global expression of a multitude of genes encoding proteins with a range of functions.
In the presence of tigecycline, the genes encoding several other adhesins also showed increased levels of expression. The cna gene, which encodes an adhesin that binds to collagen, a major component of connective tissue, was upregulated by 2.10-fold in the presence of the drug. The level of expression of this gene also increased in the other two clinical isolates analyzed by 1.2- and 1.5-fold (EMRSA-16 and EMRSA-15, respectively). The level of expression of clfB, which encodes ClfB, a protein that mediates the adherence of S. aureus to immobilized and soluble fibrinogen (14), was enhanced by fourfold in the EMRSA-16 strain used in the microarray study. These ClfB and Cna adhesins are thought to be instrumental in the pathogenesis of conditions such as septic arthritis and infective endocarditis (14, 26). In contrast to the increased levels of expression of fnbA and clfB, the level of expression of sdrD, which encodes a fibrinogen-binding protein, similar to the ClfA and ClfB proteins (15), was reduced by 2.46-fold in the presence of tigecycline. It has recently been suggested that the protease ClpX is involved in the regulation of expression of clfB (18). Analysis of the microarray data showed that in the presence of tigecycline, the expression of clpX is upregulated by 2.80-fold. This contradicts previous findings which suggested that the downregulation or deletion of clpX produced an increase in level of expression of clfB (18). At present, our evidence supports the view that the ability of S. aureus to bind to human proteins, including fibronectin and fibrinogen, is a multifactorial process involving several different surface proteins and probably as yet undiscovered adhesins, which act in synchrony to achieve attachment to host tissues or foreign surfaces within the host. Tigecycline appears to influence the expression of several genes encoding these adhesins, suggesting that the binding capacity of S. aureus will be altered during drug-induced stress.
The expression of a glycocalyx also has an influence on adherence to surfaces and biofilm formation. Microorganisms that cause invasive disease commonly produce extracellular capsular polysaccharides (29). Approximately 90% of S. aureus isolates produce 1 of 11 serotypes of capsule; serotype CP8 is produced by approximately 50% of the isolates recovered from humans (29). Capsules enhance microbial virulence by rendering the bacterium resistant to phagocytosis (29). The expression of S. aureus CP8 in vitro is highly sensitive to various environmental signals, including anaerobiosis, alkaline conditions, and the salt concentration (22, 32), and is probably heavily influenced by the in vivo environment during the infection process. The isolate used in this study is known to be serotype CP8. Tigecycline treatment downregulated the expression of the capB, capC, capD, capF, capG, and cap8H genes in the EMRSA-16 isolate used in the microarray study; and the downregulated expression of these genes may interfere with capsule production in this organism. This finding was corroborated by the gene expression data for the other two clinical isolates examined in this study, which also displayed reductions in the levels of capC and capG expression in the presence of the drug. An attempt was made to quantify the amount of capsule actually produced by this isolate in the presence and the absence of the drug; however, the rocket immunoelectrophoresis method used was not sufficiently sensitive to detect the low levels of capsule produced when cells were grown in BHI medium. The reduction in the level of capsule gene expression may be a consequence of the decrease in nonessential cell processes in an attempt to survive exposure to the drug. A welcome side effect of this is that the reduction in capsule gene expression renders the organism more sensitive to the host immune response, namely, phagocytosis, which may result in the more rapid clearance of infection.
One of the most important virulence factors that bacteria employ to colonize the host and establish infection is the formation of a biofilm. PIA plays a role in biofilm development in S. aureus by mediating the intercellular attachment of the accumulating cells to build a biofilm (10). This molecule is encoded by the ica operon, which comprises the icaA, icaD, icaB, and icaC genes. Studies have shown that the expression of PIA is altered by various environmental stimuli, including anaerobic conditions, the presence of glucose and salt, and the influence of antimicrobials, such as ethanol and antibiotics. Rachid et al. (2000) showed that the presence of a subinhibitory concentration of tetracycline enhanced the expression of icaA by ninefold in S. epidermidis (34). Contrary to these findings, our study showed that the presence of tigecycline reduced the level of expression of icaC by 2.06-fold in biofilm-associated cells. The levels of expression of the other genes in the ica operon were also reduced in the presence of tigecycline, but this downregulation was less than twofold. The icaC gene in staphylococci encodes IcaC, which is a membrane protein with a putative function in the export of the growing sugar chain (20). This suggests that in the presence of this drug, the reduced expression of IcaC may interfere with the production of PIA, therefore limiting intercellular adhesion and inhibiting the development of the three-dimensional structure of the biofilm.
Another biofilm-associated gene which is downregulated (by 2.30-fold) in the presence of tigecycline is sasF. This gene encodes a putative surface anchored protein (SasF) with significant homology to SasG. Little is known of the function of SasF; but its homologue, SasG, has been relatively well characterized and is thought to be involved in the adhesion of S. aureus to squamous epithelial cells in the anterior nares (35). A study by Corrigan et al. (2007) also suggested that SasG may be involved in biofilm formation, as isolates of S. aureus that expressed SasG formed biofilms independently of the PIA mechanism (9). The role of SasG in biofilm development has not been elucidated, but understanding of its role may be key in explaining why isolates that do not carry the ica operon have the ability to form biofilms. SasF may have a function similar to that of SasG in adherence and biofilm development, and our findings suggest that tigecycline can downregulate the expression of the gene encoding this protein, possibly reducing bacterial adhesion to surfaces and inhibiting the initial stages of biofilm formation. At the moment, tigecycline is licensed for use only in patients with severe skin and soft tissue infections. Our findings show that this drug may have therapeutic value if a diagnosis is achieved in the early stages of infection, as it downregulates the genes involved in the initial stages of biofilm development.
One other important virulence factor that affords staphylococci lethal pathogenicity is the ability to produce toxins. At present, 20 serologically distinct staphylococcal superantigens have been described; and these comprise TSST-1, the staphylococcal enterotoxins (enterotoxins A to E and G to J), and the staphylococcal enterotoxin-like toxins (2, 23). The genes encoding these virulence factors are carried by accessory genetic elements, such as phages, plasmids, and staphylococcal pathogenicity islands (SAPIs) (23, 46). TSST-1 is responsible for toxic shock associated with female menstruation, a condition characterized by high fever, diffuse erythema, arterial hypotension, and multiple organ failure (23). The toxin stimulates the massive proliferation of T cells, resulting in the release of excessive amounts of cytokines, which in turn affect a number of organ systems, including the cardiovascular system, resulting in shock (17). Previous studies have shown that the level of production of TSST-1 is reduced in cells treated with the protein inhibitors clindamycin, linezolid, and gentamicin (39). These antibiotics are recommended for use for the treatment of severe staphylococcal infections associated with the production of this potent toxin. From the findings of this study, we would also suggest that tigecycline be considered as a therapy for toxin-mediated staphylococcal infections. The presence of this antibiotic reduced the level of expression of tst in the original microarray isolate by 2.39-fold and in the second clinical isolate, EMRSA-16, by 5.52-fold. The production of the functional toxin by the isolate used for the microarray experiment was also reduced by 10-fold in the phenotypic assay. In the clinical setting, this may have a beneficial effect on the outcome of infection by attenuating the expression of TSST-1 in life-threatening cases of toxic shock.
Conclusion.
S. aureus utilizes the biofilm mode of growth to initiate and establish recalcitrant infections in a multitude of disease conditions ranging from surgical wound infections to endocarditis. Antibiotic therapy often fails to eradicate these multicellular communities from the host; however, in the study described here, we have shown that the antibiotic tigecycline does influence gene expression in survivor cells within the biofilm exposed to this agent. Exposure to a subinhibitory concentration of the drug significantly alters (greater than a twofold change) the expression of 18.6% of the genome (in the presence of the drug, 11% of the genes were upregulated and 7.6% of the genes were downregulated) in cells within the biofilm. Genes encoding a number of key virulence factors were affected. There was an increase in the levels of expression of a number of genes encoding adhesin proteins and a reduction in the levels of expression of several genes encoding biofilm-associated proteins, enzymes involved in capsule synthesis, and the gene encoding TSST-1. Although the effect of tigecycline on biofilm formation still needs to be measured, it remains that the regulation and expression of certain virulence factors involved in pathogenesis in S. aureus are markedly downregulated in the presence of the drug, which is encouraging. From our initial studies with two additional clinical isolates of MRSA, it appears that tigecycline has a similar effect on gene expression in genetically related and unrelated clinical isolates of MRSA. Further investigation with a larger panel of isolates would be required to confirm these findings and determine whether tigecycline may promote the beneficial modulation of virulence factors and prove therapeutically useful in reducing the morbidity and mortality linked with S. aureus biofilm-mediated infections.
Acknowledgments
We acknowledge Jawad Sarwar at Nabi Pharmaceuticals for kindly providing the capsule type 8 antisera and help with capsule detection. We thank Kathleen Harvey-Wood at the Royal Hospital for Sick Children, Glasgow, United Kingdom, for her assistance with the toxic shock toxin assay. Finally, we acknowledge Christopher Longshaw and Wyeth Pharmaceuticals for providing the tigecycline powder and funding for this study.
Footnotes
Published ahead of print on 26 October 2009.
REFERENCES
- 1.Andrews, J. M. 2001. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 48(Suppl. 1):5-16. [DOI] [PubMed] [Google Scholar]
- 2.Baker, M. D., and K. R. Acharya. 2004. Superantigens: structure-function relationships. Int. J. Med. Microbiol. 293:529-537. [DOI] [PubMed] [Google Scholar]
- 3.Bauer, G., C. Berens, S. J. Projan, and W. Hillen. 2004. Comparison of tetracycline and tigecycline binding to ribosomes mapped by dimethylsulphate and drug-directed Fe2+ cleavage of 16S rRNA. J. Antimicrob. Chemother. 53:592-599. [DOI] [PubMed] [Google Scholar]
- 4.Bergeron, J., M. Ammirati, D. Danley, L. James, M. Norcia, J. Retsema, C. A. Strick, W. G. Su, J. Sutcliffe, and L. Wondrack. 1996. Glycylcyclines bind to the high-affinity tetracycline ribosomal binding site and evade Tet(M)- and Tet(O)-mediated ribosomal protection. Antimicrob. Agents Chemother. 40:2226-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernardo, K., N. Pakulat, S. Fleer, A. Schnaith, O. Utermohlen, O. Krut, S. Muller, and M. Kronke. 2004. Subinhibitory concentrations of linezolid reduce Staphylococcus aureus virulence factor expression. Antimicrob. Agents Chemother. 48:546-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blickwede, M., C. Wolz, P. Valentin-Weigand, and S. Schwarz. 2005. Influence of clindamycin on the stability of coa and fnbB transcripts and adherence properties of Staphylococcus aureus Newman. FEMS Microbiol. Lett. 252:73-78. [DOI] [PubMed] [Google Scholar]
- 7.Reference deleted.
- 8.Chien, Y., A. C. Manna, S. J. Projan, and A. L. Cheung. 1999. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J. Biol. Chem. 274:37169-37176. [DOI] [PubMed] [Google Scholar]
- 9.Corrigan, R. M., D. Rigby, P. Handley, and T. J. Foster. 2007. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 153:2435-2446. [DOI] [PubMed] [Google Scholar]
- 10.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Gotz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donlan, R. M. 2002. Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8:881-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dziewanowska, K., J. M. Patti, C. F. Deobald, K. W. Bayles, W. R. Trumble, and G. A. Bohach. 1999. Fibronectin binding protein and host cell tyrosine kinase are required for internalization of Staphylococcus aureus by epithelial cells. Infect. Immun. 67:4673-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellis-Grosse, E. J., T. Babinchak, N. Dartois, G. Rose, and E. Loh. 2005. The efficacy and safety of tigecycline in the treatment of skin and skin-structure infections: results of 2 double-blind phase 3 comparison studies with vancomycin-aztreonam. Clin. Infect. Dis. 41(Suppl. 5):S341-S353. [DOI] [PubMed] [Google Scholar]
- 14.Entenza, J. M., T. J. Foster, D. Ni Eidhin, P. Vaudaux, P. Francioli, and P. Moreillon. 2000. Contribution of clumping factor B to pathogenesis of experimental endocarditis due to Staphylococcus aureus. Infect. Immun. 68:5443-5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster, T. J., and M. Hook. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6:484-488. [DOI] [PubMed] [Google Scholar]
- 16.Foster, T. J. 2004. The Staphylococcus aureus “superbug.” J. Clin. Invest. 114:1693-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fraser, J. D., and T. Proft. 2008. The bacterial superantigen and superantigen-like proteins. Immunol. Rev. 225:226-243. [DOI] [PubMed] [Google Scholar]
- 18.Frees, D., A. Chastanet, S. Qazi, K. Sorensen, P. Hill, T. Msadek, and H. Ingmer. 2004. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol. Microbiol. 54:1445-1462. [DOI] [PubMed] [Google Scholar]
- 19.Gemmell, C. G., and C. W. Ford. 2002. Virulence factor expression by Gram-positive cocci exposed to sub-inhibitory concentrations of linezolid. J. Antimicrob. Chemother. 50:665-672. [DOI] [PubMed] [Google Scholar]
- 20.Gerke, C., A. Kraft, R. Sussmuth, O. Schweitzer, and F. Gotz. 1998. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 273:18586-18593. [DOI] [PubMed] [Google Scholar]
- 21.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95-108. [DOI] [PubMed] [Google Scholar]
- 22.Herbert, S., S. W. Newell, C. Lee, K. P. Wieland, B. Dassy, J. M. Fournier, C. Wolz, and G. Doring. 2001. Regulation of Staphylococcus aureus type 5 and type 8 capsular polysaccharides by CO2. J. Bacteriol. 183:4609-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindsay, J. A., A. Ruzin, H. F. Ross, N. Kurepina, and R. P. Novick. 1998. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol. Microbiol. 29:527-543. [DOI] [PubMed] [Google Scholar]
- 24.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 25.Mah, T. F., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 26.McDevitt, D., P. Francois, P. Vaudaux, and T. J. Foster. 1994. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol. Microbiol. 11:237-248. [DOI] [PubMed] [Google Scholar]
- 27.Miro, J. M., I. Anguera, C. H. Cabell, A. Y. Chen, J. A. Stafford, G. R. Corey, et al. 2005. Staphylococcus aureus native valve infective endocarditis: report of 566 episodes from the International Collaboration on Endocarditis Merged Database. Clin. Infect. Dis. 41:507-514. [DOI] [PubMed] [Google Scholar]
- 28.Mongodin, E., O. Bajolet, J. Cutrona, N. Bonnet, F. Dupuit, E. Puchelle, and S. de Bentzmann. 2002. Fibronectin-binding proteins of Staphylococcus aureus are involved in adherence to human airway epithelium. Infect. Immun. 70:620-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Riordan, K., and J. C. Lee. 2004. Staphylococcus aureus capsular polysaccharides. Clin. Microbiol. Rev. 17:218-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parsek, M. R., and C. Fuqua. 2004. Biofilms 2003: emerging themes and challenges in studies of surface-associated microbial life. J. Bacteriol. 186:4427-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peacock, S. J., I. de Silva, and F. D. Lowy. 2001. What determines nasal carriage of Staphylococcus aureus? Trends Microbiol. 9:605-610. [DOI] [PubMed] [Google Scholar]
- 32.Pohlmann-Dietze, P., M. Ulrich, K. B. Kiser, G. Doring, J. C. Lee, J. M. Fournier, K. Botzenhart, and C. Wolz. 2000. Adherence of Staphylococcus aureus to endothelial cells: influence of capsular polysaccharide, global regulator agr, and bacterial growth phase. Infect. Immun. 68:4865-4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Projan, S. J. 2000. Preclinical pharmacology of GAR-936, a novel glycylcycline antibacterial agent. Pharmacotherapy 20:219S-223S. [DOI] [PubMed] [Google Scholar]
- 34.Rachid, S., K. Ohlsen, W. Witte, J. Hacker, and W. Ziebuhr. 2000. Effect of subinhibitory antibiotic concentrations on polysaccharide intercellular adhesin expression in biofilm-forming Staphylococcus epidermidis. Antimicrob. Agents Chemother. 44:3357-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roche, F. M., M. Meehan, and T. J. Foster. 2003. The Staphylococcus aureus surface protein SasG and its homologues promote bacterial adherence to human desquamated nasal epithelial cells. Microbiology 149:2759-2767. [DOI] [PubMed] [Google Scholar]
- 36.Rose, W. E., and M. J. Rybak. 2006. Tigecycline: first of a new class of antimicrobial agents. Pharmacotherapy 26:1099-1110. [DOI] [PubMed] [Google Scholar]
- 37.Smith, K., A. Perez, G. Ramage, D. Lappin, C. G. Gemmell, and S. Lang. 2008. Biofilm formation by Scottish clinical isolates of Staphylococcus aureus. J. Med. Microbiol. 57:1018-1023. [DOI] [PubMed] [Google Scholar]
- 38.Smith, K., A. Perez, G. Ramage, C. G. Gemmell, and S. Lang. 2009. Comparison of biofilm-associated cell survival following the in vitro exposure of methicillin resistant Staphylococcus aureus biofilms to the antibiotics clindamycin, daptomycin, linezolid, tigecycline and vancomycin. Int. J. Antimicrob. Agents 33:374-378. [DOI] [PubMed] [Google Scholar]
- 39.Stevens, D. L., Y. Ma, D. B. Salmi, E. McIndoo, R. J. Wallace, and A. E. Bryant. 2007. Impact of antibiotics on expression of virulence-associated exotoxin genes in methicillin-sensitive and methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 195:202-211. [DOI] [PubMed] [Google Scholar]
- 40.Stewart, G. R., L. Wernisch, R. Stabler, J. A. Mangan, J. Hinds, K. G. Laing, P. D. Butcher, and D. B. Young. 2002. The heat shock response of Mycobacterium tuberculosis: linking gene expression, immunology and pathogenesis. Comp. Funct. Genomics 3:348-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart, P. S. 2003. Diffusion in biofilms. J. Bacteriol. 185:1485-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wann, E. R., S. Gurusiddappa, and M. Hook. 2000. The fibronectin-binding MSCRAMM FnbpA of Staphylococcus aureus is a bifunctional protein that also binds to fibrinogen. J. Biol. Chem. 275:13863-13871. [DOI] [PubMed] [Google Scholar]
- 43.Witney, A. A., G. L. Marsden, M. T. Holden, R. A. Stabler, S. E. Husain, J. K. Vass, P. D. Butcher, J. Hinds, and J. A. Lindsay. 2005. Design, validation, and application of a seven-strain Staphylococcus aureus PCR product microarray for comparative genomics. Appl. Environ. Microbiol. 71:7504-7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiong, Y. Q., A. S. Bayer, M. R. Yeaman, W. Van Wamel, A. C. Manna, and A. L. Cheung. 2004. Impacts of sarA and agr in Staphylococcus aureus strain Newman on fibronectin-binding protein A gene expression and fibronectin adherence capacity in vitro and in experimental infective endocarditis. Infect. Immun. 72:1832-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reference deleted.


